Abstract
The transition from a specified germ cell to a population of pluripotent cells occurs rapidly following fertilization. During this developmental transition, the zygotic genome is largely transcriptionally quiescent and undergoes significant chromatin remodeling. In Drosophila, the DNA-binding protein Zelda (also known as Vielfaltig) is required for this transition and for transcriptional activation of the zygotic genome. Open chromatin is associated with Zelda-bound loci, as well as more generally with regions of active transcription. Nonetheless, the extent to which Zelda influences chromatin accessibility across the genome is largely unknown. Here we used formaldehyde-assisted isolation of regulatory elements to determine the role of Zelda in regulating regions of open chromatin in the early embryo. We demonstrate that Zelda is essential for hundreds of regions of open chromatin. This Zelda-mediated chromatin accessibility facilitates transcription-factor recruitment and early gene expression. Thus, Zelda possesses some key characteristics of a pioneer factor. Unexpectedly, chromatin at a large subset of Zelda-bound regions remains open even in the absence of Zelda. The GAGA factor-binding motif and embryonic GAGA factor binding are specifically enriched in these regions. We propose that both Zelda and GAGA factor function to specify sites of open chromatin and together facilitate the remodeling of the early embryonic genome.
Transcription factors drive cell-fate specification by binding to cis-regulatory regions and controlling gene expression. Despite the broad impact of these factors, our understanding of how they access and function at the correct sites in the genome remains incomplete. In particular, while transcription factors recognize and bind specific DNA motifs, only a small fraction of their potential binding sites are occupied at a given time point (Carr and Biggin 1999; Iyer et al. 2001; Lieb et al. 2001; Liu et al. 2006; Yang et al. 2006; Li et al. 2008). Thus, other features in addition to DNA sequence must influence where transcription factors bind in the genome. In vivo, DNA is wrapped around histone proteins to form nucleosomes, which compete with transcription factors for access to DNA-encoded information. Reduced accessibility of nucleosomal DNA can largely explain the patterns of transcription factor binding in vivo (Kaplan et al. 2011; Li et al. 2011). However, the mechanisms that create sites of accessibility are poorly understood. It has been proposed that a special class of transcription factors, termed pioneer factors, binds nucleosomal DNA and increases chromatin accessibility for other transcription factors (Zaret and Carroll 2011). This model is based on the activity of the Forkhead box (FOX) family of proteins that bind nucleosomal DNA early in hepatic development and facilitate open chromatin, making these regions accessible for subsequent transcription factor binding (Gualdi et al. 1996; Cirillo et al. 1998, 2002; Cirillo and Zaret 1999).
Although it is widely accepted that chromatin structure is reconfigured during early embryonic development, little is known about the factors that direct this process. These changes in chromatin structure accompany the transition from a specified germ cell to a population of pluripotent cells, which occurs rapidly following fertilization. At this time the zygotic genome is not transcribed, and maternally contributed mRNAs and proteins control embryonic development. Only at later cell cycles is widespread transcription initiated (Newport and Kirschner 1982; Tadros and Lipshitz 2009). The degradation of maternally deposited mRNAs is coordinated with the transcriptional activation of the zygotic genome during the maternal-to-zygotic transition (MZT). This fundamental transition is conserved among metazoans and is essential for development.
In Drosophila melanogaster, early development is characterized by a series of 13 rapid synchronous nuclear divisions. Approximately 2–3 h after fertilization, at the 14th nuclear cycle, the division cycle slows and widespread zygotic transcription initiates. This marks the end of the D. melanogaster MZT. Maternally deposited zelda (zld; also known as vielfaltig) mRNA is necessary for zygotic genome activation and development beyond the MZT (Staudt et al. 2006; Liang et al. 2008). Previous work has demonstrated that loci bound by Zelda (Zld) as early as nuclear cycle 8 are associated with regions of open chromatin and transcription factor binding at cycle 14 (Li et al. 2008; MacArthur et al. 2009; Harrison et al. 2011). Indeed, Zld binding at early time points is a robust predictor of subsequent transcription factor binding (Harrison et al. 2011), and it has recently been shown that Zld-binding sites are instrumental in regulating DNA binding by the transcription factors Dorsal (Dl), Twist (Twi), and Bicoid (Bcd) (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014). Furthermore, it has been proposed that Zld acts to potentiate transcription factor binding by determining regions of open chromatin (Harrison et al. 2011; Satija and Bradley 2012). At regions bound by Zld in wild-type embryos, histone H3 density increases when Zld is depleted (Li et al. 2014), supporting this hypothesis. Additionally, chromatin accessibility as assayed by DNase I accessibility is correlated with levels of Zld binding to the brinker (brk) and short gastrulation (sog) enhancers (Foo et al. 2014). Nonetheless, it is unknown whether Zld is required for open chromatin at all of the thousands of loci to which it binds. Furthermore, whether Zld-mediated chromatin accessibility facilitates transcription factor binding remains to be determined.
Here, we directly test the role of Zld in shaping the chromatin environment during the MZT. We used formaldehyde-assisted isolation of regulatory elements (FAIRE) (Giresi et al. 2007; McKay and Lieb 2013) to perform genome-wide profiling of open chromatin in embryos lacking maternal Zld (zldM-).
