Abstract
Early-stage oligomer formation of the huntingtin protein may be driven by self-association of the seventeen-residue amphipathic α-helix at the protein’s N-terminus (Nt17). Oligomeric structures have been implicated in neuronal toxicity and may represent important neurotoxic species in Huntington’s disease. Therefore, a residue-specific structural characterization of Nt17 is crucial to understanding and potentially inhibiting oligomer formation. Native electrospray ion mobility spectrometry-mass spectrometry (IMS-MS) techniques and molecular dynamics simulations (MDS), have been applied to study coexisting monomer and multimer conformations of Nt17, independent of the remainder of huntingtin exon 1. MDS suggests gas-phase monomer ion structures are comprised of a helix-turn-coil configuration and a helix-extended coil region. Elongated dimer species are comprised of partially-helical monomers arranged in an antiparallel geometry. This stacked helical bundle may represent the earliest stages of Nt17-driven oligomer formation. Nt17 monomers and multimers have been further probed using diethylpyrocarbonate (DEPC). An N-terminal site (N-terminus of Threonine-3) and Lysine-6 are modified at higher DEPC concentrations, which led to the formation of an intermediate monomer structure. These modifications resulted in decreased extended monomer ion conformers, as well as a reduction in multimer formation. From the MDS experiments for the dimer ions, Lys6 residues in both monomer constituents interact with Ser16 and Glu12 residues on adjacent peptides; therefore, the decrease in multimer formation could result from disruption of these or similar interactions. This work provides a structurally selective model from which to study Nt17 self-association and provides critical insight toward Nt17 multimerization and possibly, the early stages of huntingtin exon 1 aggregation.
Huntington’s disease is a fatal neurodegenerative disease caused by an expanded, glutamine-coding CAG repeat sequence in the huntingtin gene (1). The resulting expanded (>37 residue) polyglutamine (polyGln) tract in huntingtin exon 1 is responsible for aggregation (2, 3). Directly adjacent (N-terminal) to the polyGln tract is a seventeen-residue amphipathic α-helix (Nt17). This tract is the driving force behind potentially toxic oligomer formation (4, 5), anchoring to a lipid substrate (6–8), and cellular trafficking (9). In the present work Nt17, independent of the polyGln domain, was used to model the earliest stages of Nt17-driven oligomer formation of the huntingtin protein. A motivating factor in this study is that modifications of this sequence at selected side-chains may be used to glean insight to inhibiting toxic oligomerization. To date, no study has directly probed interactions of Nt17 monomers or downstream effects of monomer modification on multimer formation and structure. This study serves as a model for early stages of Nt17-mediated huntingtin aggregate nucleation and provides insight into key residues in initial Nt17 association.
Nt17 populates multiple secondary structures, ranging from 75% random-coil to mostly helical, depending on the buffer and proximity of binding partners (10–12). Jayaraman et al. suggest that Nt17 adopts a helical structure only upon interaction with a second Nt17 tract (5). Nt17 does maintain a helical structure in fibrils of huntingtin exon 1 (13), which suggests that the helical conformation of Nt17 may be involved in fibril formation. Further evidence that Nt17 is important in htt aggregation is provided by post-translational modifications within Nt17, such as phosphorylation of Thr3, Ser13 and Ser16, that retard Nt17-mediated aggregation possibly by stabilizing a random coil arrangement and thus preventing formation of helical structure (14–16). The structural heterogeneity of the system suggests the need for direct measurement of individual monomer and multimer species. High-resolution techniques, such as NMR, have been crucial to elucidating fibril structure (7, 13, 17, 18); however, labeling multiple residues, as in the case for NMR, can become cost-prohibitive. Additionally, other spectroscopic methods, such as circular dichroism, only offer a global view of protein structure, as it cannot differentiate the structure of individual, coexisting protein or peptide conformers. Systems, such as Nt17, where structural heterogeneity could be a critical determinant in oligomer formation, require structurally selective methods to fully comprehend the interplay of secondary structure content and multimer formation. The current study utilizes ion mobility spectrometry-mass spectrometry (IMS-MS) for structural characterization of peptide ions in the form of collision cross section determinations. IMS is a post-ionization, gas-phase separation technique that differentiates ion populations on the basis of a collision cross section (size). This cross section can be related to discrete secondary, tertiary, and quaternary structure through comparisons to in silico structures (19–26). If the protein is ionized at a low-energy, under native solution conditions, the gas-phase structures that are observed may be related to the solution structure (27–29). The observation of individual ion populations exhibiting different collision cross sections suggests the presence of multiple solution conformations (28, 30). Collision cross sections can also track conformational abundances as a function of solution conditions (30, 31).
IMS-MS has seen widespread use in structural characterization of carbohydrates, peptides and proteins (27, 30, 32–41). Traditional drift tube IMS (DTIMS), consisting of stacked ring electrodes providing a constant electric field (32, 42, 43) has been used in the present study. IMS-MS is used in the current study to examine native structural heterogeneity of a system whose oligomerization is predicated on formation of an amphipathic α-helix (5, 44). The conformational resolution afforded by IMS is used to determine effects of covalent modification on secondary structure stability and multimer formation. Here, IMS-MS is used to directly monitor various monomer and multimer comformers.
IMS-MS has been used extensively in aggregate (45) and early amyloid (29, 34, 46–48) characterization, primarily for the amyloid β peptide (Aβ). Specifically, IMS-MS revealed an oligomerization mechanism that occurred via multimer association for the Aβ peptide (47). Annular hexameric structures could subsequently nucleate into dodecameric oligomers that are hypothesized as precursors for amyloid fibril formation (47), which demonstrates the ability of IMS-MS to elucidate mechanistic details in aggregation phenomena. A second, complementary approach for discerning multimeric arrangement is to covalently modify solvent-accessible residues. Sites of covalent modification can be used to help determine the binding face. Covalent modifications have been widely used to probe solvent accessibility of other amyloid proteins, such as β2-microglobulin (49–51). These studies utilized diethylpyrocarbonate (DEPC) to carbethoxylate residues that have heteroatom-containing side chains. DEPC targets alcohol and amino groups, such as those found in His, Thr, Lys, and Ser. The Lys6 and Lys15 residues in Nt17 are, at least partially, solvent-inaccessible from an early aggregate state (52), which suggests the presence of an intermolecular side chain interaction. Modification at one of these residues could destabilize oligomerization.
Residue-specific label reactions and post-translational modifications can sometimes compromise the structural integrity of the native protein by altering hydrophobicity or introducing a new intramolecular interaction not present previously (53). Typically, sites of covalent modification are determined through a bottom-up approach; the protein is modified at increasing label concentrations, digested with a common enzyme, such as trypsin, and analyzed using reversed-phase liquid chromatography (49, 50, 53, 54). This work takes advantage of structural selection afforded by the IMS separation to subvert the need for enzymatic digestion.
The current study demonstrates the inherent structural heterogeneity of a sequence that is implicated in potentially toxic oligomer formation in Huntington’s disease. Rather than monitoring bulk structural populations, the present work utilized IMS to resolve and monitor relevant monomer and multimer ion structures. When combined with molecular dynamics simulations (MDS), two monomeric structural types resembling partial helices (proposed solution structures) are observed. Additionally, several multimeric species are observed, ranging from compact globular to extended helical bundle. For dimer ions, MDS suggests that the most elongated, and potentially most relevant, conformation is an arrangement of antiparallel helices stabilized by intermolecular hydrogen bonding interactions. Finally, covalent modification reveals solvent accessibility of residues near the N-terminus of the peptide at the boundary of the hydrophobic and hydrophilic faces. This modification serves to destabilize multimer formation. This is the first instance of direct, structurally-specific, monitoring of Nt17 oligomer formation. This model provides insights into the structural underpinnings of Nt17-driven huntingtin oligomerization while also providing a potential target for inhibition.
MATERIALS AND METHODS
Sample Preparation
Nt17 peptide was purchased as a lyophilized powder (Genscript, Inc.). 1.0 mg of powder was dissolved in 1.0 mL of 50/50 trifluoroacetic acid/hexafluoroisopropanol (TFA/HFIP) and separated into 200 μg aliquots. All solvent was evaporated by a stream of nitrogen, then samples were placed in a vacuum evaporator for three hours to create a peptide film. Films were stored at −20°C until use. All samples were reconstituted in 0.1 M ammonium acetate (Fisher) to yield a final concentration of 0.2 mg mL−1. All water used was purified to 18.2 MΩ resistance (Millipore).
Carbethoxylation with Diethylpyrocarbonate (DEPC)
DEPC solutions were made by diluting stock DEPC (Thermo) into LC-MS grade acetonitrile (Fisher). Typically, structural analyses avoid the use of organics as much as possible; however, DEPC has a very low solubility in water, so organic solvents, in this case, were unavoidable. Freshly reconstituted samples were labeled at 5×, 50×, and 250× (mol DEPC:mol Nt17) concentrations. In each case, 1% (v/v) acetonitrile was added to the samples. DEPC stock solutions were made such that at higher concentrations, only 1% of the final sample contained organic solvent. See Supporting Information for blanks containing 1% acetonitrile. Samples were allowed to react for 10 minutes before a 200 μL aliquot was removed for electrospray.
