Abstract
Light affects animal physiology and behavior more than simply through classical visual, image-forming pathways. Nonvisual photoreception regulates numerous biological systems, including circadian entrainment, DNA repair, metabolism, and behavior. However, for the majority of these processes, the photoreceptive molecules involved are unknown. Given the diversity of photophysiological responses, the question arises whether a single photopigment or a greater diversity of proteins within the opsin superfamily detect photic stimuli. Here, a functional genomics approach identified the full complement of photopigments in a highly light-sensitive model vertebrate, the zebrafish (Danio rerio), and characterized their tissue distribution, expression levels, and biochemical properties. The results presented here reveal the presence of 42 distinct genes encoding 10 classical visual photopigments and 32 nonvisual opsins, including 10 novel opsin genes comprising four new pigment classes. Consistent with the presence of light-entrainable circadian oscillators in zebrafish, all adult tissues examined expressed two or more opsins, including several novel opsins. Spectral and electrophysiological analyses of the new opsins demonstrate that they form functional photopigments, each with unique chromophore-binding and wavelength specificities. This study has revealed a remarkable number and diversity of photopigments in zebrafish, the largest number so far discovered for any vertebrate. Found in amphibians, reptiles, birds, and all three mammalian clades, most of these genes are not restricted to teleosts. Therefore, nonvisual light detection is far more complex than initially appreciated, which has significant biological implications in understanding photoreception in vertebrates.
Light detection (photoreception) in vertebrates is often focused on the visual system (Davies et al. 2012a), where eyes and the specialized photoreceptors they contain are critical for forming images of the external world. There are, however, many nonvisual effects of light in vertebrates, including the synchronization of biological rhythms (circadian clock photoentrainment) (Freedman et al. 1999; Chen et al. 2011; Lucas et al. 2012), seasonal or photoperiodic changes that often underpin the timing of reproduction (Follett et al. 1985), pupillary constriction (Lucas et al. 2003), DNA repair activation (Tamai et al. 2004; Hirayama et al. 2009; Gavriouchkina et al. 2010), and the cell cycle (Dekens et al. 2003; Laranjeiro et al. 2013), as well as numerous other behavioral, physiological, cellular, and molecular responses. Currently, relatively little is known about light detection and the signaling pathways that regulate these nonvisual processes, with the principle exception of the photopigment melanopsin, which has been shown to play a key role in circadian clock entrainment and pupillary constriction in mammals (Rollag et al. 2003; Hatori and Panda 2010). It is not yet clear, however, whether melanopsin is the predominant nonvisual photopigment in vertebrates or whether additional photoreceptive molecules exist. The aim of this study was to explore the diversity and complexity of opsin-based photopigments, as well as examine the extent of nonvisual photoreception in a more complete and systematic manner.
In the course of evolution, a number of gene duplication events have led to an increased repertoire of visual (image-forming) opsin family members, where the classic five classes of vertebrate visual pigment genes, a single rod (rhodopsin-like-1; rh1) and four cones classes (long-wavelength-sensitive, lws; short-wavelength-sensitive-1, sws1; short-wavelength-sensitive-2, sws2; and rhodopsin-like-2; rh2), are extended in many bony fishes (Davies et al. 2012a). In zebrafish, the complement of visual pigments has doubled through a duplication of both lws (Chinen et al. 2003) and rh1 (Morrow and Chang 2010; Davies et al. 2012a) genes and a quadrupling of the rh2 gene (Chinen et al. 2003). Similarly, in the nonvisual system, no less than five opn4 (melanopsin) genes were identified that duplicated from two ancestral melanopsin gene lineages (Bellingham et al. 2006; Davies et al. 2011). Furthermore, each opn4 gene encodes for a distinct photosensitive pigment differentially distributed throughout the neural retina of the eye (Davies et al. 2011). The zebrafish (Danio rerio), therefore, is an excellent and established model vertebrate in which to study photopigment diversity and their functional roles.
The circadian clock consists of a network of activator (e.g., clock circadian regulator, Clock; aryl hydrocarbon receptor nuclear translocator–like, Arntl) and repressor (e.g., period, Per; cryptochrome, Cry) proteins that dictate oscillatory gene expression over a period of about a day via feedback mechanisms of transcription and translation (Albrecht 2004; Tamai et al. 2005). To ensure these critical biological rhythms are synchronized to the changing light environment, photoentrainable sensory systems have evolved where light-sensitive molecules convey photic information that are linked to endogenous clocks (Panda et al. 2003). In mammals, the central circadian pacemaker is found in the suprachiasmatic nucleus (SCN) (Moore 2013) and receives light input information via melanopsin-expressing retinal ganglion cells (RGCs) in the eye (Hankins et al. 2008; Davies et al. 2010). Circadian oscillators in peripheral tissues are then synchronized (Mohawk et al. 2012; Brown and Azzi 2013) by cues emanating either from the SCN (Moore 2013), or from other signals such as temperature (Brown et al. 2002; Lahiri et al. 2005; Buhr et al. 2010) or rhythmic feeding activity (Damiola et al. 2000; Schibler et al. 2003; Challet 2013). Similar to mammals, clocks are also present in the tissues of zebrafish and other teleosts (Whitmore et al. 2000; Velarde et al. 2009; Cavallari et al. 2011; Martin-Robles et al. 2012). However, the circadian system of fish appears to be far more decentralized than in mammals (Tamai et al. 2005), as peripheral tissues are thought to be directly photoresponsive, such that their phase can be entrained by light (Whitmore et al. 2000; Tamai et al. 2005; Kaneko et al. 2006).
In the zebrafish, the majority of tissues and cells in the body contain a directly photoentrainable circadian pacemaker (Whitmore et al. 2000; Kaneko et al. 2006). Thus, these tissues and cells must contain the essential photopigments necessary for circadian photoreception and other associated physiological responses to light. Transcriptome analysis of light-regulated genes in various zebrafish tissues reveals that, in addition to the circadian system, biological processes as diverse as DNA repair, oxidative stress, mitochondrial function, and heme metabolism are directly influenced by light (Gavriouchkina et al. 2010; Weger et al. 2011). Furthermore, direct effects of light on behavior (i.e., dark photokinesis and photomotor responses) have been reported in zebrafish, even in the absence of the eyes and pineal, suggesting a role for photoreceptors in the brain (Fernandes et al. 2012; Kokel et al. 2013). It remains unknown whether a single nonvisual photopigment class (e.g., melanopsin) or multiple photopigments control these wide-ranging biological responses in parallel detection systems. Thus, further investigation is required to determine the full complement and functional roles of vertebrate photopigments.
To address these questions, this study systematically mined the zebrafish genome for opsin-like genes, with the aim of identifying and functionally characterizing the entire set of opsin genes. Such an approach will provide a significant foundation upon which critical insights linking light detection and an array of diverse molecular, biological, physiological, and behavioral processes will be determined.
Results
Direct photosensitivity in isolated zebrafish tissues
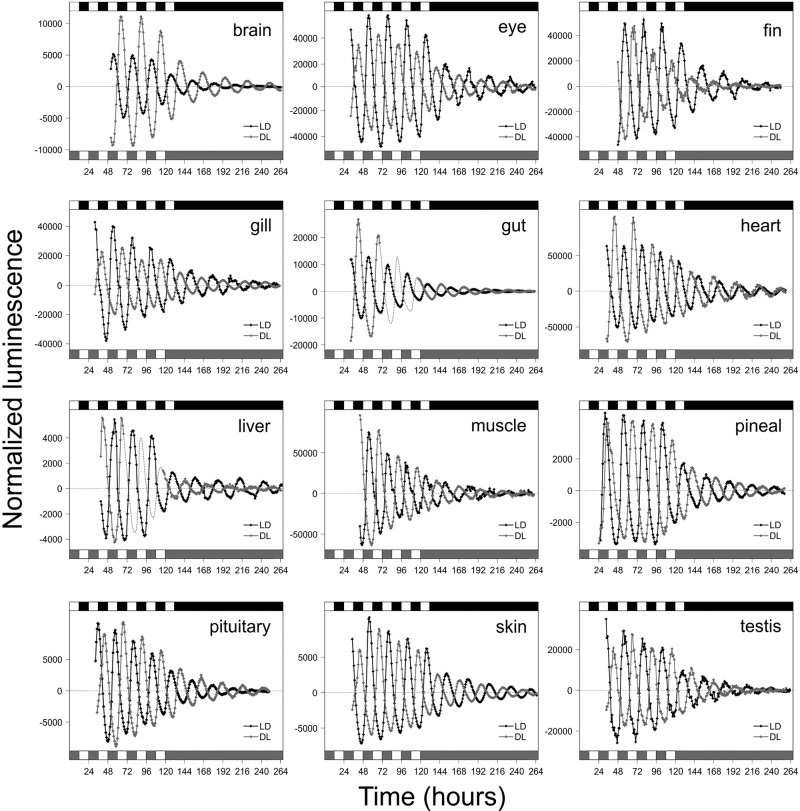
The circadian system of the zebrafish is highly decentralized. Although this phenomenon has been shown to occur in some isolated organs (Kaneko et al. 2006), whether direct clock entrainment by light actually occurs in the majority of zebrafish tissues was examined. Key tissues were dissected from transgenic per3-luciferase clock gene reporter zebrafish (Kaneko and Cahill 2005; Kaneko et al. 2006) and placed into culture while being exposed to two different lighting regimes: 12 h of light, followed by 12 h of dark (LD), or 12 h of dark, followed by 12 h of light (DL). Bioluminescence from the per3-luciferase transgene showed that exposure to the different lighting regimes synchronized their circadian clocks 12 h out of phase, which persisted in constant darkness (DD) (Fig. 1). The amplitude of per3-luciferase oscillation varied between tissues, as observed previously (Kaneko et al. 2006), but the phase difference due to the lighting regime was maintained in all tissues examined, demonstrating that these local organ clocks are being set directly by light. These results extend the phenomena of tissue photosensitivity to most, if not all, zebrafish organs and tissues.
