Abstract
Purpose:
To identify a new clinical marker based on quantitative kinetic image features analysis and assess its feasibility to predict tumor response to neoadjuvant chemotherapy.
Methods:
The authors assembled a dataset involving breast MR images acquired from 68 cancer patients before undergoing neoadjuvant chemotherapy. Among them, 25 patients had complete response (CR) and 43 had partial and nonresponse (NR) to chemotherapy based on the response evaluation criteria in solid tumors. The authors developed a computer-aided detection scheme to segment breast areas and tumors depicted on the breast MR images and computed a total of 39 kinetic image features from both tumor and background parenchymal enhancement regions. The authors then applied and tested two approaches to classify between CR and NR cases. The first one analyzed each individual feature and applied a simple feature fusion method that combines classification results from multiple features. The second approach tested an attribute selected classifier that integrates an artificial neural network (ANN) with a wrapper subset evaluator, which was optimized using a leave-one-case-out validation method.
Results:
In the pool of 39 features, 10 yielded relatively higher classification performance with the areas under receiver operating characteristic curves (AUCs) ranging from 0.61 to 0.78 to classify between CR and NR cases. Using a feature fusion method, the maximum AUC = 0.85 ± 0.05. Using the ANN-based classifier, AUC value significantly increased to 0.96 ± 0.03 (p < 0.01).
Conclusions:
This study demonstrated that quantitative analysis of kinetic image features computed from breast MR images acquired prechemotherapy has potential to generate a useful clinical marker in predicting tumor response to chemotherapy.
Keywords: dynamic contrast-enhanced breast magnetic resonance imaging, tumor response to neoadjuvant chemotherapy, quantitative image feature analysis, assessment of breast cancer prognosis
1. INTRODUCTION
Breast cancer is the most prevalent cancer and the second highest cause of cancer death in women in the United States.1 Early detection and effective treatment of breast cancer play an important role to reduce mortality rates of breast cancer patients.2,3 Although mammography is well recognized as the most cost-effective imaging modality for breast cancer screening and early detection, the performance (including both sensitivity and specificity) of screening mammography is not satisfactory,4,5 in particular, for younger women with dense breast tissues. As a result, other new imaging modalities have also been developed and tested for improving breast cancer screening and detection. Among them, dynamic contrast-enhanced breast magnetic resonance imaging (DCE-MRI) has shown superior sensitivity in detecting mammography-occult cancers and is recommended by the American Cancer Society as an adjunct screening tool to mammography for women at an elevated breast cancer risk (e.g., >20%–25% of lifetime risk) since 2007.6 Currently, breast DCE-MRI has been playing an important role in the clinical management of breast cancer in screening, diagnosis, and assessment of treatment efficacy.7,8
In order to assist radiologists to more accurately and efficiently read and interpret breast MR images, great research effort has been made in the last two decades to develop and test computer-aided detection (CAD) schemes of breast MR images.9,10 Besides the tumor morphological features (i.e., tumor volume, shape, and boundary spiculation), the quantitative analysis of the tumor kinetic features extracted from the postcontrast-enhanced MR images also demonstrated that malignant breast tumors usually have fast wash-in and wash-out ratios, as well as an increased heterogeneity in the contrast enhancement kinetics.11,12 As a result, both morphological and kinetic image features have been analyzed and used in CAD schemes for detecting and classifying suspicious breast tumors depicted on breast MR images. Recently, a number of studies demonstrated that image features computed from the background parenchymal enhancement (BPE) of breast MR images carried unique and/or supplementary information that enables to assess risk of developing breast cancer13 and increase discriminatory power in classifying between malignant and benign breast tumors depicted on breast MR images.14,15
In treating advanced stage breast cancer patients, neoadjuvant chemotherapy is commonly used before breast cancer surgery. The tumor response to the chemotherapy is typically evaluated by comparing tumor size and kinetic feature variation using breast MRI examinations taken before and after chemotherapy based on the response evaluation criteria in solid tumors (RECIST) guideline.16,17 In order to more effectively predict tumor response to the neoadjuvant chemotherapy and help clinicians (e.g., oncologists and surgeons) make optimal treatment decisions for the patients, we in this study investigated the feasibility of applying a new CAD-based quantitative breast MR image feature analysis method to assist prediction of tumor response to neoadjuvant chemotherapy. For this purpose, we developed a new CAD scheme to automatically detect the chest wall and segment breast regions and tumor regions depicted on the breast MR images. CAD scheme then computed a series of kinetic image features extracted from both the segmented breast tumor and background parenchymal tissue regions. By analysis and selection of optimal features computed from the breast MR images acquired before performing neoadjuvant chemotherapy, our goal is to identify a new potentially useful clinical marker to quantitatively predict tumor response to the chemotherapy. The details of this study design and experimental results are reported in Secs. 2 and 3.