Results
Zld establishes or maintains regions of open chromatin
We have previously shown that Zld-bound regions are highly correlated with DNA accessibility (Harrison et al. 2011). To test the function of Zld in establishing or maintaining these regions of open chromatin, we performed FAIRE on 2–3 h embryos with wild-type levels of Zld (yw) and embryos depleted for maternally contributed Zld (zldM-) (Liang et al. 2008). Obtaining embryos depleted for maternal Zld requires generating germline clones using heat-shock–induced mitotic recombination (Chou et al. 1993; Liang et al. 2008). Thus to control for any influence of this protocol on chromatin accessibility, we generated germline clones in our yw control strain in parallel with our zld mutant strain. Immunostaining confirmed the successful generation of embryos lacking Zld (Supplemental Fig. S1). We sequenced and analyzed three biological replicates of the zldM- embryos and two replicates of the yw control embryos. Regions enriched by FAIRE in our 2–3 h yw control embryos correlated well with the regions enriched in 2–4 h wild-type (OreR) embryos (cf. yw control FAIRE to OreR FAIRE; Spearman's rho = 0.75) (Fig. 1A; Supplemental Fig. S2; McKay and Lieb 2013). Differences between the two samples are likely the result of the different developmental time points at which the samples were collected, as well as any effects from the induction of germline clones.
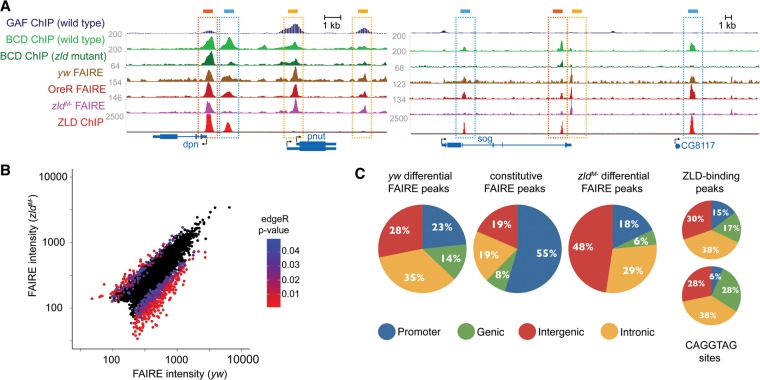
Figure 1.
Zld is required to maintain or establish discrete regions of open chromatin. (A) Normalized FAIRE-seq and ChIP-seq read profiles as labeled on the left for two genomic regions. FAIRE data for yw and zldM- embryos (this study). FAIRE data for 2–4 h OreR (wild-type) embryos from McKay and Lieb (2013). ChIP data for Zld from Harrison et al. (2011), for Bcd from Xu et al. (2014), and for GAF from Negre et al. (2011). Genes are shown at the bottom with arrows to indicate direction of transcription. Boxes indicate different classes of FAIRE peaks: blue box, differential, Zld-bound; dark orange box, constitutive, Zld-bound; and light orange box, constitutive, not Zld-bound. (B) Scatter plot of the FAIRE signal from yw embryos versus the FAIRE signal from zldM- embryos. Black dots indicate the union set of regions identified by FAIRE in both yw and zldM- embryos. Colored dots indicate differential peaks identified by edgeR. Colors represent P-values (<0.05) as indicated by the scale. (C) Pie charts showing the genomic distribution of FAIRE peaks, Zld-binding sites, and CAGGTAG motifs to promoters, genes, introns, and intergenic regions as shown.
Because active cis-regulatory regions are correlated with chromatin accessibility (Fisher et al. 2012), we focused our analysis on the high-confidence open regions. Specifically, we took the top 5000 peaks in our zldM- data set (MACS adjusted −log10 P-value = 39) and controlled for differences in data quality between the experimental and control by similarly selecting the top 5000 peaks from the yw control (MACS adjusted −log10 P-value = 18). To determine how chromatin accessibility changed when maternal Zld was removed, we took the union of the top 5000 peaks from both of our data sets (resulting in 6042 peak regions along the fly genome) and used edgeR to identify differentially accessible regions (Robinson et al. 2010). That is, FAIRE peaks that vary between the two samples (P < 0.05). We found 540 regions that had enriched FAIRE signals in our control yw embryos compared with zldM- embryos (yw differential) (Fig. 1A,B). These regions are exemplified in the cis-regulatory regions for deadpan (dpn) and sog by distinct FAIRE peaks that are present in both yw and OreR FAIRE but absent in the zldM- FAIRE (Fig. 1A; blue boxes). Conversely, there were 145 regions that were enriched in the zldM- embryos compared with the control (zldM- differential).
To determine whether Zld directly contributes to the changes in chromatin accessibility we observed between our yw control and zldM- embryos, we quantified the enrichment of the canonical Zld-binding site in our constitutive FAIRE peaks along with those regions that become more accessible (zldM- differential) and those that become less accessible (yw differential) in the absence of Zld. Specifically, we searched for occurrences of CAGGTAG—the optimal sequence recognition element for Zld—within those regions that were uniquely enriched in either yw or zldM- embryos. While only 12% of the top 5000 FAIRE peaks in our yw control contain the CAGGTAG motif, 45% of the Zld-dependent peaks (yw differential) contain CAGGTAG motifs (for full data, including the enrichment for CAGGTA, see Table 1). In contrast, of the 145 peaks that become more accessible in zldM- embryos only 0.7% contain CAGGTAG (zldM- differential) (Table 1). Thus, the Zld-binding site is selectively enriched in regions that depend on Zld for accessibility.
