Abstract
Striatin is a novel protein that interacts with steroid receptors and modifies rapid, non-genomic activity in vitro. We tested the hypothesis that striatin would in turn affect mineralocorticoid receptor function and consequently sodium, water, and blood pressure homeostasis in an animal model.
We evaluated salt sensitivity of blood pressure in novel striatin heterozygote knockout mice. When compared with wild type, striatin heterozygote exhibited a significant increase in blood pressure when sodium intake was increased from restricted (0.03%) to liberal (1.6%) sodium). Further, renal expression of mineralocorticoid receptor and its genomic downstream targets serum/glucocoticoid-regulated kinase 1 and epithelial sodium channel were increased in striatin heterozygote versus wild type mice on liberal sodium intake while the pAkt/Akt ratio, readout of mineralocoriticoid receptor's rapid, non-genomic pathway, was reduced.
To determine the potential clinical relevance of these findings, we tested the association between single nucleotide polymorphic variants of striatin gene and salt sensitivity of blood presure in 366 Caucasian hypertensive subjects. HapMap derived tagging single nucleotide polymorphisms identified an association between rs2540923 with salt sensitivity of blood pressure (OR, 6.25; 95% CI 1.7-20; P=0.01).
These data provide the first in vivo evidence in humans and rodents that associates striatin with markers of mineralocoriticoid receptor activity. The data also support the hypothesis that the rapid, non-genomic mineralocoriticoid receptor pathway (mediated via striatin) has a role in modulating the interaction between salt intake and blood pressure.
Keywords: Striatin, salt-sensitivity, blood pressure, SNP, aldosterone, mineralocorticoid receptor
Introduction
Interventional and epidemiological studies have demonstrated an association between dietary sodium consumption and increases in blood pressure (BP) 1, 2, 3. However, nearly 40% of individuals with hypertension do not demonstrate salt sensitivity (SS) of BP and even in those with SS of BP the underlying mechanism(s) remain largely unknown. To better understand the differential BP response to sodium loading, one promising approach is to study the interaction of dietary sodium intake with candidate genes involved in the regulation of salt and water homeostasis.
A primary role of aldosterone (ALDO) and the mineralocorticoid receptor (MR) is to maintain sodium and water homeostasis. Activation of this process is initiated by translocation of an activated ALDO/MR to the cell nucleus to modulate the expression of genes regulating electrolyte and fluid balance 4. It has been documented that ALDO activation of MR can also lead to rapid phosphorylation and activation of proteins in the MAPK pathways, increases in reactive oxygen species (ROS) , and intracellular calcium, among other effects 5-6. Together these effects are commonly referred to as ALDO's “non-genomic” actions. Several other steroid hormones receptors, e.g., estrogen receptor (ERα), have been shown to have both genomic and non-genomic effects 7-8.
Striatin is a newly described protein that interacts with the ERα and MR and regulates their nongenomic actions 9-10. Striatin is a highly conserved member of the WD-repeat family of proteins that possesses caveolin-1, calcium-calmodulin, and coil-coil conserved domains 11-12. Our laboratory has reported that activation of MR by ALDO increases striatin expression in kidney, heart and aortic tissue, and cultured endothelial cells 13. Our group has also described that striatin is a novel mediator for ALDO's induction of pERK and ERα's rapid action to promote phosphorylation of eNOS 14.
The objective of this study was to examine the in vivo relationship between SS of BP and striatin. Towards this goal, we generated a novel striatin heterozygous knockout mouse and demonstrated that reduction in striatin levels results in SS of BP. To translate these results to human hypertension we explored whether genetic variance within the striatin gene was associated with SS of BP in a well-phenotyped Caucasian hypertensive cohort.
Methods
Generation of Striatin Heterozygous Mouse and Experimental Protocol
The striatin heterozygous knockout strain (Strn+/−, CSD26933) was generated by the trans-NIH Knock-Out Mouse Project (KOMP). Details are available in Online Supplemental Materials.
Blood Pressure Measurements
Details are available in Online Supplemental Materials.
Plasma Hormone Measurements
Blood was collected in purple-top BD Microtainer tubes (EDTA)and plasma was separated by centrifugation. ALDO levels were measured using Coat-A-Count Radioimmunometric Assay (RIA) kit (SIEMENS, Los Angeles, CA USA) 15. Plasma renin activity (PRA) was measured by RIA assay (DiaSorin, Stillwater, MN) 16.
Western Blot Analysis
Protein was extracted from heart, kidney and adrenal tissue by homogenizing according to the Bullet Blender protocol and bead specification (Next Advance, Inc., Averill Park, NY). Briefly, 30mg of tissue was placed in a RIPA buffer (Boston Bioproducts, Worchester, MA) with protease inhibitor cocktail (SIGMA, St. Louis, Mo)(1:100). Lysates were centrifuged at 12,000g for 10 minutes at 4°C and supernatant was collected and stored at −20°C. Cell lysates were prepared with 4x-reducing Laemmli's SDS Sample Buffer (250MM Tris-HCl, 8% SDS, 40% Glycerol, 0.02% Bromophenol blue, DTT). Samples were size fractionated by electrophoresis on SDS-PAGE and proteins transferred to nitrocellulose membranes by electroblotting. Blots were incubated with primary antibody overnight at 4°C (BD biosciences: striatin; Santa Cruz: ENaC, SGK1, eNOS and cell signaling: pAkt/Akt). Blots were incubated with conjugated secondary antibody with horseradish-peroxidase for 1 hour at room temperature and analyzed using Enhanced Chemiluminescence (ECL) (Perkin-Elmer Life Sciences, Waltham, MA). Blots were reprobed for β-tubulin (Sigma) and results were normalized to correct for loading.