2. MATERIALS AND METHODS
2.A. A breast MR image dataset
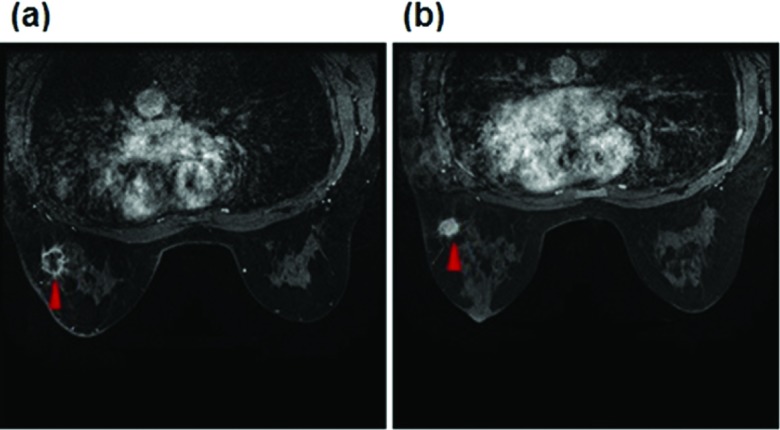
Based on our institutional review board (IRB) approved image data collection protocol, we retrospectively assembled an image dataset that includes deidentified breast MR images acquired from 68 breast cancer patients. Each patient had at least two sequential breast DCE-MRI examinations taken before and after neoadjuvant chemotherapy. Based on existing clinical records that were generated using RECIST guidelines to examine and compare breast tumor size change measured from the breast MR images acquired pre- and post-treatment of the same patient, we used the same case classification criterion as reported in the literature18 to divide the 68 patients (or cases) into two groups, which we named as (1) a “complete response” (CR) group in which the targeted or tracked breast tumors almost disappear in the postchemotherapy breast MR images and (2) a “nonresponse” (NR) group in which the tumor diameter either increased or decreased by less than 50% between pre- and postchemotherapy breast MR images. Among 68 patients, 25 were assigned in CR group in which the enhanced contrast levels inside the tumor volume were reduced to the level of normal enhanced background parenchymal tissues and 43 were categorized in NR group in which a high fraction of contrast enhancement areas remained in the original segmented tumor regions of the postchemotherapy breast MR images. In the NR group, the average tumor diameter measured based RECIST guidelines decreases by 13.3%. Figure 1 shows an example of two matched image slices acquired from pre- and post-treatment breast MRI examinations of one patient in the “NR” group. In this example, tumor morphological volume decreased based on RECIST guidelines, but the active contrast enhancement volume increased after neoadjuvant chemotherapy.
FIG. 1.
An example showing two matched breast MRI slices acquired from before (a) and after (b) neoadjuvant chemotherapy of one patient assigned in the NR group. A matched malignant tumor is marked by a red arrow in two image slices.
In this dataset, mean and standard deviations of the women’s age are 49.2 ± 11.7 and 51.1 ± 9.1 for groups of CR and NR cases, respectively, indicating that the majority of women whose breast DCE-MRI examination images were selected in this study are relatively younger, i.e., <55 yr old. In this study, only the axial view of the breast MR images acquired prechemotherapy of each patient was selected from the originally recorded image data. Then, in each breast MR image scan or examination, our CAD scheme was applied to process two MR image series that include the one acquired before injection of gadopentate dimeglumine (Gd-DTPA) contrast agent and the one of the first postcontrast agent injection. According to the different breast sizes, the number of MRI slices in each series varies for different patients, which ranges from 58 to 104. Every MRI slice is a data matrix of 512 × 512 pixels with a slice thickness of 2.6 mm and pixel size of 0.58 mm in each direction. Thus, for every patient, two sets of axial view breast MR images acquired before neoadjuvant chemotherapy treatment were involved in this study to perform the kinetic feature computation and data analysis process.
2.B. A CAD scheme
We developed and tested a new CAD scheme of breast MR images, which includes the following image processing and data analysis steps, namely, (1) detection of the chest wall depicted on breast MR images to remove all pixels (regions) behind the chest wall; (2) subtraction of two sets of matched breast MRI slices acquired in the breast MRI scans performed pre- and postinjection of contrast agent; (3) segmentation of each targeted malignant tumor and separation of its contrast-enhanced and necrotic area (without contrast enhancement); and (4) computation of relevant kinetic image features from both segmented tumor and BPE regions.
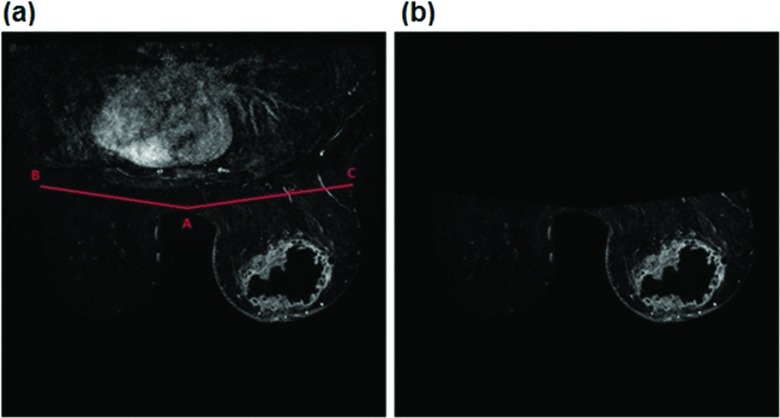
First, although many existing CAD schemes of breast MR images do not perform the task of breast area segmentation, in order to automatically compute BPE features, automated detection and segmentation of breast regions are required in our CAD scheme. A number of methods to segment breast regions from breast MR images have been reported in the previous studies.10,19–21 For example, one study reported a fuzzy c-means clustering based segmentation method, which required manually defined landmarks to distinguish between breast parenchyma and chest wall by assuming that there is an adequate retromammary fat plane.19 Another study used a straight line fitting method to separate the breast and chest wall, which may eliminate part of the fibroglandular tissue at axillary tails.20 To overcome these limitations and also develop a simple and robust approach for our application purpose, we applied a CAD scheme to detect the chest wall, which operates on the intensity and texture variation of the image processing techniques that use a thresholding approach followed by morphological operations.10 Taking Fig. 2 as an example, our CAD scheme first searches for and detects a highest point located on a concave curve of the chest skin between the left and right breasts. Next, the detected point is moved up by 5–10 pixels to define a new middle point A between the left and right breasts. Then, from the point A, our CAD scheme fits two oblique lines with 10° angles from the left and right horizontal lines, in order to transect the two breasts. The intersection points between these two (left and right) lines and the breast skin surface are defined as points B and C, respectively. Finally, these two fitted lines are approximated as the chest wall. All pixels behind these two fitted lines are removed and excluded in computing BPE features by the CAD scheme. Although we recognize that this simple line fitting method may exclude a very small fraction of breast fibroglandular tissue, its impact on computing BPE features is minimal and can be ignored.