Table 1.
Enrichment of Zld-binding motifs and in vivo Zld-binding sites (with Zld) in FAIRE peaks
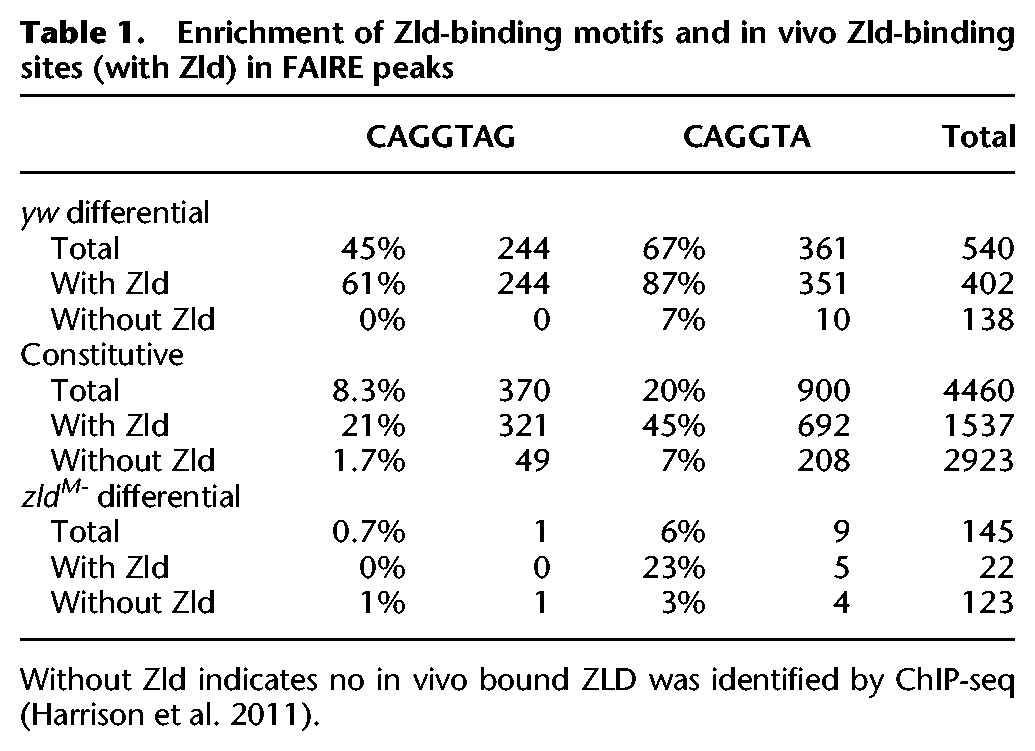
When we compared our control (yw) FAIRE peaks to regions bound by Zld, as determined by chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) (Harrison et al. 2011), we similarly identified binding enrichment specifically at those regions that depend on Zld for accessibility (yw differential). At cycle 14, 54% (2681/5000) of the control FAIRE peaks overlap with a region bound by Zld in vivo, while 77% (418/540) of the Zld-dependent peaks (yw differential) overlap with a Zld-bound region. This correlation between Zld binding and regions that require Zld for open chromatin is also evident at individual loci, such as dpn and sog (Fig. 1A, blue boxes). Furthermore, only 15% of those peaks that increase in accessibility in the absence of maternal Zld (zldM- differential) overlap with in vivo Zld-binding sites. Thus, both the Zld-binding sequence and in vivo Zld binding are enriched specifically in those regions that are more open in the control compared with zldM- embryos (yw differential). Together these data indicate that Zld is required to maintain or establish distinct regions of open chromatin.
Zld binds to promoters and enhancers of genes expressed through the MZT (Fig. 1C; Harrison et al. 2011; Nien et al. 2011). To assess which genomic regions might be particularly dependent on Zld for chromatin accessibility, we determined the distribution of FAIRE peaks that were specific to the yw control (yw differential), FAIRE peaks that were accessible in both yw and zldM- embryos (constitutive), and FAIRE peaks specific to the zldM- embryos (zldM- differential) (Fig. 1C). The observed distribution of FAIRE peaks that require Zld (yw differential) was similar to the distribution of regions bound by Zld in vivo and of CAGGTAG motifs (Fig. 1C), suggesting that Zld functions to establish or maintain chromatin accessibility at both promoters and enhancers. In contrast, the peaks specific to the zldM- (zldM- differential) showed a genomic distribution substantially different from Zld binding and CAGGTAG motifs, with a strong enrichment in intergenic regions.
Zld is not essential for chromatin accessibility at many Zld-bound loci
During the initial analysis of our FAIRE data, we identified three classes of FAIRE peaks that we had expected to identify. First, fitting with the predicted role of Zld as a pioneer-like factor, we identified Zld-bound loci that required Zld for chromatin accessibility. Second, we identified a large class of FAIRE peaks that did not bind Zld or require Zld for accessibility. Chromatin accessibility in these regions likely resulted from the activities of other proteins or from inherent DNA-sequence structure. Third, we identified a small number of regions that depend on Zld for accessibility but are not Zld bound, which are likely due to indirect effects of Zld depletion. However, our analysis unexpectedly revealed, in addition to these three classes, a large class of FAIRE peaks that overlapped Zld-bound regions but remained accessible in zldM- embryos. Thus, while Zld was required for chromatin accessibility at 540 loci, there were nearly three times as many loci (1537) that were bound by Zld but did not require Zld occupancy for accessibility.
