Abstract
The cyclin-dependent kinase inhibitor 1A (CDKN1A), p21/Cip1, is a vital cell cycle regulator, dysregulation of which has been associated with a large number of human malignancies. One critical mechanism that controls p21 function is through its degradation, which allows the activation of its associated cell cycle promoting kinases, CDK2 and CDK4. Thus, delineating how p21 is stabilized and degraded will enhance our understanding of cell growth control and offer a basis for potential therapeutic interventions. Here, we report a novel regulatory mechanism that controls the dynamic status of p21 through its interaction with Cdk5 and Abl enzyme substrate 1 (Cables1). Cables1 has a proposed role as a tumor suppressor. We found that upregulation of Cables1 protein was correlated with increased half-life of p21 protein, which was attributed to Cables1/p21 complex formation and supported by their co-localization in the nucleus. Mechanistically, Cables1 interferes with the proteasome (Prosome, Macropain) subunit alpha type 3 (PSMA3) binding to p21 and protects p21 from PSMA3-mediated proteasomal degradation. Moreover, silencing of p21 partially reverses the ability of Cables1 to induce cell death and inhibit cell proliferation. In further support of a potential pathophysiological role of Cables1, the expression level of Cables1 is tightly associated with p21 in both cancer cell lines and human lung cancer patient tumor samples. Together, these results suggest Cables1 as a novel p21 regulator through maintaining p21 stability, and support the model that the tumor suppressive function of Cables1 occurs at least in part through enhancing the tumor suppressive activity of p21.
Keywords: Cables1, p21, Cell death
Introduction
The cyclin-dependent kinase inhibitor p21 plays various critical physiological roles and regulates many cellular processes such as cell division, cell differentiation, senescence and apoptosis.1, 2 Due to its important biological functions, the expression and protein levels of p21 are tightly controlled by both transcriptional and post-transcriptional mechanisms. As an unstable protein with short half-life, p21 is degraded by both ubiquitin-dependent and -independent proteasome pathways.1 Three E3 ubiquitin ligase complexes SCFSkp2,3–5 CRL4Cdt2,6–8 and APC/CCdc20,9 have been reported to induce p21 ubiquitination and degradation at specific stages of the cell cycle. Ubiquitin-independent p21 degradation is mediated by its interaction with subunits of the proteasome such as the C8 subunit of the 20S proteasome (prosome, macrophain subunit alpha type3; PSMA3).10, 11 Such p21 degradation is also promoted by MDM2 and MDM4 but independently of their E3 ligase activities.12, 13 Thus, inhibition of these p21 degradation mechanisms is expected to result in p21 stabilization. Interestingly, an Hsp90 binding tetratricopeptide repeat (TPR) protein WISp39 stabilizes newly synthesized p21 protein by recruiting Hsp90 to prevent the proteasomal degradation of p21.14 However, the respective contributions of the various p21 stabilization and degradation mechanisms remain to be elucidated.
Cables1 (Cdk5 and Abl enzyme substrate 1) is an adaptor protein which links the Cdks to the nonreceptor tyrosine kinases and regulates the activity of Cdks by enhancing their Y15 phosphorylation. Cables1 promotes the phosphorylation of Cdk5 at Y15 by C-abl, resulting in increased Cdk5 kinase activity, which is believed to positively regulate neurite outgrowth in neurons.15 Cables1 also connects Cdk2 and Wee1, resulting in increased Cdk2 Y15 phosphorylation, decreased Cdk2 kinase activity and reduced cell growth in proliferating cells.16 It has been reported that Cables1 is also able to interact with and affect p53, p63 and p73 function.17, 18 Compared to Cables1+/+ MEFs, Cables1−/− MEFs exhibit increased growth rate, delayed senescence and decreased serum dependence.19 Furthermore, Cables1−/− mice have an increased incidence of