FIG. 2.
Illustration of chest wall detection and breast region segmentation. In this figure, the detected middle point A between the left and right breasts and two fitted oblique lines (B and C) are marked in (a), and the final segmented breast region, which is used to compute the BPE features, is shown in (b).
Second, our CAD scheme registers the segmented breast areas depicted on the sequential breast MRI scans before and after the injection of the Gd-DTPA contrast agent. Motion artifacts due to the patients’ physiologic movement (i.e., heart beating and breathing) could affect the accuracy in detection and analysis of kinetic image features computed from the sequentially scanned DCE-MRI images. However, due to the small nonrigid breast motion and the limited pixel resolution of the MR images, the majority of motion occurs in the subpixel range. As a result, using a whole pixel-based image registration (either rigid or nonrigid without known “ground-truth”)14,22 is very difficult and unreliable. Hence, for this proof-of-concept study, we simply registered the breast MR images acquired from two sequential MR imaging scans before and after injection of the contrast agent based on the order of slice order in the two scanning sequences. Specifically, two series of image sequences, before and after injection, with the same number of slices were subtracted from each other to generate a set of new contrast enhancement map based images. These new subtraction images were then used for tumor detection and kinetic feature computation.
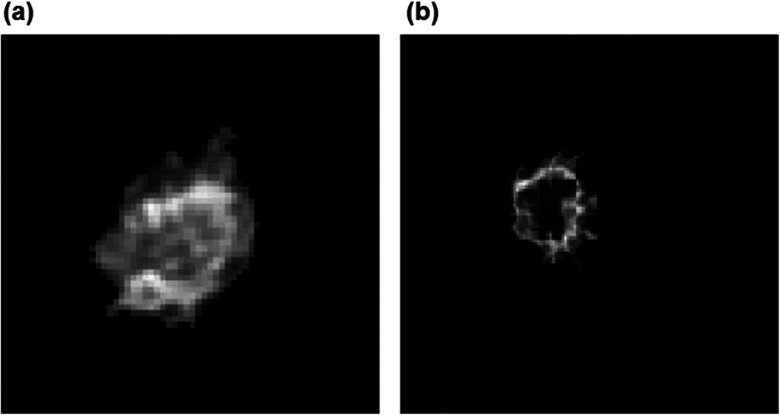
Third, from the contrast enhancement map (or subtraction) images, the CAD scheme automatically searches for and segments the contrast-enhanced regions. An algorithm including the scaling, morphological (erosion and dilation), and labeling operations is implemented in the CAD scheme. We initially rescaled our image from 0 to the maximum value of the image to 0–1 and then applied a threshold value of 0.5 to detect the tumor area. Morphological operations (i.e., erosion and dilation) were applied to modify the segmentation result. We also applied a region labeling method to group the pixels with value one into the connected blobs and deleted the isolated small blobs in order to detect the actual tumor-related blobs. Next, CAD scheme also automatically detected and extracted necrotic regions inside the segmented tumor. For example, Fig. 3 shows two breast MR image slices selected from two cases. The first breast MR image depicts a tumor with significant contrast enhancement of all pixels inside the tumor [Fig. 3(a)] and the second breast MR image displays a tumor with substantial necrotic regions inside the tumor [Fig. 3(b)].
FIG. 3.
An example of two breast tumors including (a) a tumor without necrotic area and (b) a tumor with necrotic area inside.
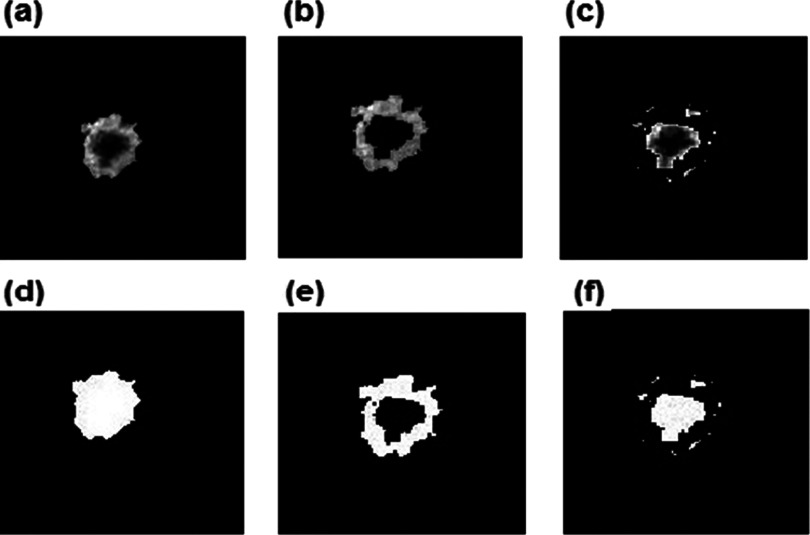
Our CAD scheme applied an Otsu’s thresholding method on the completely segmented tumor to detect and classify the active (high/significantly enhanced) and necrotic (low/poorly enhanced) areas. CAD scheme assigned all involved pixels into either active tumor or necrotic area based on the pixel contrast enhancement values, which are greater or smaller than an assigned threshold level (the threshold level was 50% of the maximum pixel value of each slice). Figure 4 shows an example of the contrast-enhanced pixels of a tumor with a large internal necrotic area. Finally, through the above steps or procedures, CAD scheme classified each pixel within the segmented breast area into one of three categories, namely, (1) breast tumor detected, (2) active contrast-enhanced pixels inside a targeted breast tumor, or (3) inactive (nonenhanced) pixels inside the tumor (necrosis).
