Abstract
RNA interference (RNAi) is a conserved eukaryotic gene regulatory mechanism that uses small non-coding RNAs to mediate post-transcriptional/transcriptional gene silencing. The fission yeast Schizosaccharomyces pombe and the filamentous fungus Neurospora crassa have served as important model systems since the beginning of RNAi studies. Studies in these two organisms and other fungi have contributed significantly to our understanding of the mechanisms and functions of RNAi in eukaryotes. In addition, surprisingly diverse RNAi-mediated processes and small RNA biogenesis pathways have been discovered in fungi. In this review, an overview is given of different fungal RNAi pathways with a focus on their mechanisms and functions.
Keywords: RNA interference, Small non-coding RNA, Saccharomyces pombe, Neurospora
INTRODUCTION
RNA interference (RNAi) was first described in 1998 in C. elegans as a phenomenon triggered by double-stranded RNA (dsRNA) that results in the silencing of the genes complementary to the dsRNA (31; 72). This process was initially thought to be a host defense mechanism against invading viruses and transposons (81); however studies in the last decade have demonstrated that RNAi pathways use small non-coding RNAs (sRNAs) to regulate diverse cellular, developmental and physiological processes. RNAi pathways are conserved in eukaryotes and among the identified sRNAs, there are three major classes: small interfering RNA (siRNA), microRNA (miRNA) and piwi-interacting RNA (piRNA) (10; 34; 54; 73; 100). Each class of sRNAs is produced differently and has diverse functions. In the siRNA and miRNA pathways, the RNase-III like Dicer enzymes cleave dsRNA/hairpin RNA to generate 21–25 nucleotide mature siRNA or miRNA duplexes. The duplex is incorporated into an effector complex, the RNA-induced silencing complex (RISC), containing an Argonaute protein with slicer activity. The passenger strand of the sRNA duplex is removed and the remaining guide RNA directs the RISC to complementary mRNA sequences, resulting in silencing either by mRNA cleavage or translational repression.
The fungi kingdom is a large and diverse group of eukaryotic organisms estimated to contain more than three million species that as a group have diverse and vital roles in the ecosystem (80). Although RNAi was first demonstrated in C. elegans, the RNAi-related phenomena were first described and studied in plants and fungi (76; 85). Quelling, the RNAi-related post-transcriptional gene silencing phenomenon in Neurospora was reported in 1992 (85). Since then, the identification of quelling components and analysis of the pathway have contributed significantly to our understanding of RNAi-mediated gene silencing at the post-transcriptional level. RNAi was first found to be required for heterochromatin formation in the fission yeast Saccharomyces pombe (105), and this system remains the best understood pathway of RNAi-mediated transcriptional gene silencing. In addition, RNAi components and their functions have also been characterized in many fungal species (7). Furthermore, many classes of sRNAs have been found in different fungal species, revealing diverse sRNA biogenesis pathways. In this review, we will focus on the mechanisms and functions of different fungal RNAi and sRNA production pathways.
QUELLING IN NEUROSPORA
The discovery of quelling
Soon after the discovery of co-suppression in plants, Macino and his colleagues discovered a similar transgene-induced gene silencing phenomenon called quelling in Neurospora crassa (76; 85). Quelling was discovered by transforming Neurospora with DNA containing albino-1 or albino-3 genes, which are involved in carotenoid biosynthesis. After transformation, white/pale yellow transformants with reduced al mRNA levels were obtained, indicating the silencing of endogenous al genes by the transgenes (9; 20; 32; 85). High copy numbers of the transgenes correlate with the quelling efficiency, suggesting that the presence of repetitive transgene causes silencing. In addition, the quelled transformants were found to spontaneously and progressively revert to wild-type or intermediate phenotypes due to reductions in copy number of the transgene.
The al quelled strains are dominant over wild-type strains, suggesting that diffusible and trans-acting silencing molecules are involved in quelling (20). In addition, a sense RNA derived from the transgene was specifically detected in quelled strains, suggesting that aberrant transcription of transgenes is involved in silencing (20). Furthermore, it was determined that quelling did not affect the levels of mRNA precursors, indicating that quelling is a post-transcriptional gene silencing (PTGS) mechanism (20). Together, these results led to the hypothesis that the production of aberrant RNA (aRNA) in the presence of multi-copy of transgenes causes PTGS.
Molecular mechanism of quelling
Macino and colleagues isolated quelling-defective (qde) mutants, which were mapped to three complementation groups, called qde-1, qde-2, and qde-3 (21). The cloning and studies of these qde genes contributed significantly to our understanding of an RNAi pathway that is well conserved across eukaryotes (13; 14; 21; 25). QDE-1, which encodes a cellular RNA-dependent RNA polymerase (RdRP), was the first eukaryotic RNAi component identified (22); it was discovered soon after the demonstration of the silencing effect of dsRNA in C. elegans (31). The requirement of an RdRP in quelling demonstrates that dsRNA is an essential intermediate of PTGS in vivo.
QDE-2 encodes an Argonaute protein that is homologous to RDE-1 in C. elegans, which is required for dsRNA-mediated silencing (11). The identification of QDE-2 provided the first evidence that RNAi in C. elegans and quelling share a common genetic mechanism.
QDE-3 belongs to the RecQ helicase family and is homologous to the mammalian Bloom’s and Werner’s syndrome proteins (23). RecQ helicases are known to be involved in homologous recombination, DNA replication and DNA repair. As expected, QDE-3 contributes to DNA repair process in Neurospora, (23; 51). The requirement of QDE-3 in quelling raises the possibility that repetitive transgenes may form aberrant DNA structures that are recognized by QDE-3 to promote aRNA and sRNA production. Interestingly, OsRecQ1, a RecQ helicase homolog in rice, is required for inverted-repeat induced RNA silencing (16). rRecQ-1, a homolog of QDE-3 in rats, is associated with the piRNA-binding complex (58).
Expression of an inverted repeat-containing transgene can bypass QDE-1 and QDE-3 to trigger gene silencing (14; 61), indicating that QDE-1 and QDE-3 function in producing aRNA and dsRNA from repetitive transgenes. In addition, these results suggest that QDE-1 is not involved in sRNA amplification. Although QDE-1 is expected to function as an RdRP to convert aRNA into dsRNA, it was not clear how aRNAs are produced and specifically recognized by QDE-1. In plants, RNA polymerases Pol IV and Pol V are required for production of the single-stranded non-coding RNA precursors of dsRNAs and siRNAs (71; 108). In the fission yeast, RNA Pol II is the RNA polymerase that produces the centromeric siRNA precursors (88). Structure of QDE-1 revealed that its catalytic core is similar to that of eukaryotic DNA-dependent RNA polymerase (DdRP) (87). In addition, QDE-1 is required for the production of rDNA-specific aRNA (61). Recently we demonstrated that QDE-1 is both DdRP and RdRP and can use either single-stranded DNA or RNA (ssDNA or ssRNA) as templates to produce DNA/RNA hybrid and dsRNA, respectively (60; 61). Together, these results suggest that QDE-1 is an RNA polymerase that produces aRNA from ssDNA in the quelling pathway (61).