For further analysis, we therefore divided the 5000 top FAIRE peaks from our yw embryos into four categories: differential, Zld-bound (402 peaks); constitutive, Zld-bound (1537 peaks); differential, not Zld-bound (138 peaks); and constitutive, not Zld-bound (2923 peaks) (Fig. 2A). Examples of these different classes are highlighted in the regulatory regions surrounding both dpn and sog (Fig. 1A). The average FAIRE signals from the differential and constitutive peaks were generally similar. However, the differential peaks showed a slightly lower level of FAIRE signal, especially those differential peaks that did not overlap Zld-bound regions (differential, not Zld-bound) (Fig. 2B). As defined, the FAIRE peaks for the differential class are substantially decreased in the zldM- embryos (Fig. 2B). Additionally, there was no detectable difference between the average FAIRE signals from constitutive regions either bound or not bound by Zld (Fig. 2B). Thus, the strength of the FAIRE signal is unlikely to explain the differences between these four classes.
Figure 2.
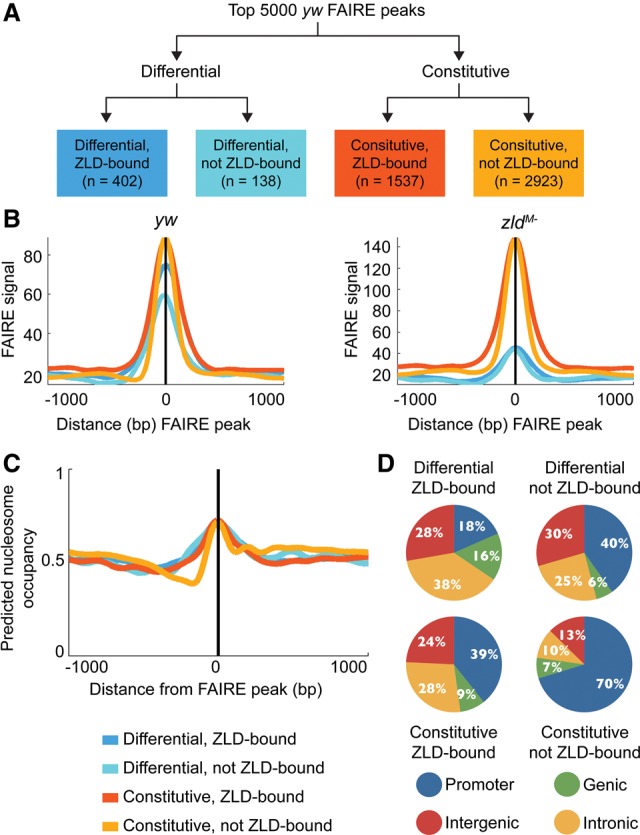
Many loci bound by Zld in wild-type embryos remain accessible in zldM- embryos. (A) Flow chart illustrating the four classes of yw FAIRE peaks used for future analysis. (B) Average FAIRE signal for each class of FAIRE peaks in both yw and zldM- embryos. (C) Predicted nucleosome occupancy based on DNA sequence surrounding FAIRE peaks in each of the indicated classes. Colors indicate the different FAIRE classes. Plots are centered on the FAIRE peak midpoint and oriented relative to the transcription start site of the nearest gene. (D) Pie charts showing the genomic distribution to promoters, genes, introns, and intergenic regions for each class of FAIRE peaks.
When we used sequence surrounding the FAIRE peaks to predict nucleosome occupancy (Kaplan et al. 2009), we did not identify a predicted region of nucleosome depletion at the FAIRE peaks in any of the four classes (Fig. 2C). On the contrary, we found that sequences underlying FAIRE peaks are predicted to have higher nucleosome occupancy, consistent with previous observations (McKay and Lieb 2013). This analysis suggests that the underlying sequence alone does not explain the open chromatin identified by FAIRE at any of these regions and that active mechanisms are likely working to create or maintain chromatin accessibility. To better define the features that determine chromatin accessibility in the early embryo, we focused our analysis on characterizing these classes of FAIRE peaks.
A large number of promoters do not require Zld for accessibility
Many factors are known to shape chromatin accessibility so we were not surprised to identify a relatively large class of FAIRE peaks that were unchanged in the absence of Zld and that did not possess Zld-binding sites (constitutive, not Zld-bound; n = 2923). When we analyzed the distribution of these regions throughout the genome, we noted they were highly enriched for promoter regions with ∼70% of these FAIRE peaks being localized to promoters (Fig. 2D). Promoters, especially of transcriptionally active genes, are known to be relatively nucleosome free in a wide variety of species (Lee et al. 2007; Schones et al. 2008; Weiner et al. 2010; Thomas et al. 2011; Valouev et al. 2011). In zebrafish, well-positioned nucleosome arrays appear at promoters during zygotic genome activation in a transcription-independent fashion, suggesting that active mechanisms may shape nucleosome occupancy at promoters during early development (Zhang et al. 2014). When we searched for motifs enriched among these constitutively open peaks, all k-mers identified were highly AT rich (Supplemental Table 1). AT-rich sequences were also enriched when we analyzed all FAIRE peaks lacking Zld compared with all FAIRE peaks with Zld bound. AT-rich sequences are associated with decreased nucleosome occupancy and have also been shown to be enriched at promoters (Sekinger et al. 2005; Hughes et al. 2012). Because the underlying sequence does not predict any inherent decrease in nucleosome occupancy at FAIRE peaks that are independent of Zld binding (constitutive, not Zld-bound) (Fig. 2C), we suggest the identification of the AT-rich sequence motif in these regions likely results from the fact that this sets of peaks is enriched for promoters and does not by itself explain the open chromatin at these regions.