endometrial cancer and a reduced survival rate in response to unopposed estrogen and an increased incidence of colorectal cancer in response to 1,2-dimethylhydrazine.20, 21 Recently, Cables1 was shown to protect p63 from proteasomal degradation to ensure deletion of cells after genotoxic stress.18 Loss of Cables1 expression is found with high frequency in many types of human cancer,16, 20, 22–24 and also enhances tumor progression in the ApcMin/+ mouse model and activates the Wnt/β-catenin signaling pathway.25 However, the mechanism that directly links the loss of Cables1 to tumorigenesis remains to be established. Here we present evidence to support a novel function of Cables1 in controlling the status of a master cell cycle regulator, p21.
Results
Cables1 regulates the protein levels of p21
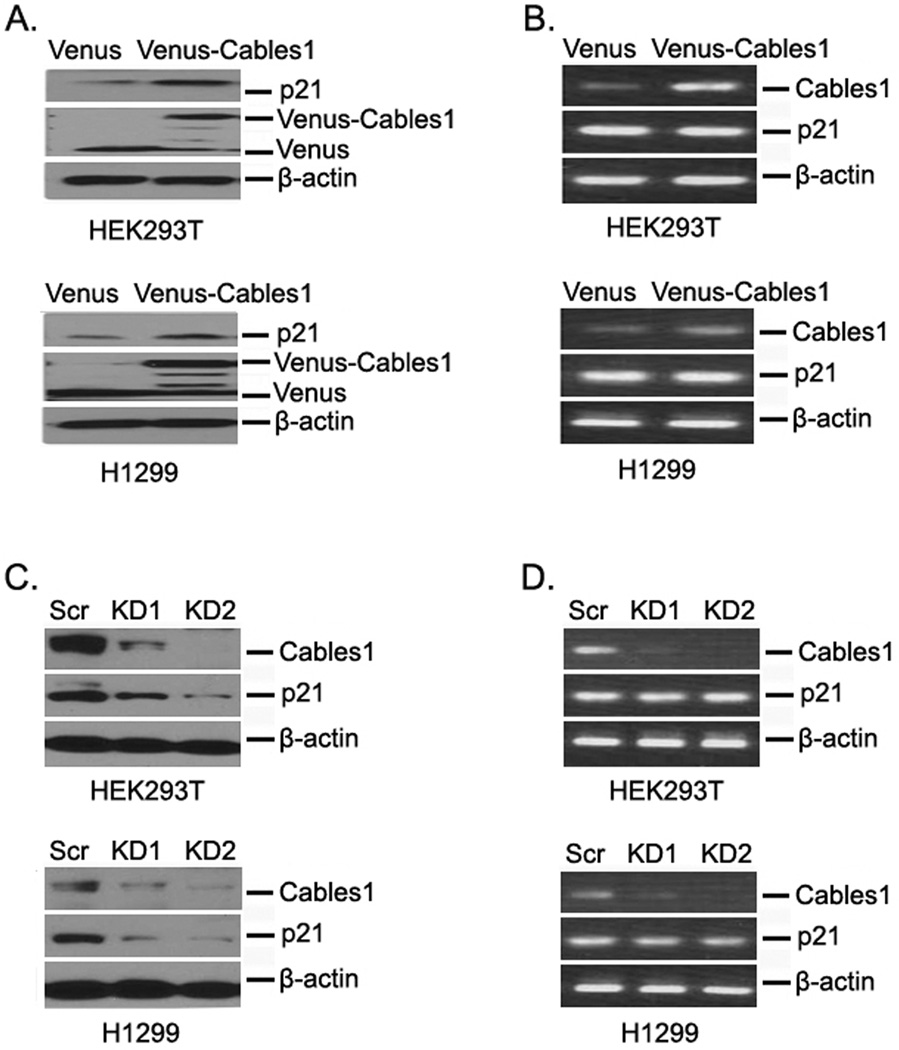
To investigate whether Cables1 affects the expression level of p21, we first overexpressed Venus control and Venus-tagged Cables1 in HEK293T cells and examined protein expression by Western blot. Compared with Venus alone, Venus-Cables1 expression significantly increased the protein levels of p21. The expression of p21 is mainly controlled by the tumor suppressor protein p531. To exclude the effect of p53 on Cables1-induced p21 expression, we also overexpressed Venus and Venus-Cables1 in p53-null H1299 cells and found that the protein levels of p21 were still increased after Venus-Cables1 overexpression (Fig 1A). These data suggest that p21 upregulation by Cables1 is p53 independent. We then examined whether p21 mRNA levels were also increased after Cables1 overexpression. The results of RT-PCR showed that there was no change in p21 mRNA levels with or without Cables1 overexpression (Figure 1B). Next, we detected p21 levels in both HEK293T and H1299 cells whose endogenous Cables1 was stably silenced by two different Cables1 shRNAs in pLKO vectors. As shown in Figure 1C, knocking down Cables1 by both shRNAs dramatically decreased the protein levels of p21 in both HEK293T and H1299 cells. Similarly, p21 mRNA levels were not altered after Cables1 silencing (Figure 1D). Thus, Cables1 regulates p21 at the protein level but not at the mRNA level.
Figure 1. Cables1 regulates protein levels of p21.
(A, B) Overexpressing Cables1 increases p21 at the protein level, but not at the mRNA level. HEK293T and H1299 cells were transfected with Venus and Venus-Cables1, then after 2 days, cells were lysed. Proteins were examined by Western blot, and mRNAs were examined by RT-PCR. (C, D) Knockdown of Cables1 decreases p21 at the protein level, but not at the mRNA level. HEK293T and H1299 cells were transfected with pLKO.1 Cables1 shRNA vectors and selected with puromycin (2 µg/ml) for 7 days, then cells were lysed. Proteins were examined by Western blot, and mRNAs were examined by RT-PCR.
Cables1 interacts with p21
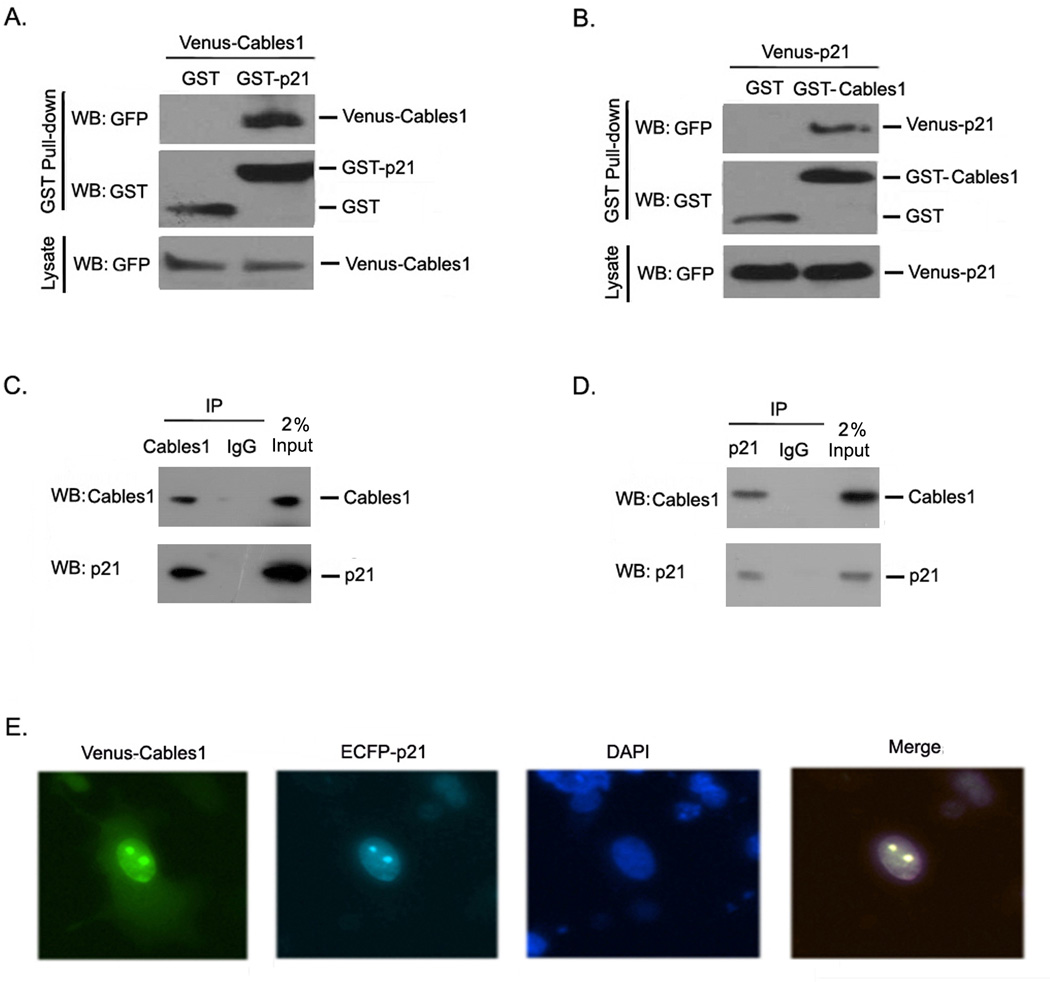
To elucidate how Cables1 regulates p21 protein expression, we examined whether the two proteins interact with each other. We co-transfected GST or GST-p21 and Venus-Cables1 into HEK293T cells and performed a GST pull-down assay. As shown in Figure 2A, Venus-p21 was detectable in the GST-Cables1 complex, but not in the GST complex. We also reversed the fusions and co-transfected GST or GST-Cables1 and Venus-p21 into HEK293T cells. Venus-p21 was detectable only in the GST-p21 complex, not in the GST complex (Figure 2B). To further confirm the interaction of Cables1 with p21, we investigated their endogenous interaction. Endogenous Cables1 and p21 were separately immunoprecipitated from HEK293T cells and reciprocal protein detection was performed. As shown in Figure 2C and 2D, both Cables1 and p21 were detectable in their individual immunoprecipitated complexes but not in the control IgG complexes. Additionally, both Cables1 and p21 were co-localized in the nucleus (Figure 2E). From these data, we conclude that Cables1 and p21 interact with each other.