How is QDE-1 recruited to ssDNA substrates to produce aRNA? QDE-1 associates with RPA-1, which is a subunit of Replication Protein A (RPA), a ssDNA binding complex involved in DNA replication, recombination and repair (60; 78). Deletion of rpa-3 in Neurospora abolished quelling and DNA damage-induced production of qiRNAs (see below) (60). In addition, the interaction between QDE-1 and RPA requires QDE-3. QDE-1 can use ssDNA templates to produce dsRNA, an activity that is strongly promoted by RPA as it prevents the formation of DNA/RNA hybrids. These results suggest that RPA has two roles in the production of aRNA: It recruits QDE-1 to ssDNA with the aid of QDE-3, and it blocks the formation of DNA/RNA hybrids. This mechanism explains how aRNAs are produced and are specifically recognized by an RdRP in the quelling pathway. Repeat-induced gene silencing has been reported in many systems, including fungi, plants, C. elegans, Drosophila and mammals (46). The mechanism of how repetitive DNA sequences trigger the production of sRNA remains largely unclear. The involvement of QDE-3 and RPA in quelling suggests that DNA repeats may result in some aberrant DNA structures during the DNA replication or recombination processes that may then be recognized by RNA polymerases to produce aRNAs and dsRNA.
Two partially redundant Neurospora Dicer proteins (DCL-1 and DCL-2) process dsRNA into about 25-nt small RNAs in an ATP-dependent manner (14). The deletion of both dicer genes completely abolishes quelling and Dicer activity. QDE-2 associates with siRNA duplexes and forms an inactive RISC complex (11; 12). In order to activate the RISC complex, the passenger strand of the siRNA duplex must be removed. We showed that QDE-2 and its slicer activity are required for single-stranded siRNA production, indicating that QDE-2 slices the passenger strand of siRNA to produce a nicked siRNA duplex (70); this reaction is analogous to the slicing of an mRNA substrate. This provides the first in vivo evidence that the Argonaute is involved in generating single-stranded siRNA. However, QDE-2 alone is not sufficient to remove the passenger strand. Biochemical purification of QDE-2 led to the identification of a QDE-2 interacting protein called QIP; this putative exonuclease is required for the removal of the passenger strand of siRNAs (70). Together, these results indicate that both the cleavage and removal of passenger strand of siRNA duplex are essential steps in RISC activation. A Drosophila ribonuclease, C3PO (component 3 promoter of RISC), was later found to promote RISC activation by removing siRNA passenger strand cleavage products (68), indicating that the RISC activation mechanism is similar in Neurospora and animals.
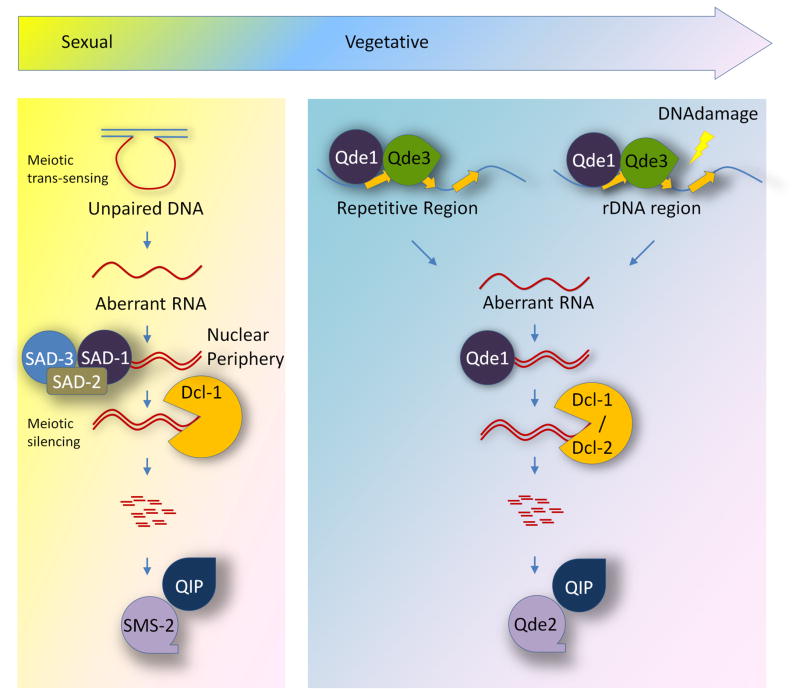
The current model for the Neurospora quelling pathway is depicted in Figure 1. Repetitive DNA sequences form aberrant DNA structures after encountering DNA replication stress. These aberrant DNA structures are recognized by QDE-3 and RPA, which recruit QDE-1 to the ssDNA locus. QDE-1 acts first as a DdRP to produce aRNAs, which are then converted into dsRNAs by the DdRP activity of QDE-1. The long dsRNA precursors are processed into siRNA duplexes by Dicer, and siRNAs are then loaded onto the RISC. QDE-2 cleaves and removes the passenger strand with the help of QIP to form an active RISC associated with single-stranded siRNA guide strand that targets complementary mRNA substrates for degradation. Although the mechanism of quelling downstream of dsRNA is well understood, how repetitive DNA sequences trigger the formation of aberrant DNA structures, which are specifically recognized by QDE-3, RPA and QDE-1, is not known and should be a major focus of future studies.
Figure 1.
Models for RNAi-related pathways in Neurospora. During vegetative stage (right), both repetitive transgene (quelling) and ribosomal DNA loci (qiRNA) lead to the production of aRNAs by QDE-1 and QDE-3. Single-stranded aRNAs are then converted to dsRNA precursors by QDE-1. dsRNA are processed by DCLs into sRNAs, which are then loaded onto QDE-2/QIP-based RISC to execute gene silencing. During meiosis (left), unpaired DNA can be sensed and results in the production of aRNAs. aRNAs are converted to dsRNA by SAD1, SAD2, and SAD3 in the nuclear periphery. dsRNA are processed by DCL-1 into sRNAs, which are then loaded onto an SMS-2/QIP-based RISC to execute gene silencing.
qiRNA, a type of sRNA induced by DNA damage
Quelling triggered by repetitive transgenes occurs in the vegetative Neurospora culture under normal growth conditions. Intriguingly, we recently uncovered a class of QDE-2 interacting sRNA, designated qiRNA, which is specifically induced after Neurospora is treated with a DNA damaging agent (61). qiRNAs mostly originate from the ribosomal DNA (rDNA) locus, which is the only highly repetitive sequence in the wild-type Neurospora genome. These sRNAs are ~21–23 nt in length and generally have a 5′ uridine. qiRNA biogenesis requires QDE-1, QDE-3 and the Dicers, indicating that qiRNAs are specific sRNA species made by the RNAi machinery upon DNA damage (61). qiRNAs originate from aRNA precursors. In fact, aRNAs ranging in size from 500 bp to 2 kb from the intergenic spacer regions accumulate in the dcl double mutant. The production of aRNA is abolished in the qde-3 mutant, indicating that the RecQ helicase QDE-3 is required for the biogenesis of DNA-damage-induced aRNA. In addition, QDE-1 is essential for aRNA production, but the conventional RNA polymerases (Pol I, Pol II and Pol III) are not, suggesting that QDE-1 acts as a DdRP to produce the DNA damage-induced aRNAs.
DNA damage results in cell arrest, decrease of DNA replication and protein synthesis. In mutants that are defective in qiRNA synthesis, DNA damage-induced reduction of protein synthesis is blunted (61). QDE-3 plays a role in DNA damage repair (23; 51). Furthermore, qde-1 and dcl mutants also exhibit increased sensitivity to DNA damage agents, suggesting that qiRNAs contribute to DNA damage checkpoints by inhibiting protein synthesis (61). In Arabidopsis, RNAi components are enriched in the nucleolus, and rDNA-derived siRNAs facilitate heterochromatin formation (83). Furthermore, the Drosophila RNAi-deficient mutants have disorganized nucleoli and rDNA loci (82). These studies suggest that rDNA-derived sRNAs have a shared role in eukaryotes to maintain genome integrity.