FIG. 4.
The result of the detection process for tumor, enhanced, and dead area shown as image slices: (a) tumor detection, (b) enhanced area detection, and (c) necrotic area detection with their corresponding binary images (d)–(f), respectively.
Finally, the CAD scheme computed a set of kinetic image features from the mapped images generated by the subtraction between the baseline MRI scan (before injection of contrast agent) and the first sequential postcontrast injection MRI scan. A total of 39 features were computed, which is divided into five groups. These groups include image features computed from (1) the entire tumor area, (2) the active contrast-enhanced tumor area, (3) the tumor necrotic area, (4) the entire background parenchymal region of two breasts, and (5) the absolute value of bilateral BPE feature difference computed from the left and right breasts. Table I summarizes these 39 image features in the five feature groups. Apart from the kinetic image features that were directly computed from the pixel value distribution of the subtraction images, several tumor morphological features were also included in Table I. For example, the maximum value of , where (cx, cy) represents the central point of the tumor in each slice and (i, j) represents the extracted tumor pixels, was defined as the maximum value of tumor diameter. Tumor shape factor was defined as the number of pixels in the tumor surface divided by the tumor volume. For the last two groups of the BPE features, CAD used the same method as reported in our previous study23 to sort all pixels assigned in BPE region from the highest to the lowest pixel values based on the computed contrast enhancement values. CAD then selected a small percentage (e.g., 1% and 5% as shown in Table I) of sorted pixels with higher contrast enhancement values from the segmented BPE regions and computed the kinetic BPE image features.
TABLE I.
Summary of computed kinetic image features in five groups.
| Feature group | Feature number | Description |
|---|---|---|
| Tumor area | 1–7 | Volume, average intensity, maximum pixel intensity, standard deviation, and skewness of tumor pixel intensity, maximum value of tumor radius, and shape factor |
| Enhanced area | 8–11 | Volume, average intensity, standard deviation, and skewness of contrast-enhanced pixel intensity |
| Necrotic area | 12–16 | Volume, average intensity, standard deviation, and skewness of low-enhanced pixel intensity, ratio of necrotic volume over tumor volume |
| Background parenchymal areaa | 17–34 | Average intensity, standard deviation, skewness, maximum pixel intensity, average value of top 1%, and average value of top 5% of pixel values |
| Absolute bilateral difference of BP areab | 35–39 | Average intensity, standard deviation, skewness, average value of top 1%, and average value of top 5% of pixel values |
These features are computed from three different regions—background parenchymal region of the whole (left and right) breast regions, left breast and right breast.
Absolute bilateral feature difference of BPE between the left and right breasts.
2.C. Feature analysis and performance assessment
In this study, we used two approaches to perform image feature analysis and examine performance or discriminatory power of predicting breast tumor response to the neoadjuvant chemotherapy. In the first approach, we used an area under a receiver operating characteristic (ROC) curve (AUC) as an evaluation index to assess the discriminatory power of each image feature. ROC curves and AUCs were computed using a publically available ROC curve fitting program (rockit, http://www-radiology.uchicago.edu/krl/, University of Chicago). We also computed and compared the correlation coefficients of individual image features to identify nonredundant image features. Based on the above image feature analysis procedures, we selected a small set of highly performing and nonredundant image features. We then tested a number of image feature fusion methods (e.g., average, weighted combination, and selection of the maximum or minimum feature value)24 to generate a new classification score and used this score as a potential breast MR image based marker for predicting tumor response to neoadjuvant chemotherapy.
In the second approach, we built and tested a statistical machine learning classifier based method to select optimal features and predict tumor response to neoadjuvant chemotherapy. All experiments in this approach were performed using a publicly available weka data mining and machine learning software package.25 In the weka Explorer window, we chose a specific machine learning classifier (or function), namely, the “AttributeSelectedClassifier,” which integrates an artificial neural network (ANN) as a base classifier and a wrapper subset evaluator (WSE) to guide feature or attribute selection. This integration takes a search algorithm and evaluator next to the base classifier, which makes the feature selection process transparent and the base classifier operate only in a reduced optimal feature space.26 The classifier was trained using a standard leave-one-case-out (LOCO) method.27 In each of the LOCO training/testing iterations, one case was selected as an independent testing case and the remaining cases in the dataset were used to form a training dataset. The AttributeSelectedClassifier was applied to all training cases to search for optimal features from the entire feature pool and train an ANN based on the WSE results. The trained ANN was then applied to the testing case and generates a classification score for the testing case. As a result, in each LOCO training and testing iteration cycle, different image features may be selected from the initial pool of 39 features and used to build a corresponding ANN.
To reduce the potential training bias and yield higher robust levels of the classifier performance, we also took two additional measures in training and testing each ANN classifier. (1) Due to the imbalanced case numbers in the two classes (25 CR cases vs 43 NR cases), we applied a well-examined synthetic minority oversampling technique called SMOTE28 to add a set of synthetic data in the CR case group to generate a more balanced training dataset of two class cases. Using this technique, we doubled the test cases in the CR class from 25 to 50 and increased the number of cases to 93 in the training dataset. (2) Each ANN uses a common three-layer structure. The first layer is an input layer that has ni neurons connecting to the number of NF = 5 selected input features. The second layer is a hidden layer that has the number of nh = (NF + NC)/2 hidden neurons, where NC is the number of classes (NC = 2 for “CR” and NR classes in this study). The third layer is an output layer with one output neuron that produces a probability based classification score ranging from 0 to 1 indicating the likelihood of the case being a NR class. Each ANN was trained using the default parameters of weka program. In brief, the number of ANN training seed, momentum, and learning rate are 0, 0.2, and 0.3, respectively. The ANN training iteration number is 500.