Figure 1.
Light-entrainable circadian clocks are present in adult zebrafish (Danio rerio) tissues, specifically brain, eye, fin, gill, gut, heart, liver, muscle, pineal, pituitary, skin, and testis. Bioluminescence was monitored throughout the experiment and showed alternating cycles of per3 expression when tissues were exposed either to a forward cycle of 12 h of light, 12 h of dark (LD; black) or to an inverse cycle of 12 h of dark, 12 h of light (DL; gray). Note that open and solid boxes represent periods of light and darkness, respectively. In gut and liver experiments, a gray dotted line represents interpolated cycles of bioluminescence where data are partly absent. All tissues experienced LD or DL conditions for 5 to 6 d prior to transference into constant darkness for 7 d.
Discovery of diverse pigment genes in the zebrafish genome
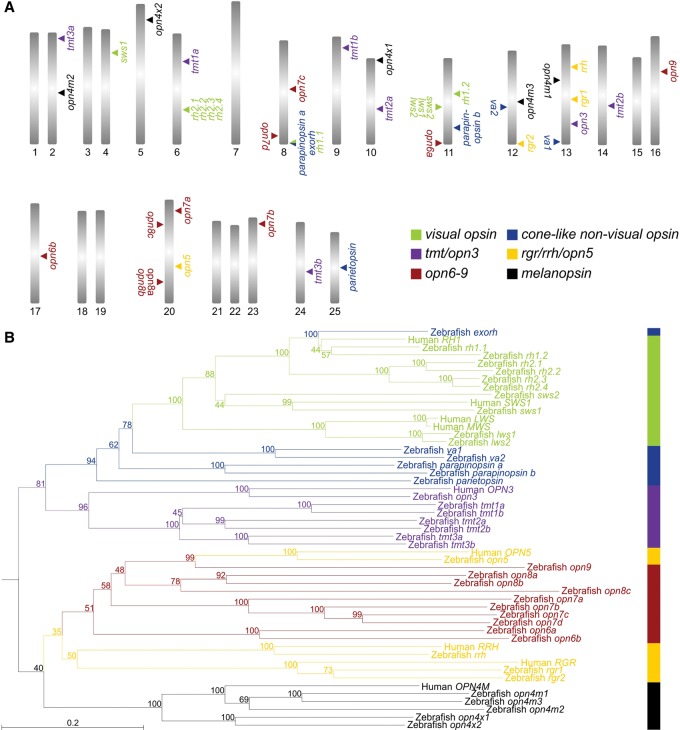
Systematic mining of the zebrafish genome identified many opsin-like genes that might contribute to tissue light sensitivity. Additionally, reverse transcription–polymerase chain reaction (RT-PCR) amplification, cloning, and sequencing of full-length coding regions confirmed their presence in the zebrafish eye. Together with the isolation of known visual and nonvisual opsins, our functional genomics approach led to the identification of several additional members of known opsin gene families, such as melanopsin (opn4m1 [ZFIN: opn4a], opn4m2 [ZFIN: opn4.1], and opn4m3 [ZFIN: opn4b] [mammalian-like class]; opn4x1 [ZFIN: opn4xa] and opn4x2 [ZFIN: opn4b] [Xenopus-like class]) (Davies et al. 2011); retinal pigment epithelium (RPE) retinal G protein receptor (rgr; two members: rgr1 [ZFIN: rgra] and rgr2 [ZFIN: rgrb]); and teleost multiple tissue (tmt) opsin (six members renamed based on their phylogenetic position: tmt1a [ZFIN: tmtops2a] and tmt1b [ZFIN: tmtops2b]; tmt2a [ZFIN: tmtops3b] and tmt2b [ZFIN: tmtops3a]; and tmt3a [ZFIN: tmtopsa] and tmt3b [ZFIN: tmtopsb]) (Fig. 2; Supplemental Figs. S1–S3; Fischer et al. 2013). Four new classes of photopigment genes were also discovered that consist of 10 unique and distinct genes that are most closely related phylogenetically to neuropsin (opn5) (Tarttelin et al. 2003), retinal pigment epithelium-derived rhodopsin homolog (rrh; also known as peropsin) (Bellingham et al. 2003b), and rgr opsin (Jiang et al. 1993) classes (Fig. 2). Specifically, these new opsins clade between the opn5 and rrh branches into four discrete groups (Fig. 2; Supplemental Fig. S1) that have distinct gene structures from either opsin class, although some intron positions are conserved (Supplemental Fig. S3). Collectively, these data show that the new opsin pigment genes are not simply opn5-like and arose from a series of duplication events of the opn5 gene, but are distinct gene classes in their own right. Indeed, the most closely related pigment gene to opn5 (historically presented as a direct sister clade to rrh and rgr opsin genes, but now shown to be incorrectly phylogenetically placed) is opn9, whose protein is structurally distinct from its sister class by the presence of a third extracellular loop that is extended by 80 amino acids. This is a unique feature of Opn9 pigments alone (Supplemental Fig. S3). These four distinct gene classes are herein named opn6 (opsin 6, group member a and opsin 6, group member b), opn7 (opsin 7, group member a; opsin 7, group member b; opsin 7, group member c; and opsin 7, group member d), opn8 (opsin 8, group member a; opsin 8, group member b; and opsin 8, group member c), and opn9 (opsin 9), respectively, based on their relative phylogenetic positions (Fig. 2; Supplemental Figs. S1–S3).
Figure 2.
Opsin diversity in the zebrafish genome. (A) Mapping of opsins shows that the zebrafish genome contains 42 distinct pigment genes across 17 chromosomes. (B) Phylogenetic analysis of pigment genes in human (Homo sapiens) and zebrafish (D. rerio) genomes, showing the presence of 10 novel opsin genes. Outgroups are not shown. The scale bar indicates the number of nucleotide substitutions per site and the degree of internal branching is expressed as a percentage at each node. For a more extensive tree and GenBank accession numbers (KT008391–KT008432), including ZFIN (http://www.zfin.org) gene nomenclature where appropriate, please see Supplemental Figure S1. Opsin classes are subdivided into six main groups that contain visual opsins (green), cone-like nonvisual opsins (blue), opn3/tmt opsins (purple), rgr/rrh/opn5 opsins (yellow), opn4 opsins (black), and the new opsins (red). Specifically, these are subdivided into extra-ocular rhodopsin (exorh), rhodopsin-like-1 (rh1) rod opsin, rhodopsin-like-2 (rh2) cone opsin, short-wavelength-sensitive-2 (sws2) cone opsin, short-wavelength-sensitive-1 (sws1) cone opsin, long-wavelength-sensitive (lws) cone opsin, vertebrate ancient (va) long opsin, parapinopsin, parietopsin, encephalopsin/panopsin (opn3), teleost multiple tissue (tmt) opsin, neuropsin (opn5), novel opsins (opn6, opn7, opn8, and opn9), retinal pigment epithelium (RPE) rhodopsin homolog (rrh; also known as peropsin), RPE retinal G protein receptor (rgr), mammalian-like melanopsin (opn4m), and Xenopus-like melanopsin (opn4x).
When comparing teleosts, it was evident that the genome of the Japanese pufferfish (Takifugu rubripes) contained corresponding orthologs for all except two of the new zebrafish opsin genes, namely, opn6b and opn7d (Supplemental Fig. S1). The pufferfish genome does contain a sequence corresponding to opn6b within scaffold 102, but a likely recent mutational event has introduced a premature stop codon in exon 2 that renders this a pseudogene. Investigation of the opn7 subclass identified only three out of the four zebrafish orthologs in the pufferfish genome (Supplemental Fig. S1). Phylogenetic analysis showed that both zebrafish opn7a and opn7b have direct orthologs in the pufferfish genome, with the third pufferfish opn7 ortholog branching prior to the zebrafish opn7c and opn7d genes (Supplemental Fig. S1). Thus, it would appear that the gene duplication that gave rise to opn7c and opn7d in the zebrafish genome is an independent event.
Collectively, these data show that the zebrafish possesses a complement of 42 opsin genes (GenBank accession numbers KT008391–KT008432) (Supplemental Fig. S1) of which >75% are classed as nonvisual pigments (Fig. 2; Supplemental Figs. S1, S2, S3) and are conserved in other vertebrate classes (Fig. 3). In the zebrafish karyotype, 17 out of 25 chromosomes contain at least one opsin gene, with some chromosomes containing as many as five or six opsins (Fig. 2A).
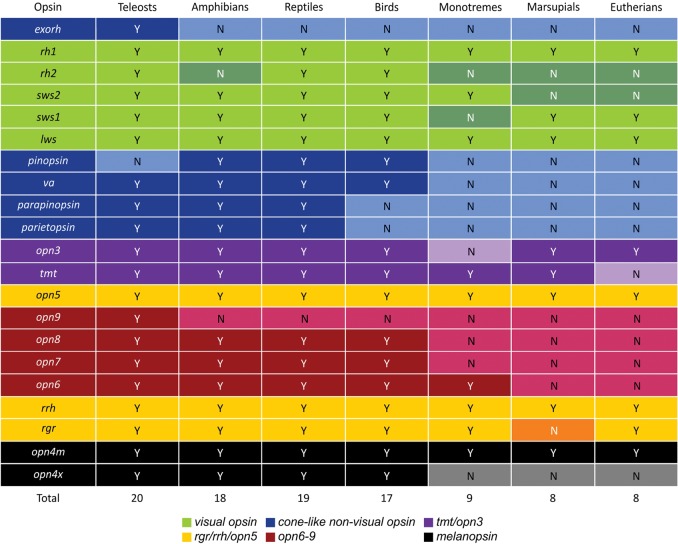
Figure 3.