Both quelling and qiRNA pathways appear to require the same components, suggesting that they are mechanistically similar (60; 61). The repetitive nature of transgenes and the rDNA locus is very likely to be the common trigger for quelling and qiRNA production. The production of qiRNA is induced by DNA damage, suggesting that replication stress or double-stranded breaks trigger formation of aberrant DNA structures. Although quelling occurs under normal growth conditions, it is likely that the transgene integration loci are fragile DNA sites with elevated levels of replication stress (28; 35). If so, quelling should also be the result of DNA damage or replication stress at the transgene integration loci trigged by presence of repetitive DNA sequences.
MEIOTIC SILENCING BY UNPAIRED DNA
Meiotic silencing by unpaired DNA (MSUD) was originally discovered during studies on meiotic transvection of the ascospore maturation 1 gene (asm-1) in Neurospora (3; 93; 94). Unlike quelling, which takes place in the vegetative stage, MSUD occurs during meiosis. Neurospora is a haploid ascomycete and becomes transiently diploid during meiosis following the fusion of two haploid nuclei from opposite mating types. MSUD occurs in prophase I of meiosis when unpaired DNA sequences are present and leads to the silencing of all homologous genes in the diploid ascus cell. It has been proposed that trans-sensing and meiotic silencing are two related but distinct steps in MSUD (53; 84). The trans-sensing step should occur in the nucleus, where the presence of unpaired DNA during meiosis is sensed and likely triggers the production of unpaired DNA-specific aRNA, whereas the meiotic silencing step will convert the aRNA into dsRNA and sRNA, resulting the sRNA mediated gene silencing. Although, the mechanism of trans-sensing is still largely unclear, DNA methylation appears to influence trans-sensing without affecting silencing (84). Recently, MSUD was also found to operate in the homothallic fungus Gibberella zeae (96).
A breakthrough in understanding the mechanism of MSUD was achieved by forward genetic screens for mutants that suppress meiotic silencing (94). The first gene identified that is required for MSUD is sad-1 (suppressor of ascus dominance-1), which encodes an RdRP and is a paralog of QDE-1 (93; 94). SAD-2 was the second component of MSUD identified (5; 95). SAD-1 and SAD-2 co-localize in the perinuclear region and interact physically. A functional sad-2 gene is required for the proper localization of SAD-1, indicating that SAD-2 might function to recruit SAD-1 to the perinuclear region. The fact that SAD-1 encodes a putative cellular RdRP indicates that MSUD is an RNAi-related phenomenon. In addition, sms-2 (suppressor of meiotic silencing-2), an Argonaute protein homolog in Neurospora, is also required for MSUD (59). Although a sms-2 mutant did not show obvious defects during vegetative growth, the homozygous crosses of sms-2 mutants are completely barren. In contrast, QDE-2 is not required for MSUD, indicating that there are two parallel RNAi pathways functioning separately in the vegetative and meiotic stages (59).
MSUD was further established as an RNAi-related pathway from studies of the role of Neurospora Dicer proteins in meiotic silencing (1). Unlike the single mutants of sad-1 and sad-2, the dcl-1 single mutants did not function as dominant suppressors of meiotic silencing (1; 94; 95). However, when DCL-1 is inactivated at a later stage of the sexual cycle, meiotic silencing is suppressed, indicating a critical role for DCL-1 in MSUD, and therefore a requirement for dsRNA and sRNA in meiotic silencing. In addition, DCL-1 co-localizes with other MSUD components, including SAD-1, SAD-2 and SMS-2, in the nuclear periphery (1; 95). Interestingly, DCL-2, the major Dicer in the quelling pathway is not required for MUSD (14). QIP, the exonuclease that is required for the RISC activation in the quelling pathway, is also involved in MSUD (42; 60; 109). Like DCL-1, QIP is localized in the perinuclear region with other meiotic silencing components.
Recently, a putative RNA/DNA helicase, SAD-3, was demonstrated to be required for meiotic silencing (42). SAD-3 is homologous to S. pombe Hrr1, which is a component of the RNA-directed RNA polymerase complex required for the RNAi-mediated heterochromatin formation in fission yeast (75). This link raises the possibility that MUSD and the RNAi-induced heterochromatin assembly may be mechanistically related. SAD-3 is also found in the perinuclear region and associates with SAD-1, SAD-2 and SMS-2 (42). The fact that all the known components involved in MUSD are localized in the perinuclear region and associate with each other suggests that they form a large meiotic silencing complex in this region. It is likely that this complex function by converting the aRNA produced in the nucleus into dsRNA and then sRNA to trigger gene silencing.
A simple model of the MSUD pathway is shown in Figure 1: During meiosis, the pairing of the homologous chromosomes leads to the detection of unpaired DNA region and production of aRNA transcripts. In the perinuclear region, aRNA transcripts are converted into dsRNA by SAD-1, with the help of SAD-2 and SAD-3. DCL-1 process dsRNA into sRNAs, which are then loaded onto an SMS-2/QIP-based RISC to execute post-transcriptional silencing of homologous genes. Despite the identification of many components involved in the meiotic silencing step of the MUSD pathway, little is known about the trans-sensing mechanism. First, what is the mechanism that scans and senses the unpaired DNA in homologous chromosomes during meiosis? DNA methylation is known to be involved at this step (84), suggesting that chromatin structures are important. Second, what is the DdRP that produces the aRNA and how is the specificity achieved? In quelling, QDE-1 serves as both DdRP and RdRP (60). It will be interesting to test whether QDE-1/SAD-1 has a similar role in MUSD. In addition, some genes appear to be immune to MSUD, such as rDNA clusters and the mating type genes mat A and mat a, which have very different in DNA sequences (92; 94). Thus, there must be a mechanism that protects certain genes from silencing during meiosis in Neurospora. Finally, since the discovery of meiotic silencing in Neurospora, unpaired homologous DNA-triggered silencing during meiosis has been described in C. elegans, Drosophila and mammals (53). Although the silencing mechanisms in Drosophila and mammals appear to be independent of RNAi, it is still not clear whether MUSD in Neurospora is mechanistically similar to the silencing phenomenon in animals.
RNAI-MEDIATED HETEROCRHOMATIN FORMATION IN SCHIZOSACCHAROMYCES POMBE
Heterochromatin was initially considered to be transcriptionally inert. Heterochromatin in the fission yeast S. pombe is distributed in three different loci: telomeres, centromeres and the mating-type loci. The centromeres of fission yeast are similar to those of humans in their structure and epigenetic modifications. The central core region sequence is unique for every centromere, and this region is where the kinetochore binds. It is flanked by two types of repeated DNA sequences, the innermost (imr) and outermost (otr) repeats. The outermost pericentromeric repeats are further comprised of the dg and dh sequences; these sequences are coated with histone H3 that is methylated at lysine 9 (H3K9). These specialized chromatin modifications provide docking sites for chromodomain proteins that maintain the transcriptionally silent status of the heterochromatin (29; 65).
A direct link between heterochromatin formation and an RNAi pathway was first established in the fission yeast in 2002, when Volpe et al. (104; 105) showed that RNAi components are required for the heterochromatin formation in the centromeric regions. In addition, transcription is detected in the heterochromatin region and the heterochromatic RNAs accumulate in mutants of the RNAi pathway including those in the RNA-dependent RNA polymerase (rdp1), dicer (dcr1) and argonaute (ago1). The transcripts arising from the heterochromatin regions are mapped to both strands of DNA, indicating a dsRNA precursor. Reduced H3K9 methylation and increased H3K4 methylation are also observed in these mutants. Furthermore, small RNAs derived from the centromeric region are detected, and Rdp1 is enriched at the centromere (105). Together, these results demonstrated the requirement of the S. pombe RNAi pathway in heterochromatin formation.