Ion Mobility Spectrometry-Linear Ion Trap Mass Spectrometry (IMS-MS)
Samples were analyzed on a hybrid (home-built) IMS-linear ion trap (LIT) mass spectrometer (Thermo), which has been described in detail elsewhere (55, 56). Briefly, the 1-m-long drift tube was constructed of stainless steel ring electrodes separated by Delrin spacers. IMS is a post-ionization technique that separates analytes based on mobility (K) through an inert buffer gas. The mobility of an ion is related to its drift velocity (vd) according to Equation 1:
| (1) |
where E is the electric field applied along the axis of the drift tube. vd is related to drift time (tD) and drift tube length (L) according to Equation 2:
| (2) |
Drift velocities can be related to collision cross sections according to Equation 3:
| (3) |
where z is numerical charge, e is the elementary charge, kb is the Boltzmann constant, and T is the temperature of the buffer gas. mI and mb are the masses of the ion and buffer gas (helium), respectively. P is the pressure (maintained at ~2.61 Torr for these experiments) and N is the neutral number density of the buffer gas at STP. Ions were produced via nano-electrospray ionization (nESI). The nESI voltage was maintained at 1.7 kV, with a 0.5 μL min−1 solution flow rate. The source temperature was maintained at room temperature, approximately 25°C. nESI emitters were pulled from untreated fused silica capillary. A linear voltage drop was applied along the length of the drift tube to give a constant field of 1000 V m−1. Ions are trapped at the front of the drift tube after electrospray by an ion funnel (57), where they are introduced at 20.0 ms intervals. The potential on a second gate located near the drift tube exit was lowered to allow ions into the mass spectrometer (Thermo LTQ Velos Pro, Thermo Scientific, San Jose, CA). The delay between the front gate and the back gate is scanned to create the IMS-MS spectrum. Each mass spectrum was collected for 30 seconds at each delay time setting, with a 400 ms maximum ion trap injection time. Four microscans were averaged to generate the mobility-resolved mass spectra. All mass spectra were collected from 150 to 2000 m/z.
Molecular Dynamics
MDS experiments required the construction of the [M+2H]2+ monomer and [M+3H]3+ dimer peptide ions. The charge sites for these ions were selected from experimental mass spectra. The ε-nitrogens of residues Lys9 and Lys15 were protonated for the doubly-charged monomer, while both Lys9 residues and a single Lys15 residue were charged for the triply-charged dimer ion. Although a tentative selection, these residues are chosen as the charge site locations because upon carbethoxylation of Lys6, no reduction in [M+2H]2+ ion production is observed (see Results and Discussion). One linear starting structure was employed for each charge state. Each initial structure was optimized at a quantum chemical level of HF/6-31G(d) and used for calculating molecular electrostatic potential (MEP) values and charge fitting performed with the R.E.D. Server (58–61). To generate the initial extended structures for MDS, the AMBER force field FF12SB was employed. The method used for annealing was similar to that described previously (62). Energy-minimized conformers were subjected to simulated annealing for 40 ps using the AMBER12 package (63). First the conformers were subjected to dynamics at 1000 K. Next, they were subjected to a gradual temperature gradient (to 50 K) and were subsequently energy-minimized. For each ion conformer study, 1000 cycles of annealing were conducted for 40 ps and the final, energy-minimized structure from one cycle was subjected to dynamics at 1000 K for the next cycle. The 1000 cycles of annealing were conducted with no non-bonded cutoffs to generate 1000 candidate structures. The collision cross sections of the 1000 conformers were calculated using the Mobcal (64) software employing the trajectory method (TM) (65). The potential energies of the 1000 conformers were plotted as a function of collision cross section. Matching (<1% difference in collision cross section), low-energy ion structures were selected for comparisons to experimental data. Structures were illustrated using VMD ver. 1.9.2. The accuracy of mobility measurements for this instrument is estimated to be ~1.5 5o 2% based on comparisons to previous experiments (66). Additionally, the high reproducibility of the drift time measurement (see Tables 1 and 2) ensures a good comparison between IMS dataset features and structures from MDS as demonstrated in numerous studies (28, 31, 67, 68).
Table 1.
Calculated collision cross-sections for selected Nt17 conformers.
| Speciesa | m/zb | td (ms)c | Ω (Å2)d |
|---|---|---|---|
| Nt17 | 988.1 | 11.2 | 399.8 ± 0.8 |
| 12.4 | 442.7 ± 0.1 | ||
| Nt17 x 2 | 1317 | 11.6 | 622 ± 0.1 |
| 12.6 | 666 ± 5 | ||
| 13.2 | 709 ± 1 | ||
| Nt17 x 3 | 1481 | 12.6 | 874 ± 1 |
| Nt17 x 4 | 1581 | 12.0 | 1074 ± 1 |
Species are identified as monomer (Nt17), dimer (Nt17 x 2), trimer (Nt17 x 3), and tetramer (Nt17 x 4).
measured m/z value.
measured drift time in ms.
calculated collision cross section with standard deviation (n = 3 replicates).
Table 2.
Calculated collision cross-sections for selected Nt17 modified conformers.
| Speciesa | m/zb | td (ms)c | Ω (Å2)d |
|---|---|---|---|
| Nt17 N/Thr3C | 1024 | 11.6 | 413.75 ± 0.09 |
| 12.8 | 457.2 ± 0.1 | ||
| Nt17 N/Thr3C, Lys6C | 1060 | 11.6 | 413.73 ± 0.10 |
| 12.2 | 435.5 ± 0.1 | ||
| 13.0 | 464.4 ± 0.1 | ||
| Nt17 x 2 N/Thr3C | 1365 | 13.0 | 696.3 ± 0.2 |
| 13.6 | 729.0 ± 0.2 | ||
| Nt17 x 2 N/Thr3C, Lys6C | 1389 | 13.4 | 718.1 ± 0.2 |
Species are identified as monomer (Nt17), dimer (Nt17 x 2), with residue modifications listed.
measured m/z value.
measured drift time in ms.
calculated collision cross section with standard deviation (n = 3 replicates).
RESULTS & DISCUSSION
Nt17 Adopts Several Monomeric and Multimeric Conformations
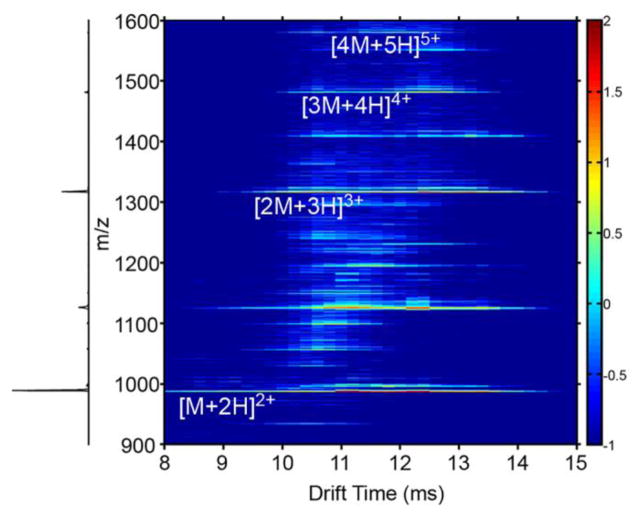
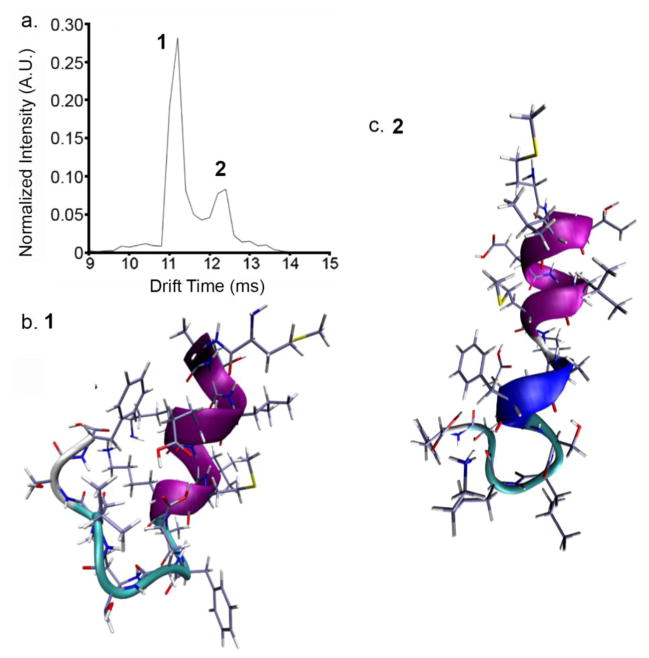
IMS-MS distributions produced upon electrospray of the unlabeled sample are shown in Figure 1. Multimers up to, and including, a tetramer are observed. The mass spectrum is dominated by the doubly-charged monomer ([M+2H]2+) ions, which primarily adopt two conformations that are mobility resolved (Figure 1b.). The first conformer, arriving at 11.0 ms, is of higher abundance (~4 fold) than the other as demonstrated in the drift time distributions shown in Figure 3a. The second, less-intense, feature corresponding to doubly-charged monomer ions exhibits a longer drift time (~12.4 ms). From Equation 3, collision cross sections for the compact and more elongated [M+2H]2+ ions are determined to be 399.8 ± 0.8 and 442.7 ± 0.1 Å2 (see Table 1).