After completing a LOCO training and testing process of the ANN-based classifier, we removed 25 synthetic CR cases and assessed classification performance using the original 68 cases in our dataset. The AUC value was used as a performance prediction index. We also applied an operation threshold (T = 0.5) on the ANN-generated classification scores to divide 68 testing cases into two predicted (CR and NR) case groups and then assessed the prediction accuracy (including the sensitivity and specificity) of applying this ANN-based classifier to the testing dataset.
3. RESULTS
Table II summarizes and compares a number of basic patient characteristics and commonly used breast MR image features between the two chemotherapy response groups (CR and NR). There is no statistically significant difference of each of these image features between the CR and NR case groups (p > 0.05 computed using DeLong’s test). The result indicates that the conventional feature assessment based on tumor volume and average tumor contrast enhancement using breast MR images acquired prechemotherapy has lower discriminatory power to predict tumor response to neoadjuvant chemotherapy.
TABLE II.
Comparisons of patient and tumor characteristics between the CR and NR groups.
| CR (N = 25) | NR (N = 43) | p-value | |
|---|---|---|---|
| Mean patient’s age | 9.2 ± 11.7 | 51.1 ± 9.1 | 0.53 |
| Mean tumor volume | 5077.20 ± 5778 | 8453.93 ± 15137 | 0.20 |
| Mean tumor necrotic volume | 2383.6 ± 3139.1 | 4478.7 ± 9045.2 | 0.27 |
| Mean ratio between necrosis and total tumor volumes | 41% ± 13% | 43% ± 16% | 0.58 |
| Mean tumor diameter (RECIST) | 440.3 ± 160.0 | 439.9 ± 145.0 | 0.99 |
| Mean average tumor enhancement | 475.5 ± 126.0 | 451.5 ± 126.0 | 0.46 |
| Mean tumor enhancement STD | 122.0 ± 16.6 | 122.4 ± 18.9 | 0.92 |
| Mean enhancement area average | 585.6 ± 144.8 | 568.9 ± 154.0 | 0.66 |
After applying ROC analysis on each of the 39 features (as listed in Table I) using rockit program, 10 features yielded AUC > 0.6 in classifying between the CR and NR case groups (as shown in Table III). Table IV shows the correlation coefficients among these ten “highly performed” features. From the comparison result, we then selected five low-redundancy image features from tumor and background parenchymal regions with correlation lower than 0.5 (i.e., F3, F8, F15, F17, and F35—the feature description is given in Table I). When using a maximum score based fusion process to fuse (combine) these five features (using the maximum value among the five selected features after data normalization) to classify between the CR and NR case groups, classification performance significantly increased to AUC = 0.85 ± 0.05 as compared to the performance of each individual feature displayed in Table III (p < 0.05).
TABLE III.
Kinetic image features of tumor and background parenchymal regions with AUC > 0.6 (the individual feature description is given in Table I).
| Feature | AUC | Feature | AUC |
|---|---|---|---|
| F2 | 0.679 ± 0.064 | F12 | 0.626 ± 0.071 |
| F3 | 0.683 ± 0.071 | F15 | 0.606 ± 0.075 |
| F8 | 0.604 ± 0.072 | F17 | 0.671 ± 0.068 |
| F9 | 0.654 ± 0.069 | F18 | 0.611 ± 0.073 |
| F10 | 0.778 ± 0.066 | F35 | 0.713 ± 0.065 |
TABLE IV.
Correlation coefficients between each pair of ten kinetic image features (the individual feature description is given in Table I).