Summary table showing the evolution of all opsin classes in gnathostome vertebrates (teleosts to eutherians), where many opsins present in some classes (yes [Y]) were lost in others (no [N]), especially at the base of the mammalian lineage. For further information relating to opsin class subdivision into six main groups, see Figure 2: (1) visual opsins (green), (2) cone-like nonvisual opsins (blue), (3) opn3/tmt opsins (purple), (4) rgr/rrh/opn5 opsins (yellow), (5) opn4 opsins (black), and (6) the new opsins (red).
Opsin mRNA profiling using the NanoString nCounter system
To examine the expression pattern of all 42 opsin genes, a digital barcode assay developed by NanoString Technologies (http://www.nanostring.com) was used (Fig. 4; Supplemental Fig. S4). This hybridization-based protocol is highly sensitive and can quantify the expression of hundreds of genes in a single RNA sample (Geiss et al. 2008). To determine the relative levels of opsin transcript expressed in different zebrafish tissues, multiple organs were dissected from adult zebrafish (same as in Fig. 1) between zeitgeber time (ZT) 3 and ZT6 (where ZT0 is defined as the onset of light and the digits refer to the number of hours post-ZT0).
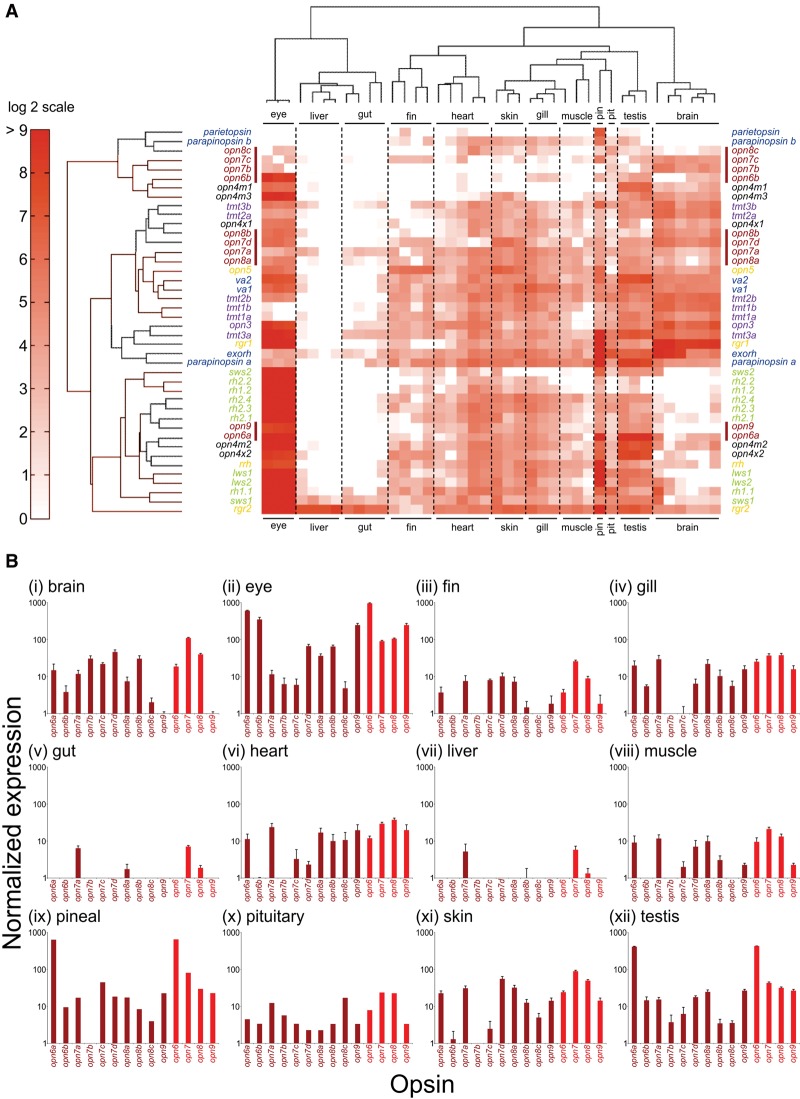
Figure 4.
Gene expression profiling of zebrafish opsin transcripts using the NanoString nCounter assay. Adult zebrafish tissues (i, brain; ii, eye; iii, fin; iv, gill; v, gut; vi, heart; vii, liver; viii, muscle; ix, pineal [pin]; x, pituitary [pit]; xi, skin, and xii, testis) were dissected and processed as described in the Methods. (A) A heat map shows relative opsin gene expression on a log2 scale, with red and white indicating high and low expression, respectively. Each column represents an independent tissue sample, and each row represents a different opsin, which are color-coded by class as in Figure 2. A red bar further highlights the novel opsins. Hierarchical clustering of both the opsins (by Euclidean distance) and tissues (by correlation) organizes the data set into similar expression patterns. The widespread yet complex expression patterns in all tissues, except the liver and gut, are readily discernable. (B) Novel opsin gene expression (dark red) in different tissues, measured by NanoString, presented as bar charts on a log10 scale. Relative transcript data are shown for each distinct new opsin gene, as well as a total for each novel opsin class (i.e., opn6, opn7, opn8, and opn9) (light red).
The results from this analysis are presented on a log2 scale as a heat map, with abundantly expressed genes shown in red and less abundantly expressed genes shown in white (Fig. 4A). In contrast with circadian clock genes (arnt1a, clocka, cry1a, per1b, and per2) being detected in all tissue samples (Supplemental Fig. S5), the opsin gene classes exhibited diverse, partially overlapping expression profiles across all tissues, as revealed by hierarchical clustering (Fig. 4). Indeed, only rgr2 and, to a lesser extent, sws1 [ZFIN: opn1sw1] showed consistently significant expression across all the tissue samples (Fig. 4; Supplemental Fig. S4). Several patterns of tissue-specific opsin gene expression emerged. First and perhaps not surprisingly, the eye, as a known photosensitive organ, expressed the greatest number of highly abundant opsin gene classes, with most transcripts being the visual opsins of the classical photoreceptors. However, the eye also expressed a number of nonvisual opsin genes, including all five opn4 submembers (Davies et al. 2011), two vertebrate ancient (va) long opsins (va1 [ZFIN: valopa] and va2 [ZFIN: valopb]), encephalopsin/panopsin (opn3), rrh, rgr1, and rgr2, as well as a number of new opsins, including opn6a, opn6b, and opn9 (Fig. 4; Supplemental Fig. S4). The second well-characterized, light-sensitive organ in zebrafish, the pineal complex, also expressed a large set of the opsin genes, including pigment genes already known to be present in the pineal in fish and other vertebrates, such as extra-ocular rhodopsin [ZFIN: exorh] (Bellingham et al. 2003a), parietopsin (Su et al. 2006), parapinopsin a (Blackshaw and Snyder 1997; Koyanagi et al. 2004), as well as a subset of visual opsins (Koyanagi et al. 2004; Kawano-Yamashita et al. 2007), namely, rh1.1 [ZFIN: rho], lws1 [ZFIN: opn1lw1], lws2 [ZFIN: opn1lw2], and sws2 [ZFIN: opn1sw2]. In addition, tmt3a, opn4m2, opn4x2, and opn6a were present at high levels in the pineal, as well as some genes thought to be retinal pigment epithelium (RPE) specific, such as rgr1, rgr2, and rrh (Fig. 4; Supplemental Fig. S4; Sun et al. 1997; Hao and Fong 1999). Third, the brain abundantly expressed a number of opsins, including rgr1 (and rgr2 to a lesser degree), extra-ocular rhodopsin, opn3, all six tmt genes, va1, va2, parapinopsin a, opn5, rrh, all five opn4 genes, and all of the novel opsins (especially opn6a, all four opn7 genes, opn8a, and opn8b) except for opn9 (Fig. 4; Supplemental Fig. S4). While many of the genes expressed in brain are also abundant in the eye, several opsins are restricted to either tissue. For example, a subset of visual (e.g., rh1.2 [ZFIN: rhol], rh2.2 [ZFIN: opn1mw2], rh2.4 [ZFIN: opn1mw4], and lws1) and nonvisual (e.g., opn9) opsins are excluded from the brain, while tmt1a and tmt1b are found in brain, but not consistently in the eye. Fourth, most other tissues (e.g., fin, gill, heart, muscle, skin, and testis), with the exception of the liver and gut, exhibited moderate to high levels of expression for numerous opsin genes, including most of the new opsins. Indeed, nearly the entire opsin repertoire is expressed in varying amounts in the testis (41 of 42 genes), heart (38 of 42 genes), skin (38 of 42 genes), and gill (36 of 42 genes) (Fig. 4; Supplemental Fig. S4). Uniquely, profiling of the zebrafish transcriptome using the NanoString system has provided a novel and complete analysis of opsin photopigment expression across a wide range of tissues (Fig. 4; Supplemental Fig. S4), all of which have been shown to be photoentrainable (Fig. 1).