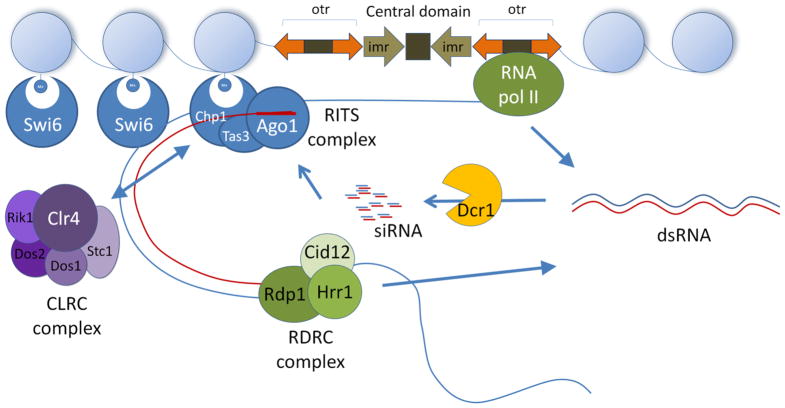
In fission yeast, heterochromatin at the centromeric regions is formed through a self-enforcing loop orchestrated by the RNAi components and chromatin-modifying components (7; 37; 73; 103) (Figure 2). This self-enforcing loop is brought about by the transcription of the pericentromeric nascent transcripts by RNA Pol II during the S phase of the cell cycle. Rdp1 converts the nascent transcripts into dsRNA, which Dcr1 recognizes and process into siRNA. The siRNA is then loaded onto Ago1 to form the RNA-induced transcriptional silencing (RITS) complex (101; 102).
Figure 2.
Model for RNAi-mediated heterochromatin formation in S. pombe. siRNA guided RITS complex targets the centromeric nascent transcript synthesized by RNA polymerase II during the S phase of the cell cycle. RITS associates with RDRC complex, where the RdRP activity of the RDRC complex generates more dsRNA. Dcr1 processes the dsRNA into siRNA, feeding into the loop. CLRC complex associates with the RITS complex, which comes into close proximity with the chromatin. Clr4 methylates H3K9 and allows Swi6 (HP1 homolog) to dock at the modified chromatin, thereby forming heterochromatin.
RNA polymerase II appears to be the DdRP responsible for the production of centromeric siRNA in S. pombe. The role of Pol II in this process was supported by the identification of two point mutations in the RNA Pol II subunits (Rpb2 and Rpb7)(52). These mutations impair the Pol II elongation step during transcription resulting in reduced levels of heterochromatic transcripts and siRNA production. Genome-wide transcriptional analyses suggested that these defects are not due to an indirect effect caused by changes in global gene expression. Furthermore, neither mutation altered the Pol II association with the centromeric DNA repeats, suggesting that the defects in siRNA production in these mutants are not due to impairment of Pol II recruitment to the centromeric DNA.
The identification of the RITS complex provided a mechanistic link between siRNA and heterochromatin assembly (101). The RITS complex consists of Ago1, Chp1 and Tas3. Chp1 is a chromodomain-containing protein that binds the histone H3K9me, which recruits the RITS to the pericentromeric region. The active Ago1 bound to a single-stranded guide siRNA also reinforces this interaction through homology-dependent association with the centromeric nascent RNA transcripts.
The production of the nascent transcripts provides a template for Rdp1 (105). Rdp1 is part of the RNA-dependent RNA polymerase complex (RDRC) that also contains Hrr1 and Cid12 (75). Both Rdp1 and Hrr1 associate with heterochromatin and centromeric (cen) RNA transcripts (cen RNA). Rdp1 generates dsRNA in a primer-independent manner from single-stranded cen RNA, and it has been proposed that the RITS complex acts as a priming complex for RDRC to localize to the centromeric DNA. Dicer cleaves the dsRNA into siRNA and greatly enhances the activity of RDRC in siRNA production in vitro (24). Although Dcr1 was previously shown to be mostly cytoplasmic (43), a recent study showed that Dicer is also present in the nucleus, suggesting that Dcr1-mediated siRNA production occurs in cis with transcription(30). Dcr1 may shuttle between the nucleus and cytoplasm, and the retention of Dcr1 in the nucleus is essential for the production of centromeric siRNAs.
The recruitment of RDRC to the centromeric region is dependent on Clr4 (75). Clr4, a histone methyltransferase, is recruited to the pericentromeric region where it methylates H3K9. The methylated chromatin further recruits the HP1 homolog, Swi6, Chp1, Chp2 and Clr4 to the region (86; 111). siRNA production is greatly reduced in the Clr4 mutants. Clr4 associates with Rik1, Dos1, Dos2 and Cul4 in a complex called CLRC; all of the components in this complex are required for heterochromatin assembly (44; 45; 48; 66). The recruitment of Swi6 to the heterochromatin allows the recruitment of cohesion; whereas the binding of Clr4 and Chp1 stabilize CLRC and RITS at the heterochromatin. Together, this allows the reinforcement of silencing signal and spreading of heterochromatin (Figure 2).
Two models have been previously proposed to explain how RITS is recruited to specific chromosome regions. In the first model, the RITS is guided by siRNA to the centromeric DNA through direct base pairing, which would require unwinding of the DNA double helix. In the second model, the siRNA is targeted to the nascent transcript produced from the pericentromeric region. RNA immunoprecipitation demonstrated that RITS and RDRC are associated with the non-coding cen RNA and that this association requires Clr4 methyltransferase and Dcr1 (75). These results suggest that recruitment of RITS to the centromeric regions is not direct but rather is mediated through the siRNA-targeted binding of AGO1 to the nascent transcripts (7; 8; 107). This RNA platform model was tested in vivo by tethering the Tas3 (a component of the RITS complex) to the RNA transcript of normally active ura4+ gene. In this system, recruitment of RITS to the RNA led to transcriptional silencing of ura4+ (4; 7). This study demonstrated that the association of RITS with the nascent transcript is sufficient to trigger H3K9me and transcriptional gene silencing.
The formation of heterochromatin in the centrometric regions is cell-cycle dependent (16; 56). The DNA replication of the heterochromatin is known to occur in early S phase. This is also precisely the time during the cell cycle when pericentromeric dg/dh transcripts are detected. In addition, Pol II localization to the centromeric regions is detected in the early S phase when the level of H3K9 methylation is at its lowest level. In S phase, RNAi components accumulate at the pericentromeric region and H3K9 methylation marks are detected. By the end of replication, the epigenetic marks of heterochromatin are faithfully passed on to the sister chromatids. The H3K9me3 levels further increase as the cell cycle progresses and peak during the G2 phase of the cell cycle. Thus, the RNAi components in fission yeast aid in the formation of heterochromatin and also assist the cell in completing DNA synthesis in heterochromatic regions (110). Co-transcriptional RNAi releases the RNA Pol II from the centromeric regions, which allows the completion of DNA replication and subsequent spreading of histone modifications.