| F2 | F3 | F8 | F9 | F10 | F12 | F15 | F17 | F18 | |
|---|---|---|---|---|---|---|---|---|---|
| F3 | 0.737 | ||||||||
| F8 | 0.086 | 0.319 | |||||||
| F9 | 0.878 | 0.920 | 0.147 | ||||||
| F10 | 0.718 | 0.744 | 0.041 | 0.863 | |||||
| F12 | −0.047 | 0.382 | 0.872 | 0.163 | 0.130 | ||||
| F15 | −0.112 | −0.343 | −0.595 | −0.181 | −0.077 | −0.620 | |||
| F17 | 0.415 | 0.519 | 0.168 | 0.476 | 0.301 | 0.117 | −0.064 | ||
| F18 | 0.407 | 0.510 | 0.207 | 0.470 | 0.282 | 0.158 | −0.136 | 0.976 | |
| F35 | 0.290 | 0.442 | 0.533 | 0.431 | 0.413 | 0.560 | −0.329 | 0.293 | 0.320 |
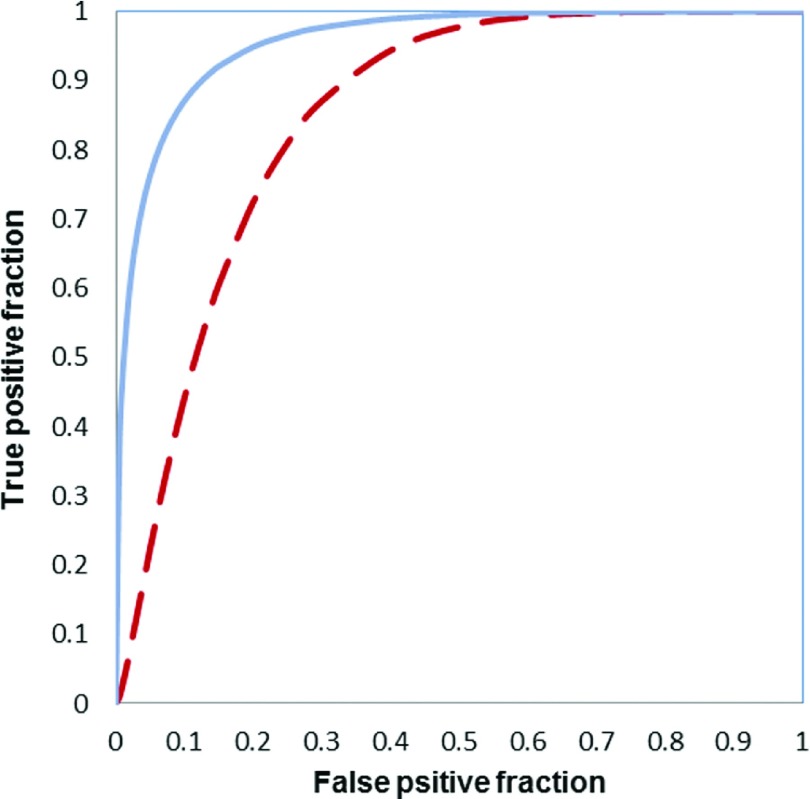
Table V lists 11 features that were selected at least 10% times in 68 LOCO training/testing iteration cycles (discarding the synthetic cases generated by SMOTE algorithm). The top five selected features (with the highest percentage of the selection) were average contrast enhancement and standard deviation of contrast enhancement inside an entire tumor region (F2 and F4), standard deviation of contrast enhancement in the active tumor region (excluding necrotic region) (F10), average pixel value of necrotic regions (F13), and ratio of necrotic volume over tumor volume (F16). The ANN-based classifier yielded an AUC value of 0.96 ± 0.03, which is significant higher than AUC = 0.85 ± 0.05 yielded using a simple feature fusion method (p < 0.01). Figure 5 shows and compares two ROC curves generated using the classification scores of the ANN-based classifier and feature fusion method. In addition, Table VI displays a confusion matrix that was obtained by applying an operating threshold of 0.5 on the classification scores generated by the ANN-based classifier. The overall prediction accuracy was 94% with a sensitivity of 88% (22/25) at a specificity of 98% (42/43).
TABLE V.
A list of ten image features that were selected in ≥10% of LOCO training and testing iteration cycles to test 68 testing cases in our dataset.
| Feature | Percentage (%) | Feature | Percentage (%) |
|---|---|---|---|
| F2 | 88 (60/68) | F10 | 100 (68/68) |
| F3 | 44 (30/68) | F13 | 87 (59/68) |
| F4 | 100 (68/68) | F14 | 15 (10/68) |
| F5 | 12 (8/68) | F16 | 44 (30/68) |
| F6 | 21 (14/68) | F17 | 10 (7/68) |
| F9 | 13 (9/68) |
FIG. 5.
Two ROC curves generated using the classification scores of the ANN-based classifier (solid curve) and feature fusion method (dashed curve).
TABLE VI.
A confusion matrix of prediction scores generated using an ANN-based classifier that was trained using five selected image features.
| Prediction result | CR | NR |
|---|---|---|
| Actual cases | ||
| CR | 22 | 3 |
| NR | 1 | 42 |
4. DISCUSSION
In this study, we developed and tested a new type of CAD scheme that aims to predict tumor response to neoadjuvant chemotherapy. We also demonstrated that similar to the use of the tumor texture features computed from breast MR images,18 using quantitative kinetic image features computed from both tumor and background parenchymal enhancement could also generate a new clinical marker with a high discriminatory power to predict breast tumor response to neoadjuvant chemotherapy using the breast MR images acquired before the neoadjuvant chemotherapy. If the result of this study can be validated in future large prospective studies, this new clinical marker has potential to assist clinicians (i.e., oncologists and breast surgeons) in selecting an optimal or personalized cancer treatment plan for different individual patients with different prognosis status. This will result in the improvement of cancer treatment efficacy and benefits the patients.
Since our new CAD scheme is not another conventional CAD scheme to detect breast lesions depicted on breast MR images, it has a number of unique characteristics. First, our scheme applies a simple but robust algorithm to segment the breast region by removing all pixels behind the chest wall. Although this easy-to-implement approach may eliminate a small fraction of background parenchymal tissues, it does not have significant impact in computing the BPE features. Second, our CAD scheme not only segments the entire tumor regions but it also detects and segments necrotic regions inside the tumor. As a result, we are able to compute a number of new image features to better describe the tumor characteristics. Third, we applied an AttributeSelectedClassifier function in weka software package to train and test an ANN-based machine learning classifier by integrating the “optimal” feature selection process within the LOCO training and testing iterations to predict tumor response to the chemotherapy.
From the experimental results, we can also make a number of new observations in supporting the advantages and potential clinical utility of applying a quantitative image feature analysis method to predict tumor response to chemotherapy. First, in current clinical practice, tumor size and average contrast enhancement are two important parameters visually assessed from breast MR images for cancer diagnosis and prognosis assessment. However, using quantitative image feature analysis methods, we are able to identify a number of new image features that have higher discriminatory power to predict tumor response to chemotherapy. For example, we observed that the heterogeneity of tumor contrast enhancement represented by the standard deviation of the contrast enhancement on the active tumor region (F10) had the highest discriminatory power (or AUC value as shown in Table III), which was also one of the two features that were selected in all LOCO training/testing cycles to build the ANN-based classifier (100% in Table V). Although this is an important phenotype based clinical marker, which correlates well with biologic characteristics of the tumor, it cannot be accurately and reliably evaluated using a visual or subjective evaluation method.