Localization of new opsin gene expression in the eye and brain by R-ISH
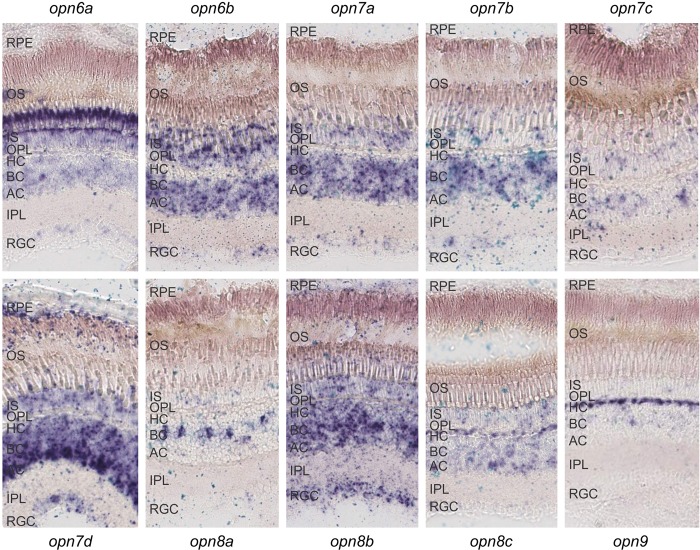
Quantitative gene expression analysis using the NanoString approach demonstrated that the novel opsins are abundant in both the eye and the brain of the zebrafish. As these are complex tissues containing many different cell types, RNA in situ hybridization (R-ISH) on tissue sections was used to explore the precise cellular pattern of expression in these structures (Figs. 5, 6). In all cases, diffuse ocular expression was found in the inner nuclear layer (INL) with mRNA transcripts detected in bipolar cells (opn8a), horizontal cells (opn8c and opn9), and amacrine cells (opn7d, but lower expression and in fewer cells for opn7c, opn8b, and opn8c) (Fig. 5). The RPE expresses opn7d only, with the RGC layer revealing the presence of several new opsin transcripts, in particular opn8b, but not opn7c, opn8a, opn8c, and opn9. Interestingly, expression was high in the outer nuclear layer (ONL) of the photoreceptors for opn6a, opn6b, and opn7a, with perhaps low levels of diffuse expression for opn7b, opn7c, opn7d, opn8b, and opn8c, but no expression for opn8a or opn9 (Fig. 5). Collectively, these and previously presented data (Fig. 4; Supplemental Fig. S4) show that the new opsins are not expressed in a pattern that is correlated with gene class.
Figure 5.
The localization of all 10 novel opsin transcripts in the adult zebrafish retina (sampled at ZT3) by RNA in situ hybridization, showing that each gene exhibits a discrete, partly overlapping expression profile. (RPE) Retinal pigment epithelium, (OS) outer segments, (IS) inner segments, (OPL) outer plexiform layer, (HC) horizontal cell layer, (BC) bipolar cell layer, (AC) amacrine cell layer, (IPL) inner plexiform layer, and (RGC) retinal ganglion cell layer.
Figure 6.
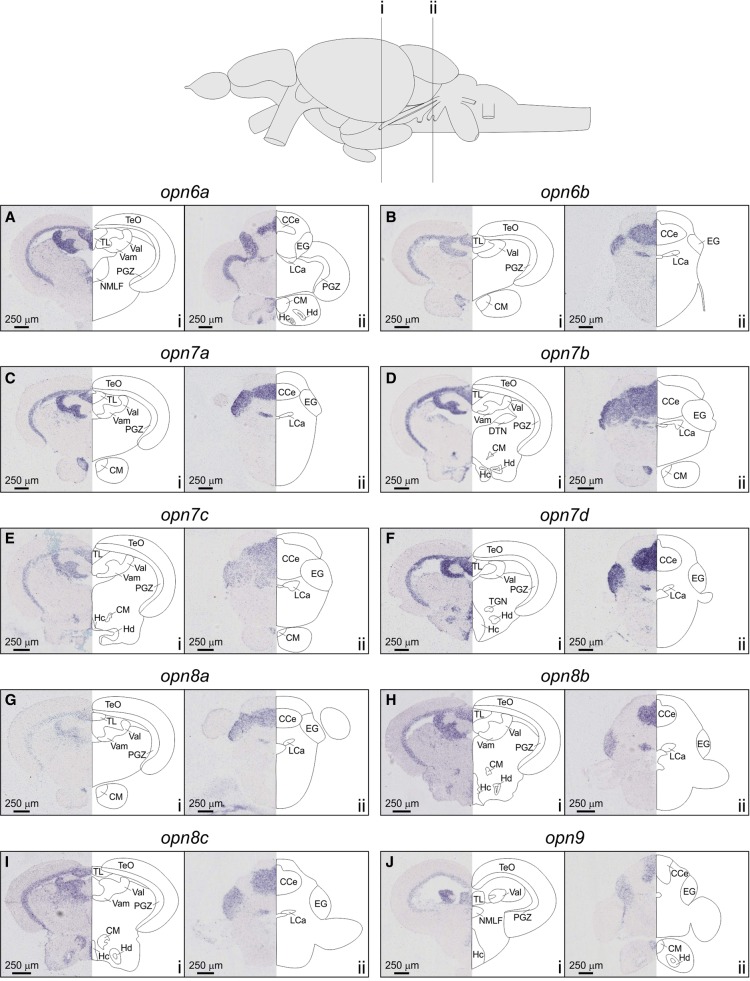
RNA in situ hybridization of novel opsin expression in the adult zebrafish brain collected at ZT3 (opn6a, A; opn6b, B; opn7a, C; opn7b, D; opn7c, E; opn7d, F; opn8a, G; opn8b, H; opn8c, I; and opn9, J). Two areas of the brain were analyzed in transverse section through the mesencephalon (i) and cerebellum (ii), where new opsin transcripts were located to the periventricular gray zone (PGZ), the optic tectal torus longitudinalis (TL), the corpus mamillare (CM), the granular layers of both the medial and lateral valvula cerebelli (Vamgra and Valgra, respectively), the granular layer of the corpus cerebellum (CCegra), the eminentia granularis (EG), the granular layer of the lobus caudalis cerebelli (LCagra), and two hypothalamic nuclei (caudal nucleus [Hc] and dorsal nucleus [Hd]).
The eye is the most photoreceptive organ in vertebrates; however, there is increasing evidence to support the idea that the brain is also an important tissue involved in light detection. R-ISH determined the location of expression for each new opsin in the adult zebrafish brain with tissues harvested at ZT3, with all 10 novel opsins expressed in the periventricular gray zone (PGZ) and torus longitudinalis (TL) of the optic tectum (TeO) (Fig. 6). The expression of the new opsin genes were also identified in the corpus mamillare (CM), the granular layers of both the medial and lateral valvula cerebelli (Vamgra and Valgra, respectively), the granular layer of the corpus cerebellum (CCegra), the eminentia granularis (EG), and the granular layer of the lobus caudalis cerebelli (LCagra; all genes except opn9) (Fig. 6). Interestingly, particular novel opsin transcripts were detected in many other specific areas of the brain, such as the hypothalamus (e.g., opn6a, opn7b, opn7c, opn7d, opn8b, opn8c, and opn9 in the caudal nucleus [Hc]; and opn6a, opn7b, opn7d, opn8b, opn8c, and opn9 in the dorsal nucleus [Hd]) (Fig. 6). As is evident from these data, multiple regions of the adult zebrafish brain express all 10 new opsins, either specifically or in many cases through coexpression in the same areas. Furthermore, these new opsins greatly overlap with the expression of multiple clock genes (cry1a, per2, and per3) (Moore and Whitmore 2014) and c-fos [ZFIN: fosab] (a marker of neuronal activity) in the presence of light (Moore and Whitmore 2014). Thus, the zebrafish brain is likely to be a particularly important photoreceptive organ, more than previously appreciated and perhaps only second to the eye concerning the degree of light sensitivity.