Recent advances in our understanding of the RNAi-mediated heterochromatin formation have demonstrated how RNAi and chromatin modifying components are recruited specifically to the centromeric loci and have revealed the time frame in which these processes occur. However, several recent studies have raised questions regarding the initial trigger that results in heterochromatin formation at the centromeric regions. Halic et al. proposed that the production of “primal” RNAs (priRNAs) are initiators of this process (39). These priRNAs are thought to be the products of degraded transcripts, are generated independently of Dcr1 and RDRC and are loaded onto Ago1. The primed Ago1-priRNAs complexes target the centromeric loci and further recruit Clr4, which methylates H3K9 and initiates the RNAi-mediated heterochromatin formation. Results of another study led to an alternative hypothesis that the CLRC complex is recruited to the centromeric regions independently of RNAi components (91). Consistent with this hypothesis, over-expression of Clr4 in a clr4 ago1 double mutant was able to induce de novo H3K9me2 at the centromeric regions, suggesting RNAi may be dispensable for initiation of heterochromatin formation but critical in maintaining the heterochromatic status. Forcibly tethering the Clr4 methyltransferase to a euchromatic region is also sufficient to establish heterochromatin formation in the absence of RNAi components (50). Although a tremendous amount of progress has been made, further studies will be needed to fully understand the mechanism of RNAi-mediated heterochromatin formation in the fission yeast.
RNAI IN BUDDING YEASTS
Although the RNA silencing pathway is highly conserved throughout the fungal kingdom, some budding yeasts such as Saccharomyces cerevisiae lack homologs of Argonaute, Dicer and RdRP. However, sequence homologs of Argonaute but not other RNAi components are present in some of the budding yeasts, such as S. castellii, Kluyveromyces polysporus and Candida albicans (27). These yeasts use non-canonical Dicer proteins to produce siRNAs, which mostly map to transposable elements and subtelomeric repeats. Remarkably, the expression of the S. castellii argonaute and dicer genes in S. cerevisiae activates an RNAi pathway that inhibits transposon transposition. These results suggest that RNAi pathway is an ancient host defense mechanism that has been lost in some of fungal species.
MIRNA-LIKE SRNA IN NEUROSPORA
miRNAs are endogenous ~21 nt RNA duplexes processed by Dicers from single-stranded RNA precursors with a hairpin structure. miRNAs have been discovered in animals, plants and algae and regulate physiological and developmental processes by targeting mRNA for cleavage or translational repression (2; 6; 38; 57; 63; 64; 69; 74). miRNA was thought to be absent in fungi until our recent identification of miRNA-like small RNAs (milRNAs) in Neurospora (62). There are at least 25 Neurospora milRNA-producing loci identified. Several lines of evidence suggest that milRNAs in Neurospora regulate gene expression similar to animal and plant miRNAs. First, the mRNA of predicated target genes of milR-1 are up-regulated in the dcl and qde-2 mutants. Second, the expression of a reporter gene containing an milR-1 target site is increased at in the qde-2 strain. Third, QDE-2 specifically associates with the predicted milR-1 target mRNAs. Interestingly, milRNAs do not appear to play critical roles in Neurospora growth or developmental processes because qde-2 or milRNA knock-out strains grow normally. Future studies are needed to establish the physiological importance of milRNAs in Neurospora. Although currently no miRNA-like sRNA has been experimentally confirmed in other fungal species, a study of the wheat pathogen Mycosphaerella graminicola found 65 potential milRNA-producing loci that produce sRNAs (36).
Unlike the animal and plant miRNAs, the Neurospora milRNAs are produced by at least four different biogenesis pathways that utilize different combinations of components, including Dicers, QDE-2, QIP and an RNase III domain-containing protein, MRPL3 (62). Of the four miRNAs that were investigated in detail, only the biogenesis of milR-3 is mechanistically similar to that of miRNAs in plants; it requires only Dicer for the generation of pre-milRNA and milRNA (49). The production of milR-4 is only partially dependent on Dicers, indicating the involvement of other nucleases in milRNA biogenesis. milR-1 is the most abundant small RNA-producing locus in the Neurospora genome, and production of milR-1 requires Dicers, QDE-2 and catalytically active QIP. The pri-milR-1 is first processed by Dicer to produce pre-milR-1. Afterwards, QDE-2 binds to pre-milR-1 and recruits the exonuclease QIP to process the pre-milR-1 into mature milR-1. In this pathway, the Argonaute QDE-2 functions as a scaffold for milR-1 maturation.
Although biogenesis of milR-1, milR-3 and milR-4 are completely or partially dependent on Dicers, the biogenesis of milR-2 is completely independent of Dicer. Instead, the production of milR-2 requires catalytically active QDE-2. Our results suggest that QDE-2 binds to the pre-milRNA and cleaves the passenger strand using its slicer activity. The milR-2 biogenesis pathway is the first known example of a Dicer-independent – but Argonaute-dependent – mechanism for small RNA production. Soon after our study of milR-2 biogenesis, miR-451 in both mouse and zebrafish was shown to be produced by a similar mechanism (15; 19), suggesting that this is a conserved sRNA biogenesis pathway. The surprisingly diverse milRNA pathways in Neurospora shed important light on the biogenesis and evolution of eukaryotic miRNAs.
RNAi AS A HOST-DENFENSE MECHANISM IN FUNGI
Transposon control in Neurospora
Quelling results in the silencing of repetitive transgenes in Neurospora, suggesting that quelling is a mechanism that silences transposons. In most Neurospora strains, there is no functional transposon due to a process called repeat induced point mutation (RIP) that can mutate repetitive sequences during meiosis (33). A functional LINE-like transposon was previously identified in an African strain (55), and Nolan et al. introduced the Tad transposon into laboratory Neurospora strains and showed that QDE-2 and Dicer, but not QDE-1 and QDE-3, are required for the suppression of transposon replication (77). These results suggest that transposition may generate inverted repeats that lead to the production of dsRNA, resulting in the silencing of transposon.
An antiviral mechanism in Cryphonectria parasitica and Aspergillus nidulans
Virus infection often leads to the production of siRNAs derived from viral double-stranded RNA, suggesting a role for RNAi as part of the innate immunity mechanism against viral replication (26). Plants, arthropods, nematodes and some animals rely on the RNAi pathway to protect them from viral infection. To counter the effects of RNAi, viruses often express viral suppressors of RNA silencing (VSRs) that can inhibit RNAi. In fungi, a similar RNAi-based antiviral mechanism was first observed in the Ascomycete filamentous fungus, Cryphonectria parasitica (90; 97; 112; 113). C. parasitica provides a rich resource for study of basic aspects of virus-fungus interactions since it can support the replication of five virus families and is a well-established experimental system for the study of mycoviruses.
Like Neurospora, C. parasitica has two dicer-like genes, dcl1 and dcl2. Hypovirus or mycoreovirus infection of the dcl2 mutant results in a severe growth phenotype and elevated viral RNA levels (90), indicating that DCL2 is the major Dicer in C. parasitica. Consistently, 21–22 nt virus-derived sRNAs were abolished in the dcl2 mutant (113). Although there are four Argonaute-like genes in C. parasitica, only agl2 is required for the antiviral defense response (97). Together, these observations demonstrate that RNAi acts as a defense mechanism against viruses.
The mycovirus Cryphonectria hypovirus 1 (CHV1) expresses p29, a papain-like protease similar to the plant potyvirus-encoded suppressor of RNA silencing HC-Pro (17; 89; 98; 99). Deletion of p29 results in the reduction of viral RNA levels. p29 suppresses the hairpin RNA-induced, virus-induced and agroinfiltration-induced RNA silencing, indicating that it is a suppressor of RNA silencing. When C. parasitica is infected by a mutant hypovirus without p29, the mRNAs of dcl2 and agl2 accumulate to high levels, suggesting that p29 represses RNAi by inhibiting the expression of RNAi genes.