mRNA Analysis
Total mRNA was extracted from tissue using RNeasy mini kit (QIAGEN Sciences, Germantown, MD). cDNA was synthesized from 1.5μg RNA with first-strand cDNA synthesis kit (GE Healthcare, Piscataway, NJ). PCR amplification reactions were performed in duplicate using the ABI Prism 7000 sequence detection system (Life Technologies, Foster City, CA) and using the comparative threshold cycle method to determine mRNA levels. The gene expression data was normalized to 18S rRNA levels. PCR amplification to detect SGK1, ENaC and 18S rRNA levels was performed with TaqMan gene expression assays. Data are represented as fold increases relative to the measurement in WT mice.
Zona Glomerulosa Cells Stimulation
Adrenal glands were excised during sacrifice and zona glomerulosa (ZG) cells were isolated as previously reported 17. ZG cell suspensions were made by diluting pellets to obtain 1 to 2×105 cells/0.5ml of modified Kreb-Ringer bicarbonate (KRGBA). Cells were then incubated in duplicate, vehicle or in the presence of angiotensinogen II (10−7 M) for 1 hour at 37°C under % CO2-95% O2 atmosphere. ALDO levels were measured using Coat-A-Count Radioimmunometric Assay (RIA) kit as listed above.
HyperPATH Cohort and Study Protocol
Our analysis consisted of 366 hypertensive Caucasian subjects from the Hypertensive Pathotype (HyperPATH) Cohort, a cohort designed to determine the genetic underpinnings of hypertension, as previously extensively reported). A detailed description of the study protocol and cohort methods can be found in online supplementary materials.
Human Study Genotyping
Statistical Analysis
Animal Study
All values are reported as means ± SEM unless otherwise indicated, and corrections for multiple comparisons are made where appropriate. Within each genotype, paired Student's t-tests were used to determine the significance of the increase in systolic BP when intake was changed from ResS to LibS diets. Student's non-parametric t-test was utilized when comparing various parameters between WT and Strn+/− mice on either a LibS or ResS diet. A difference was considered statistically significant if a two-tailed was ≤ 0.05. Because the biomarkers came from different mice on the two diets, non-parametric tests were used and the nominal p value was adjusted for multiple comparisons (p≤0.025). All studies were completed with the individual performing the study blinded as to genotype and diet which the tissue or samples were obtained.
Human Study
Given the large number of SNPs identified via HapMap, haplotypes were constructed from the genotyping of this cohort using the Haploview program 4.1 20. All subsequent analysis is described in supplemental methods.
Results
Striatin Modulates Rapid, Non-Genomic MR Activity
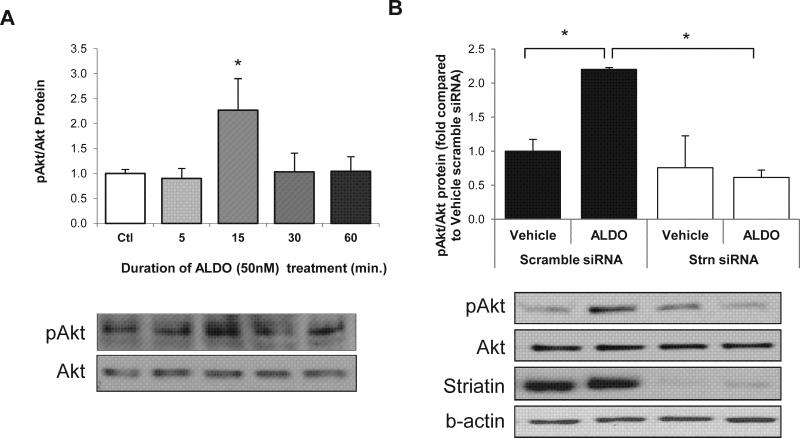
We first examined the in vitro rapid effects of ALDO (50 nM) in EA.hy926 cells, an endothelial cell line. Our results show that increases in pAkt/Akt protein ratio is time dependent, with the highest increase in activation at 15 minutes and rapid return to baseline thereafter (Figure 1A). Next we tested whether decreasing striatin levels through the use of siRNA technology would affect pAkt/Akt activation. In EA.hy926 cells, striatin siRNA reduced striatin levels and inhibited the rapid effects of ALDO/MR signaling (Figure 1B).
Figure 1.
Aldosterone rapid non-genomic actions in EA.hy926 cells. A) EA.hy926 cells treated acutely with 50 nM ALDO. B.) Cells transfected with 1 nM striatin siRNA or scramble siRNA were stimulated with ALDO for 15 minutes. pAkt/Akt levels were assessed by Western blot. Data represent means ± SEM (n=4 in duplicate).