Second, we observed that tumor necrosis was an important tumor characteristic that may also affect tumor response to chemotherapy. The kinetic features computed from the entire tumor regions and only the active tumor regions (after excluding the necrotic regions) are not equal or highly correlated. Their performance in predicting tumor response to the chemotherapy is also different. For example, to more accurately evaluate tumor size (volume) and tumor density heterogeneity, we should use the features computed from only the active tumor regions.
Third, since previous studies have shown that BPE features enabled to help predict cancer risk13 and improve tumor diagnosis,14,15 in this study we observed that BPE features (i.e., F17 and F35 as shown in Fig. 5) may also have potential to help improve performance of predicting tumor response to chemotherapy. However, no BPE features were selected by WSE in LOCO training/testing iteration cycles for more than 10% of times (cycles). This is probably due to limited characteristics of our small testing dataset. Further investigation using large datasets is needed.
In summary, this is a preliminary proof-of-concept study to demonstrate the feasibility of developing a new clinical marker based on quantitative breast MR image feature analysis to predict tumor response to neoadjuvant chemotherapy. The study also has a number of limitations. First, the size of the dataset used is small. Thus, the robustness of the classifier performance needs to be validated in future studies with large and diverse image datasets. Second, the classification target of this study is limited to the CR and NR case groups based on RECIST guidelines. This tumor response criterion may not highly correlate to the actual prognosis of the patients assessed using other evaluation criteria (i.e., progression-free survival or overall survival). Hence, whether and how applying this new quantitative breast MR image feature analysis concept and scheme to predict patients’ prognosis using other evaluation criteria needs to be investigated in future studies. Third, a nonrigid (or deformable) image registration algorithm was not applied in our CAD scheme to register MR images acquired from the pre- and postcontrast enhancement scans. Although there is no ground-truth to evaluate the accuracy of breast MR image registration, how to identify and compute kinetic image features that are invariant or less sensitive to the small nonrigid motion (in the subpixel range) of the breasts during the sequential breast MRI scans needs to be investigated further. Fourth, due to the small dataset, only a simple machine learning classifier of ANN with a set of fixed configuration (e.g., the number of hidden neurons) and training parameters was trained and tested in this study. As we reported in our other CAD studies using a large dataset of digital mammograms,29,30 once a larger and diverse dataset of breast MR images is available, other better and more “deep learning” based machine learning methods and tools can be developed and tested for this purpose. Last, in this study, we used RECIST criteria to divide the cases into two classes (CR and NR). How the CR defined by RECIST relates to the actual pathologic complete response was unknown (not investigated) in this study, which may be more important in the clinical practice for oncologists to select optimal treatment strategy for the individual patients.
Despite these limitations, we believe that as a first study that aims to develop a new quantitative image marker based on diagnostic breast MR images to predict tumor response to neoadjuvant chemotherapy, the results are encouraging. From the study results, we concluded that (1) it is feasible to compute and identify a number of quantitative breast MR image features that have relatively higher discriminatory power to predict tumor response to the chemotherapy (i.e., ten features listed in Table III), and (2) using a machine learning classifier to combine multiple image features has potential to further significantly improve classification performance than using the individual features. Therefore, this study provided useful data and evidence to support future studies to identify and optimize more accurate and reliable clinical markers based on quantitative image feature analysis to predict prognosis of cancer patients and help develop a more effective personalized cancer treatment paradigm.
ACKNOWLEDGMENTS
This work is supported in part by Grant Nos. R01 CA160205 and R01 CA197150 from the National Cancer Institute, National Institutes of Health. The authors would also like to acknowledge the support from the Peggy and Charles Stephenson Cancer Center, University of Oklahoma.