In addition to their roles in silencing viral replication by degrading viral RNAs, RNAi components also promote viral RNA recombination in C. parasitica, which leads to the production of hypovirus defective-interfering (DI) RNAs (97; 112). The DI RNAs are a result of deletion events during recombination; DI RNAs suppress parental RNA accumulation causing symptoms to attenuate. Importantly, the DI RNAs do not accumulate in agl1, agl2 or dcl2 mutants, indicating that these RNAi components are also involved in hypoviral RNA recombination and DI RNA production. The RNAi components may contribute to viral recombination by providing 5′ and 3′ fragments of viral RNA to the viral RdRP, but future studies are needed to determine such a mechanism.
Studies of the RNAi pathway in Aspergillus nidulans also provided insights into the fungal-virus interaction. A. nidulans has one Argonaute and one Dicer gene; both required for dsRNA-triggered gene silencing in the organism (41). Although there are two RdRP genes in the genome, neither is required for dsRNA-induced gene silencing, suggesting the absence of an sRNA amplification step by RdRPs (41). Infection with the Aspergillus virus 1816 suppresses the dsRNA-induced RNA silencing (40), indicating the existence of an RNA silencing suppressor encoded by this virus. In addition, the virus 341-derived siRNA was detected at a high level in the Argonaute mutant, indicating that this virus is targeted by the RNAi machinery (40).
dsRNA/viral infection-induced transcriptional response in Neurospora and C. parasitica
Innate immunity, triggered after an organism detects dsRNA or foreign particles, is an important part of the eukaryotic host-defense mechanism. The mechanism activates a signaling cascade, which in turn activates the transcription of effector genes that defend against the foreign invader. In Neurospora, a similar transcriptional response is observed upon the induction of dsRNA expression, which induces the expression of key RNAi components, QDE-2 and DCL2, and other putative anti-viral genes are up-regulated (18). The transcriptional response is retained in the dcl1/2 double mutant, indicating that the response is induced by dsRNA not siRNA. Furthermore, the known RNAi components in Neurospora are not required for this response. Interestingly, dsRNA significantly induces the expression of QDE2 both transcriptionally and post-transcriptionally. In the dcl double mutant, despite the induction of qde-2 transcription, the QDE-2 protein levels stay constant after dsRNA expression. This suggests that siRNAs are important for stabilizing the QDE2 protein. The induction of QDE-2 by dsRNA is required for efficient RNAi since mutants that lack this response have lower RNAi efficiency.
Genome-wide analyses identified around 60 genes that are activated by dsRNA. In addition to RNAi components, many of these genes are homologs of antiviral and interferon-stimulated genes, suggesting that the dsRNA-induced transcriptional response is part of an ancient host defense response. Because Neurospora does not encode homologs of the known mammalian dsRNA sensors, the transcriptional program triggered by dsRNA should represent a novel signaling cascade.
Interestingly, in C. parasitica, both the expression of dcl2 and agl2 transcripts are strongly induced upon viral infection and expression of hairpin RNA (112; 113), indicating that this antiviral transcriptional activation response is conserved in filamentous fungi. In addition, the deletion of p29, also results in up-regulation of both dcl2 and agl2. Moreover, by using a GFP reporter fused with the promoter of agl2, it was demonstrated that the induction of agl2 by viral infection is regulated at the transcriptional level (97). Interestingly, the viral infection-induced dcl-2 expression is blocked in the agl2 mutant, suggesting that AGL2 plays a regulatory role in this gene activation pathway. These results suggest that the transcriptional regulation of RNAi components is important for the host antiviral response.
RNA SILENCING AND SMALL RNA STUDIES IN OTHER FILAMENTOUS FUNGI
Gene silencing and small RNAs in Mucor circinelloides
Mucor circinelloides, which causes invasive maxillofacial zygomycosis, is an important system for studying RNAi in the zygomycete clade. Transformation with hairpin RNA-producing constructs and self-replicative plasmids show that this organism can silence gene expression. Silencing results in the accumulation of two size classes of siRNAs (21 and 25 nt). M. circinelloides has two redundant dicer (dcl1 and dcl2) and two rdrp genes (rdrp1 and rdrp2); DCL-2 plays the major role in the RNA silencing pathway and is induced both by sense and hairpin transgenes. Secondary sRNA were detected, suggesting an sRNA amplification step in the RNA silencing pathway.
Detailed examination of sRNA profile in this fungus revealed four classes of endogenous small RNA (esRNA). Interestingly, the esRNAs detected in Mucor are derived from exons and can regulate mRNA accumulation. The largest class of these exons-derived siRNAs (ex-siRNA) are dependent on DCL-2 and RDRP1, suggesting that mRNAs from these loci are converted into dsRNA by RdRP1 and processed by DCL-2. The second class of ex-siRNAs requires DCL-2 and RdRP2 for biogenesis. The third group requires both RdRP1 and RdRP2, and the two Dicers have redundant roles in their biogenesis. The last group requires DCL1, but not DCL2, and the two RdRPs are both required. These results indicate that different RNAi components are used to produce different ex-siRNAs.
RNAi studies in the human fungal pathogen Cryptococcus neoformans
Cryptococcus neoformans, which causes fatal meningoencephalitis, has Argonaute, Dicer, and RdRP genes (47; 67). The RNAi pathway in Cryptococcus has recently been shown to act in a process named sex-induced silencing (SIS) (106). This silencing process, which is similar to quelling, also requires multiple copies of transgenes. In addition, the efficiency of SIS increases dramatically during the sexual cycle, which is correlated with increased expression of known RNAi components. SIS and SIS-associated sRNA production is abolished in RNAi mutants, indicating the role of RNAi in this silencing process during the sexual cycle. Examination of the sRNA profile revealed that some of the sRNAs, which were not derived from the trangenes, mapped to repetitive transposable elements and presumptive centromeric regions, suggesting that the RNAi pathway is a genome defense mechanism. Supporting this notion, in the rdp1 mutant, retrotransposons were found to be highly expressed with higher transposition rates.
Small RNAs from Magnaporthe oryzae
Magnaporthe oryzae is a model organism for study of pathogen-host interactions because it is a primary plant pathogen of rice and wheat. A recent sRNA analysis revealed that a large variety of sRNAs map to loci including the tRNA loci, rRNA loci, coding regions and intergenic loci (79). In the mycelia, small RNAs mostly come from intergenic region and repetitive elements, such as LTR retrotransposons. However, small RNAs in the appressoria are enriched for tRNA-derived RNA fragments (tRFs). The differential distribution of small RNAs in different tissues suggests roles of these sRNAs in the regulation of growth and development of this pathogen.
SUMMARY POINTS.
Neurospora crassa has multiple RNAi-related silencing pathways including quelling, qiRNA, and MSUD, which are triggered by multiple copies of transgenes, DNA damage, and unpaired DNA, respectively.
Schizosaccharomyces pombe requires an RNAi pathway for heterochromatin formation.
There are multiple small RNA biogenesis pathways in fungi. miRNA-like sRNAs have been found in Neurospora with distinct biogenesis pathways that require combinations of different components.
RNAi is an important fungal host-defense mechanism against transposon and viral invasion.
Acknowledgments
We thank the members of the Liu laboratory for discussion and comments. This work was supported by grants from the National Institutes of Health and the Welch Foundation to Yi Liu (I-1560).