Figure 1.
False color IMS-MS distribution of Nt17 ions. Colored intensity is presented on a log scale.
Figure 3.
Nt17 ribbon structures calculated from MDS. Pink regions are α-helical. Blue regions correspond to a 3–10 helix. a.) Extracted tD distribution for [M+2H]2+ ions. b.) Compact conformer (1) at 11.0 ms. c.) Extended conformer (2) at 12.4 ms. See Table 1 for assigned collision cross-sections
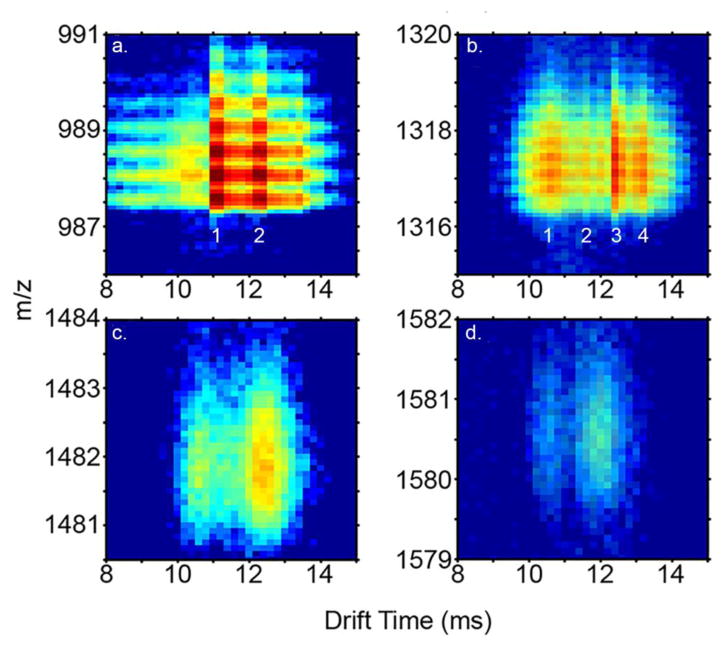
At least four distinct dataset features corresponding to triply-charged dimer ([2M+3H]3+) ions are observed in the two-dimensional (2D) IMS-MS datasets (Figure 1c). A broad feature (tD ~1 ms FWHM) is observed at a drift time of ~10.6 ms as shown in the 2D drift time distribution in Figures 2b – e, as well as the tD distribution in Figure 4. A dataset feature was also observed at 12.0 ms, but was not reliably resolved within all replicate analyses; it was not possible to reliably ascertain a collision cross section and so this feature is not considered further in this work. Narrower features are observed at tD 11.6 and 12.6 ms (Figure 4a). The dataset feature at 12.6 represents the most intense peak for the [2M+3H]3+ ions, exhibiting a peak intensity that is ~2 fold greater than the other dataset features, which demonstrate similar intensity levels. A separate dataset feature representing a shoulder on the most intense peak is observed at a tD of 13.2 ms. The collision cross sections for the partially-resolved [2M+3H]3+ ions are 568.1 ± 0.3, 620.3 ± 0.1, 671 ± 5, and 708 ± 1 Å2 for the various conformations in order of increasing size. The broad feature at 10.6 ms appears in essentially all multimeric tD distributions, up to and including the tetramer (See Supporting Information Figure S1). In each spectrum, the feature has the same peak shape. This may suggest the feature is derived from higher-order (n>4) multimer species that dissociate after the drift separation. In support of this argument, a calculated collision cross-section for a triply-charged dimer ion occurring at 10.6 ms is 568 Å2. This value is ~10% smaller than all of the smallest dimer configurations sampled by MDS (see Conformations of Multimeric Ions results section) and matches closely (~2.3% larger) to a spherical compact arrangement (45). In separate experiments, 2D IMS-MS spectra were acquired with an an ion activation potential was elevated to 120 V (as opposed to 2 V under standard conditions). This bias was applied across a small activation region at the exit portion of the drift tube. A large increase in the 10.6 ms feature was observed (see Supporting Information Figure S2) which is consistent with the dissociation of higher-order multimers at the rear of the drift separation. That said, we cannot entirely rule out the possibility that this species could be indicative of even greater structural homogeneity possibly comprised of unstructured monomers as supported by some CD studies (5). Early-stage oligomers would be expected to consist of arrangements of elongated helices, as observed in fibril structures (5, 13, 69). Unfortunately, the multimer from which the 10.6 ms feature may have arisen is not distinguished in the 2D distribution and cannot be reliably identified. These results are described in the Supporting Information section.
Figure 2. Expanded regions of the.
IMS-MS distribution (Figure 1) showing monomer (a.), dimer (b.), trimer (c.), and tetramer (d.) ions. Discrete features corresponding to separate conformations are labeled sequentially in monomer and dimer distributions.
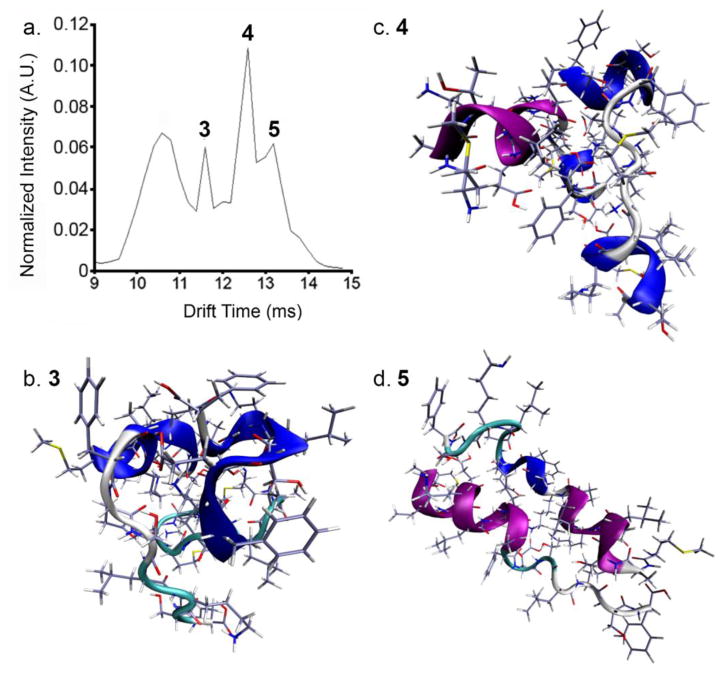
Figure 4.
Nt17 dimer structures calculated from MD simulation. Color schemes are the same as in Figure 3 a.) Extracted arrival time distribution. ‘3’, ‘4’, and ‘5’ correspond to relevant dimer structures, depicted in panels b., c., and d., respectively.
Several higher-order multimeric structures ([3M+4H]4+ and [4M+5H]5+ ions, Figures 1D and E) are also evident in the 2D spectrum. For the former ions, the same broad dataset feature centered at a drift time of 10.6 ms was observed as in the other conformations. A narrower yet still relatively broadfeature corresponding to a collision cross section of 886 Å2 is observed at a longer drift time (12.4 ms). For the [4M+5H]5+ ions, a broad dataset feature is observed to have collision cross-section of 1073 Å2. In addition to these ions, at long data collection times, [5M+6H]6+ and [6M+7H]7+ ions are observed in 2D datasets. However, these ions are of such low intensity that the poor ion statistics associated with signal dispersion in two dimensions do not allow the determination of “peaks” for which collision cross sections can be assigned.
Compact and elongated Nt17 [M+2H]2+ Ion Structures from Molecular Dynamics Simulations
Solution studies have proposed that Nt17 exists primarily as a random coil in a native-like environment (13). Separate studies, both solution (70, 71) and simulation (12, 72, 73), have suggested the presence of helical monomers in solution. The IMS data presented here (Figure 2 and Figure 3) suggests the presence of two dominant coexisting solution structures for peptide monomers.