Figure 2. Cables1 interacts with p21.
(A, B) Exogenous Cables1 interacts with p21. HEK293T cells were co-transfected with the indicated vectors. GST pull-downs were performed from cell lysates and proteins were detected by Western blot. (C–D) Endogenous Cables1 interacts with p21. Endogenous Cables1 and p21 were immunoprecipitated from HEK293T cells and detected reciprocally. (E) Cables1 co-localizes with p21 in the nucleus. HEK293T cells were transfected with Venus-Cables1 and ECFP-p21. Two days after transfection, cells were monitored under a fluorescence microscope.
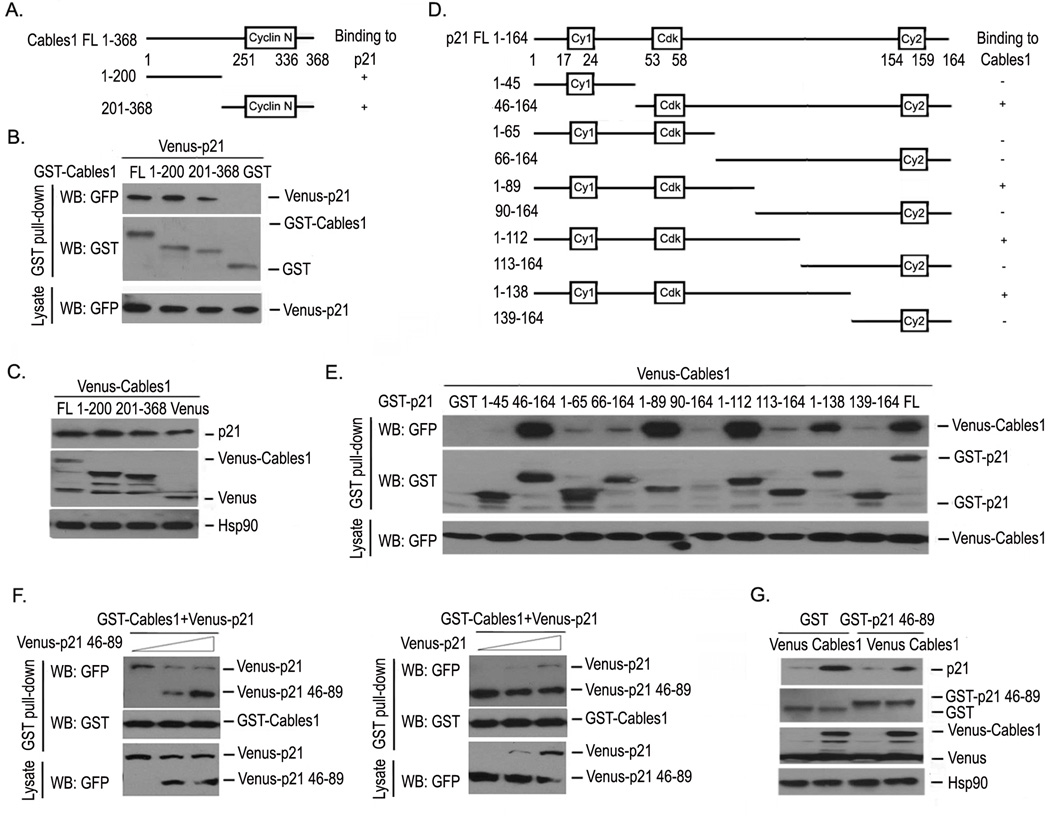
To determine the binding specificity and explore which regions in Cables1 mediate the interaction of Cables1 with p21, we overexpressed GST, GST-Cables1 truncations 1–200, 201–368, full-length (FL) and Venus-p21 in HEK293T cells, and performed GST pull-down and Western blot assays. Venus-p21 was present in the complexes for both full-length and truncated GST-Cables1, but not in the GST complexes (Figure 3B). This result indicates that the regions of Cables1 responsible for its interaction with p21 are located in both Cables1 truncations 1–200 and 201–368. Next, we asked whether both of these truncations were able to increase p21 protein levels through their interaction. As shown in Figure 3C, overexpression of Venus-Cables1 truncations 1–200, 201–368 and full-length all significantly increased p21 protein level when compared with Venus alone.
Figure 3. Interacting regions of Cables1 and p21.
(A, B) The interacting regions of Cables1 with p21 were mapped. The indicated vectors were transfected into HEK293T cells. GST pull-down and Western blot were performed after 2 days. (C) Both Cables1 truncations 1–200 and 201–368 increased p21 protein levels. Proteins were detected in HEK293T cells overexpressing Venus, Venus-Cables1 truncations 1–200, 201–368 and full length Cables1. (D, E) The interacting regions of p21 with Cables1 were mapped. The indicated vectors were transfected into HEK293T cells, then Venus-Cables1 was detected in each GST pull-down complex. (F) The p21 46–89 fragment interferes with p21 for Cables1 binding. HEK293T cells were co-transfected with GST-Cables1, Venus-p21 and Venus-p21 46–89 fragment. GST pull-down and Western blot were performed after 2 days. (G) The p21 46–89 fragment inhibits Cables1-induced increase in p21 protein. HEK293T cells were co-transfected with Venus or Venus- Cables1 and GST or GST-p21 46–89 fragment. Cells were lysed after 2 days and proteins were examined.
To map the regions of p21 which interact with Cables1, we generated 10 p21 truncations (Figure 3D) and examined their interaction with Cables1 by detecting Venus-Cables1 levels in the GST-p21 truncation complexes from overexpressing lysates. Figure 3E shows the presence of Venus-Cables1 in the complexes with GST-p21 truncations 46–164, 1–89, 1–112, 1–138 and full-length, but not in the other truncation complexes. It is likely that the region between amino acids 46–89 is involved in the Cables1 interaction. We then expressed p21 truncation 46–89 to detect whether it could block the interaction of Cables1 with p21 and inhibit the Cables1-induced increase of endogenous p21 protein. HEK293T cells were co-transfected with GST-Cables1, Venus-p21 and the Venus-p21 46–89 fragment, then a GST pull-down assay and Western blot were carried out. The level of Venus-p21 in the GST-Cables1 complex gradually decreased when the expression of Venus-p21 46–89 fragment was dose-dependently increased, and vice versa (Figure 3F). As shown in Figure 3G, compared with GST control, the GST-p21 46–89 fragment decreased the p21 protein level in Venus-Cables1 overexpressing lysates. These results suggest the regions of p21 that mediate its interaction with Cables1 are located in the region 46–89.