Spectral sensitivity of zebrafish novel opsins
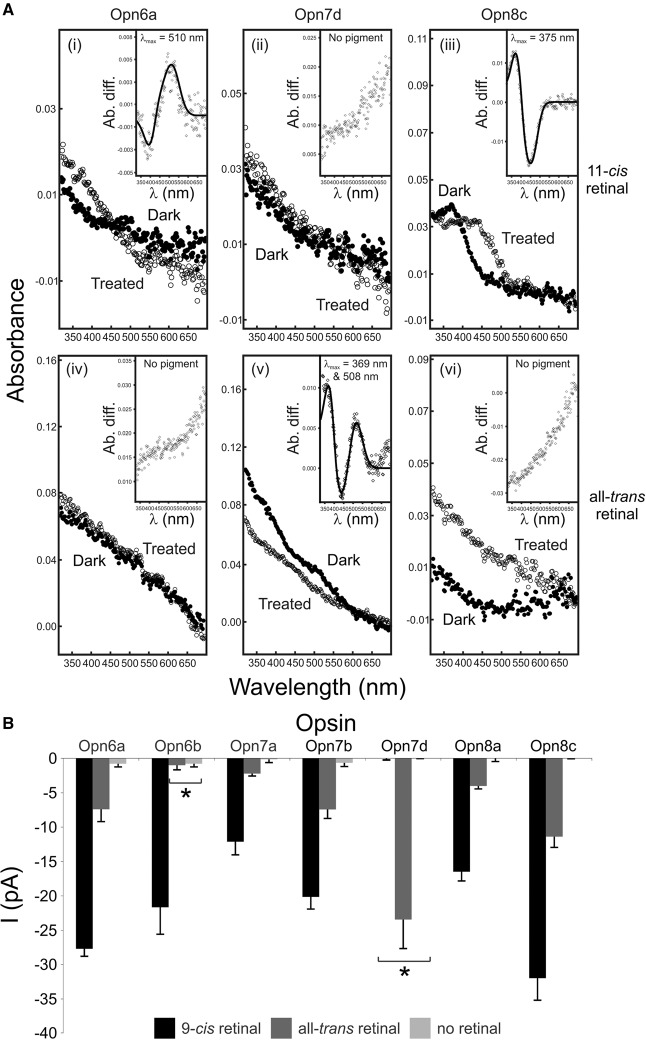
The visual pigments of zebrafish have been shown to be sensitive to wavelengths that range from the UV to the red end of the visible spectrum, with photosensitive molecules that form a stable interaction with the 11-cis retinal chromophore only (monostable) (Chinen et al. 2003). Typically, nonvisual pigments can be monostable (e.g., pinopsin [Nakamura et al. 1999] and va opsin [Sato et al. 2011]) or bistable (e.g., neuropsin [Yamashita et al. 2010] and parapinopsin [Koyanagi et al. 2004]), as they form stable opsin and chromophore complexes with multiple retinal isomers (11-cis and all-trans retinoids). To explore whether the new opsins are monostable or bistable, UV-visible (UV-vis) spectrophotometry was used to determine their particular absorbance characteristics. Spectral analyses show that the novel opsins appear to be monostable, with each subclass being biochemically very different in the chromophores utilized and in the shape and absorbance maxima of their spectra (Fig. 7A). Specifically, Opn6 binds 11-cis retinal alone to form a monophasic (one main peak) dark spectrum with a λmax at 510 nm. In contrast, Opn7 is a biphasic (two main peaks) pigment that binds all-trans retinal with λmax values at 369 nm and 508 nm, and Opn8 pigments are UV-sensitive (λmax = 375 nm), monophasic, and interact with 11-cis retinal only. The in vitro regeneration of Opn9 was not successful, most likely due to technical difficulties rather than an intrinsic lack of pigment function (Davies et al. 2010). Interestingly, pigments that are most similar to the new opsins (e.g., neuropsin [Yamashita et al. 2010] and peropsin [Nagata et al. 2010]) are known to be bistable, binding to 11-cis and all-trans retinal isomers, accompanied by a spectral switch between UV-sensitive and medium wavelength-sensitive states. Thus, it is likely that the ancestral novel opsin was biochemically similar. However, a series of gene duplications has resulted in a group of photosensitive proteins in which each subclass of pigment has maintained a distinct range of biochemical properties, collectively forming a functionally diverse group of photopigments (Fig. 7A).
Figure 7.
Functional analyses of representative zebrafish new opsins by UV-visible spectroscopy (to determine spectral sensitivity) and whole-cell electrophysiology (to assay phototransduction cascade activation). (A) Diverse absorption spectra of regenerated zebrafish Opn6a (i,iv), Opn7d (ii,v), and Opn8c (iii,vi) photopigments with 11-cis (i–iii) and all-trans (iv–vi) retinal chromophores. For all photopigments, dark (closed circles; i–iii [11-cis retinal] and iv–vi [all-trans retinal]) and light-bleached or acid-treated (open circles) spectra are shown. (Inset) Difference spectra were fitted with a Govardovskii A1-template to determine the λmax value, with x- and y-axes showing the wavelength (λ [nm]) and absorbance difference (Ab. diff.), respectively. (B) Quantification of novel opsin-dependent, light-evoked currents (mean ± SEM) by whole-cell patch-clamp electrophysiology. Recordings were measured from Neuro-2a cells expressing representative zebrafish new photopigments (Opn6a, Opn6b, Opn7a, Opn7b, Opn7d, Opn8a, and Opn8c) preincubated with 9-cis retinal (n = 5–7 cells; black) or all-trans retinal (n = 5–7 cells; dark gray) and exposed to a 10-sec light pulse (420 nm; 8 × 1014 photons/cm2/s1). Data show that these new pigments encode functional photopigments in vitro that interact differently with multiple chromophores, with some pigments being monostable (Opn6b and Opn7d) and others being bistable (Opn6a, Opn7a, Opn7b, Opn8a, and Opn8c). Compared with controls (cells with no retinal; n = 5–7 cells; light gray), all light-evoked currents were significantly greater (P < 0.05, Student's t-test), except for Opn6b preincubated with all-trans retinal and Opn7d preincubated with 9-cis retinal (shown by an asterisk).
Electrophysiological characterization of zebrafish novel opsins
Investigation of the amino acid sequences for each novel opsin demonstrates the presence of all of the critical residues required for the correct folding of the protein structure and subsequent function as a photopigment, including the conserved chromophore binding residue at Lys296 (Supplemental Table S1; Davies et al. 2011). However, unlike many conventional visual pigments, spectrophotometric studies with nonvisual pigments often fail to generate pigments that can be biochemically characterized (Davies et al. 2010). To overcome this limitation and test whether the broader family of new opsins is able to form functional pigments in the presence of light and a retinal chromophore, a sensitive heterologous cell expression system was used, as previously described for melanopsin and a number of nonvisual photopigments (Melyan et al. 2005; Davies et al. 2011). In this system, whole-cell patch clamping measures light-evoked currents, resulting from opsins expressed in a neuroblastoma cell line (Neuro-2a), by recording in the presence of 9-cis, all-trans, or no retinal. The Neuro-2a cell expression system is devoid of endogenous retinal processing (turnover) and allows for the chromophore dependence of each pigment to be determined directly. Expression of the novel opsins resulted in a chromophore-dependent activation of the native calcium signaling pathway induced through Gq/G11 in Neuro-2a cells (Fig. 7B). In this manner, light-evoked functional currents were successfully recorded for the majority of the representative new pigments tested, including Opn6a, Opn7b, Opn7d, Opn8a, and Opn8c (Fig. 7B). Some of these pigments behave more like classical photopigments, primarily working with only cis forms of the chromophore (Opn6b), whereas others (Opn6a, Opn7a, Opn7b, Opn8a, and Opn8c) appear to utilize both cis and trans isomers. Where both chromophore types were stably bound, the light-evoked currents were three- to fivefold larger in the presence of 9-cis retinal compared with those activated by all-trans retinal, a common result for electrophysiological work with bistable pigments (Tsukamoto et al. 2005; Davies et al. 2011, 2012b), where the techniques used are able to determine whether a particular chromophore type is utilized, but not the binding affinity of the interaction. In the case of Opn7d, the pigment induced a significant light-responsive current only in the presence of all-trans-retinal. This pigment, therefore, is monostable, but is uniquely different from classical visual pigments that only stably interact with 11-cis retinal. For Opn7d, this profile of light-responsive chromophore biochemistry in the patch-clamp recordings is entirely consistent with the data obtained by UV-vis spectroscopy, where a functional photopigment formed in the presence of all-trans retinal only. Similarly, the binding of chromophore and activation of retinal-dependent, light-evoked currents between these two functional approaches were identical for Opn6a and Opn8c when cis retinal isomers were used (Fig. 7). For all-trans experiments, however, both Opn6a and Opn8c activated retinal-dependent, light-evoked currents, but failed to generate pigments by the less sensitive technique of UV-vis spectrophotometry. Of the novel opsin classes as a whole, Opn9 failed to elicit retinal-dependent light responses with both assay systems. It remains, therefore, to be resolved whether Opn9 can form a native functional photopigment or has some other orphan function.
Discussion
Exposure to light has a major impact on the behavior, physiology, and pathophysiology of most vertebrates, including mammals (Munch and Bromundt 2012; Golombek et al. 2013). The mechanisms that mediate the detection of nonvisual photic cues, however, remain largely unexplored. The zebrafish (D. rerio) represents an ideal model system in which to explore the process of nonvisual photoreception, not least because most, if not all, tissues and cells in these animals appear to be directly light sensitive (Whitmore et al. 2000; Kaneko et al. 2006). In this study, it has been shown that endogenous peripheral circadian pacemakers, contained within a number of varied zebrafish tissues, can be re-entrained by shifted light-dark cycles, even when these tissues are explanted and maintained in culture. The cells in these tissues, therefore, must contain the necessary photopigments and signaling pathways required to perform this functional role. The question arises as to which photopigments are present in these tissues to mediate their direct photosensitivity.
Functional genomic analyses of the zebrafish genome have revealed many more candidate photopigments than initially predicted. In fact, 42 distinct opsin genes were identified, comprising 10 known visual (Morrow et al. 2011; Davies et al. 2012a) and 32 nonvisual opsins, which includes four new photopigment gene classes (opn6, opn7, opn8, and opn9) that have duplicated to yield 10 distinct novel opsin genes with a paired set that arose by a gene duplication event specific to the zebrafish branch. They are most similar in sequence to neuropsin (encoded by opn5), which has been shown to be a bistable UV-responsive photopigment (Yamashita et al. 2010) and is expressed in the mammalian retina, brain, spinal cord, and even ears (Kojima et al. 2011). Of these new classes of photopigment genes, only opn9 appears restricted to the teleosts, with opn6, opn7, and opn8 present in bony fishes, amphibians, reptiles, and birds. In the mammalian lineage, opn6 is present in monotremes, but is absent in representative genomes of both marsupials and eutherian mammals. The broad phylogenetic range of these opsins underpins the idea that nonvisual photoreception is a far more common and biologically significant process than was perhaps previously appreciated. Studies that focus on the visual system have suggested that the loss of visual pigments in mammals compared with nonmammalian species reflects their migration from day to night with an accompanying change in sleep/wake behavior from one that is diurnal to one that is mesopic (Davies et al. 2012a; Gerkema et al. 2013) or nocturnal (Menaker and Tosini 1996; Gerkema et al. 2013). A similar effect due to light restriction during the so-called “mesopic-bottleneck” (Davies et al. 2012a) or “nocturnal-bottleneck” (Menaker and Tosini 1996; Gerkema et al. 2013) may be the underlying cause of the nonvisual opsin loss observed in mammals. The loss of nonvisual pigment genes, however, appears to occur more gradually than that of the visual opsin genes and may reflect a prolonged period of light sensitivity diminution. Thus, it follows that early mammals may have progressively adapted to twilight first, then subsequently to the nocturnal realm, instead of a sudden evolutionary switch from diurnality to nocturnality.