Figure 3a shows the tD distribution of the doubly-charged monomer. Figure 3b and 3c depict energy-minimized compact (Structure 1) and elongated (Structure 2) structures of Nt17 obtained from MDS. These structures represent the lowest energy conformers that are within 1% of the experimental collision cross-sections of the two [M+2H]2+ dataset features. It should be noted here, that all structures presented in the present study represent gas-phase conformations that best fit to experimental gas phase conformers arising from different solution structures (30). That is, although the monomer structures are modeled in the gas-phase, they may not completely resemble solution conformers. Both monomeric conformers are helical. The high degree of helical propensity for both structures is consistent with proposed and experimentally observed solution structures contained within a huntingtin peptide aggregate (13, 74). The primary difference between the compact and elongated monomer structures is the propensity for the random coil region (residues 11 to 17) in Structure 1 to turn back on itself and potentially associate with the helix (Figure 3b). In the elongated structure, the portion C-terminal to the helical region is extended away from the rest of the peptide. Additionally, Structure 2 contains a 3–10 helix spanning residues Ala10 to E12. This helix could prohibit C-terminal association as is observed in Structure 1. In Structure 1, residue Lys6 is in relative proximity to Ser16. Interestingly, Lys6 is potentially involved in intermolecular interaction from an early aggregate state (52). In solution, such proximity could lead to charge-dipole interactions between the ε-amino group on Lys6 and the primary alcohol group on Ser16. Previous experiments have shown that phosphorylation and S16D mutations decrease aggregation kinetics through inhibition of Nt17-driven oligomerization (14, 15). One hypothesis for the inhibition of oligomer formation could be a charge-dipole interaction formed between Lys6 and residue Ser16, though the proximity could be due to charge solvation of the neighboring Lys9 in the gas phase. Still, if this is reminiscent of a solution structure, this interaction could prevent the formation of the extended helices observed in short huntingtin peptide oligomers and mature huntingtin fibrils (5, 13). It is intriguing that both of these structures match closely to the extended solution conformer found by Kelley et al (74). Indeed, the calculated collision-cross section of the elongated structure modeled here is within ±3% of the proposed solution helix.
Conformations of Multimeric Ions
Simulated annealing was performed on a dimeric species to gain some insight to possible gas-phase structures that arise from solution conformations. Figure 4a shows the tD distribution of all dimer species. Figure 4b, c, and d depict the results from the simulated annealing experiment (Structures 3, 4, and 5, respectively). Structure 3 corresponds to a dimer ion species that would have a tD of 11.2 ms (622 Å2), while Structures 4 and 5 depict ions that would have a tD of 12.6 (666.3 Å2) and 13.2 ms (709.9 Å2), respectively. Structure 3 is composed of two partial 3–10 helices that are arranged skew to one another, which could represent the first step in transition to structures representative of Structures 4 and 5. The next, more elongated, conformer, Structure 4, consists of two partially helical monomers that are arranged offset to one another. The α-helix in one monomer ranges from the N-terminus to Ala10, where a random coil region leads into a 3–10 helix. This monomer constituent resembles Structure 2. The second monomer exhibits two 3–10 helices: one from the N-terminus to Glu5, the other from K15 to F17. It should be stressed here that the structures obtained from simulation are gas-phase structures and the actual degree of similarity to solution structures is unknown. Still, reliable estimates of solution conformation have been presented from gas-phase structures (27, 30, 31). It is also noteworthy that elements of secondary structure similar to those found in the monomer species (and contained in a fibril (5, 13)) are observed for the dimers.
Structure 5 depicts a structure obtained from MDS that is consistent with the most elongated conformation of the Nt17 dimer. In this model, both peptides are partially helical, with the α-helix spanning from the N-terminus to Ala10 in one monomer, and an extended α-helix-coil-3-10 helix motif in the second monomer. Interestingly, the helices are arranged antiparallel to one another. Lys6 interactions stabilize the dimer structure through hydrogen bonding; at one end, Lys6 associates with Ser16, while at the other end Lys6 interacts with Glu12 (See Supporting Information Figure S3 for hypothetical structure depicting the interactions). Previous solution docking simulations have alluded to a stabilizing Lys6-Ser16 interaction in antiparallel Nt17 helices (10). Additionally, solution-based deuterium exchange measurements revealed an increased role of Lys6 in Nt17 multimer structure (52). Finally, S13D and S16D mutations in Nt17 prevented aggregation of huntingtin exon 1 synthetic models via the Nt17-mediated pathway (14). The dimer conformations resemble the assembly of two elongated monomer structures (Structure 5). Considering structure comparisons above, as well as the native nESI conditions, these analyses may well provide information regarding Nt17 binding face in the earliest stages of oligomer formation.
For the [3M+4H]4+ ions, the larger ions (Figure 1D) with collision cross sections of 873 Å2 and 902 Å2 are ~22% and 24% larger, respectively, than expected values for globular structures based on a fit to experimental data (45). The larger ions show a percent difference that is near that (~21%) expected for helical bundles of this size but well below that (~51%) determined for helices joined end on. The calculated cross section of the elongated trimer is near that of insulin A chain packed as a helical bundle (874 Å2 vs. 840 Å2 for insulin chain A) (45). The two peptides are similar in length (17 amino acid residues versus 21 for insulin chain A), suggesting the possibility of a similar helical arrangement for this smaller peptide. The [4M+5H]5+ ion conformers (Figure 1E) display a similar trend. The elongated (1073 Å2) ion has a collision cross section that is ~26% larger than the expected value for a globular multimeric structure based on a fit to experimental data. This value is slightly above the percent difference for helical bundles (~16), and well below that expected for the percent difference increase (~75%) for helices arranged end on. Additionally, the calculated cross section of the Nt17 tetramer, 1073 Å2, is in the same range as helical bundled insulin A chain (approximately 1010 Å2) The lack of a polyGln domain in the current study should be considered. In a fibril, polyGln β-sheet structure can impinge upon the Nt17 region at the last two Nt17 residues (17). In the present study, the C-terminal portion of Nt17 are disordered, so structural transition to β-sheet is a possibility. Finally, some higher-order oligomers were transiently observed ([5M+6H]6+ and [6M+7H]7+); however, signal levels were not sufficient to determine collision cross sections for these ions and are therefore not discussed here.
Carbethoxylation Introduces a New Monomer Structure
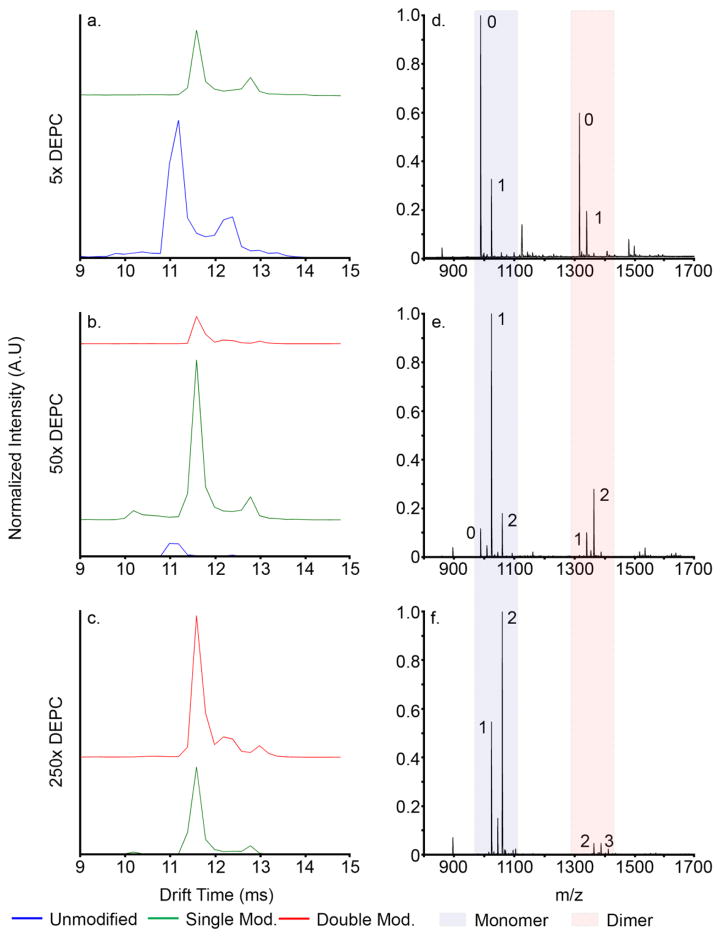
Covalent modification with DEPC was performed to determine effects of charged side chain modification on Nt17 multimerization. Figure 5 shows IMS-MS distributions of [M+2H]2+ peptide ions at various states of modification after ten minutes of incubation with DEPC (for 2D IMS-MS distributions depicting the course of modification, see Supporting Information Figures S2 and S3). Drift times and collision cross sections for each species are presented in Table 2. At 5× label (Figure 5a), two distinct unlabeled [M+2H]2+ ion conformers are observed as well as two distinct species for the singly-labeled ions. The unmodified peptide and singly-modified peptide ion conformers are observed at shorter and longer drift times, respectively. The first modification increases the collision cross section of [M+2H]2+ peptide conformers by ~14 Å2 (Figure 5a). Doubly-modified peptide ion conformers are not observed in the 5× trials but were observed as significant species at higher label concentrations. Additionally, at the 5× label concentration, total monomeric peptide ions accounts for approximately 50% of the total peptide signal, with ~36% of the total peptide signal attributed to unlabeled monomer. The rest of the peptide signal is comprised of multimeric structures (see Figure 4D – F for range of dimer m/z values as well as the Supporting Information for modified dimer 2D IMS-MS distributions). Total dimer, trimer, and tetramer species comprise ~38%, ~9%, and ~3% of the total peptide signal, respectively. Each multimer exists in several states of covalent modification. It is noted that this technique is unable to determine the origin of multimers; that is, it cannot distinguish between species that are modified before or after association. For this reason, the effect of covalent modification on formation of multimeric species was calculated as a percent of total peptide species.