Cables1 affects the half-life of p21 protein
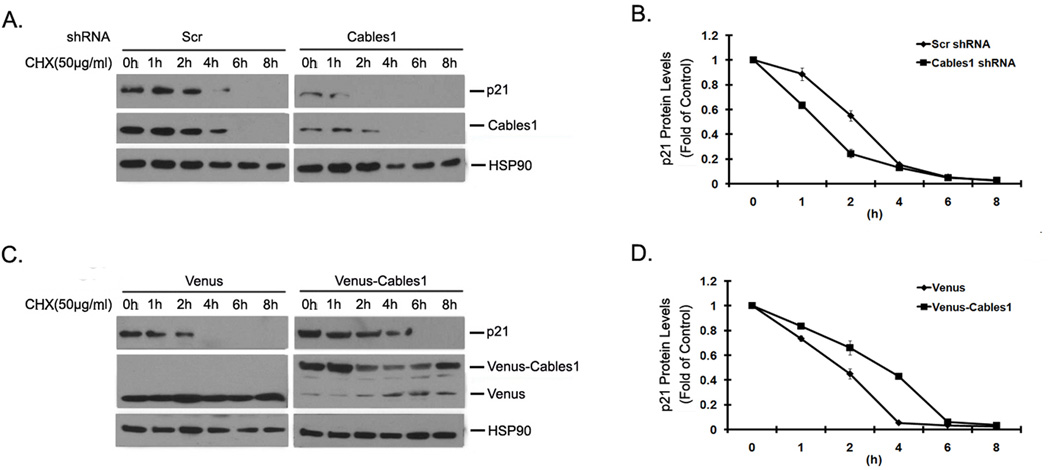
The protein level of p21 is controlled by multiple mechanisms1. To understand the mechanism by which Cables1 increases p21 protein level, we addressed whether Cables1 directly affects the half-life of p21 protein in cells. HEK293T cells with exogenous Venus and Venus-Cables1 were treated with the protein synthesis inhibitor cyclohexamine (CHX) at the indicated time-points, and the levels of endogenous p21 protein were examined. The Western blot results are shown in Figure 4A, and their quantification in Figure 4B. Compared with Venus alone, Venus-Cables1 clearly increased the half-life of p21. We also compared the half-life of endogenous p21 protein in HEK293T cells with or without knocking-down endogenous Cables1. As shown in Figure 4C and D, after CHX treatment, the half-life of p21 in Cables1-silenced cells was significantly shorter than that in scramble control cells. These results demonstrate that Cables1 stabilizes p21 by blocking its protein turnover in cells.
Figure 4. Cables1 affects the half-life of p21 protein.
(A) Knockdown of Cables1 decreases the half-life of p21 protein. HEK293T cells were expressedwith pLKO-Cables1 shRNA vectors and selected with puromycin (2 µg/ml) for 7 days, then cells were treated with CHX (50 µg/ml) for the indicated times. Then cells were lysed and proteins were examined. (B) Quantified relative p21 protein levels are shown. (C) Overexpression of Cables1 increases the half-life of p21 protein. Overexpressing Venus-Cables1 HEK293T cells were treated with CHX (50 µg/ml) at the indicated times. Then cells were lysed and proteins were examined. (D) Quantified relative p21 protein levels are shown.
Cables1 interferes with PSMA3 to bind to and inhibit p21 degradation
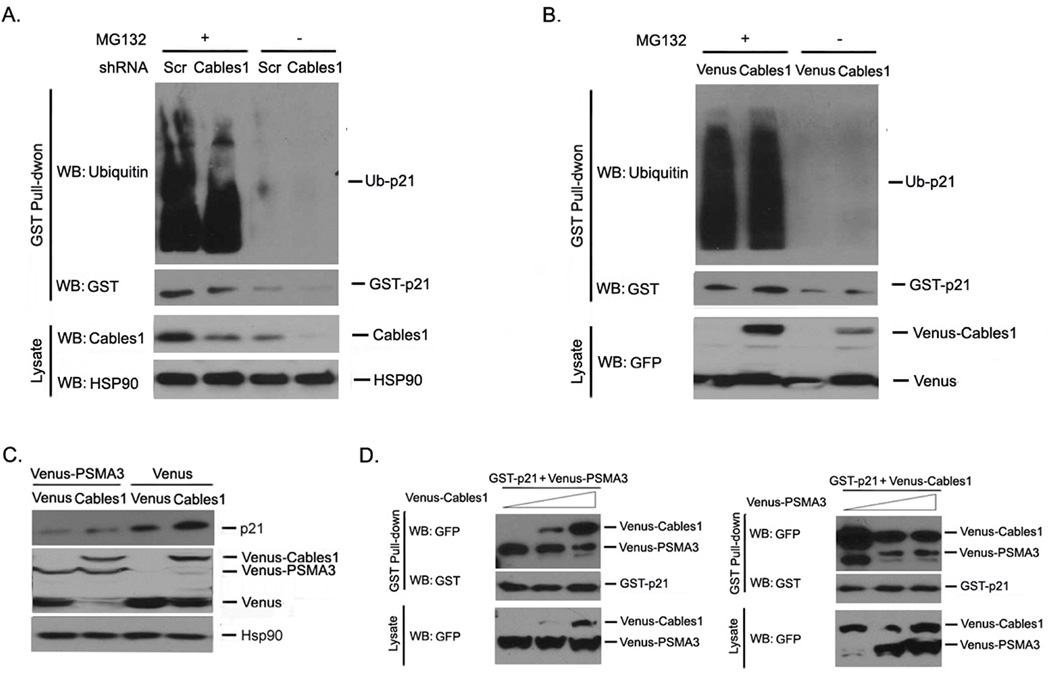
The degradation of p21 is mediated by both ubiquitin-dependent and -independent proteasomal pathways1. To address whether stabilization of p21 by Cables1 occurs through a ubiquitin-dependent or -independent mechanism, we treated both Venus and Venus-Cables1 cells with or without the proteasome inhibitor MG132 and examined p21 ubiquitination. In comparison to Venus alone, Venus-Cables1 did not alter the levels of ubiquitinated p21 (Figure 5A). We also examined p21 ubiquitination in HEK293T cells with or without knocking-down endogenous Cables1. Similarly, after MG132 treatment, there was no difference in the levels of ubiquitinated p21 between Cables1-silenced and non-silenced cells (Figure 5B).