Generation of Strn+/− Mice
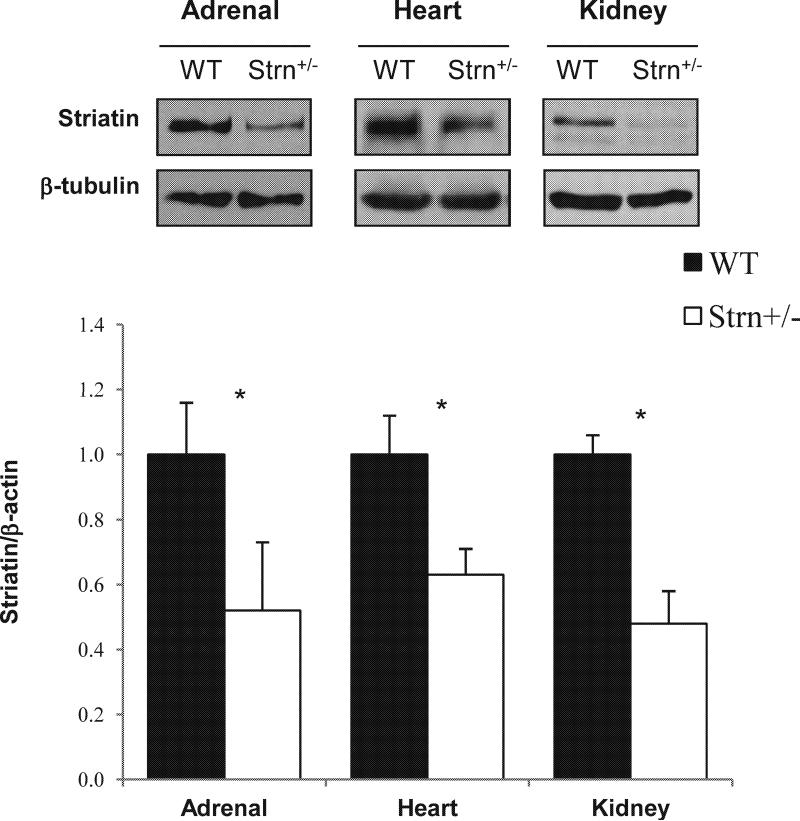
To investigate the physiological relevance of rapid, non-genomic MR activation striatin gene targeting was performed by the International Knockout Mouse Consortium (IKMC) in JM8 embryonic stem cell line on a C57BL/6N genetic background (Supplemental Materials). Immunoblotting analysis of Strn+/− mouse tissues (adrenal, heart and kidney) confirmed that targeted allele effectively reduced striatin protein levels (Figure 2); striatin protein levels were reduced approximately 50% in Strn+/− mice as compared to WT mice.
Figure 2.
Strn gene targeting in mice. Western blot analysis of striatin expression in adrenal, heart and kidney tissue. Gels for striatin and β-actin are presented for each tissue/genotype. Data represent mean ± SD, N=4. *p<0.01, comparing WT to Strn+/− mice.
Effects of High Salt Diet on BP in Strn+/− Mice
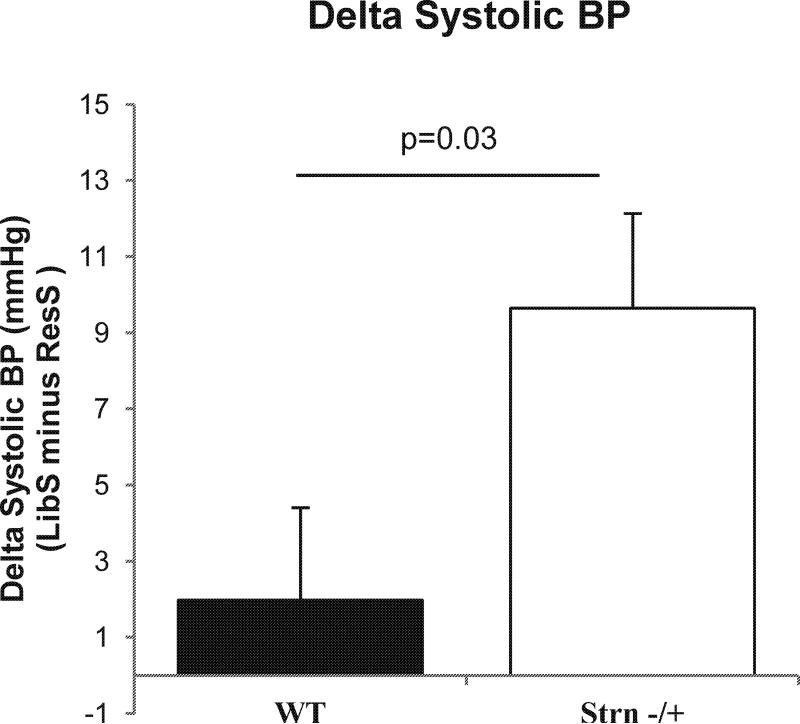
WT and Strn+/− mice had similar body weights when consuming a ResS diet (30.5 ± 1.5 and 30.5 ± 1.5 g, respectively) or when consuming a LibS diet (31.1 ± 0.8 and 29.7 ± 1.0 g, respectively). Strn+/− mice had significantly higher BPs on LibS when compared to ResS diet (ResS 98 ± 3 vs. LibS 107 ± 4 mmHg, p=0.001), whereas WT mice did not exhibit SS BP (ResS 102 ± 2 vs. LibS (104 ± 2 mmHg, p=0.26). Furthermore, the rise in blood pressure was significantly different comparing WT to Strn+/− mice (Δ 2.0 ± 2 and 9.6 ± 2, p = 0.04) (Figure 3).