Acronyms
- RNAi
RNA interference
- PTGS
post-transcriptional gene silencing
- sRNA
small non-coding RNA
- QDE-2
Quelling-deficient 2
- MSUD
Meiotic Silencing by Unpaired DNA
- RITS
RNA-induced transcriptional silencing complex
- milRNA
microRNA-like small RNA
- dsRNA
double-stranded RNA
Glossary
- RNA interference
A conserved eukaryotic gene regulatory mechanism that uses small RNA to mediate gene silencing
- Quelling
An RNAi-related gene silencing mechanism in Neurospora, which is induced by repetitive transgenic sequences. Quelling occurs in the vegetative growth stage
- Heterochromatin
Chromosome regions that are tightly packed throughout the cell cycle and is mostly genetically inactive
- RNA-induced transcriptional silencing
An RNAi-related mechanism that triggers or maintains the formation of heterochromatin, resulting in the downregulation of gene expression at the level of transcription
- Meiotic Silencing by Unpaired DNA
An RNAi-related gene silencing mechanism that occurs with the presence of unpaired homologous DNA sequences during meiosis
- microRNAs
A type of small RNAs that are generated from precursor RNAs with stem-loop structures. microRNAs can silence gene expression at post-transcriptional and translational levels
References
- 1.Alexander WG, Raju NB, Xiao H, Hammond TM, Perdue TD, et al. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol. 2008;45:719–27. doi: 10.1016/j.fgb.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996;86:103–13. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- *4.Bühler M, Verdel A, Moazed D. Tethering RITS to a Nascent Transcript Initiates RNAi- and Heterochromatin-Dependent Gene Silencing. Cell. 2006;125:873–86. doi: 10.1016/j.cell.2006.04.025. This study demonstrate that the targeting of RITS complex via RNA to a chromosomal locus is sufficient to induce transcriptional gene silencing. [DOI] [PubMed] [Google Scholar]
- 5.Bardiya N, Alexander WG, Perdue TD, Barry EG, Metzenberg RL, et al. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics. 2008;178:593–6. doi: 10.1534/genetics.107.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–8. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 8.Cam HP, Chen ES, Grewal SIS. Transcriptional Scaffolds for Heterochromatin Assembly. Cell. 2009;136:610–4. doi: 10.1016/j.cell.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli A, Cogoni C, Morelli G, Macino G. Molecular characterization of upstream regulatory sequences controlling the photoinduced expression of the albino-3 gene of Neurospora crassa. Mol Microbiol. 1994;13:787–95. doi: 10.1111/j.1365-2958.1994.tb00471.x. [DOI] [PubMed] [Google Scholar]
- 10.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. This study identified the Argonaute protein QDE-2 as a factor required for qelling, indicating that the dsRNA-induced gene silencing in animals and quelling in Neurospora share a common mechanism. [DOI] [PubMed] [Google Scholar]
- 12.Catalanotto C, Azzalin G, Macino G, Cogoni C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–5. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalanotto C, Nolan T, Cogoni C. Homology effects in Neurospora crassa. FEMS Microbiol Lett. 2006;254:182–9. doi: 10.1111/j.1574-6968.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- 14.Catalanotto C, Pallotta M, ReFalo P, Sachs MS, Vayssie L, et al. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24:2536–45. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. This study dicovered that the siRNA production and heterochromatin formation are regulated by the cell cycle and suggested a role for RNAi in cell cycle. [DOI] [PubMed] [Google Scholar]
- 17.Choi GH, Pawlyk DM, Nuss DL. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology. 1991;183:747–52. doi: 10.1016/0042-6822(91)91004-z. [DOI] [PubMed] [Google Scholar]
- 18.Choudhary S, Lee H-C, Maiti M, He Q, Cheng P, et al. A Double-Stranded-RNA Response Program Important for RNA Interference Efficiency. Molecular and Cellular Biology. 2007;27:3995–4005. doi: 10.1128/MCB.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. Embo J. 1996;15:3153–63. [PMC free article] [PubMed] [Google Scholar]
- 21.Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci U S A. 1997;94:10233–8. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–9. doi: 10.1038/20215. This study identified the first eukaryotic RNAi component and suggested that double-stranded RNA is an endogenous trigger for post-transcriptional gene silenicng. [DOI] [PubMed] [Google Scholar]
- 23.Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–4. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 24.Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. Coupling of Double-Stranded RNA Synthesis and siRNA Generation in Fission Yeast RNAi. Molecular Cell. 2007;27:449–61. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Dang Y, Yang Q, Xue Z, Liu Y. RNA interference in fungi: pathways, functions and applications. Eukaryot Cell. 2011 doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding SW, Lu R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr Opin Virol. 2011;1:533–44. doi: 10.1016/j.coviro.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, et al. RNAi in Budding Yeast. Science. 2009;326:544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durkin SG, Glover TW. Chromosome fragile sites. Annual review of genetics. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 29.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:377–92. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 30.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Bühler M. Nuclear Retention of Fission Yeast Dicer Is a Prerequisite for RNAi-Mediated Heterochromatin Assembly. Developmental Cell. 2010;18:102–13. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 32.Fulci V, Macino G. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol. 2007;10:199–203. doi: 10.1016/j.mib.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends in Genetics. 2004;20:417–23. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glover TW. Common fragile sites. Cancer letters. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Goodwin SB, Ben M’Barek S, Dhillon B, Wittenberg AHJ, Crane CF, et al. Finished Genome of the Fungal Wheat Pathogen Mycosphaerella graminicola Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis. PLoS Genet. 2011;7:e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Current opinion in genetics & development. 2010;20:134–41. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–7. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halic M, Moazed D. Dicer-Independent Primal RNAs Trigger RNAi and Heterochromatin Formation. Cell. 2010;140:504–16. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond TM, Andrewski MD, Roossinck MJ, Keller NP. Aspergillus Mycoviruses Are Targets and Suppressors of RNA Silencing. Eukaryotic Cell. 2008;7:350–7. doi: 10.1128/EC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond TM, Keller NP. RNA Silencing in Aspergillus nidulans Is Independent of RNA-Dependent RNA Polymerases. Genetics. 2005;169:607–17. doi: 10.1534/genetics.104.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammond TM, Xiao H, Boone EC, Perdue TD, Pukkila PJ, Shiu PK. SAD-3, a Putative Helicase Required for Meiotic Silencing by Unpaired DNA, Interacts with Other Components of the Silencing Machinery. G3: Genes, Genomes, Genetics. 2011;1:369–76. doi: 10.1534/g3.111.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi A, Da-Qiao D, Tsutsumi C, Chikashige Y, Masuda H, et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes to Cells. 2009;14:217–25. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 44.Hong E-JE, Villén J, Moazed D. A Cullin E3 Ubiquitin Ligase Complex Associates with Rik1 and the Clr4 Histone H3-K9 Methyltransferase and is Required for RNAi-Mediated Heterochromatin Formation. RNA Biology. 2005;2:106–11. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 45.Horn PJ, Bastie J-N, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes & Development. 2005;19:1705–14. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh J, Fire A. Recognition and silencing of repeated DNA. Annual review of genetics. 2000;34:187–204. doi: 10.1146/annurev.genet.34.1.187. [DOI] [PubMed] [Google Scholar]
- 47.Idnurm A, Bahn Y-S, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the Model Pathogenic Fungus Cryptococcus Neoformans. Nat Rev Micro. 2005;3:753–64. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 48.Jia S, Noma K-i, Grewal SIS. RNAi-Independent Heterochromatin Nucleation by the Stress-Activated ATF/CREB Family Proteins. Science. 2004;304:1971–6. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 49.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annual review of plant biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 50.Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, et al. Synthetic Heterochromatin Bypasses RNAi and Centromeric Repeats to Establish Functional Centromeres. Science. 2009;324:1716–9. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato A, Akamatsu Y, Sakuraba Y, Inoue H. The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr Genet. 2004;45:37–44. doi: 10.1007/s00294-003-0459-3. [DOI] [PubMed] [Google Scholar]
