In Table 4 and Supplemental Table 1 of the published paper entitled “Genetic Diagnosis of Charcot-Marie-Tooth Disease in a Population by Next-Generation Sequencing” [1], we classified two variants as likely pathogenic. These two variants (NM_001126131.1:c.[1491G>C(;)2243G>C]), identified in the POLG gene, were carried by one patient (ID 62). As it was not possible to examine whether the two variants were in cis or trans, that is, situated on the same or different alleles, these two variants should have been reported as uncertain pathogenic variants.
This change also influences the Results, where the prevalence of identified variants is corrected to 8 likely and 17 uncertain pathogenic variants.
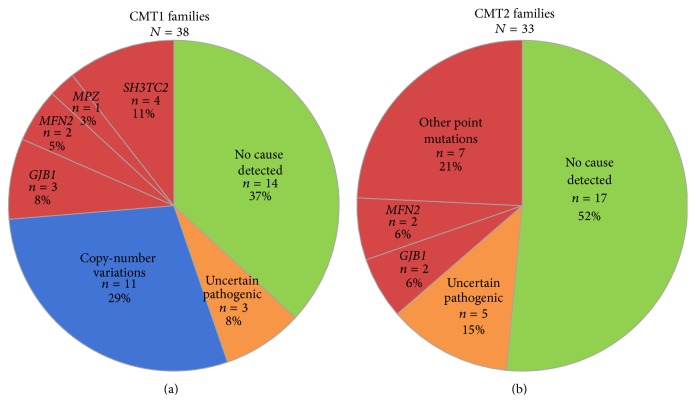
In Figure 1, the number of families with likely pathogenic variants is reduced to n = 7 (9%) and the number for families with uncertain pathogenic variants is increased to n = 10 (12%). And in Figure 2(b), the number of CMT2 families in the category “other point mutations” is reduced to n = 7 (21%) and the number of families with uncertain pathogenic variants is increased to n = 5 (15%). See corrected Figures 1 and 2(b).
Figure 1.
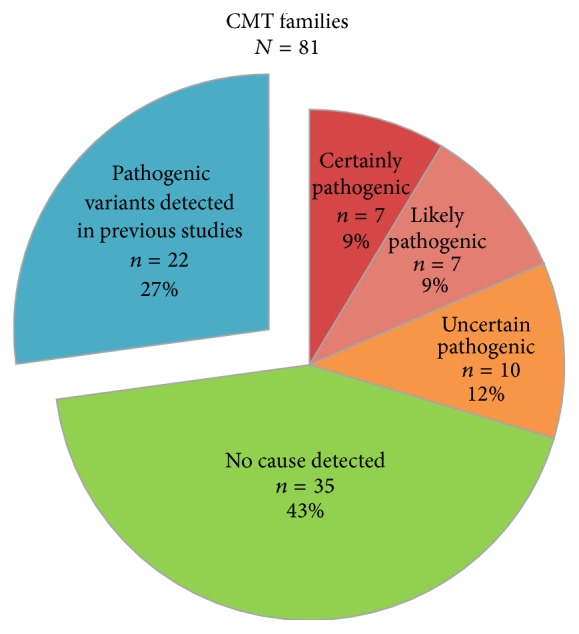
Identified variants in 81 Norwegian CMT families from the general population. Our previous studies identified copy-number variations in 12 CMT families and pathogenic point mutations in 10 CMT families [2, 14, 19]. The remaining 59 CMT families were investigated by next-generation sequencing.
Figure 2.
Frequencies of certain and likely pathogenic variants in CMT1 and CMT2 families from the Norwegian general population.
References
- 1.Høyer H., Braathen G. J., Busk Ø. L., et al. Genetic diagnosis of Charcot-Marie-Tooth disease in a population by next-generation sequencing. BioMed Research International. 2014;2014:13. doi: 10.1155/2014/210401.210401 [DOI] [PMC free article] [PubMed] [Google Scholar]