Figure 3.
Change in systolic blood pressure in response to changes in sodium intake. Delta is calculated as systolic BP on 1.6% Na+ minus systolic BP on 0.03% Na+ diet. Data represent means ± SEM (n=13 per genotype). P value for student t-test comparing WT and Strn+/−.
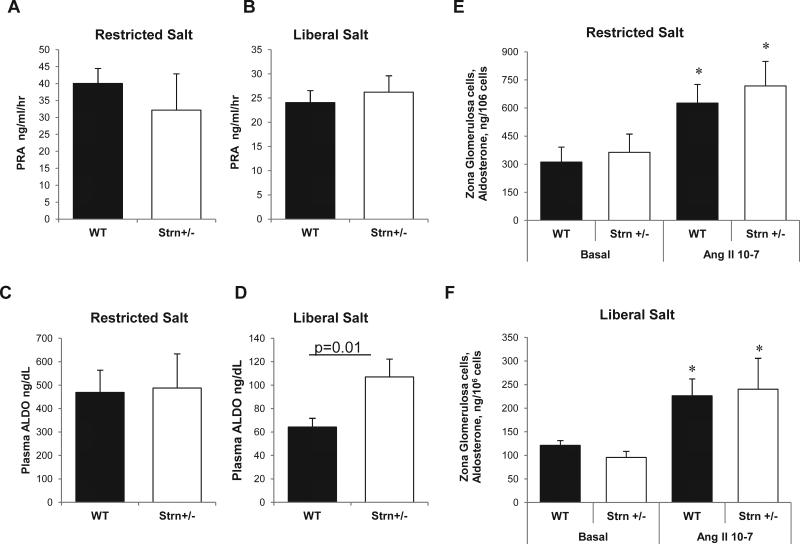
Next we assessed the impact of dietary sodium intake on the level of activation of the reninangiotensin-aldosterone system by measuring the levels of PRA and ALDO at the time of sacrifice. While PRA levels were significantly reduced on the LibS versus ResS diets, there were no differences between genotypes (Figure 4A, B). Interestingly, ALDO levels were significantly higher on a LibS diet in Strn+/− mice compared to WT (Figure 4C, D). Furthermore, we measured ALDO production in response angiotensin II (AngII) stimulation ex vivo in an isolated glomerulosa cell system. Similar to plasma measurements, ALDO production was significantly lower on LibS versus ResS diets. AngII stimulation induced a ~2-fold increase in ALDO on ResS diets, but there were no significant differences between genotypes (Figure 4C).
Figure 4.
Effect of dietary Na+ on ALDO production in vivo and ex vivo in WT and Strn+/− mice. PRA levels on (A) ResS or (B) LibS diets (n=4-8/group). Plasma ALDO levels on a (C) ResS or (D) LibS diets (n=4-8/group). ALDO production is assessed in media from isolated zona glomerulosa cells (ZG) ZG in the presence or absence of ANGII 10−7 M for 2 hours on (E) ResS and (F) LibS diets. ALDO. Black bar indicates WT, open bar indicates Strn+/−. Data expressed as mean ± SEM; * indicates p<0.001.
Heart and Kidney Tissue Analysis
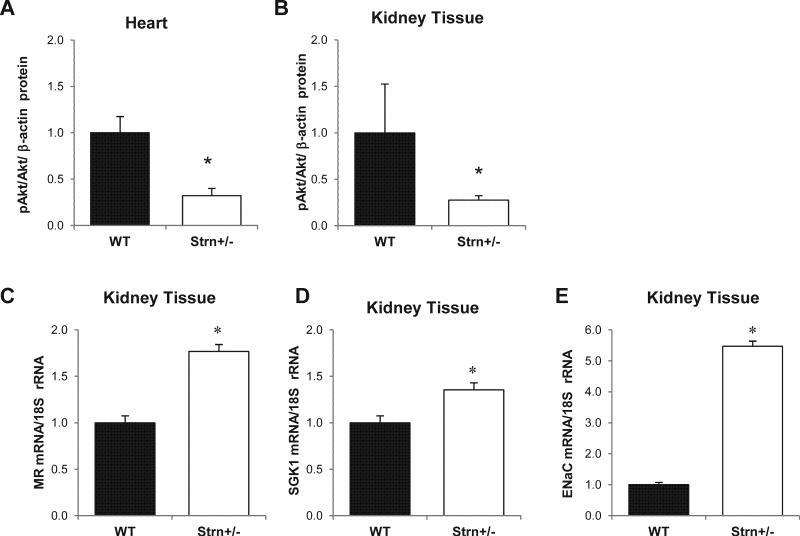
Heart weight and heart/body weight ratios did not differ between WT and Strn+/− mice whether on a ResS diet (4.8 ± 0.2 and 4.9 ± 0.2 mg/g, respectively) or on a LibS diet (4.4 ± 0.2 and 4.2 ± 0.2 g, respectively). However, as anticipated from the cell based studies, phosphorylation of Akt in heart tissue was significantly (p<0.05) reduced (~70%) in Strn+/− mice as compared with WT animals on the same diet (Figure 5A).