Zld-mediated chromatin accessibility is associated with early gene expression
Our analysis of the top FAIRE peaks demonstrated that there were two classes of accessible chromatin regions bound by Zld: those that required Zld for accessibility (differential, ZLD-bound; n = 402) and those that did not (constitutive, Zld-bound; n = 1537). To begin to differentiate between these two classes of Zld-bound loci, we used ChIP-seq data to determine the relative strength of Zld binding at nuclear cycle 14 (Harrison et al. 2011). Constitutive peaks with Zld bound (constitutive, Zld-bound) had on average threefold lower Zld ChIP-signals than those that were dependent on Zld for accessibility (differential, Zld-bound) (Fig. 3A). This shows that regions highly occupied by Zld in vivo are more dependent on Zld for establishing or maintaining chromatin accessibility than those less-occupied sites (see also Supplemental Fig. S3). We have previously shown that levels of Zld binding are correlated with the timing of gene expression; regions with higher Zld ChIP-seq signal are expressed earlier in embryonic development (Harrison et al. 2011). This suggests that regions that depend on Zld for accessibility may be enriched near genes that require Zld for expression.
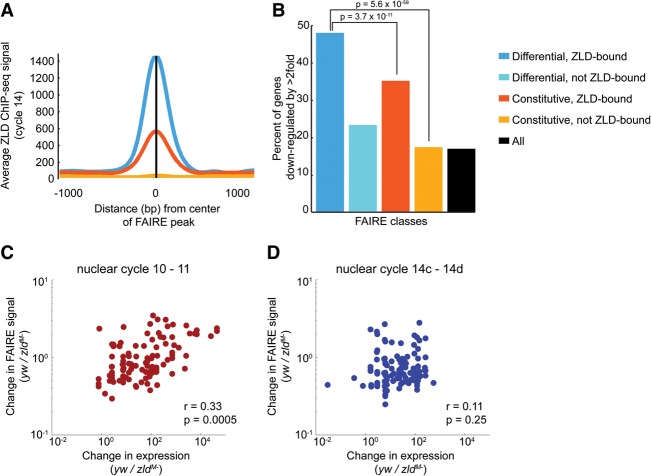
Figure 3.
Zld-mediated chromatin accessibility is correlated with early Zld-dependent gene expression. (A) Average ChIP-seq signal for Zld at cycle 14 for each class of FAIRE peaks. (B) FAIRE-enriched sites were associated with the nearest gene. For each associated gene, the expression change when maternal Zld was depleted was identified by RNA-seq. The percentage of genes that show a greater than twofold decrease in expression is plotted for each class of FAIRE peaks. Hypergeometric P-values are shown. (C,D) Correlation between Zld-mediated changes in gene expression and FAIRE signal for 83 genes expressed at nuclear cycles 10–11 (C) or 91 genes expressed at late nuclear cycle 14 (D). r = Spearman's correlation coefficient.
To test the functional impact of Zld-mediated chromatin accessibility on gene expression, we performed RNA-seq from single embryos and compared stage 5 zldM- to yw controls. We then associated FAIRE peaks with the nearest gene and identified what percentage of these FAIRE-associated genes require Zld for robust expression. Because Zld is known to act as a transcriptional activator (Liang et al. 2008), we focused on genes whose expression decreased by greater than twofold in the zldM- embryos. The analysis shows that 48% of genes associated with Zld-dependent FAIRE peaks (differential, Zld-bound) had a greater than twofold decrease in expression in the absence of Zld compared with only 18% for all FAIRE peaks (Fig. 3B). Those regions that do not depend on Zld for accessibility (constitutive, Zld-bound) were also enriched near genes that require Zld for expression, but to a lesser degree than those genes near regions that require Zld for open chromatin (differential, Zld-bound). However, the large amount of maternally supplied RNA in the early embryo masked any changes in expression of genes transcribed both maternally and zygotically. Thus, our RNA-seq only identified changes in gene expression for those genes that are exclusively zygotically expressed. For this reason, the actual numbers of down-regulated genes in the zldM- embryos may be larger than reported. To independently verify these results, we repeated our gene expression analysis using published microarray data (Liang et al. 2008) and similarly demonstrated that those regions that require Zld for accessibility were enriched near genes that require Zld for expression (Supplemental Fig. S4).