Figure 5. Cables1 interferes with PSMA3 to bind to and inhibit p21 degradation.
(A, B) Cables1 does not alter p21 ubiquitination. HEK293T cells were co-transfected with the indicated vectors. GST pull-down and Western blot were performed after 2 days. (C) PSMA3 decreases p21 protein levels. HEK293T cells were co-transfected with Venus-PSMA3 and Venus or Venus-Cables1. After 2 days, cells were lysed and proteins were examined. (D) Cables1 competes with PSMA3 to bind to p21. HEK293T cells were co-transfected with GST-p21, Venus-PSMA3 and Venus-Cables1. GST pull-down and Western blot were performed after 2 days. Left panel showed the effect of Cables1 overexpression on the p21/PSMA3 association while right panel showed the effect of PSMA3 on the p21/Cables1 interaction.
It has been reported that the proteasome subunit PSMA3 mediates ubiquitin-independent p21 degradation by directly interacting with and recruiting p21 into the proteasome for degradation10, 11. To investigate whether PSMA3 is involved in the action of Cables1, we introduced PSMA3 with Venus or Venus-Cables1 into HEK293T cells and examined endogenous p21 protein levels in cells. As shown in Figure 5C, exogenous PSMA3 decreased endogenous p21 protein levels in both Venus and Venus-Cables1 cells. Next, we examined whether Cables1 and PSMA3 interferes with each other for the binding of p21. HEK293T cells were co-transfected with GST-p21, Venus-PSMA3 and Venus-Cables1, then GST pull-down assay and Western blot were carried out. The levels of Venus-PSMA3 in the GST-p21 complexes gradually decreased when the expression of Venus-Cables1 was dose-dependently increased, and vice versa (Figure 5D). Taken together, these data suggest that Cables1 stabilizes p21 by inhibiting PSMA3-dependent but ubiquitin-independent p21 degradation.
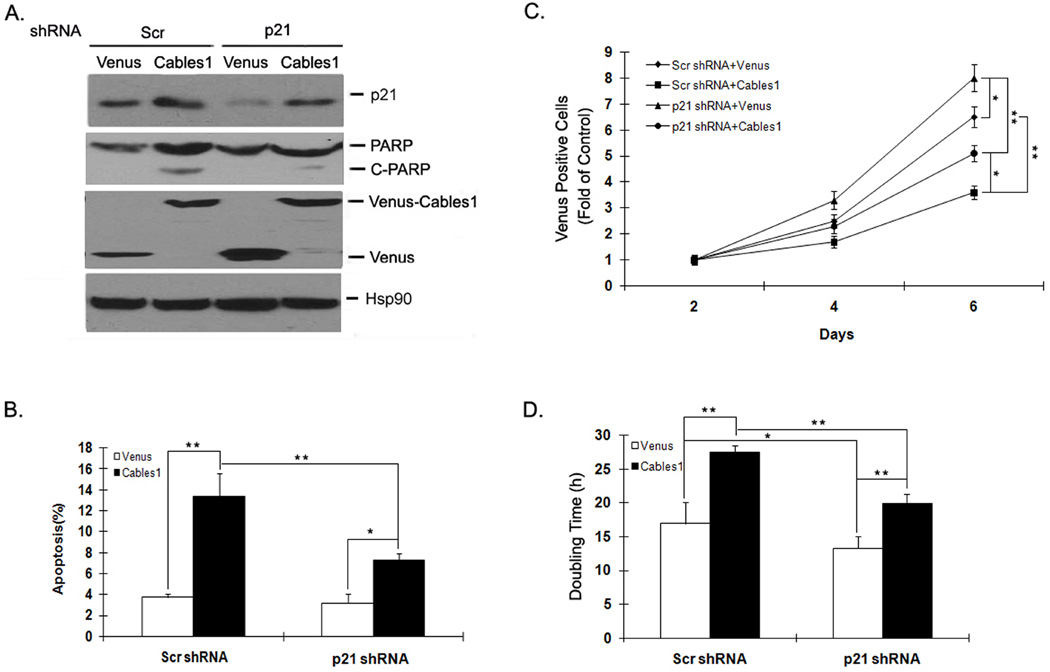
Cables1 induces apoptosis and inhibits growth partially through p21
In proliferating cells, Cables1 was reported to induce cell apoptosis and inhibit cell growth.16, 26 To explore the role of p21 in these Cables1 biological functions, we used shRNA to silence p21 expression in both Venus and Venus-Cables1 cells and examined the degree of cell apoptosis and growth. The level of apoptosis in Venus positive cells was determined by detecting Annexin V-positive cells and the protein levels of cleaved PARP by Western blot. As shown in Figure 6A and B, the levels of cell apoptosis and PARP cleavage were greater in Venus-Cables1 than in Venus alone cells. Levels of p21 protein were significantly decreased in both Venus and Venus-Cables1 cells by the expression of p21 shRNA, while apoptosis and PARP cleavage were partially decreased in Venus-Cables1 cells after p21 was silenced. We also counted the number of Venus-positive cells to monitor cell growth. As shown in Figure 6C, Venus-Cables1 cells had lower cell numbers than Venus alone either with or without p21 silencing, and knocking down p21 partially reversed this Cables1-induced decrease in cell number. Furthermore, the tendency toward lower cell number was enhanced with increasing culture time. Analyzing the cell doubling time from these data showed the following order of cell doubling times: Venus-Cables1 plus scrambled shRNA (27 h) > Venus-Cables1 plus p21 shRNA (20 h) > Venus plus scrambled shRNA (17 h) > Venus plus p21 shRNA (13 h) (Figure 6D). These data suggest that knockdown of p21 only partially reverses Cables1-mediated cell growth inhibition, and that p21 is probably one of several downstream effector molecules to mediate Cables1 tumor suppressor action.
Figure 6. Cables1 induces apoptosis and inhibits growth partially through p21.
(A, C) Knockdown of p21 partially reverses Cables1-induced cell apoptosis and growth inhibition. Venus or Venus-Cables1 WT was co-transfected into HEK293T cells with or without pLKO.1 p21 shRNA. After 2 days, cells were lysed and proteins were detected. Cells were also stained with Annexin V-PE and the apoptosis of Venus-positive cells was analyzed by flow cytometry. After every 2 days, Venus-positive cells were counted under the fluorescence microscope. (B, D) Quantified data are shown. * and ** represent P<0.05 and P<0.01, respectively.