Figure 5.
Molecular studies using heart and kidney tissue. pAkt/Akt ratio protein expression in heart (A) and kidney (B). mRNA expression using real-time PCR in kidney cortical tissue: MR (C), SGK1 (D) and ENaC (E). * p<0.01 relative to WT-ResS. Data represent mean ± SD, N=4.
Similar to the heart kidney, weight and kidney to body weight ratios did not differ between WT and Strn+/− mice whether on a ResS diets (5.8 ± 0.3 and 5.3 ± 0.2 mg/g, respectively) or on a LibS diets (5.5 ± 0.2 and 5.7 ± 0.2 mg/g, respectively) while the pAkt/Akt ratio in kidneys obtained from mice on a LibS diet was significantly lower (p=0.01) in the Strn+/− mice compared to the WT mice (Figure 5B). In contrast MR transcript levels on a LibS diet were significantly increased (~1.7 fold; p=0.01) in Strn+/− versus WT mice (Figure 5C). This increased MR expression levels was associated with differences in expression of known MR activated downstream genes: Strn+/− mice had significantly (p<0.01) higher mRNA transcript levels of serum/glucocoritcoid regulated kinase (SGK1) (~1.4 fold) and epithelial sodium channel (ENaC) (>5 fold) as compared to WT animals on the same diet (Figure 5D, E).
Human Study Participant Characteristics
To assess the relevance of the findings in mice to humans, the association between known SNP variation in the striatin gene and salt sensitive BP in humans was assessed. Study participant characteristics are summarized in Supplemental Materials (Table S2). There were 366 Caucasian individuals from the HyperPATH cohort examined. Genotyping had a completion rate of ≥ 95% and all SNPs were in Hardy-Weinberg equilibrium. Of the 40 HapMap derived SNPs genotyped in the STRN gene, 21 were removed before the start of the analysis (7 displayed monomorphism in our population and 14 had a minor allele frequency of less than 0.03). The remaining 19 SNPs captured 100% of the common HapMap Caucasian variation in this region and were contained in 3 distinct haplotype blocks (Table S3).
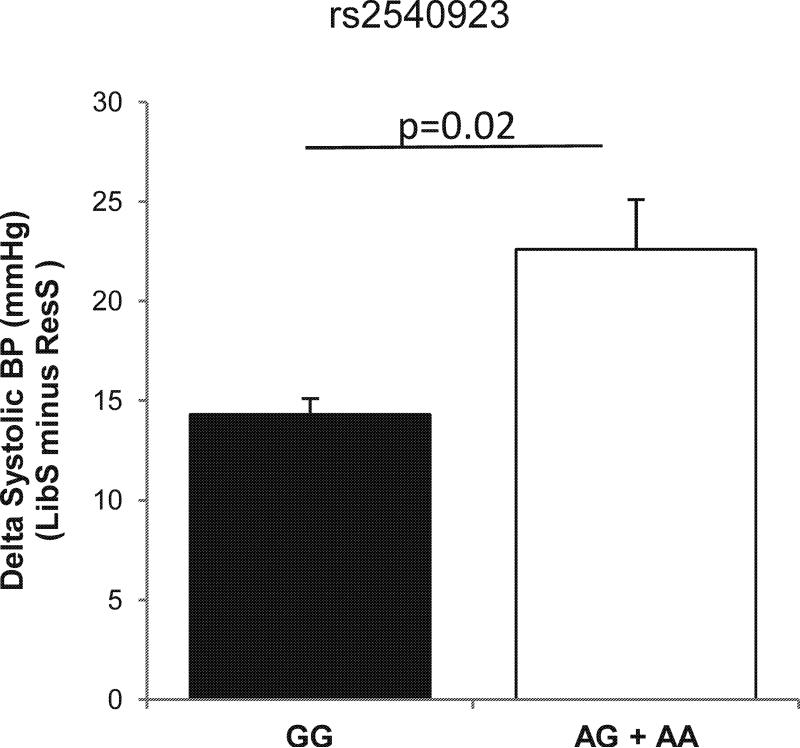
A haplotype analysis (Table 1) was first conducted to determine the association of each of the three haplotype blocks with salt sensitive BP. The global block analysis was significantly associated with SS of BP (pglobal=0.04). This association was entirely driven by haplotype number 3 rs2540923A|rs888083G|rs11678303G|rs6744560A|rs7562109A| rs10490658A|rs7573966A (p= 0.002) and specifically the first SNP within that haplotype (rs2540923). The relationship of the rs2540923 SNP with SS of BP was then assessed (Figure 6). Adjusting for age, gender, and BMI, minor allele carriers (AG/AA) for rs2540923 displayed 6 fold increased odds ratio for SS of BP versus major allele homozygotes (OR 6.25, 95% CI 1.70 – 20). There were no genotype associated differences in ALDO levels either on the LibS or ResS intakes.