Figure 5.
tD distributions of covalently modified Nt17 [M+2H]2+ monomer at 5x (a.), 50x (b.), and 250x (c.) DEPC. tD distributions are normalized to total peptide ion counts. Blue trace: unlabeled Nt17; green trace: singly modified Nt17; red trace: doubly modified. d. – f., mass spectra at each DEPC concentration showing monomer m/z range (light blue box) and dimer m/z range (light red box). The number of covalent modifications is shown next to each peak of interest in the mass spectra.
Overall monomer signal increases at the 50× label concentration (Figure 5e), to approximately 66% of the total peptide signal. At this concentration, a second modification is evident at m/z 1060 (Figure 5b and Supporting Information). The second modification has less effect on collision cross section; the largest increase in collision cross section was ~7 Å2 for the most extended conformer. Notably, a new conformer is observed at a tD of 12.2 ms (653 Å2) in the doubly-modified [M+2H]2+ ion tD distribution. It is noted that the size of this is closer to the more compact arrangement (Figure 3b, Structure 2). No additional collapsed monomeric structures are observed, and the small amount of organic solvent added to the solution to initiate the labeling was not sufficient to induce denaturation of the peptide (less than 1% by volume for each concentration); evidence for new structures was only observed once the peptide was doubly modified. Supporting Information Figure S6 shows monomer and dimer distributions under solvent conditions with 1% (v/v) acetonitrile. When compared to Figure 1b and c, no significant change in monomeric or multimeric conformation is evident. Thus, the appearance of the compact conformer at 12.2 ms must arise from the modification of peptide, and not from addition of a small amount of organic solvent. Under these conditions, the majority of the monomer signal stems from the singly modified peptide; approximately 53% of the total peptide signal is attributed to this species alone. Multimer signal also decreases, falling to ~27%, ~5%, and ~2% total dimer, trimer, and tetramer at the 50× label concentration level, respectively.
At the 250× label concentration (Figure 5c and f), almost no residual unlabeled monomer exists. The base peak of the mass spectrum is the doubly modified monomer. Total monomer increased in this experiment to ~89% of the total peptide signal, while dimeric, trimeric, and tetrameric signal dropped to ~8%, ~2%, and ~0.5%, respectively. The appearance of the doubly-modified monomer does correlate with a sharp decline in multimer formation. One possible explanation is that the covalent modification induces instability in the complex leading to a shift in equilibrium to the monomeric state. These results could have implications in posttranslational modification studies of huntingtin exon 1, where modifications of hydrophilic residues have been shown to alter aggregation kinetics and morphologies, either by introducing a monomeric structural change or through inhibition of residue-specific interactions (14–16, 75).
Lysine-6 is Modified at Elevated DEPC Concentrations
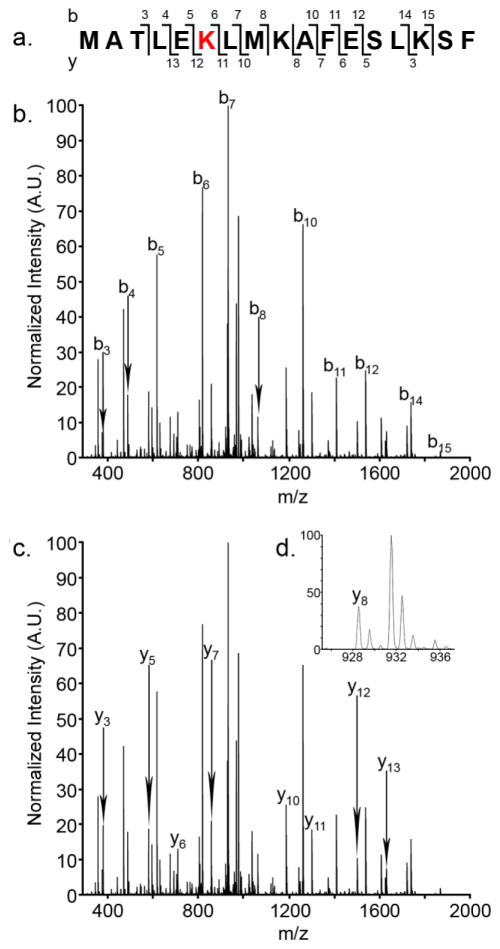
Figure 6 shows a mobility-selected MS/MS spectrum of the new, doubly modified, intermediate monomer conformer at 12.2 ms. The front and back gates in the drift tube have been set to allow only species with an arrival time distribution from 12.0 to 12.6 ms. Mass selection and fragmentation was performed in the mass spectrometer. Figure 6 shows the sequence coverage obtained with MS/MS analysis. The Nt17 peptide contains several potential sites of modification, including the N-terminus, and residues Thr3, Lys6, Lys9, Ser14, Lys15, and Ser16. Singly modified peptides from MS/MS analysis are modified either at the N-terminus or at residue Thr3. Unfortunately, the b1 and b2 ions are not present in the observed ion sequence; therefore, the first modification could not be unambiguously identified. The N-terminus would be the most likely site of modification, because even in an aggregated state, it is solvent-exposed and heteroatom sites are very labile (5, 13, 52). It would appear that the modification of residues N-terminal to Leu4 does not introduce any structural rearrangement (Figure 5b, e, and h); this modification has little to no bearing on gas-phase conformation, other than a slight increase in collision cross-section owing to the larger carbethoxy moiety.
Figure 6.
Mobility selected MS/MS analysis of the intermediate, doubly modified conformer. a.) Sequence coverage by ion. b.) b-ion series. c.) y-ion series. d.) Enhanced region showing the y8 ion for clarity.
At elevated DEPC concentrations, Lys6 is modified. MS/MS studies confirm that all three doubly modified monomeric conformers are modified at the same residues. The b7 ion is the most abundant ion in the MS/MS spectrum. A fragmentation product ion corresponding to unmodified Lys6 was not observed, suggesting that covalent modification is not heterogeneous. For these conformers, no other lysine residues were modified; carbethoxylation occurred exclusively on Lys6, and as such, resulted directly in the newly observed conformer at 12.2 ms (Figure 5 and Figure S4 F and I). Lys6 is contained within the conserved monomeric ion α-helix in the N-terminal portion of Nt17 from MDS (Figure 3b and c, Structures 1 and 2). Additionally, MDS suggests that Lys6 could be directly involved in two stabilizing interactions in the Nt17 dimer; both Glu12 and Ser16 are potential binding partners for this residue. The most dramatic decrease in total multimer formation correlates with the emergence of the doubly-modified conformer at tD 12.2 ms, which suggests this structural transition increases destabilization of multimeric species, either by introducing a new intermolecular interaction that destabilizes the α-helix, or by preventing hydrophilic Lys6 interactions, such as those suggested by MDS.
Interpretation of Structural Findings
Simulations of pathogenic and non-pathogenic Nt17-polyGln-polyPro reveal that Nt17 has the propensity to form helical bundles early in the aggregation process (5, 13). Additional simulation and CD spectropolarimetry has shown 36% helicity in the Nt17 region at room temperature.(12) The monomeric extended helix conformer has been implicated in studies containing the amyloidogenic polyGln tract as the most prevalent structure, with helices ranging from residues 4 – 12 to 2 – 17 (5, 13, 72, 74). In contrast, solid state NMR studies on fully aggregated Nt17Glu35Pro10Lys2 showed that β-sheet character from the amyloid core penetrated to Ser16; however, helical nature was conserved over the region containing Lys6 (17). This is not out of the realm of possibility, as in light of the results obtained here, the random nature of the C-terminal portion of Nt17 suggested by MDS could very easily transition to a β-sheet conformation. Additonally, in recent studies a helix-breaking Pro-Gly segment was inserted between Phe17 and Gln18 in a huntingtin exon 1 mimic (76). Kinetics of aggregation were not changed upon insertion of the helix-breaker, which suggests penetration of the helix into the polyGln structure, or vice-versa, does not affect aggregation kinetics, and that the structure of Nt17 is mostly independent of polyGln structure. Previous simulations on the Nt17 region alone show a two-helix bundle is a dominant structure at room temperature; residues Met1 – Met8 are contained in an α-helix with a loop from residues Lys9 - Phe11 (74). In the same study, a second, straight helix conformer is evidenced at room temperature. The current study provides evidence for two relevant monomeric Nt17 structures stemming from solution species, which are mostly consistent with the findings of Kelley et al (74). A strong helical propensity is observed for the first eleven N-terminal residues; however, where Kelley et al. demonstrate a helix-turn-helix structure, referred to in their work as a two-helix bundle, the current study shows that the most abundant ion conformer contains a helical N-terminus with a disordered C-terminus. The second structure presented in the current study could most closely resemble the extended helix structure observed by Kelley et al.. The data presented in the study by Kelley et al., and the work presented here, contrasts previously published work that states no stable helix is evident in N-terminal huntingtin monomers (5, 77). It should be noted that measurements in these referenced studies are based on circular dichroism spectropolarimetry, which only reports on average characteristics. It was assumed that α-helix character was a result of transient oligomers present in solution upon reconstitution. While this was a valid conclusion based on the measurements and solution conditions, in light of the data presented here, this conclusion may be partially correct, as both the matching monomeric and multimeric MDS conformations contain a degree of helicity. Indeed, crystal structures of huntingtin model peptides with an extended polyGln tract show a large amount of helicity in the Nt17 region (71).