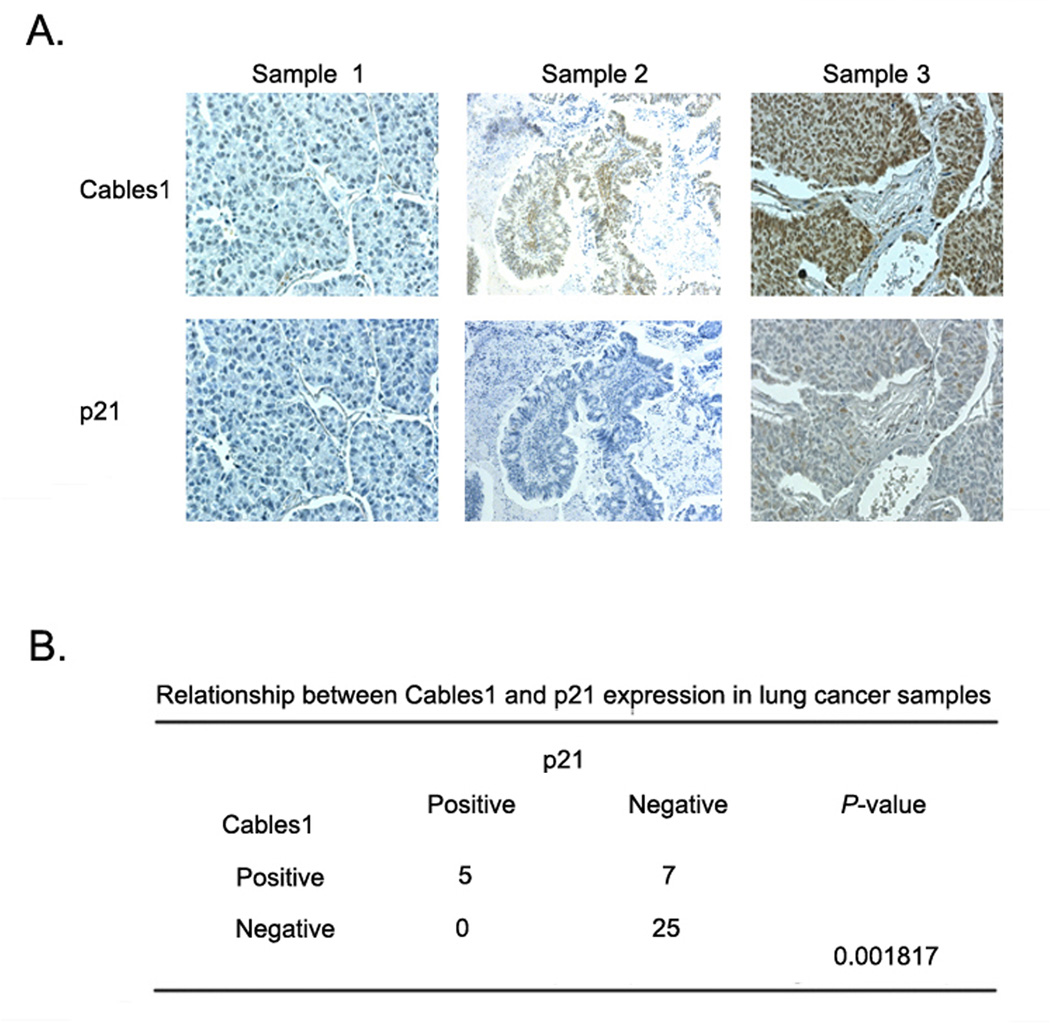
Cables1 protein expression is correlated with that of p21 in lung cancer patient tumor samples
Our results clearly demonstrate that p21 is regulated by Cables1 in cell culture. To determine whether this is also the case in tumor tissues, we compared the levels of Cables1 and p21 in 37 human lung cancer samples by immunostaining with specific antibodies against Cables1 and p21. Positive Cables1 staining was present in 12 (32%) out of 37 samples, and positive p21 staining was present in 5 (14%) out of 37 samples. The expression of p21 was highly correlated with the expression of Cables1 (Figure 7B). Results of three representative samples are shown in Figure 7A. Sample 1 showed negative staining of both Cables1 and p21. Sample 2 showed positive Cables1 staining but negative staining of p21. Sample 3 showed positive staining of both Cables1 and p21. Thus, these results in human lung cancer specimens support our proposed mechanism based on cell-culture experiments that expression of Cables1 is able to enhance the protein expression of p21. Loss of Cables1 may lead to p21 degradation and enhanced tumorigenesis in many lung cancer patients.
Figure 7. Expression of Cables1 and p21 proteins in lung cancer samples.
The levels of Cables1 and p21 in 37 human lung cancer samples were examined by immunostaining with specific antibodies. (A) Three representative samples are shown where either Cables1 or p21 staining were negative or positive. (B) Summary of staining results from 37 tumor tissue samples. Correlation between Cables1 and p21 expression was analyzed using Fisher’s exact test.
Discussion
In the present study, we have identified Cables1 as a novel p21 regulator that maintains its protein stability and suppresses its degradation. Our results showed that overexpression of exogenous Cables1 increased p21 at the protein level, while silencing of endogenous Cables1 had an inverse effect to decrease p21 at the protein level.
Our immunoprecipitation data indicated that Cables1 and p21 exist in the same protein complex, and can interact with each other. The cyclin N domain of Cables1 is sufficient to interact with p21 and increase p21 protein levels after their co-transfection. The region of p21 that mediates its interaction with Cables1 was not located in the two cyclin-binding motifs (Cy1, Cy2), although Cables1 contains a cyclin N domain at the C-terminus. This suggests that the mechanism by which Cables1 stabilizes p21 may be different from the known mechanisms by which some cyclins bind and stabilize p21. It has been reported that cyclin D1 stabilizes p21 by interaction with its C-terminal cyclin-binding motif (Cy2),27 while cyclin E stabilizes p21 by interaction with its N-terminal cyclin-binding motif (Cy1).28 The physiological role of p21 stabilization by cyclins is to promote assembly of cyclin-CDK complexes.27 In our case, it is possible that p21 stabilization by Cables1 may mediate the function of Cables1 as a tumor suppressor. This is partially supported by our data showing that silencing of p21 was able to partially reverse the effect of Cables1 on inducing cell apoptosis and inhibiting cell growth.