Haplotype analyses for association with salt sensitive blood pressure in individuals with hypertension
| BLOCK 1 | Pglobal = 0.04* | ||
|---|---|---|---|
| Haplotype |
P value |
Frequency |
|
| GAGCGAA | 0.194 | 0.31 | |
| GGGAGAC | 0.974 | 0.52 | |
| AGGAAAA | 0.002* | 0.04 | |
| GGGCGAA | 0.369 | 0.03 | |
| GAGAGGA | 0.969 | 0.09 | |
| GGAAAAA | 0.725 | 0.01 | |
| rs2540923|rs888083|rs11678303|rs6744560|rs7562109|rs10490658|rs7573966 | |||
| BLOCK 2 | Pglobal=0.14 | ||
|---|---|---|---|
| Haplotype |
P value |
Frequency |
|
| CGA | 0.43 | 0.21 | |
| GGA | 0.98 | 0.02 | |
| CAA | 0.65 | 0.7 | |
| GAA | 0.01* | 0.05 | |
| CGG | 0.18 | 0.02 | |
| GAG | 0.69 | 0.61 | |
| rs17020071 |rs17497197|rs10490657 | |||
The primary SNP and allele driving the significant findings are in bold.
P value < 0.05
Figure 6.
SNP and SS of BP associations in a hypertensive population. Data represent mean ± SD.
Discussion
Knocking down the striatin gene in mice and polymorphic variants of the striatin gene in humans is associated with SS of BP. In the mice SS is not related to an alteration in ALDO production but may be related to alteration in its metabolism and effectiveness. MR expression in the kidney is increased as are the levels of expression of two downstream targets of MR activation---SGK1 and especially ENaC. Importantly, both in the kidney and the heart, a signature for a defect in ALDO's rapid, non-genomic pathway---decrease phosphorylation of Akt---is present. From this and a previous study, we have documented that dietary sodium restriction (that increases ALDO) and direct ALDO administration increases MR expression. Thus, on a liberal salt diet one would expect a decrease in expression of MR, SGK1 and ENaC. This did not occur in strn+/− mice, suggesting that striatin plays a role, as yet undefined, in modulating the normal effect of salt intake on this cascade. The net result is an inappropriately increased activity of sodium retaining mechanisms on a liberal salt diet and SS of BP.
Our previous studies documented in EA.hy926 cells, endothelial cell cultures that: 1) striatin coimmunoprecipitates with MR in vitro and in vivo 2) ALDO stimulates rapid phosphorylation of ERK1/2 that is prevented by knocking down striatin with siRNA; 3) ALDO stimulates striatin synthesis; and 4) knocking down striatin in vitro does not modify the transcript levels of classical genomic targets of ALDO, e.g. SGK1 and ENaC. In the present study, the cell culture data extend the molecules involved in striatin associated effects to include Akt 13, 21. Furthermore, the in vivo studies document that knocking down striatin levels has the same effect in the heart and kidney as in endothelial cells---reduction of phosphorylation of Akt.
To date the data supporting a role for striatin as a critical intermediate in the non-genomic effects of steroid hormones in general, and ALDO in particular has come from in vitro studies. The current study extends this support to the in vivo state and suggests that striatin is involved in regulating the organism's BP response to sodium intake 22. The mechanism(s) by which alterations in striatin leads to SS of BP does not appear to be related to genotype driven differences in PRA levels in humans (minor allele carriers of rs2540923) or Strn+/− mice or potassium levels in humans. Also the biosynthesis in ALDO from glomerulosa cells does not differ by genotype in the mice either with a change in sodium diet or in response to incubation with angiotensin II. Thus, striatin does not appear to be involved in mediating the external signals regulating ALDO secretion or its de novo production by glomerulosa cells. However, the effectiveness of ALDO seems to be increased as documented by three changes in the kidneys of the Strn+/− mice: a significant increase in renal MR mRNA expression associated with increased SGK1 and ENaC mRNA expressions. Thus, the data from this study suggest that excess MR activity underlies the SS of BP phenotype observed in Strn+/− mice.
Excess MR Expression could potentially come from the increased circulating ALDO levels in the Strn+/− mice studied on the LibS diet. Given the absence of an effect on the production of ALDO in the Strn+/− mice, altered metabolism may be the mechanism for the increased circulating ALDO. How this effect occurs, is uncertain. These results raise the possibility that the excess volume expansion leading to SS of BP in Strn+/− mice is secondary to increased renal expression of MR leading to increased SGK1 and ENaC and thus increased ENaC-mediated reabsorption of sodium in the renal collecting tubules.
An increasing body of data supports the concept that hypertension is not a disease but a heterogeneous syndrome, with a number of pathways leading to an increase in BP and more specifically to salt sensitive hypertension. Thus, GWAS studies have been nearly uniformly unsuccessful in identify risk genes and/or alleles, likely because of the heterogeneity of the patient population used. Thus, it is not surprising that there was no signal on 2p22.2 in the CHARGE and GlobalBP GWAS studies given the low frequency of the risk allele, the heterogeneity of the population and the lack of control of environmental factors critical for identifying the intermediate phenotype identified in this study.