Trimeric and tetrameric species have been observed that appear to have conformers similar in size to bundles of elongated helices (45). These structural assignments are in good agreement with prior work that states Nt17 associates via α-helical interactions and maintains its helical nature in mature amyloid fibrils (5, 13, 17, 74). Because the data in the current study seem to indicate that elongated, helical, structures are conserved in the gas phase, it is noted that the elongated trimer and tetramer conformers observed here are assumed to contain a degree of helicity. This can also be argued from the nature of Nt17 to form extended helices in amyloid fibrils as well as the similarity of collision cross sections to calculated helical bundle peptides of similar length.
Covalent Modifications Alter Structures
Multiple PTM’s have been implicated in accelerating or inhibiting huntingtin exon 1 aggregation (14–16, 78, 79), while some have been implicated in promoting new morphologies from pre-aggregated species (80). Phosphorylation of Thr3 leads to decreased toxicity but increased aggregation rates (16). Carbethoxylation of heteroatoms, while not a physiological PTM, is a means to increase hydrophobicity and introduces steric hindrance in the helical region. Excess of label reagent can destabilize secondary structures (54). Previous studies have used carbethoxylation by DEPC as a means to probe solvent accessibility (49, 50, 54). Mendoza et al. use the reagent to track lysine, threonine, and histidine residues in isoforms of β2 microglobulin (49, 50). In the present work, however, the modification of a single, hydrophilic residue led to a new monomer conformer and in concert, reduced overall multimerization. This occurs with a decrease in the elongated monomer conformer. The total contribution of the elongated conformer drops from 10% to approximately 7% as the new conformer appears. This may suggest that the new conformer arises at the expense of the formation of the elongated species. Notably, the majority of the doubly modified conformer still exists primarily as the most compact conformation.
Previous work highlights the importance of Lys6 and its protection from an aggregated state in a huntingtin exon 1 model, indicating that Lys6 is involved in a stabilizing intra- or intermolecular interaction (52). In previous studies, which utilized top-down deuterium exchange techniques, it was not possible to determine the exact origin of monomeric peptide. Results from the present study suggest that Lys6 is available in a monomeric state and may be involved in stabilizing multimer interactions, which would account for previous reports of decreased accessibility (52). Modification of Lys6 in the peptide’s multimeric state could reduce stabilizing interactions, and as a result, favor dissociation of the multimer species. Additionally, the modification could present a monomer structure that is not amenable for interaction mediated through Lys6 due to altered intramolecular interactions, once again preventing multimerization.
Implication of New Monomeric Structure
In some cases, post-translational modification can have accelerating effects on multimerization. For example, phosphorylation of Thr3 in huntingtin exon 1 increased aggregation rate, as well as decreased toxicity, in Drosophila models; Aiken et al. show that Thr3 phosphorylation occurs in vivo and may represent a therapeutic strategy for Huntington’s disease remediation (16). Two explanations for neuroprotection are proposed. Increased hydrophobicity of Thr3 due to modification either increases the rate of aggregation through the more toxic oligomeric state, thus reducing the amount of available toxic oligomer and leading to the neuroprotective fibrillar form. Conversely, modification of Thr3 destabilizes helical propensity and intermolecular association, thus preventing an oligomeric state to form entirely, leading to formation of fibrils that are not mediated through an oligomeric precursor. It should be noted that Lys6 lies on the boundary of the hydrophilic and hydrophobic faces, so the interpretation provided by Aiken et al. for Thr3 could also be extended to Lys6. That is, the data in the current study would support the latter interpretation for neuroprotection proposed by Aiken et al.; that is, an altered helix state does not allow for extended Nt17 multimers to form, promoting aggregation via a pathway that is not modulated by Nt17. Alternatively, it must be noted that Nt17-self association is not the only potential mechanism by which huntingtin may form fibrils. Nt17 may interact with the polyGln tract, leading to increased fibrillization kinetics (the domain cross-talk model) (12, 81). In this model, hydrophobic residues in Nt17 are sequestered in polyGln, stabilizing polyGln structure and driving fibrillization. Alteration of hydrophilic residues could increase the propensity for domain cross-talk by altering the boundary between hydrophilic and hydrophobic domains. This could lead to an increase in aggregation kinetics, perhaps through bypass of the oligomeric state. However, since the model in the current study did not contain a polyGln tract, no comment on fibril formation or polyGln interaction can be made, nor can any conclusion be drawn regarding the effects of β-sheet formation in Nt17 as a result of extended polyGln segments; however, it can be concluded that modification of Nt17 at a critical boundary lysine residue reduces multimerization, which could lead to altered aggregation pathways.
Multiple studies have reported on the propensity of Nt17 to form an α-helix upon association with another Nt17 tract (5, 13) or a lipid bilayer (7, 82). Solid state NMR studies have shown that Nt17 is mostly helical in amyloid fibrils and retains its helicity even after advanced fibrillization, and that helicity increases with increased association (13, 17). Indeed, formation of an amphipathic α-helix is a critical step in exon 1 nucleation (5, 44, 70). Tam et al. have shown that an amphipathic helix from residues 4 – 12 are necessary for advanced huntingtin fibrillization, which represents a more extended helix (70). In the present study, a large portion of the monomer conformation appears to exist in an extended helical state in the gas phase and, upon labeling, is destabilized. It would appear that antiparallel association (Figure 4C) of pre-elongated peptides is possible; however, it is unclear which (if either) conformation is the most disease-relevant. One interpretation is that destabilization of the pre-existing helix abrogates bundled multimer structure formation that resembles Nt17 nucleation in full-length huntingtin exon 1. It could be that the more compact monomer conformer may not contribute as significantly to Nt17-mediated nucleation as the extended state due to the C-terminal coil region folding back into, and potentially interacting with, the helical region.
Implication of Lysine Modification
Depletion of the most elongated dimer conformers was observed upon modification of Lys6, which implies that Lys6 could be involved in more elongated helical associations of Nt17. Additionally, simulations of gas-phase dimer interactions show the involvement of Lys6 in stabilizing hydrogen bond interactions with oxygen-containing residues. Multiple solution simulations show that Lys6 is contained in the helical portion of Nt17, whether in its monomeric form (17) or bound to a lipid substrate (6, 7, 82). While the current study did not probe aggregation kinetics, the study does suggest that modifying the ε-amino group on Lys6 inhibits inter-helical association. Modification of both the N-terminal residue and Lys6 could work synergistically, by introducing steric hindrances that prohibit association and by altering secondary structure, to reduce multimer formation. Currently, it is not clear as to the mechanism of inhibition; however, Lys6 appears to be critical in Nt17 association.
Huntingtin Nt17 peptide monomer and multimer structures have been analyzed by IMS-MS and MDS. Direct observation of monomer structure reveals two populated solution states that correlate well with prior simulation and experiment, both of which could contain partial α-helices. Several multimeric conformations are also observed, particularly for the dimer structure, each containing a degree of helicity. The most elongated dimer structure may consist of an arrangement of antiparallel α helices, and could be the most disease-relevant conformation. This arrangement most closely resembles association of Nt17 in full-length huntingtin exon 1. MDS revealed intermolecular association in the elongated dimer ions is stabilized through Lys6-Glu12 and Lys6-Ser16 hydrogen bond interactions. Carbethoxylation of an N-terminal residue and Lys6 may destabilize the helical structure as a new compact conformer is observed in the doubly-modified spectrum. Additionally, this modification drastically reduced multimer formation, which may suggest the critical role of Lys6 in the earliest stages of Nt17-mediated huntingtin exon 1 aggregation.