Zld influences expression of both a small number of genes expressed early in the MZT, as well as a large number of genes expressed at nuclear cycle 14. To determine if Zld-mediated accessibility might specifically drive expression of either one of these sets of genes, we used a high-temporal resolution data set to examine the relationship between Zld-mediated chromatin accessibility and the timing of gene expression (Lott et al. 2011). Genes were grouped based on their timing of initial expression (Li et al. 2014), and we analyzed the correlation between Zld-mediated changes in accessibility and Zld-mediated changes in expression. Specifically, we determined the change in FAIRE signal between control and zldM- embryos and plotted this against the change in RNA levels, as determined by RNA-seq, for the gene with the nearest associated transcription start site. There was a significant correlation between Zld-mediated chromatin accessibility and Zld-mediated gene expression for genes that initiate expression during nuclear cycles 10–11 (Spearman's rho = 0.33, P = 0.0005) but not for genes that are expressed during late cycle 14 (Spearman's rho = 0.11, P = 0.25) (Fig. 3C,D). Furthermore, during nuclear cycles 8–12 RNA polymerase II (Pol II) is bound only to the promoters of the early expressed genes (Chen et al. 2013; Li et al. 2014), and we demonstrated that this promoter localized Pol II is preferentially found at promoters that require Zld for chromatin accessibility (differential, Zld-bound) (Supplemental Fig. S5). Only later, at cycles 13–14, was Pol II associated with promoters from every FAIRE class (Supplemental Fig. S5; Chen et al. 2013). Thus Zld-mediated chromatin accessibility likely drives the very earliest expressed zygotic genes. Later in embryonic development, Zld may activate gene expression by a different mechanism.
H3K18ac and H4K8ac are enriched around Zld-bound regions that require Zld for accessibility
We recently mapped the location of nine histone modifications at four different developmental times spanning the MZT using ChIP-seq (Li et al. 2014) and demonstrated that acetylation of three different lysines on two different histones (H3K18, H3K27, and H4K8) correlated with Zld binding, especially early in development (Li et al. 2014). Thus we speculated that Zld-mediated chromatin accessibility might correlate with specific histone modifications. To further investigate the connection between chromatin accessibility and early gene expression, we examined the relationship between chromatin accessibility and histone modifications during the MZT. We analyzed the enrichment of acetylation and methylation marks around our four classes of FAIRE peaks (Fig. 4; Supplemental Fig. S6). Prior to widespread zygotic genome activation (at cycles 8 and 12), H3K18ac and H4K8ac were enriched on nucleosomes surrounding the Zld-bound regions that require Zld for accessibility (differential, Zld-bound) (Fig. 4). Similar enrichment was not evident at constitutively open regions or those that lack Zld binding (constitutive, Zld-bound; constitutive, not Zld-bound, differential, not Zld-bound) (Fig. 4). Thus, Zld-mediated chromatin accessibility, and not Zld-binding in general, is correlated with early histone acetylation. We previously demonstrated that these early acetylation marks are associated with early expressed genes (Li et al. 2014), strengthening the connection between regions that require Zld for accessibility and early gene expression. Furthermore, H3K18ac levels are decreased in zldM- embryos (Li et al. 2014), demonstrating that Zld is required for H3K18 acetylation at specific regions. Together with our FAIRE data, this suggests that Zld may potentiate chromatin accessibility and early gene expression by increasing histone acetylation.
Figure 4.
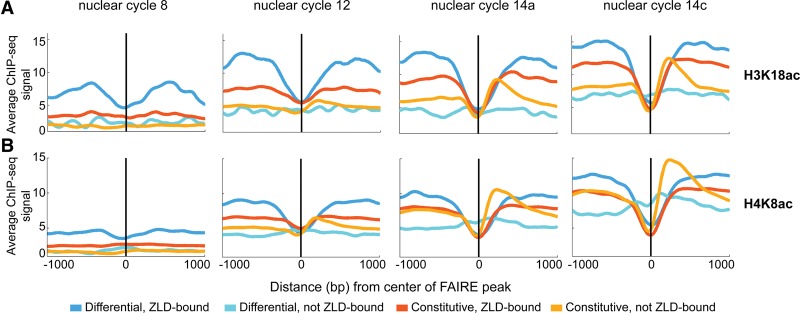
Early histone acetylation is enriched around loci that require Zld for chromatin accessibility. (A,B) Distribution of ChIP-seq signals for H3K18ac (A) and H4K8ac (B) surrounding individual classes of FAIRE peaks at specific nuclear cycles during the early stages of embryonic development. Colors indicate the different FAIRE classes. Plots are centered on the FAIRE peak and oriented relative to the transcription start site of the nearest gene.
Regions that require Zld for chromatin accessibility also require Zld for Bcd binding
One mechanism by which Zld may activate gene expression is by facilitating transcription factor binding. We have previously demonstrated that early Zld binding is predictive of where transcription factors will later bind (Harrison et al. 2011), and it has been recently reported that Zld promotes binding of the transcription factors Twi, Dl, and Bcd to DNA (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014). To determine if Zld enabled binding of these proteins by creating open regions of chromatin, we reanalyzed a published ChIP-seq data set identifying Bcd-binding sites in both wild-type embryos and those lacking maternal zld (Xu et al. 2014). Previous analysis had shown that Bcd binding is partially redistributed when Zld is absent. Specifically, Bcd binding in the mutant was decreased at a large number of sites that overlap with Zld-bound regions (Xu et al. 2014). Comparing our four classes of peaks with the Bcd ChIP-seq data set demonstrated that Bcd binds preferentially to Zld-bound loci, both those that require Zld for accessibility and those that do not (Fig. 5A), and the average Bcd ChIP-signal at these two classes of Zld-bound regions is approximately equivalent (Fig. 5A). We identified a preferential reduction in Bcd binding at the differential peaks with Zld bound (differential, Zld-bound) compared with the constitutively open peaks with Zld bound (constitutive, Zld-bound) upon Zld depletion (Fig. 5B). This is demonstrated by the regulatory regions of dpn and sog (Fig. 1A). In these examples, promoter proximal FAIRE peaks do not require Zld for accessibility or Bcd binding (dark orange box), while neighboring enhancers lose both chromatin accessibility and Bcd binding in the absence of Zld (blue box). This shows that Zld-dependent Bcd binding is correlated with Zld-mediated chromatin accessibility and not just with Zld binding. Together this suggests that Zld facilitates transcription factor binding through the establishment or maintenance of open chromatin.