Most proteins degraded by the proteasome require ubiquitin conjugation for targeting via the association of ubiquitin with the 19S lid of the proteasome.29 However, p21 protein can be degraded by both ubiquitin-dependent and -independent proteasomal pathways.1 Here, we demonstrate that p21 stabilization by Cables1 occurs through a ubiquitin-independent pathway based on our data showing that Cables1 did not alter the ubiquitination of p21. Ubiquitin-independent degradation of p21 appears to involve the interaction of its C-terminus with PSMA3.10, 11 Our results showed that PSMA3 decreased the stabilization of p21 by Cables1, and Cables1 interferes with PSMA3 for binding to p21. Although Cables1 and PSMA3 bind to different regions of p21, their binding may induce a conformational change in p21, leading to masking of the other binding site. Inhibition of p21 degradation by masking of the PSMA3 binding site may represent a general mechanism of p21 stabilization. For example, cyclin D1 stabilizes p21 by interaction with the Cy2 domain of p21, which prevents p21 association with and subsequent degradation by PSMA3.27 However, the respective contribution of the various stabilization and degradation mechanisms operating on p21 requires further validation. Additionally, Cables1−/− MEFs showed decreased p16 and p27 comparing with Cables1+/+ MEFs.19. This reported requirement of Cables1 for p16 and p27 proteins supports a broad regulatory role of Cables1 in the control of cell division. Because of their central importance in determining cell fate, these CDK inhibitors are regulated by different mechanisms at transcriptional, translational, and/or posttranslational levels. Whether Cables1 regulates p16 and p27 in a mechanism analogous to p21 remains to be established. Our findings provide strong a rationale to further examine the relationship between Cables1 and other p21-like CDK regulatory proteins in the future, including members of the INK4 family (p15, p16, p18, and p19) and Cip/Kip members (p21, p27, and p57).
Loss of Cables1 expression is observed with high frequency in many types of human cancer.16, 20, 22–24 In human colorectal cancer tissues, Cables1 expression was seen in 35% of samples.23 Zirbes, et al. showed that 67% of colorectal tumor samples were p21-staining positive with significantly better survival.30 In human non-small cell lung cancer tissues, Tan, et al. showed that 55% of samples were Cables1-staining positive without any relationship to clinicopathologic parameters except histologic tumor type,24 while p21 expression was detected in 35% samples and was associated with longer survival time and was found more frequently in stage I or II compared with stage III disease.31 In this report, we examined the expression of Cables1 and p21 in 37 human lung cancer tissues by immunostaining and found that Cables1 was lost in 68% of samples, which showed significant correlation with co-loss of p21 expression. Thus, it is possible that during tumorigenesis, Cables1 suppresses the growth of tumors not only through decreasing the activity of CDK2 to reduce cell cycle progression and enhancing the function of p53 families to induce cell apoptosis,16,17, 18 but also through stabilizing the CDK inhibitor p21 to inhibit cell proliferation.
In summary, we have identified Cables1 as a novel p21 regulator that protects p21 from proteasomal degradation. The tumor suppressive function of Cables1 is mediated partially via regulating the protein level of p21. This enhanced understanding of the mechanism by which Cables1 controls cell cycle progression may allow the identification of strategies to manipulate its tumor suppressor function for future therapeutic discovery.
Material and Methods
Cells and reagents
HEK293T and H1299 cells were maintained in DMEM with 10% fetal bovine serum and 100 units penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. Anti-GST and Hsp90 antibodies were from Santa Cruz Biotechnology. Anti-p21 and PARP antibodies were from Cell Signaling Technologies. Anti-Cables1 and GFP antibodies were from Abcam. Anti-β-actin and other chemical regents were from Sigma.
Plasmids and transfection
Cables1 and p21 cDNAs were amplified by PCR and cloned into the Gateway expression vectors (Invitrogen). pLKO.1 Cables1 shRNA1 with target sequence (5’-GCACTTACTTACTACTGGAAA-3’), pLKO.1 Cables1 shRNA2 with target sequence (5’-CCTGGGAGACTTTATGGACTA-3’), pLKO.1 p21 shRNA with target sequence (5’-GTCACTGTCTTGTACCCTTGT-3’) and scrambled shRNA were purchased from OpenBiosystems. Site-directed mutagenesis was performed essentially following the manufacturer’s protocol (Stratagene). Transfections were performed using FuGene HD (Roche).
Protein interaction assays
GST pull-down assay
Cells were lysed in GST pull-down lysis buffer (1% Nonidet P-40, 150 mM NaCl, 100 mM Hepes, 5 mM Na4P2O7, 5 mM NaF, 2 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 10 mg/L aprotinin, 10 mg/L leupeptin). Cleared cell lysates were incubated with glutathione-conjugated sepharose or the appropriate antibody and Protein G conjugated sepharose for 2 hours at 4°C. Then the resin was washed 3 times with GST pull-down lysis buffer and boiled in 6X SDS sample buffer for Western blot analysis.
Co-immunoprecipitation (Co-IP) assay
Cells were lysed in Co-IP lysis buffer (1% Nonidet P-40, 150 mM NaCl, 100 mM Hepes, 5 mM Na4P2O7, 5 mM NaF, 2 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 10 mg/L aprotinin, 10 mg/L leupeptin). Cleared cell lysates were incubated with Protein A or G conjugated sepharose (GE Healthcare) and the appropriate antibody for 2 hours to overnight at 4°C. Following incubation, the resin was washed 3 times with Co-IP lysis buffer and protein samples were eluted by boiling in 6X SDS sample buffer for Western blot analysis.
Western blot
Proteins were separated on 12.5% SDS-PAGE gels and transferred to PVDF membranes. Membranes were blocked with 5% BSA and incubated with the indicated primary antibodies. Corresponding horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) were used against each primary antibody. Proteins were detected using West-Pico or West-Dura enhanced chemiluminescent detection reagents (Pierce) and a Kodak imaging system or films.
RT-PCR
Total RNA was extracted from cells and the reverse transcription reaction was performed with the first strand cDNA synthesis kit (Invitrogen). The newly synthesized cDNAs were amplified by PCR with Cables1 primers (5’-CTCCGGAGATGTCGAGCTCTCTCAGGTTC-3’; 5’-GCTCCCTGGGTGCCGGCTGCCCGGCCTATGGAG-3’), p21 primers (5’CTTTGTCACCGAGGCGCCAGTGGAGGGTG-3’; 5’- CAGGTCCACCTGGCGCCCCTTCAGCCTGC-3’), and β-actin primers (5’-GCTCGTCGTCGACAACGGCTC--3’; 5’-CAAACATGATCTGGGTCATCTTCTC--3’). Aliquots of product were electrophoresed on 1.5% agarose gels, and PCR fragments were visualized by ethidium bromide.
Apoptosis assay
Cells were harvested and washed with PBS 3 times, fixed with 70% ethanol, stained with Annexin V-PE (BD), and finally analyzed with Guava flow cytometer (Millipore) to determine the percentage of apoptotic cells.