Supplementary Material
Acknowledgments
Funding statement: This work was supported by the National Institutes of Health grants R15NS090380. J. Arndt was supported by the WV Higher Education Policy Commission/Division of Science and Research
Abbreviations
- IMS-MS
Ion mobility-mass spectrometry
- Nt17
N-terminal 17-residue amphipathic α-helix of the huntingtin protein
- MD
Molecular dynamics
- DEPC
diethyl pyrocarbonate
- tD
drift time
Footnotes
Additional IMS-MS distributions and mass spectra are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.The Huntington’s Disease Collaborative Research Group . A NOVEL GENE CONTAINING A TRINUCLEOTIDE REPEAT THAT IS EXPANDED AND UNSTABLE ON HUNTINGTONS-DISEASE CHROMOSOMES. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Penney JB, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington’s disease. Ann Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- 3.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 4.Jayaraman M, Mishra R, Kodali R, Thakur AK, Koharudin LMI, Gronenborn AM, Wetzel R. Kinetically Competing Huntingtin Aggregation Pathways Control Amyloid Polymorphism and Properties. Biochemistry. 2012;51:2706–2716. doi: 10.1021/bi3000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayaraman M, Kodali R, Sahoo B, Thakur AK, Mayasundari A, Mishra R, Peterson CB, Wetzel R. Slow Amyloid Nucleation via alpha-Helix-Rich Oligomeric Intermediates in Short Polyglutamine-Containing Huntingtin Fragments. J Mol Biol. 2012;415:881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Côté S, Wei G, Mousseau N. Atomistic mechanisms of huntingtin N-terminal fragment insertion on a phospholipid bilayer revealed by molecular dynamics simulations. Proteins: Structure, Function, and Bioinformatics. 2014 doi: 10.1002/prot.24509. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 7.Michalek M, Salnikov ES, Werten S, Bechinger B. Membrane Interactions of the Amphipathic Amino Terminus of Huntingtin. Biochemistry. 2013;52:847–858. doi: 10.1021/bi301325q. [DOI] [PubMed] [Google Scholar]
- 8.Burke KA, Kauffman KJ, Umbaugh CS, Frey SL, Legleiter J. The Interaction of Polyglutamine Peptides With Lipid Membranes is Regulated by Flanking Sequences Associated with Huntingtin. J Biol Chem. 2013 doi: 10.1074/jbc.M112.446237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng ZQ, Li AM, Holmes BB, Marasa JC, Diamond MI. An N-terminal Nuclear Export Signal Regulates Trafficking and Aggregation of Huntingtin (Htt) Protein Exon 1. J Biol Chem. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalek M, Salnikov ES, Bechinger B. Structure and Topology of the Huntingtin 1 17 Membrane Anchor by a Combined Solution and Solid-State NMR Approach. Biophys J. 2013;105:699–710. doi: 10.1016/j.bpj.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giulia Rossetti PC, Laio Alessandro, Carloni Paolo. Conformations of the Huntingtin N-term in aqueous solution from atomistic simulations. FEBS Letters. 2011:3086–3089. doi: 10.1016/j.febslet.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 12.Williamson TE, Vitalis A, Crick SL, Pappu RV. Modulation of Polyglutamine Conformations and Dimer Formation by the N-Terminus of Huntingtin. J Mol Biol. 2010;396:1295–1309. doi: 10.1016/j.jmb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivanandam VN, Jayaraman M, Hoop CL, Kodali R, Wetzel R, van der Wel PCA. The Aggregation-Enhancing Huntingtin N-Terminus Is Helical in Amyloid Fibrils. J Am Chem Soc. 2011;133:4558–4566. doi: 10.1021/ja110715f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra R, Hoop CL, Kodali R, Sahoo B, van der Wel PCA, Wetzel R. Serine Phosphorylation Suppresses Huntingtin Amyloid Accumulation by Altering Protein Aggregation Properties. J Mol Biol. 2012;424:1–14. doi: 10.1016/j.jmb.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu XF, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aiken CT, Steffan JS, Guerrero CM, Khashwji H, Lukacsovich T, Simmons D, Purcell JM, Menhaji K, Zhu YZ, Green K, LaFerla F, Huang L, Thompson LM, Marsh JL. Phosphorylation of Threonine 3 IMPLICATIONS FOR HUNTINGTIN AGGREGATION AND NEUROTOXICITY. J Biol Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoop CL, Lin H-K, Kar K, Hou Z, Poirier MA, Wetzel R, van der Wel PCA. Polyglutamine amyloid core boundaries and flanking domain dynamics in huntingtin fragment fibrils determined by solid-state NMR. Biochemistry. 2014 doi: 10.1021/bi501010q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kar K, Hoop CL, Drombosky KW, Baker MA, Kodali R, Arduini I, van der Wel PCA, Horne WS, Wetzelt R. beta-Hairpin-Mediated Nucleation of Polyglutamine Amyloid Formation. J Mol Biol. 2013;425:1183–1197. doi: 10.1016/j.jmb.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyttenbach T, von Helden G, Bowers MT. Gas-Phase Conformation of Biological Molecules: Bradykinin. J Am Chem Soc. 1996;118:8355–8364. [Google Scholar]
- 20.Counterman AE, Clemmer DE. Cis–Trans Signatures of Proline-Containing Tryptic Peptides in the Gas Phase. Anal Chem. 2002;74:1946–1951. doi: 10.1021/ac011083k. [DOI] [PubMed] [Google Scholar]
- 21.Taraszka JA, Counterman AE, Clemmer DE. Large anhydrous polyalanine ions: substitution of Na+ for H+ destabilizes folded states. Int J Mass Spectrom. 2001;204:87–100. [Google Scholar]
- 22.Mao Y, Woenckhaus J, Kolafa J, Ratner MA, Jarrold MF. Thermal Unfolding of Unsolvated Cytochrome c: Experiment and Molecular Dynamics Simulations. J Am Chem Soc. 1999;121:2712–2721. [Google Scholar]
- 23.Hall Z, Politis A, Robinson CV. Structural modeling of heteromeric protein complexes from disassembly pathways and ion mobility-mass spectrometry. Structure. 2012;20:1596–1609. doi: 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Politis A, Park AY, Hyung S-J, Barsky D, Ruotolo BT, Robinson CV. Integrating Ion Mobility Mass Spectrometry with Molecular Modelling to Determine the Architecture of Multiprotein Complexes. PLoS ONE. 2010;5:e12080. doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Ion Mobility–Mass Spectrometry Reveals Long-Lived, Unfolded Intermediates in the Dissociation of Protein Complexes. Angew Chem Int Ed. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 26.Gidden J, Kemper PR, Shammel E, Fee DP, Anderson S, Bowers MT. Application of ion mobility to the gas-phase conformational analysis of polyhedral oligomeric silsesquioxanes (POSS) Int J Mass Spectrom. 2003;222:63–73. [Google Scholar]
- 27.Wyttenbach T, Bowers MT. Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility-Mass Spectrometry Environment. J Phys Chem B. 2011;115:12266–12275. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- 28.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. Number of Solution States of Bradykinin from Ion Mobility and Mass Spectrometry Measurements. J Am Chem Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-beta protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi HL, Pierson NA, Valentine SJ, Clemmer DE. Conformation Types of Ubiquitin M+8H (8+) Ions from Water: Methanol Solutions: Evidence for the N and A States in Aqueous Solution. J Phys Chem B. 2012;116:3344–3352. doi: 10.1021/jp210797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE. Characterizing Intermediates Along the Transition from Polyproline I to Polyproline II Using Ion Mobility Spectrometry-Mass Spectrometry. J Am Chem Soc. 2014 doi: 10.1021/ja505899g. [DOI] [PubMed] [Google Scholar]
- 32.Hudgins RR, Woenckhaus J, Jarrold MF. High resolution ion mobility measurements for gas phase proteins: correlation between solution phase and gas phase conformations. Int J Mass Spectrom. 1997;165–166:497–507. [Google Scholar]
- 33.Clemmer DE, Hudgins RR, Jarrold MF. Naked Protein Conformations: Cytochrome c in the Gas Phase. J Am Chem Soc. 1995;117:10141–10142. [Google Scholar]
- 34.Bleiholder C, Do TD, Wu C, Economou NJ, Bernstein SS, Buratto SK, Shea JE, Bowers MT. Ion Mobility Spectrometry Reveals the Mechanism of Amyloid Formation of A beta(25-35) and Its Modulation by Inhibitors at the Molecular Level: Epigallocatechin Gallate and Scyllo-inositol. J Am Chem Soc. 2013;135:16926–16937. doi: 10.1021/ja406197f. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Bendiak B, Siems WF, Gang DR, Hill HH. Carbohydrate Structure Characterization by Tandem Ion Mobility Mass Spectrometry (IMMS)2. Anal Chem. 2013;85:2760–2769. doi: 10.1021/ac303273z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloniecki M, Jablonowska A, Poznanski J, Langridge J, Hughes C, Campuzano I, Giles K, Dadlez M. Ion Mobility Separation Coupled with MS Detects Two Structural States of Alzheimer’s Disease A beta 1-40 Peptide Oligomers. J Mol Biol. 2011;407:110–124. doi: 10.1016/j.jmb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Bornschein RE, Hyung SJ, Ruotolo BT. Ion Mobility-Mass Spectrometry Reveals Conformational Changes in Charge Reduced Multiprotein Complexes. J Am Soc Mass Spectrom. 2011;22:1690–1698. doi: 10.1007/s13361-011-0204-y. [DOI] [PubMed] [Google Scholar]
- 38.Trimpin S, Tan B, Bohrer BC, O’Dell DK, Merenbloom SI, Pazos MX, Clemmer DE, Walker JM. Profiling of phospholipids and related lipid structures using multidimensional ion mobility spectrometry-mass spectrometry. Int J Mass Spectrom. 2009;287:58–69. [Google Scholar]