Figure 5.
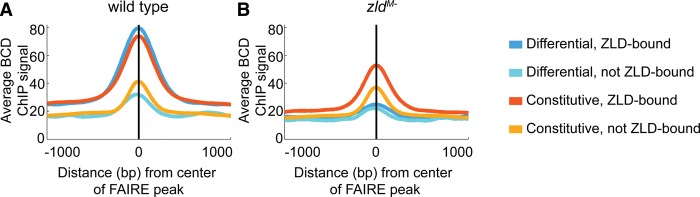
In the absence of Zld, Bcd binding is preferentially lost at those regions that lose chromatin accessibility. (A,B) Average Bcd binding in wild-type embryos (A) and embryos lacking maternal Zld (B) for each class of FAIRE peaks. Colors indicate the different FAIRE classes.
Collective binding by a large number of transcription factors does not distinguish between Zld-bound FAIRE classes
In addition to being associated with regions of open chromatin, another feature correlated with Zld-bound loci is occupancy by a large number of transcription factors (Li et al. 2008; MacArthur et al. 2009; Roy et al. 2010; Kvon et al. 2012). These regions have been termed highly occupied target (HOT) regions and have been identified in a wide variety of organisms (Moorman et al. 2006; The ENCODE Project Consortium 2007; Li et al. 2008; MacArthur et al. 2009; Gerstein et al. 2010; Roy et al. 2010; Negre et al. 2011). Thus it was possible that Zld-bound regions that do not require Zld for accessibility are maintained in an open chromatin state by binding of a large number of additional factors. To determine if this was the case, we analyzed the number of transcription factors bound in vivo to each of the four classes of FAIRE peaks. As expected, we showed that FAIRE peaks overlapping Zld-bound regions were enriched for binding by more than 10 transcription factors (constitutive, Zld-bound and differential, Zld-bound) (Fig. 6A). On average there were 2.5 times as many transcription factors present at Zld-bound FAIRE peaks than at FAIRE peaks that were not Zld bound (Fig. 6A). Thus, among regions of chromatin accessibility, Zld binding is correlated with those loci that are bound by a large number of transcription factors. Nonetheless, we found no obvious difference in the number of transcription factors bound when we compared Zld-bound peaks that depend on Zld for accessibility (differential, Zld-bound) to those that do not require Zld (constitutive, Zld-bound). This was also the case when we controlled for the fact that constitutive, Zld-bound peaks are enriched for promoters compared with differential, Zld-bound peaks by further subdividing each class by genome annotation (Supplemental Fig. 7). Thus, these data demonstrate that binding by a large number of transcription factors is not what functionally distinguishes these classes of Zld-bound regions and that some other feature must allow the chromatin to remain accessible in the absence of Zld at those regions that remain constitutively open.
Figure 6.
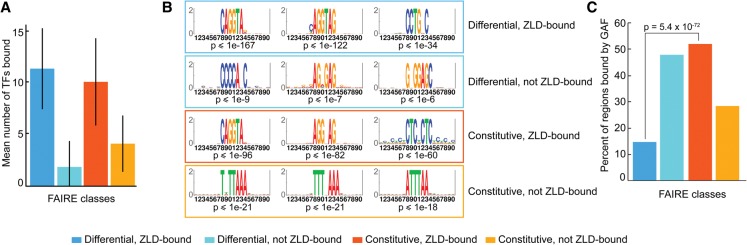
GAF likely functions with Zld to define chromatin accessibility in the early embryo. (A) Mean number of transcription factors (TFs) bound to each class of FAIRE peaks. Colors indicate the different FAIRE classes. Error bars, SD. (B) Position weight matrices for the motifs enriched in the FAIRE peaks for each class compared with all of the top 5000 FAIRE peaks identified in yw embryos. Hypergeometric P-values are shown below each motif. The top three motifs for each class are shown (for additional motifs, see Supplemental Table 1). (C) Percentage of FAIRE peaks in each class that overlap a GAF-binding site as identified by ChIP-seq in 0- to 8-h embryos (Negre et al. 2011). Hypergeometric P-value is shown.