- 52.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA Polymerase II Is Required for RNAi-Dependent Heterochromatin Assembly. Science. 2005;309:467–9. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 53.Kelly WG, Aramayo R. Meiotic silencing and the epigenetics of sex. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2007;15:633–51. doi: 10.1007/s10577-007-1143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 55.Kinsey JA. Tad, a LINE-Like Transposable Element of Neurospora, Can Transpose Between Nuclei in Heterokaryons. Genetics. 1990;126:317–23. doi: 10.1093/genetics/126.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA Interference Guides Histone Modification during the S Phase of Chromosomal Replication. Current Biology. 2008;18:490–5. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 58.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 59.Lee DW, Pratt RJ, McLaughlin M, Aramayo R. An argonaute-like protein is required for meiotic silencing. Genetics. 2003;164:821–8. doi: 10.1093/genetics/164.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee HC, Aalto AP, Yang Q, Chang SS, Huang G, et al. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–7. doi: 10.1038/nature08041. This study suggested that DNA-damage is an important trigger for small RNA production and proposed that the RNA-dependent RNA polymerase also functions as an RNA polymerase to produce aberrant RNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *62.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–14. doi: 10.1016/j.molcel.2010.04.005. This study discovered the miRNA-like small RNAs in Neurospora and uncovered surprisingly diverse biogenesis mechanisms for small RNA production. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 64.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 65.Lejeune E, Bayne EH, Allshire RC. On the connection between RNAi and heterochromatin at centromeres. Cold Spring Harbor symposia on quantitative biology. 2010;75:275–83. doi: 10.1101/sqb.2010.75.024. [DOI] [PubMed] [Google Scholar]
- 66.Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two Novel Proteins, Dos1 and Dos2, Interact with Rik1 to Regulate Heterochromatic RNA Interference and Histone Modification. Current Biology. 2005;15:1448–57. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 67.Lin X, Heitman J. The Biology of the Cryptococcus neoformans Species Complex. Annual Review of Microbiology. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Ye X, Jiang F, Liang C, Chen D, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–3. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. The Plant Cell. 2002;14:1605–19. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Current opinion in cell biology. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 72.Mello CC, Conte D. Revealing the world of RNA interference. Nature. 2004;431:338–42. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 73.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–9. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- *75.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. This study discovered the critical link between siRNA production production process and heterocrhomatin assembly machinery. [DOI] [PubMed] [Google Scholar]
- 76.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. The Plant Cell. 1990;2:279–85. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nolan T, Braccini L, Azzalin G, De Toni A, Macino G, Cogoni C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Research. 2005;33:1564–73. doi: 10.1093/nar/gki300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nolan T, Cecere G, Mancone C, Alonzi T, Tripodi M, et al. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008;36:532–8. doi: 10.1093/nar/gkm1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nunes C, Gowda M, Sailsbery J, Xue M, Chen F, et al. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Brien HE, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R. Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol. 2005;71:5544–50. doi: 10.1128/AEM.71.9.5544-5550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 84.Pratt RJ, Lee DW, Aramayo R. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics. 2004;168:1925–35. doi: 10.1534/genetics.104.031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *85.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–53. doi: 10.1111/j.1365-2958.1992.tb02202.x. This study describe the discovery of quelling in Neurospora and is the one of the first known RNAi-related phenomena. [DOI] [PubMed] [Google Scholar]
- 86.Sadaie M, Iida T, Urano T, Nakayama J-i. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 2004;23:3825–35. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schramke V, Sheedy DM, Denli AM, Bonila C, Ekwall K, et al. RNA-interference-directed chromatin modification coupled to RNA polymerase II transcription. Nature. 2005;435:1275–9. doi: 10.1038/nature03652. [DOI] [PubMed] [Google Scholar]
- 89.Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot Cell. 2006;5:896–904. doi: 10.1128/EC.00373-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci U S A. 2007;104:12902–6. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shanker S, Job G, George OL, Creamer KM, Shaban A, Partridge JF. Continuous Requirement for the Clr4 Complex But Not RNAi for Centromeric Heterochromatin Assembly in Fission Yeast Harboring a Disrupted RITS Complex. PLoS Genet. 2010;6:e1001174. doi: 10.1371/journal.pgen.1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shiu PK, Glass NL. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr Opin Microbiol. 2000;3:183–8. doi: 10.1016/s1369-5274(00)00073-4. [DOI] [PubMed] [Google Scholar]
- 93.Shiu PK, Metzenberg RL. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics. 2002;161:1483–95. doi: 10.1093/genetics/161.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *94.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–16. doi: 10.1016/s0092-8674(01)00609-2. This study demonstrated that meiotic silencing in Neurospora is triggered by unpaired DNA and is an RNAi-related phenomenon. [DOI] [PubMed] [Google Scholar]
- 95.Shiu PK, Zickler D, Raju NB, Ruprich-Robert G, Metzenberg RL. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc Natl Acad Sci U S A. 2006;103:2243–8. doi: 10.1073/pnas.0508896103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Son H, Min K, Lee J, Raju NB, Lee YW. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 2011;115:1290–302. doi: 10.1016/j.funbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 97.Sun Q, Choi GH, Nuss DL. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci U S A. 2009;106:17927–32. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suzuki N, Chen B, Nuss DL. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. Journal of virology. 1999;73:9478–84. doi: 10.1128/jvi.73.11.9478-9484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki N, Maruyama K, Moriyama M, Nuss DL. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. Journal of virology. 2003;77:11697–707. doi: 10.1128/JVI.77.21.11697-11707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annual review of cell and developmental biology. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *101.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, et al. RNAi-Mediated Targeting of Heterochromatin by the RITS Complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. This study identified the RITS complex, which is the link between the RNAi machinery and heterochromatin assembly factors in S. pombe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS letters. 2005;579:5872–8. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 103.Volpe T, Martienssen RA. RNA interference and heterochromatin assembly. Cold Spring Harbor perspectives in biology. 2011;3:a003731. doi: 10.1101/cshperspect.a003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, et al. RNA interference is required for normal centromere function infission yeast. Chromosome Research. 2003;11:137–46. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- *105.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. This is the first study that demonstrated the requrement of RNAi components in heterochromatin formation in S. pombe. [DOI] [PubMed] [Google Scholar]
- 106.Wang X, Hsueh Y-P, Li W, Floyd A, Skalsky R, Heitman J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes & Development. 2010;24:2566–82. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.White SA, Allshire RC. RNAi-Mediated Chromatin Silencing in Fission Yeast. Current Topics in Microbiology and Immunology. 2008;320:157–83. doi: 10.1007/978-3-540-75157-1_8. [DOI] [PubMed] [Google Scholar]
- 108.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–48. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiao H, Alexander WG, Hammond TM, Boone EC, Perdue TD, et al. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics. 2010;186:119–26. doi: 10.1534/genetics.110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–8. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–8. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 112.Zhang X, Nuss DL. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proceedings of the National Academy of Sciences. 2008;105:16749–54. doi: 10.1073/pnas.0807225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. Journal of virology. 2008;82:2613–9. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]