Immunohistochemistry assay
Formalin-fixed, paraffin-embedded human lung cancer tissue array slides (ABXIS and Biochain) were stained with anti-pCables1 T44, T150 (21st Century) and pAkt S473 (Epitomics) antibodies using a microwave-enhanced avidin-biotin staining method. For the quantitation of protein expression, the following formula was used: IHC score = % positive cells × intensity score. The intensity was scored as follows: 0, negative; 1, weak; 2, moderate; and 3, intense. An IHC score of 100 or greater was considered as positive.
Statistical analysis
A student’s t-test was used to compare individual data points among each group. Correlation was analyzed using Fisher’s exact test. A P value of less than 0.05 was set as the criterion for statistical significance.
Acknowledgement
We would like to thank past and present members of the Fu/Khuri laboratory for many stimulating discussions. We thank Drs Zukerberg LR and Rueda BR (Massachusetts General Hospital) for sharing reagents. This work was supported by the National Institutes of Health Grant P01 CA116676 (to F.R.K. and H.F.), Georgia Cancer Coalition (to H.F. and F.R.K.), the Chinese National Natural Science Foundation No. 31271444 and No. 81201726 (to Z.S.), the Foundation for Research Cultivation and Innovation of Jinan University No. 21612407 (to Z.S.) and the Specialized Research Fund for the Doctoral Program of Higher Education No. 20124401120007 (to Z.S.).
Footnotes
Disclosure statement
The authors declare no conflict of interest.
References
- 1.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Current opinion in oncology. 2013;25:52–58. doi: 10.1097/CCO.0b013e32835b639e. [DOI] [PubMed] [Google Scholar]
- 3.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 6.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem. 2008;283:13707–13713. doi: 10.1074/jbc.M710030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday MJ. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. Embo J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 12.Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, Xiao Z, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol Cell Biol. 2008;28:1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J Biol Chem. 2004;279:16000–16006. doi: 10.1074/jbc.M312264200. [DOI] [PubMed] [Google Scholar]
- 14.Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, et al. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell. 2005;17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 15.Zukerberg LR, Patrick GN, Nikolic M, Humbert S, Wu CL, Lanier LM, et al. Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron. 2000;26:633–646. doi: 10.1016/s0896-6273(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 16.Wu CL, Kirley SD, Xiao H, Chuang Y, Chung DC, Zukerberg LR. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 2001;61:7325–7332. [PubMed] [Google Scholar]
- 17.Tsuji K, Mizumoto K, Yamochi T, Nishimoto I, Matsuoka M. Differential effect of ik3-1/cables on p53- and p73-induced cell death. J Biol Chem. 2002;277:2951–2957. doi: 10.1074/jbc.M108535200. [DOI] [PubMed] [Google Scholar]
- 18.Wang N, Guo L, Rueda BR, Tilly JL. Cables1 protects p63 from proteasomal degradation to ensure deletion of cells after genotoxic stress. EMBO Rep. 2010;11:633–639. doi: 10.1038/embor.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirley SD, Rueda BR, Chung DC, Zukerberg LR. Increased growth rate, delayed senescense and decreased serum dependence characterize cables-deficient cells. Cancer Biol Ther. 2005;4:654–658. doi: 10.4161/cbt.4.6.1732. [DOI] [PubMed] [Google Scholar]
- 20.Zukerberg LR, DeBernardo RL, Kirley SD, D'Apuzzo M, Lynch MP, Littell RD, et al. Loss of cables, a cyclin-dependent kinase regulatory protein, is associated with the development of endometrial hyperplasia and endometrial cancer. Cancer Res. 2004;64:202–208. doi: 10.1158/0008-5472.can-03-2833. [DOI] [PubMed] [Google Scholar]
- 21.Kirley SD, D'Apuzzo M, Lauwers GY, Graeme-Cook F, Chung DC, Zukerberg LR. The Cables gene on chromosome 18Q regulates colon cancer progression in vivo. Cancer Biol Ther. 2005;4:861–863. doi: 10.4161/cbt.4.8.1894. [DOI] [PubMed] [Google Scholar]
- 22.Dong Q, Kirley S, Rueda B, Zhao C, Zukerberg L, Oliva E. Loss of cables, a novel gene on chromosome 18q, in ovarian cancer. Mod Pathol. 2003;16:863–868. doi: 10.1097/01.MP.0000084434.88269.0A. [DOI] [PubMed] [Google Scholar]
- 23.Park do Y, Sakamoto H, Kirley SD, Ogino S, Kawasaki T, Kwon E, et al. The Cables gene on chromosome 18q is silenced by promoter hypermethylation and allelic loss in human colorectal cancer. Am J Pathol. 2007;171:1509–1519. doi: 10.2353/ajpath.2007.070331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan D, Kirley S, Li Q, Ramnath N, Slocum HK, Brooks JS, et al. Loss of cables protein expression in human non-small cell lung cancer: a tissue microarray study. Hum Pathol. 2003;34:143–149. doi: 10.1053/hupa.2003.26. [DOI] [PubMed] [Google Scholar]
- 25.Arnason T, Pino MS, Yilmaz O, Kirley SD, Rueda BR, Chung DC, et al. Cables1 is a tumor suppressor gene that regulates intestinal tumor progression in Apc(Min) mice. Cancer Biol Ther. 2013;14:672–678. doi: 10.4161/cbt.25089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto H, Friel AM, Wood AW, Guo L, Ilic A, Seiden MV, et al. Mechanisms of Cables 1 gene inactivation in human ovarian cancer development. Cancer Biol Ther. 2008;7:180–188. doi: 10.4161/cbt.7.2.5253. [DOI] [PubMed] [Google Scholar]
- 27.Coleman ML, Marshall CJ, Olson MF. Ras promotes p21(Waf1/Cip1) protein stability via a cyclin D1-imposed block in proteasome-mediated degradation. Embo J. 2003;22:2036–2046. doi: 10.1093/emboj/cdg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Nie L, Maki CG. Cdk2-dependent Inhibition of p21 stability via a C-terminal cyclin-binding motif. J Biol Chem. 2005;280:29282–29288. doi: 10.1074/jbc.M407352200. [DOI] [PubMed] [Google Scholar]
- 29.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 30.Zirbes TK, Baldus SE, Moenig SP, Nolden S, Kunze D, Shafizadeh ST, et al. Prognostic impact of p21/waf1/cip1 in colorectal cancer. Int J Cancer. 2000;89:14–18. doi: 10.1002/(sici)1097-0215(20000120)89:1<14::aid-ijc3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 31.Komiya T, Hosono Y, Hirashima T, Masuda N, Yasumitsu T, Nakagawa K, et al. p21 expression as a predictor for favorable prognosis in squamous cell carcinoma of the lung. Clin Cancer Res. 1997;3:1831–1835. [PubMed] [Google Scholar]