REFERENCES
- 1.Siegel R. L., Miller K. D., and Jemal A., “Cancer statistics, 2015,” Ca-Cancer J. Clin. 65, 5–29 (2015). 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 2.Tabar L., Vitak B., and Chen H. H., “Beyond randomized controlled trials: Organized mammographic screening substantially reduces breast carcinoma mortality,” Cancer 91, 1724–1731 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Smith R. A. et al. , “Cancer screening in the United States, 2015: A review of current American Cancer Society guidelines and current issues in cancer screening,” Ca-Cancer J. Clin. 65, 30–54 (2015). 10.3322/caac.21261 [DOI] [PubMed] [Google Scholar]
- 4.Fenton J. J., Wheeler J., and Carney P. A., “Reality check: Perceived versus actual performance of community mammographers,” Am. J. Roentgenol. 187, 42–46 (2006). 10.2214/AJR.05.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlin L. and Hall F. M., “More mammography muddle: Emotions, politics, science, costs and polarization,” Radiology 255, 311–316 (2010). 10.1148/radiol.10100056 [DOI] [PubMed] [Google Scholar]
- 6.Saslow D., Boetes C., and Burke W., “American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography,” Ca-Cancer J. Clin. 57, 75–89 (2007). 10.3322/canjclin.57.2.75 [DOI] [PubMed] [Google Scholar]
- 7.Klijn J. G. M., “Early diagnosis of hereditary breast cancer by magnetic resonance imaging: What is realistic?,” J. Clin. Oncol. 28, 1441–1445 (2010). 10.1200/JCO.2009.26.3467 [DOI] [PubMed] [Google Scholar]
- 8.Graham L. J., Shupe M. P., and Schneble E. J., “Current approaches and challenges in monitoring treatment 405 response in breast cancer,” J. Cancer 5, 58–68 (2014). 10.7150/jca.7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W., Giger M. L., Lan L., and Bick U., “Computerized interpretation of breast MRI: Investigation of enhancement-variance dynamics,” Med. Phys. 31, 1076–1086 (2004). 10.1118/1.1695652 [DOI] [PubMed] [Google Scholar]
- 10.Wu S., Weinstein S., Conant E., Schnall M., and Kontos D., “Automated chest wall line detection for 410 whole-breast segmentation in sagittal breast MR images,” Med. Phys. 40, 042301 12pp. (2013). 10.1118/1.4793255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie K., Chen J. H., Yu J. H., Chu Y., Nalcioqlu O., and Su M. Y., “Quantitative analysis of tumor morphology and texture features for diagnostic prediction in breast MRI,” Acad. Radiol. 15, 1513–1525 (2008). 10.1016/j.acra.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karahaliou A., Vassiou K., Arikidis N. S., Skiadopoulos S., Kanavou T., and Costaridou L., “Assessing heterogeneity of tumor enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis,” Br. J. Radiol. 83, 296–306 (2010). 10.1259/bjr/50743919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King V., Brooks J. D., and Bernstein J., “Background parenchymal enhancement at breast MR imaging and breast cancer risk,” Radiology 260, 50–60 (2011). 10.1148/radiol.11102156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q., Li L., Zhang J., Shao G., and Zheng B., “A computerized image biomarker to assist diagnosis of breast cancer using breast magnetic resonance images: A preliminary assessment,” J. Digital Imaging 27, 152–160 (2014). 10.1007/s10278-013-9617-4 [DOI] [Google Scholar]
- 15.Yang Q., Li L., Zhang J., Shao G., and Zheng B., “A new quantitative image analysis method for improving breast cancer diagnosis using DCE-MRI examinations,” Med. Phys. 42, 103–109 (2015). 10.1118/1.4903280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Therasse P. et al. , “New guidelines to evaluate the response to treatment in solid tumors,” JNCI, J. Natl. Cancer Inst. 92, 205–216 (2000). 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 17.Eisenhauer E. A. et al. , “New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1),” Eur. J. Cancer 45, 228–247 (2009). 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 18.Ahmed A., Gibbs P., Pickles M., and Turnbull L., “Texture analysis in assessment and prediction of chemotherapy response in breast cancer,” J. Magn. Reson. Imaging 38, 89–101 (2013). 10.1002/jmri.23971 [DOI] [PubMed] [Google Scholar]
- 19.Koenig M., Laue H., Boehler T., and Peitgen H., “Automatic segmentation of relevant structures in DCE MR mammograms,” Proc. SPIE 6514, 65141S (2007). 10.1117/12.708558 [DOI] [Google Scholar]
- 20.Su M. Y., Nie K., and Chen J. H., “Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI,” Med. Phys. 35(12), 5253–5262 (2008). 10.1118/1.3002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J. H., Moon W. K., and Shen Y. W., “Comparative study of density analysis using automated whole breast ultrasound and MRI,” Med. Phys. 38(1), 382–389 (2011). 10.1118/1.3523617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Y. C., Huang Y. H., Huang C. S., Chen J. H., and Chang R. F., “Computerized breast tumors detection using kinetic and morphologic analysis for dynamic contrast-enhanced MRI,” J. Magn. Reson. Imaging 32, 514–522 (2014). 10.1016/j.mri.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 23.Yang Q., Li L., Zhang J., Shao G., and Zheng B., “A computerized global MR image feature analysis 440 scheme to assist diagnosis of breast cancer: A preliminary assessment,” Eur. J. Radiol. 83, 1086–1091 (2014). 10.1016/j.ejrad.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lederman D., Zheng B., Wang X., Wang X. H., and Gur D., “Improving breast cancer risk stratification using resonance-frequency electrical impedance spectroscopy through fusion of multiple classifiers,” Ann. Biomed. Eng. 39, 931–945 (2011). 10.1007/s10439-010-0210-4 [DOI] [PubMed] [Google Scholar]
- 25.Witten I., Frank E., and Hall M. A., Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed. (Morgan Kaufmann Publishers, Elsevier, Burlington, MA, 2011), http://www.cs.waikato.ac.nz/ml/weka/. [Google Scholar]
- 26.Kohavi R. and John G. H., “Wrappers for feature subset selection,” Artif. Intell. 97, 273–324 (1997). 10.1016/S0004-3702(97)00043-X [DOI] [Google Scholar]
- 27.Li Q. and Doi K., “Reduction of bias and variance of evaluation of computer-aided diagnostic schemes,” Med. Phys. 33, 868–875 (2006). 10.1118/1.2179750 [DOI] [PubMed] [Google Scholar]
- 28.Chawla N. V., Bowyer K. W., Hall L. O., and Kegelmeyer W. P., “SMOTE: Synthetic minority over-sampling technique,” J. Artif. Intell. Res. 16, 321–357 (2002). 10.1613/jair.953 [DOI] [Google Scholar]
- 29.Tan M., Pu J., and Zheng B., “A new and fast image feature selection method for the developing an optimal mammographic mass detection scheme,” Med. Phys. 41, 081906 12pp. (2014). 10.1118/1.4890080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan M., Qian W., Pu J., Liu H., and Zheng B., “A new approach to develop computer-aided detection schemes of digital mammograms,” Phys. Med. Biol. 60, 4413–4427 (2015). 10.1088/0031-9155/60/11/4413 [DOI] [PMC free article] [PubMed] [Google Scholar]