To explore the complement of opsins transcribed in the zebrafish, NanoString nCounter and R-ISH approaches quantified the expression levels of all opsins (visual and nonvisual) in a wide range of zebrafish tissues and located novel opsin transcripts in both ocular and cranial tissues. All of the major tissues and organs of the body contain multiple identified opsin mRNAs, although this study did not assess the relative pigment protein complement and levels. Perhaps not surprisingly, the retina contains the greatest diversity of pigments, with not only all of the visual opsins, but also the majority of nonvisual opsins, including the newly identified novel opsins. For example, opn9 shows distinct and very high transcript levels within the horizontal cells of the retina, whereas opn8a shows clear expression within a subset of bipolar cells. Several of the other new opsins show distinct expression within the RGCs (opn6b, opn7d, and opn8b). The complexity of expression was far greater than expected; this strongly suggests that these novel opsins play very specific roles within the organization, physiology, and cell biology of the retina.
Recently, a study demonstrated that zebrafish larvae lacking both eyes and a pineal gland are still able to respond to light stimuli, most likely via the Opn4m1 photopigment, supporting the presence of functionally relevant deep brain photoreceptors (Fernandes et al. 2012). Consistently, the adult zebrafish brain demonstrates a similar level of opsin complexity, as well as in the retina. Indeed, out of the 10 novel opsins, the brain highly expresses all subclass members compared with internal controls, except opn9, where the expression level is very low, but still evident. Specifically, certain neural areas, such as the PGZ, TL, VAL, VAM, CM, and EG, remarkably coexpress all 10 new opsins, with the hypothalamus being more conservative in the number of novel opsins expressed. The zebrafish brain is directly light responsive (Whitmore et al. 2000), a fact shown by the acute induction of clock genes cry1a and per2 in culture (Moore and Whitmore 2014). Most, but not all, areas of the adult brain do demonstrate the acute induction of these genes in response to light pulses (Moore and Whitmore 2014), and this correlates with the expression pattern of numerous opsins. Indeed, there is great overlap in the expression and location between genes that are photoinduced and those that are fundamental to the clock machinery, as well as the photosensitive opsin molecules themselves. The cell-type–specific nature of opsin expression within the brain certainly suggests that light directly regulates refined neural and behavioral responses, at least in teleosts. Interestingly, higher vertebrates possess deep brain photoreceptors that express a number of photopigments. For example, VA and OPN5 photopigments expressed in the avian hypothalamus are likely to play a key role in orchestrating the timing of reproduction in birds by directly monitoring light levels as the day length alters from one season to the next (Halford et al. 2009; Nakane et al. 2010; Davies et al. 2012c). Collectively, therefore, many vertebrates may express an array of cranial photopigments with functional roles in directly regulating endogenous pacemakers that control photoentrainment (Whitmore et al. 1998, 2000) and sleep (Zhdanova 2006; Rihel et al. 2010), swimming, photokinesis and phototaxis (via deep brain photoreceptors) (Fernandes et al. 2012), and the timing of reproduction (via the axis that connects the hypothalamus, pituitary gland, and the gonads) (Marino and Jensen 2008).
Other tissues, such as fin, gill, heart, muscle, pineal, pituitary, skin, and testis, also contain a surprisingly large number of photopigments, an observation that supports studies demonstrating that they are directly light-responsive structures (Whitmore et al. 2000; Kaneko et al. 2006). In particular, the zebrafish heart contains the full complement of new opsins apart from opn6b. Zebrafish skin expresses opn5 (neuropsin), consistent with the identification of this photopigment in the ears of mice and humans (Kojima et al. 2011), and several novel opsins (indeed all except opn6b and opn7b) that may have a functional role in regulating body pigmentation (Shiraki et al. 2010). Like neuropsin, many of these new photopigments show responsiveness at wavelengths <380 nm (e.g., Opn7d and Opn8c), which might be expected for a tissue that is exposed to direct sunlight and, thus, dangerous UV rays. Indeed, the spectral sensitivities of the new opsins, with a range of monophasic and biphasic characteristics and differential binding to 11-cis retinal and all-trans retinal, suggest that they may have crucial roles as UV sensors in processes such as the activation of the DNA repair cascade (Hirayama et al. 2009), the cell cycle, and stress pathway responses (Tamai et al. 2012). In contrast, the two deepest internal organs, the gut and the liver, express the fewest number of opsins. The most highly expressed gene in both these tissue types is rgr2, followed by a few novel opsins (e.g., opn7a and opn8a to a lesser degree). Since the circadian clock in both of these tissues is directly photoentrainable (Peyric et al. 2013), the restricted repertoire of opsins in the gut and liver suggests that these pigments alone are sufficient for the circadian light response and may regulate intestinal movements and digestion.
In summary, zebrafish tissues possess a remarkable number and diversity of opsin photopigments, including four new classes of opsins and 10 subclass members. This is not simply a teleost-specific phenomenon, as many vertebrate genomes also contain these novel opsins, including both lower and higher gnathostomes. These novel opsins can form functional photopigments with unique chromophore handling properties and spectral sensitivities. Furthermore, they show broad expression profiles across most of the tissues analyzed in this model vertebrate. The diversity of opsin photopigments almost certainly has its evolutionary origins in the whole-genome duplication events, although their conservation in the zebrafish genome indicates that they must have evolved under significant positive selective pressures. Retention of pigment multiplicity probably confers a number of advantages for survival of this freshwater teleost in a dynamic photic environment. Interestingly, the broader vertebrate lineage retains many of the 42 pigments identified in this study, suggesting the future discovery of a complex array of novel photoreceptive processes. The unique discoveries presented here form the foundation for a greater exploration of direct tissue photoreception, which is an important step, as a complete appreciation of the breadth and significance of nonvisual light responsiveness is currently limited.
Methods
Fish care
Adult wild-type zebrafish (AB, AB/TL, and the per3-luciferase transgenic line) (Kaneko and Cahill 2005; Kaneko et al. 2006) were raised according to standard protocols used by the University College London (UCL) Fish Facility at 28.5°C on a 14 h light:10 h dark (14L:10D) cycle. All procedures adhered to the United Kingdom Animals (Scientific Procedures) Act 1986 and local Animal Ethics Committee guidelines (UCL).
Bioluminescence assay
Adult per3-luciferase transgenic zebrafish (Kaneko and Cahill 2005; Kaneko et al. 2006) were dissected to investigate direct photoreception in multiple tissues. Specifically, small tissue pieces of brain, fin, gill, gut, liver, muscle, skin, and testis were placed into 96-well dishes, while eye, heart, and pineal and pituitary glands were placed in whole-organ culture and incubated in Leibovitz (L15) media, supplemented with 15% fetal calf serum, penicillin/streptomycin (50 U/mL), gentamicin (50 mg/mL), and 0.5 mM beetle luciferin (Promega). Subsequently, plates were exposed for up to 6 d to two different lighting regimes comprising (1) a forward light cycle consisting of 12 h of light, followed by 12 h of dark (LD), or (2) a reverse light cycle comprising 12 h of dark, followed by 12 h of light (DL), before being transferred into constant darkness (DD) for up to 7 d. A bioluminescence reporter assay measured gene activity during the entire 2 wk of the experiment (LD or DL into DD) on a Packard TopCount NXT scintillation counter at 28°C.
Functional genomics and gene discovery
Visual and nonvisual zebrafish opsins were used as bait to mine the zebrafish genome (Zv9 assembly) (http://www.ensembl.org/Danio_rerio/Info/Index) for opsin-like genes, prior to obtaining pigment gene orthologs from database repositories of other vertebrate species (for further information, see Supplemental Material).
Phylogenetic analysis
A codon-matched nucleotide sequence alignment of all 42 zebrafish opsin coding regions was generated, comparing these sequences to pigment gene sequences derived from a variety of gnathostome vertebrates ranging from teleosts to primates. A neighbor-joining phylogenetic tree (Saitou and Nei 1987) was generated by using the MEGA version 5 computer package (Tamura et al. 2011) and applying a maximum composite likelihood (MCL) model algorithm (for further information, see Supplemental Material) (Tamura and Nei 1993).
NanoString nCounter assay
Dissected tissues (outlined above) from adult AB and AB/TL zebrafish strains between ZT3 and ZT6 (where ZT0 is lights on) were isolated (n ≥ 3, where each sample [n] contained one or more whole or tissue pieces from multiple animals), homogenized in TRIzol Reagent (Life Technologies) and stored at −20°C. Following the manufacturer's instructions (Life Technologies), total RNA was extracted, and gene-specific probes were designed to the coding region of all 42 opsins and five clock components by the manufacturer (NanoString Technologies). In addition, probes to eukaryotic translation elongation factor 1 alpha 1, like-1 (eef1al1), ribosomal protein L13a (rpl13a), and ubiquitin B (ubb) acted as three internal reference genes. Scientific Support Services (UCL Genomics) performed the NanoString nCounter assay, using RNA samples diluted to the appropriate concentration. Opsin and clock gene expression levels (where raw data were initially normalized to internal positive and negative controls for each lane using a medium level of stringency) were normalized to the geometric mean (Vandesompele et al. 2002) of the reference genes for each tissue studied. The hierarchical cluster analyses were performed in MATLAB (Release 2012a), and the bar graphs were produced in Microsoft Excel 2010.