There are limitations to these studies. First, in the human gene association study the sample size is relatively small which could play a role in driving our positive findings. However the study is strengthened by the strict environmental control imposed (medication washout, calculated diets, standardization in procedures and use of a central laboratory) 23. SNP rs2540923 is a tagging SNP and therefore likely is a marker in linkage disequilibrium with the functional SNP. Given the nature of the phenotype and the fact that this SNP is not in the coding region of the gene, suggests that the functional SNP is likely in the regulatory region of the gene. Given the similarity between the mouse and human phenotypes, it is likely that the functional SNP leads to a decrease in the expression of striatin, although there is no direct data to support this hypothesis. Second our cohort was restricted to Caucasians and additional studies are required to determine whether this phenotype will be observed in other racial groups. Thirdly, in contrast to the mice studies, the human studies did not demonstrate a difference in ALDO levels on the LibS diet. This lack of difference could be related to the level of expression of striatin associated with the polymorphic variants in the gene and/or other differences in the protocols used. Fourth, in the mice studies, while the molecular data are suggestive of the mechanism involved in the SS of BP, the evidence is somewhat indirect, particularly because of the differential effects on SGK1 levels between the culture endothelial cells and the Strn+/− mice. In addition protein levels were not assessed for MR, SGK1 or ENaC in this study, but others have suggested a correlation between changes in protein and mRNA of these molecules. However, it has previously been suggested that the non-genomic MR pathway may interact with genomic mechanisms to provide an integrated cellular response 5. Such would be more likely observed in vivo than in vitro. Finally all generalized knockout models suffer from the possibility that the observed phenotype is due to an effect in another disrupted gene or an effect in another tissue.
Perspectives
Striatin is a novel protein that interacts with and modulates the rapid, non-genomic activity of steroid receptors, including the mineralocorticoid receptor (MR), however its role in sodium, water, and blood pressure (BP) homeostasis is unknown. In the present study, we provide direct support for a role of striatin in modulating the organism's response to salt intake and suggest that an alteration in the MR and its downstream targets in the kidney may be the responsible mechanism(s). However, further studies are needed to elucidate the pathophysiological effects of striatin gene variants deficiency and whether striatin SNPs can be exploited to identify individuals who would benefit from sodium restriction and/or specific anti-hypertensive therapies.
Supplementary Material
Novelty and Significance.
What is New?
Striatin is a key intermediate in the non-genomic actions of aldosterone. Polymorphic variants in its gene in humans and knocking-down the gene in mice are associated with increased salt sensitivity of blood pressure (BP).
What is Relevant?
Salt sensitive hypertension is a major, heterogeneous subset of the hypertensive population whose cause(s) is (are) largely unknown. Identifying the underling mechanisms in the homogeneous sub-groups of this subset will aid in the development of more precise and specific treatment and prevention strategies. Variants in the expression of the striatin gene may underlie one of these homogeneous sub-groups.
Summary
In a novel striatin heterozygous knockout mouse, we demonstrated that reduction in striatin levels results in salt sensitivity of blood pressures and on a liberal salt diet was associated with increased plasma aldosterone levels and renal tissue expression of mineralocorticoid receptor, serum/glucocorticoid regulated kinase and epithelial sodium channel. By using a well-phenotyped Caucasian hypertensive cohort, we documented that genetic variance within the striatin gene was associated with salt sensitivity of blood pressures. Taken together, this study supports a role for striatin in modulating the response to salt intake and proposes that an alteration in mineralocorticoid receptor and its downstream targets may be possible mechanism.
Acknowledgments
We would like to thank all other investigators and staff of the HyperPATH Protocol as well as the Center for Clinical Investigation, Brigham and Women's Hospital's staff and participants at each protocol site, including the Clinical Investigation Centre, INSERM CIC 9201, Hôpital Européen Georges Pompidou, Paris France. Additionally, we would like to thank the Tissue Culture Facility at the University of North Carolina Lineberger Comprehensive Cancer Center for providing the EA.hy926 cells.
Source(s) of Funding
This work was supported by the National Institute of Health National Heart, Lung and Blood Institute Grants R01HL11476 (GHW), R01HL104032 (LHP), P50HL055000 (HyperPATH Cohort), R01HL096518 (JRR) and a NIH T32 training grant T32HL007609-27.