- 39.Shvartsburg AA, Li FM, Tang KQ, Smith RD. Characterizing the structures and folding of free proteins using 2-D gas-phase separations: Observation of multiple unfolded conformers. Anal Chem. 2006;78:3304–3315. doi: 10.1021/ac060283z. [DOI] [PubMed] [Google Scholar]
- 40.Ridenour WB, Kliman M, McLean JA, Caprioli RM. Structural Characterization of Phospholipids and Peptides Directly from Tissue Sections by MALDI Traveling-Wave Ion Mobility-Mass Spectrometry. Anal Chem. 2010;82:1881–1889. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campuzano I, Bush MF, Robinson CV, Beaumont C, Richardson K, Kim H, Kim HI. Structural Characterization of Drug-like Compounds by Ion Mobility Mass Spectrometry: Comparison of Theoretical and Experimentally Derived Nitrogen Collision Cross Sections. Anal Chem. 2011;84:1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- 42.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 43.Wu C, Siems WF, Asbury GR, Hill HH. Electrospray Ionization High-Resolution Ion Mobility Spectrometry–Mass Spectrometry. Anal Chem. 1998;70:4929–4938. doi: 10.1021/ac980414z. [DOI] [PubMed] [Google Scholar]
- 44.Mishra R, Jayaraman M, Roland BP, Landrum E, Fullam T, Kodali R, Thakur AK, Arduini I, Wetzel R. Inhibiting the Nucleation of Amyloid Structure in a Huntingtin Fragment by Targeting alpha-Helix-Rich Oligomeric Intermediates. J Mol Biol. 2012;415:900–917. doi: 10.1016/j.jmb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Counterman AE, Valentine SJ, Srebalus CA, Henderson SC, Hoaglund CS, Clemmer DE. High-order structure and dissociation of gaseous peptide aggregates that are hidden in mass spectra. J Am Soc Mass Spectrom. 1998;9:743–759. doi: 10.1016/S1044-0305(98)00052-X. [DOI] [PubMed] [Google Scholar]
- 46.Gessel MM, Wu C, Li HY, Bitan G, Shea JE, Bowers MT. A beta(39-42) Modulates A beta Oligomerization but Not Fibril Formation. Biochemistry. 2012;51:108–117. doi: 10.1021/bi201520b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to beta-sheet in amyloid fibril formation. Nat Chem. 2011;3:172–177. doi: 10.1038/nchem.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. Amyloid beta-protein: Monomer structure and early aggregation states of A beta 42 and its Pro(19) alloform. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 49.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Structure of the Preamyloid Dimer of beta-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendoza VL, Baron-Rodriguez MA, Blanco C, Vachet RW. Structural Insights into the Pre-Amyloid Tetramer of beta-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stocks BB, Rezvanpour A, Shaw GS, Konermann L. Temporal Development of Protein Structure during S100A11 Folding and Dimerization Probed by Oxidative Labeling and Mass Spectrometry. J Mol Biol. 2011;409:669–679. doi: 10.1016/j.jmb.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 52.Arndt JR, Brown RJ, Burke KA, Legleiter J, Valentine SJ. Lysine residues in the N-terminal huntingtin amphipathic α-helix play a key role in peptide aggregation. J Mass Spectrom. 2015;50:117–126. doi: 10.1002/jms.3504. [DOI] [PubMed] [Google Scholar]
- 53.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spec Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendoza VL, Vachet RW. Protein Surface Mapping Using Diethylpyrocarbonate with Mass Spectrometric Detection. Anal Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donohoe GC, Maleki H, Arndt JR, Khakinejad M, Yi J, McBride C, Nurkiewicz TR, Valentine SJ. A New Ion Mobility–Linear Ion Trap Instrument for Complex Mixture Analysis. Anal Chem. 2014;86:8121–8128. doi: 10.1021/ac501527y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khakinejad M, Kondalaji SG, Maleki H, Arndt JR, Donohoe GC, Valentine SJ. Combining Ion Mobility Spectrometry with Hydrogen-Deuterium Exchange and Top-Down MS for Peptide Ion Structure Analysis. J Am Soc Mass Spectrom. 2014;25:2103–2115. doi: 10.1007/s13361-014-0990-0. [DOI] [PubMed] [Google Scholar]
- 57.Kim T, Tolmachev AV, Harkewicz R, Prior DC, Anderson G, Udseth HR, Smith RD, Bailey TH, Rakov S, Futrell JH. Design and Implementation of a New Electrodynamic Ion Funnel. Anal Chem. 2000;72:2247–2255. doi: 10.1021/ac991412x. [DOI] [PubMed] [Google Scholar]
- 58.Wang FBJP, Cieplak P, Dupradeau FY. R.E.D. Python: Object oriented programming for Amber force fields. Université de Picardie - Jules Verne, Sanford|Burnham Medical Research Institute; 2013. [Google Scholar]
- 59.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A WELL-BEHAVED ELECTROSTATIC POTENTIAL BASED METHOD USING CHARGE RESTRAINTS FOR DERIVING ATOMIC CHARGES - THE RESP MODEL. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 60.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA. GENERAL ATOMIC AND MOLECULAR ELECTRONIC-STRUCTURE SYSTEM. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- 61.Gordon MS, Schmidt MW. Advances in electronic structure theory: GAMESS a decade later. In: Dykstra CE, Frenking G, Kim KS, Scuseria GE, editors. Theory and Applications of Computational Chemistry: the first forty years. Elsevier; 2005. pp. 1167–1189. [Google Scholar]
- 62.Wyttenbach T, vonHelden G, Bowers MT. Gas-phase conformation of biological molecules: Bradykinin. J Am Chem Soc. 1996;118:8355–8364. [Google Scholar]
- 63.Case DAD, TA, Cheatham TE, III, Simmerling CL, Wang L, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. Amber 12. University of California; San Francisco: 2012. [Google Scholar]
- 64.Jarrold Research Group, I. U. MOBCAL – A Program to Calculate Mobilities. [Google Scholar]
- 65.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. The J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 66.Donohoe G, Khakinejad M, Valentine S. Ion Mobility Spectrometry-Hydrogen Deuterium Exchange Mass Spectrometry of Anions: Part 1. Peptides to Proteins. J Am Soc Mass Spectrom. 2015;26:564–576. doi: 10.1007/s13361-014-1045-2. [DOI] [PubMed] [Google Scholar]
- 67.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea J-E. Structure of the 21–30 fragment of amyloid β-protein. Protein Science. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, Shea J-E. Amyloid β-protein monomer structure: A computational and experimental study. Protein Science. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bugg CW, Isas JM, Fischer T, Patterson PH, Langen R. Structural Features and Domain Organization of Huntingtin Fibrils. J Biol Chem. 2012;287:31739–31746. doi: 10.1074/jbc.M112.353839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, Frydman J. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–U1298. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary Structure of Huntingtin Amino-Terminal Region. Structure. 2009;17:1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dlugosz M, Trylska J. Secondary Structures of Native and Pathogenic Huntingtin N-Terminal Fragments. J Phys Chem B. 2011;115:11597–11608. doi: 10.1021/jp206373g. [DOI] [PubMed] [Google Scholar]
- 73.Rossetti G, Cossio P, Laio A, Carloni P. Conformations of the Huntingtin N-term in aqueous solution from atomistic simulations. Febs Letters. 2011;585:3086–3089. doi: 10.1016/j.febslet.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 74.Kelley NW, Huang X, Tam S, Spies C, Freedman J, Paned VS. The Predicted Structure of the Headpiece of the Huntingtin Protein and Its Implications on Huntingtin Aggregation. J Mol Biol. 2009;388:919–927. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Rourke JG, Gateau JR, Ochoa J, Song W, Risk T, Reverted D, Lee J, Moneys AM, Palos J, Me L, Vashishtha M, Apostol BL, Nicholson TP, Illes K, Zhu YZ, Dasso M, Bates GP, Difiglia M, Davidson B, Wanker EE, Marsh JL, Lima CD, Steffan JS, Thompson LM. SUMO-2 and PIAS1 Modulate Insoluble Mutant Huntingtin Protein Accumulation. Cell Reports. 2013;4:362–375. doi: 10.1016/j.celrep.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kokona B, Rosenthal ZP, Fairman R. Role of the Coiled-Coil Structural Motif in Polyglutamine Aggregation. Biochemistry. 2014;53:6738–6746. doi: 10.1021/bi500449a. [DOI] [PubMed] [Google Scholar]
- 77.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJL, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Atwal RS, Desmond CR, Caron N, Maiuri T, Xia JR, Sipione S, Truant R. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7:453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 79.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O’Rourke JG, Khashwji H, Lukacsovich T, Zhu YZ, Lau AL, Massey A, Hayden MR, Zeitlin SO, Finkbeiner S, Green KN, LaFerla FM, Bates G, Huang L, Patterson PH, Lo DC, Cuervo AM, Marsh JL, Steffan JS. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitomi Y, Nomura T, Kurosawa M, Nukina N, Furukawa Y. Post-aggregation Oxidation of Mutant Huntingtin Controls the Interactions between Aggregates. J Biol Chem. 2012;287:34764–34775. doi: 10.1074/jbc.M112.387035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Nat Acad Sci. 2013;110:20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagarajan A, Jawahery S, Matysiak S. The Effects of Flanking Sequences in the Interaction of Polyglutamine Peptides with a Membrane Bilayer. The J Phys Chem B. 2013 doi: 10.1021/jp407900c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.