RNA in situ hybridization
R-ISH on adult zebrafish eye and brain sections (collected at ZT3) was performed as previously described (for further information, see Supplemental Material) (Davies et al. 2011). Slides were imaged with an automated SCN400 Slide Scanner (Leica) at the IQPath Facility (UCL Institute of Neurology).
UV-visible spectroscopy of recombinant novel opsin proteins
Each of the new opsin full-length coding sequences was cloned into the pMT4 mammalian expression vector and repeatedly regenerated with excess 11-cis retinal or all-trans retinal (Sigma) under dark conditions as previously described (for further information, see Supplemental Material) (Davies et al. 2011). A Shimadzu UV-visible spectrophotometer (UV-2550) (Shimadzu) recorded triplicate absorbance spectra in complete darkness, before applying light or acid treatment to each sample and subsequent rerecording. A difference spectrum was calculated by subtracting the treated spectral trace from that measured in the dark, which was then fitted to a modified Govardovskii rhodopsin A1-template (Govardovskii et al. 2000) using Microsoft Excel to determine the λmax as previously shown (Davies et al. 2007, 2009, 2011). For the biphasic difference spectrum of the Opn7d pigment regenerated with all-trans retinal, the λmax value for each peak was determined separately and followed the same analytical procedure as for all monophasic pigments. The two partial traces were then merged together to generate the overall biphasic spectrum.
Expression of novel opsins for electrophysiology
Constructs comprising new opsin sequences cloned in pMT4 were transiently transfected into Neuro-2a cells with GeneJuice reagent (Novagen), according to the manufacturer's instructions (for further information, see Supplemental Material). For each transfection, a control pSIREN-DNR-DsRed-Express vector (Clontech) was cotransfected with each novel opsin plasmid to allow for identification of cells expressing a particular pigment gene prior to electrophysiological analysis. The addition of retinoic acid (20 μM; Tocris) induced cell differentiation 24 h after transfection, prior to conducting electrophysiology experiments after a further 48–60 h in complete darkness.
Whole-cell electrophysiology
Transfected cells were maintained and whole-cell patch-clamp recordings were measured as previously described (for further information, see Supplemental Material) (Melyan et al. 2005; Davies et al. 2011). Briefly, Neuro-2a cells were voltage-clamped at holding potentials of −50 mV, and currents were recorded using Axopatch 200B (Axon Instruments), where recordings were subsequently filtered (1 kHz) and sampled (20 kHz). A slit monochromator, using a Cairn Optoscan Xenon Arc lamp source, produced specific wavelengths of light, where all stimuli lasted 10 sec with a half-bandwidth of 20 nm. Irradiance levels were determined using an optical power meter (Macam Photometrics) and transformed into photon flux values. Clampfit (Axon Instruments) determined each response magnitude as defined by measuring the peak sustained. Statistical assessments comprised multiple, independent Student's t-tests (two-tailed distribution with two-sample unequal variance [heteroscedastic] parameters) between light-evoked currents generated in the presence of either 9-cis retinal or all-trans retinal compared with no retinal controls (for further information, see Supplemental Material).
Data access
All sequence data from this study have been submitted to the NCBI GenBank repository (GenBank; http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KT008391–KT008432 (for further details, see Supplemental Fig. S1).
Supplementary Material
Acknowledgments
We thank visiting scholar Dr. Leonardo Lima (University of Sao Paolo, Brazil) for helping to collect tissue samples, Dr. Simone Sharma (University College London [UCL] Genomics, Scientific Support Services, UK) for help with the NanoString assay, Angela Richard-Londt for imaging the slides (UCL Institute of Neurology, IQPath Facility, UK), and Dr. Helen A. Moore for providing adult zebrafish brain sections. We thank Dr. Rosalie Crouch (Medical University of South Carolina, USA) and Prof. David Hunt (University of Western Australia, Australia) for the generous provision of 11-cis retinal and 1D4 antibody, respectively. We also thank Prof. David Hunt for supporting the salary of W.I.L.D. during the initial bioinformatics study. A grant awarded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) to M.W.H., and a Future Fellowship (FT110100176) and Discovery Project grant (DP140102117) awarded to W.I.L.D. by the Australian Research Council (ARC) supported this work.
Author contributions: W.I.L.D., T.K.T., R.G.F., D.W., and M.W.H. contributed to the conception and design of the study and wrote the paper. W.I.L.D. performed the bioinformatics, RNA in situ hybridization experiments, and spectral studies. T.K.T. performed the bioluminescence assays and gene expression studies. L.Z. and J.K.F. performed the electrophysiology. J.R. performed the cluster analysis and generated the heat map. J.K.F. helped with the RNA in situ hybridization experiments and electrophysiology. W.I.L.D., T.K.T, L.Z., J.K.F., J.R., and M.W.H. analyzed the data.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.189886.115.
References
- Albrecht U. 2004. The mammalian circadian clock: a network of gene expression. Front Biosci 9: 48–55. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Tarttelin EE, Foster RG, Wells DJ. 2003a. Structure and evolution of the teleost extraretinal rod-like opsin (errlo) and ocular rod opsin (rho) genes: Is teleost rho a retrogene? J Exp Zool B Mol Dev Evol 297: 1–10. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Wells DJ, Foster RG. 2003b. In silico characterisation and chromosomal localisation of human RRH (peropsin)—implications for opsin evolution. BMC Genomics 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham J, Chaurasia SS, Melyan Z, Liu C, Cameron MA, Tarttelin EE, Iuvone PM, Hankins MW, Tosini G, Lucas RJ. 2006. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol 4: e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Snyder SH. 1997. Parapinopsin, a novel catfish opsin localized to the parapineal organ, defines a new gene family. J Neurosci 17: 8083–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Azzi A. 2013. Peripheral circadian oscillators in mammals. Handb Exp Pharmacol 45–66. [DOI] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. 2002. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583. [DOI] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallari N, Frigato E, Vallone D, Frohlich N, Lopez-Olmeda JF, Foa A, Berti R, Sanchez-Vazquez FJ, Bertolucci C, Foulkes NS. 2011. A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol 9: e1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E. 2013. Circadian clocks, food intake, and metabolism. Prog Mol Biol Transl Sci 119: 105–135. [DOI] [PubMed] [Google Scholar]
- Chen SK, Badea TC, Hattar S. 2011. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476: 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen A, Hamaoka T, Yamada Y, Kawamura S. 2003. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WL, Cowing JA, Carvalho LS, Potter IC, Trezise AE, Hunt DM, Collin SP. 2007. Functional characterization, tuning, and regulation of visual pigment gene expression in an anadromous lamprey. FASEB J 21: 2713–2724. [DOI] [PubMed] [Google Scholar]
- Davies WL, Carvalho LS, Tay BH, Brenner S, Hunt DM, Venkatesh B. 2009. Into the blue: Gene duplication and loss underlie color vision adaptations in a deep-sea chimaera, the elephant shark Callorhinchus milii. Genome Res 19: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WL, Hankins MW, Foster RG. 2010. Vertebrate ancient opsin and melanopsin: divergent irradiance detectors. Photochem Photobiol Sci 9: 1444–1457. [DOI] [PubMed] [Google Scholar]
- Davies WI, Zheng L, Hughes S, Tamai TK, Turton M, Halford S, Foster RG, Whitmore D, Hankins MW. 2011. Functional diversity of melanopsins and their global expression in the teleost retina. Cell Mol Life Sci 68: 4115–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WI, Collin SP, Hunt DM. 2012a. Molecular ecology and adaptation of visual photopigments in craniates. Mol Ecol 21: 3121–3158. [DOI] [PubMed] [Google Scholar]
- Davies WI, Tay BH, Zheng L, Danks JA, Brenner S, Foster RG, Collin SP, Hankins MW, Venkatesh B, Hunt DM. 2012b. Evolution and functional characterisation of melanopsins in a deep-sea chimaera (elephant shark, Callorhinchus milii). PLoS One 7: e51276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WI, Turton M, Peirson SN, Follett BK, Halford S, Garcia-Fernandez JM, Sharp PJ, Hankins MW, Foster RG. 2012c. Vertebrate ancient opsin photopigment spectra and the avian photoperiodic response. Biol Lett 8: 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekens MP, Santoriello C, Vallone D, Grassi G, Whitmore D, Foulkes NS. 2003. Light regulates the cell cycle in zebrafish. Curr Biol 13: 2051–2057. [DOI] [PubMed] [Google Scholar]
- Fernandes AM, Fero K, Arrenberg AB, Bergeron SA, Driever W, Burgess HA. 2012. Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr Biol 22: 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RM, Fontinha BM, Kirchmaier S, Steger J, Bloch S, Inoue D, Panda S, Rumpel S, Tessmar-Raible K. 2013. Co-expression of VAL- and TMT-opsins uncovers ancient photosensory interneurons and motorneurons in the vertebrate brain. PLoS Biol 11: e1001585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follett BK, Foster RG, Nicholls TJ. 1985. Photoperiodism in birds. Ciba Found Symp 117: 93–105. [DOI] [PubMed] [Google Scholar]
- Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, Foster R. 1999. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science 284: 502–504. [DOI] [PubMed] [Google Scholar]