Footnotes
Conflict(s) of Interest/Disclosure(s)
NONE
References
- 1.Hurwitz S, Fisher ND, Ferri C, Hopkins PN, Williams GH, Hollenberg NK. Controlled analysis of blood pressure sensitivity to sodium intake: interactions with hypertension type. J Hypertens. 2003;21:951–959. doi: 10.1097/00004872-200305000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Williams GH, Hollenberg NK. Non-modulating hypertension. A subset of sodium-sensitive hypertension. Hypertension. 1991;17:I81–85. doi: 10.1161/01.hyp.17.1_suppl.i81. [DOI] [PubMed] [Google Scholar]
- 3.Nabika T, Nara Y, Ikeda K, Endo J, Yamori Y. Genetic heterogeneity of the spontaneously hypertensive rat. Hypertension. 1991;18:12–16. doi: 10.1161/01.hyp.18.1.12. [DOI] [PubMed] [Google Scholar]
- 4.Viengchareun S, Le Menuet D, Martinerie L, Munier M, Pascual-Le Tallec L, Lombes M. The mineralocorticoid receptor: insights into its molecular and (patho)physiological biology. Nucl Recept Signal. 2007;5:e012. doi: 10.1621/nrs.05012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev. 2003;83:965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- 6.Funder JW. Non-genomic actions of aldosterone: role in hypertension. Curr Opin Nephrol Hypertens. 2001;10:227–230. doi: 10.1097/00041552-200103000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Bernelot Moens SJ, Schnitzler GR, Nickerson M, Guo H, Ueda K, Lu Q, Aronovitz MJ, Nickerson H, Baur WE, Hansen U, Iyer LK, Karas RH. Rapid estrogen receptor signaling is essential for the protective effects of estrogen against vascular injury. Circulation. 2012;126:1993–2004. doi: 10.1161/CIRCULATIONAHA.112.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grossmann C, Gekle M. New aspects of rapid aldosterone signaling. Mol Cell Endocrinol. 2009;308:53–62. doi: 10.1016/j.mce.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Ueda K, Lu Q, Baur W, Aronovitz MJ, Karas RH. Rapid estrogen receptor signaling mediates estrogen-induced inhibition of vascular smooth muscle cell proliferation. Arterioscler Thromb Vasc Biol. 2013;33:1837–1843. doi: 10.1161/ATVBAHA.112.300752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Q, Pallas DC, Surks HK, Baur WE, Mendelsohn ME, Karas RH. Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor alpha. Proc Natl Acad Sci U S A. 2004;101:17126–17131. doi: 10.1073/pnas.0407492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moqrich A, Mattei MG, Bartoli M, Rakitina T, Baillat G, Monneron A, Castets F. Cloning of human striatin cDNA (STRN), gene mapping to 2p22-p21, and preferential expression in brain. Genomics. 1998;51:136–139. doi: 10.1006/geno.1998.5342. [DOI] [PubMed] [Google Scholar]
- 12.Castets F, Rakitina T, Gaillard S, Moqrich A, Mattei MG, Monneron A. Zinedin, SG2NA, and striatin are calmodulin-binding, WD repeat proteins principally expressed in the brain. J Biol Chem. 2000;275:19970–19977. doi: 10.1074/jbc.M909782199. [DOI] [PubMed] [Google Scholar]
- 13.Pojoga LH, Coutinho P, Rivera A, Yao TM, Maldonado ER, Youte R, Adler GK, Williams J, Turchin A, Williams GH, Romero JR. Activation of the mineralocorticoid receptor increases striatin levels. Am J Hypertens. 2012;25:243–249. doi: 10.1038/ajh.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutinho P, Vega C, Pojoga LH, Rivera A, Prado GN, Yao TM, Adler G, Torres-Grajales M, Maldonado ER, Ramos-Rivera A, Williams JS, Williams G, Romero JR. Aldosterone's rapid, nongenomic effects are mediated by striatin: a modulator of aldosterone's effect on estrogen action. Endocrinology. 2014;155:2233–2243. doi: 10.1210/en.2013-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pojoga LH, Williams JS, Yao TM, Kumar A, Raffetto JD, do Nascimento GR, Reslan OM, Adler GK, Williams GH, Shi Y, Khalil RA. Histone demethylase LSD1 deficiency during high-salt diet is associated with enhanced vascular contraction, altered NO-cGMP relaxation pathway, and hypertension. Am J Physiol Heart Circ Physiol. 2011;301:H1862–1871. doi: 10.1152/ajpheart.00513.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuengsamarn S, Garza AE, Krug AW, Romero JR, Adler GK, Williams GH, Pojoga LH. Direct renin inhibition modulates insulin resistance in caveolin-1-deficient mice. Metabolism. 2013;62:275–281. doi: 10.1016/j.metabol.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braley LM, Menachery AI, Yao T, Mortensen RM, Williams GH. Effect of progesterone on aldosterone secretion in rats. Endocrinology. 1996;137:4773–4778. doi: 10.1210/endo.137.11.8895346. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins PN, Lifton RP, Hollenberg NK, Jeunemaitre X, Hallouin MC, Skuppin J, Williams CS, Dluhy RG, Lalouel JM, Williams RR, Williams GH. Blunted renal vascular response to angiotensin II is associated with a common variant of the angiotensinogen gene and obesity. J Hypertens. 1996;14:199–207. doi: 10.1097/00004872-199602000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins PN, Hunt SC, Jeunemaitre X, Smith B, Solorio D, Fisher ND, Hollenberg NK, Williams GH. Angiotensinogen genotype affects renal and adrenal responses to angiotensin II in essential hypertension. Circulation. 2002;105:1921–1927. doi: 10.1161/01.cir.0000014684.75359.68. [DOI] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Pojoga LH, Yao TM, Opsasnick LA, Garza AE, Reslan OM, Adler GK, Williams GH, Khalil RA. Dissociation of hyperglycemia from altered vascular contraction and relaxation mechanisms in caveolin-1 null mice. J Pharmacol Exp Ther. 2014;348:260–270. doi: 10.1124/jpet.113.209189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Good DW. Nongenomic actions of aldosterone on the renal tubule. Hypertension. 2007;49:728–739. doi: 10.1161/01.HYP.0000259797.48382.b2. [DOI] [PubMed] [Google Scholar]
- 23.Underwood PC, Sun B, Williams JS, Pojoga LH, Raby B, Lasky-Su J, Hunt S, Hopkins PN, Jeunemaitre X, Adler GK, Williams GH. The association of the angiotensinogen gene with insulin sensitivity in humans: a tagging single nucleotide polymorphism and haplotype approach. Metabolism. 2011;60:1150–1157. doi: 10.1016/j.metabol.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.