- Gavriouchkina D, Fischer S, Ivacevic T, Stolte J, Benes V, Dekens MP. 2010. Thyrotroph embryonic factor regulates light-induced transcription of repair genes in zebrafish embryonic cells. PLoS One 5: e12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Gerkema MP, Davies WI, Foster RG, Menaker M, Hut RA. 2013. The nocturnal bottleneck and the evolution of activity patterns in mammals. Proc Biol Sci 280: 20130508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek DA, Casiraghi LP, Agostino PV, Paladino N, Duhart JM, Plano SA, Chiesa JJ. 2013. The times they're a-changing: effects of circadian desynchronization on physiology and disease. J Physiol Paris 107: 310–322. [DOI] [PubMed] [Google Scholar]
- Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. 2000. In search of the visual pigment template. Vis Neurosci 17: 509–528. [DOI] [PubMed] [Google Scholar]
- Halford S, Pires SS, Turton M, Zheng L, Gonzalez-Menendez I, Davies WL, Peirson SN, Garcia-Fernandez JM, Hankins MW, Foster RG. 2009. VA opsin-based photoreceptors in the hypothalamus of birds. Curr Biol 19: 1396–1402. [DOI] [PubMed] [Google Scholar]
- Hankins MW, Peirson SN, Foster RG. 2008. Melanopsin: an exciting photopigment. Trends Neurosci 31: 27–36. [DOI] [PubMed] [Google Scholar]
- Hao W, Fong HK. 1999. The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J Biol Chem 274: 6085–6090. [DOI] [PubMed] [Google Scholar]
- Hatori M, Panda S. 2010. The emerging roles of melanopsin in behavioral adaptation to light. Trends Mol Med 16: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Miyamura N, Uchida Y, Asaoka Y, Honda R, Sawanobori K, Todo T, Yamamoto T, Sassone-Corsi P, Nishina H. 2009. Common light signaling pathways controlling DNA repair and circadian clock entrainment in zebrafish. Cell Cycle 8: 2794–2801. [DOI] [PubMed] [Google Scholar]
- Jiang M, Pandey S, Fong HK. 1993. An opsin homologue in the retina and pigment epithelium. Invest Ophthalmol Vis Sci 34: 3669–3678. [PubMed] [Google Scholar]
- Kaneko M, Cahill GM. 2005. Light-dependent development of circadian gene expression in transgenic zebrafish. PLoS Biol 3: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Hernandez-Borsetti N, Cahill GM. 2006. Diversity of zebrafish peripheral oscillators revealed by luciferase reporting. Proc Natl Acad Sci 103: 14614–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano-Yamashita E, Terakita A, Koyanagi M, Shichida Y, Oishi T, Tamotsu S. 2007. Immunohistochemical characterization of a parapinopsin-containing photoreceptor cell involved in the ultraviolet/green discrimination in the pineal organ of the river lamprey Lethenteron japonicum. J Exp Biol 210: 3821–3829. [DOI] [PubMed] [Google Scholar]
- Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y. 2011. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS One 6: e26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokel D, Dunn TW, Ahrens MB, Alshut R, Cheung CY, Saint-Amant L, Bruni G, Mateus R, van Ham TJ, Shiraki T, et al. 2013. Identification of nonvisual photomotor response cells in the vertebrate hindbrain. J Neurosci 33: 3834–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Kawano E, Kinugawa Y, Oishi T, Shichida Y, Tamotsu S, Terakita A. 2004. Bistable UV pigment in the lamprey pineal. Proc Natl Acad Sci 101: 6687–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, Foulkes NS. 2005. Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol 3: e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laranjeiro R, Tamai TK, Peyric E, Krusche P, Ott S, Whitmore D. 2013. Cyclin-dependent kinase inhibitor p20 controls circadian cell-cycle timing. Proc Natl Acad Sci 110: 6835–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas RJ, Hattar S, Takao M, Berson DM, Foster RG, Yau KW. 2003. Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science 299: 245–247. [DOI] [PubMed] [Google Scholar]
- Lucas RJ, Lall GS, Allen AE, Brown TM. 2012. How rod, cone, and melanopsin photoreceptors come together to enlighten the mammalian circadian clock. Prog Brain Res 199: 1–18. [DOI] [PubMed] [Google Scholar]
- Marino CL, Jensen BH. 2008. Zebrafish Danio rerio, ovaries contract just prior to lights on. J Behav Neurosci Res 6: 10–14. [Google Scholar]
- Martin-Robles AJ, Aliaga-Guerrero M, Whitmore D, Pendon C, Munoz-Cueto JA. 2012. The circadian clock machinery during early development of Senegalese sole (Solea senegalensis): effects of constant light and dark conditions. Chronobiol Int 29: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Melyan Z, Tarttelin EE, Bellingham J, Lucas RJ, Hankins MW. 2005. Addition of human melanopsin renders mammalian cells photoresponsive. Nature 433: 741–745. [DOI] [PubMed] [Google Scholar]
- Menaker M, Tosini G. 1996. The evolution of vertebrate circadian systems. Hokkaido University Press, Sapporo, Japan. [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. 2012. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY. 2013. The suprachiasmatic nucleus and the circadian timing system. Prog Mol Biol Transl Sci 119: 1–28. [DOI] [PubMed] [Google Scholar]
- Moore HA, Whitmore D. 2014. Circadian rhythmicity and light sensitivity of the zebrafish brain. PLoS One 9: e86176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JM, Chang BS. 2010. The p1D4-hrGFP II expression vector: a tool for expressing and purifying visual pigments and other G protein-coupled receptors. Plasmid 64: 162–169. [DOI] [PubMed] [Google Scholar]
- Morrow JM, Lazic S, Chang BS. 2011. A novel rhodopsin-like gene expressed in zebrafish retina. Vis Neurosci 28: 325–335. [DOI] [PubMed] [Google Scholar]
- Munch M, Bromundt V. 2012. Light and chronobiology: implications for health and disease. Dialogues Clin Neurosci 14: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Koyanagi M, Tsukamoto H, Terakita A. 2010. Identification and characterization of a protostome homologue of peropsin from a jumping spider. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 196: 51–59. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Kojima D, Imai H, Terakita A, Okano T, Shichida Y, Fukada Y. 1999. Chimeric nature of pinopsin between rod and cone visual pigments. Biochemistry 38: 14738–14745. [DOI] [PubMed] [Google Scholar]
- Nakane Y, Ikegami K, Ono H, Yamamoto N, Yoshida S, Hirunagi K, Ebihara S, Kubo Y, Yoshimura T. 2010. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc Natl Acad Sci 107: 15264–15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. 2003. Circadian light input in plants, flies and mammals. Novartis Found Symp 253: 73–82; discussion 82–78, 102–109, 281–104. [PubMed] [Google Scholar]
- Peyric E, Moore HA, Whitmore D. 2013. Circadian clock regulation of the cell cycle in the zebrafish intestine. PLoS One 8: e73209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihel J, Prober DA, Schier AF. 2010. Monitoring sleep and arousal in zebrafish. Methods Cell Biol 100: 281–294. [DOI] [PubMed] [Google Scholar]
- Rollag MD, Berson DM, Provencio I. 2003. Melanopsin, ganglion-cell photoreceptors, and mammalian photoentrainment. J Biol Rhythms 18: 227–234. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sato K, Yamashita T, Ohuchi H, Shichida Y. 2011. Vertebrate ancient-long opsin has molecular properties intermediate between those of vertebrate and invertebrate visual pigments. Biochemistry 50: 10484–10490. [DOI] [PubMed] [Google Scholar]
- Schibler U, Ripperger J, Brown SA. 2003. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms 18: 250–260. [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kojima D, Fukada Y. 2010. Light-induced body color change in developing zebrafish. Photochem Photobiol Sci 9: 1498–1504. [DOI] [PubMed] [Google Scholar]
- Su CY, Luo DG, Terakita A, Shichida Y, Liao HW, Kazmi MA, Sakmar TP, Yau KW. 2006. Parietal-eye phototransduction components and their potential evolutionary implications. Science 311: 1617–1621. [DOI] [PubMed] [Google Scholar]
- Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. 1997. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci 94: 9893–9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai TK, Vardhanabhuti V, Foulkes NS, Whitmore D. 2004. Early embryonic light detection improves survival. Curr Biol 14: R104–105. [PubMed] [Google Scholar]
- Tamai TK, Carr AJ, Whitmore D. 2005. Zebrafish circadian clocks: cells that see light. Biochem Soc Trans 33: 962–966. [DOI] [PubMed] [Google Scholar]
- Tamai TK, Young LC, Cox CA, Whitmore D. 2012. Light acts on the zebrafish circadian clock to suppress rhythmic mitosis and cell proliferation. J Biol Rhythms 27: 226–236. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ. 2003. Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett 554: 410–416. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Terakita A, Shichida Y. 2005. A rhodopsin exhibiting binding ability to agonist all-trans-retinal. Proc Natl Acad Sci 102: 6303–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde E, Haque R, Iuvone PM, Azpeleta C, Alonso-Gomez AL, Delgado MJ. 2009. Circadian clock genes of goldfish, Carassius auratus: cDNA cloning and rhythmic expression of Period and Cryptochrome transcripts in retina, liver, and gut. J Biol Rhythms 24: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, Dolle D, Lahiri K, Vallone D, Ettwiller L, Geisler R, et al. 2011. The light responsive transcriptome of the zebrafish: function and regulation. PLoS One 6: e17080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. 1998. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci 1: 701–707. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Sassone-Corsi P. 2000. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404: 87–91. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y. 2010. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc Natl Acad Sci 107: 22084–22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova IV. 2006. Sleep in zebrafish. Zebrafish 3: 215–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.