Constitutively open Zld-bound loci contain GAGA motifs and are preferentially bound by GAGA factor
Because binding by a large number of transcription factors did not functionally distinguish between the two Zld-bound FAIRE classes, we proposed that binding by a specific transcription factor might maintain chromatin accessibility at constitutive, Zld-bound peaks in the absence of Zld. To determine potential factors, we identified motifs enriched in the DNA sequences underlying the FAIRE peaks. For both classes of peaks with Zld bound (differential and constitutive), we showed, as expected, that the most highly enriched motif was the canonical Zld-binding motif, CAGGTA (Fig. 6B; Supplemental Table 1). For the differential Zld-bound peaks, the next most highly enriched k-mers were overlapping or related to this canonical Zld-binding site (Supplemental Table 1; ten Bosch et al. 2006; Liang et al. 2008; Harrison et al. 2011; Nien et al. 2011; Struffi et al. 2011). However, for the constitutively open Zld-bound peaks, there was a strong enrichment for GAGA motifs (Fig. 6B; Supplemental Table 1), suggesting that these regions may be occupied by GAGA factor (GAF; encoded by the gene Trithorax-like [Trl]) in addition to Zld (Wilkins and Lis 1997). Importantly, GAF binding is strongly correlated with chromatin accessibility, and GAGA motifs are enriched in HOT regions (Lu et al. 1993; Kvon et al. 2012; Slattery et al. 2014). Because GAF-binding motifs are known to be enriched at promoters, it was possible that the identification of GAF-binding motifs in constitutive, Zld-bound peaks was a function of the fact that 39% of these peaks were in promoter regions compared with only 18% for the differential, Zld-bound peaks (Fig. 2D). Thus, we directly determined the enrichment of the GAF-binding motif in the constitutive, Zld-bound versus the differential, Zld-bound peaks for promoters, intergenic regions, intronic regions, and coding regions. We found that in all cases there was a significant enrichment of the GAF-binding motif in the constitutive, Zld-bound peaks. This enrichment was most significant at the intronic and intergenic peaks. Comparing GAGA motif enrichment in constitutive versus differential Zld-bound FAIRE peaks, we demonstrated that at intronic regions, 62% of constitutively accessible Zld-bound regions contained GAGA motifs, while only 25% of the introns that depended on Zld for accessibility (differential, Zld-bound) contained the motif (P = 10−15). In intergenic regions, 20% of constitutively accessible Zld-bound regions had GAGA motifs in comparison to none of the differential, Zld-bound intergenic peaks (P < 10−8). These analyses demonstrate that the GAF-binding motif is specifically enriched in the constitutive, Zld-bound regions independently of where in the genome these peaks are located.
Consistent with this sequence enrichment, GAF binding, as assayed by ChIP-seq in 0- to 8-h embryos (Negre et al. 2011), is enriched in those regions that are constitutively accessible and bound by Zld (constitutive, Zld-bound). Specifically, 52% (796/1537) of constitutively open Zld-bound regions overlap GAF peaks, compared with only 15% (59/402) of regions that depend on Zld for open chromatin (Fig. 6C, P < 0.0001). Thus both the GAF-binding site and embryonic GAF binding are specifically enriched at Zld-bound loci that remain accessible in the absence of Zld. Furthermore, only 28% (829/2923) of constitutively open regions that do not bind Zld overlap GAF-binding sites, indicating that in addition to GAF, other transcription factors may be required for open chromatin. Together, these data suggest that a subset of Zld-bound regions that do not change in accessibility in the zldM- embryos remain open due to co-occupancy by GAF.
Discussion
We have used FAIRE to identify regions of open chromatin in the early embryo and determine the role of Zld in establishing or maintaining chromatin accessibility. We demonstrate on a genome-wide level that Zld is instrumental in defining specific regions of open chromatin. Furthermore, this Zld-mediated chromatin accessibility dictates both transcription factor binding and early gene expression. Unexpectedly, we discovered that most open chromatin regions to which Zld is bound do not absolutely require Zld for chromatin accessibility. At these regions, Zld may function redundantly with GAF to determine the chromatin state. We suggest that Zld directly mediates the very earliest gene expression by facilitating chromatin accessibility. At cycle 14, when thousands of genes are transcribed, Zld and GAF may coordinate to determine both regions of open chromatin and levels of gene expression.
Zld-mediated chromatin accessibility facilitates transcription factor binding and early gene expression
Zld is known to be instrumental in regulating expression of both the very first set of zygotic genes transcribed after fertilization, as well as a large set of genes transcribed at cycle 14. Zld is already bound to thousands of loci at cycle 10, including those that will not be activated until four nuclear cycles later during the major wave of genome activation (Harrison et al. 2011). This suggests that early Zld binding is poising genes for later activation. Nonetheless, it remains unclear what differentiates the small subset of Zld-bound loci that are transcribed early from the hundreds of Zld-bound genes activated at cycle 14. Here we demonstrate regions that require Zld for chromatin accessibility are correlated with the subset of genes transcribed prior to cycle 14 and with histone acetylation (Figs. 3, 4; Chen et al. 2013; Li et al. 2014). However, not all Zld-bound regions are equally dependent on Zld for chromatin accessibility. We therefore propose that Zld is essential for creating regions of open chromatin that drive expression of the subset of earliest expressed genes. This may be mediated, in part, by local histone acetylation. At cycle 14, other factors likely function with Zld to determine chromatin accessibility.
Zld-mediated transcriptional activation may be potentiated by the subsequent binding of additional transcription factors. Early Zld binding is a robust predictor of where multiple additional transcription factors will later bind (Harrison et al. 2011). More recently, it has been shown that Zld is required for the DNA binding of three different transcription factors: Twi, Dl, and Bcd (Yanez-Cuna et al. 2012; Foo et al. 2014; Xu et al. 2014). Additionally, transgenic versions of the brk and sog enhancers show a correlation between the number of Zld-binding sites and both Dl binding and DNase I accessibility (Foo et al. 2014). Thus, prior work has clearly demonstrated a role for Zld in mediating transcription factor binding, but the mechanism by which Zld served this function was unclear. Here we demonstrate that Bcd binding is lost in zldM- embryos preferentially at those regions that depend on Zld for chromatin accessibility. These data show that Zld potentiates transcription factor binding through the establishment or maintenance of open chromatin, and this is likely to be important for Zld-mediated transcriptional activation (Fig. 7).
Figure 7.
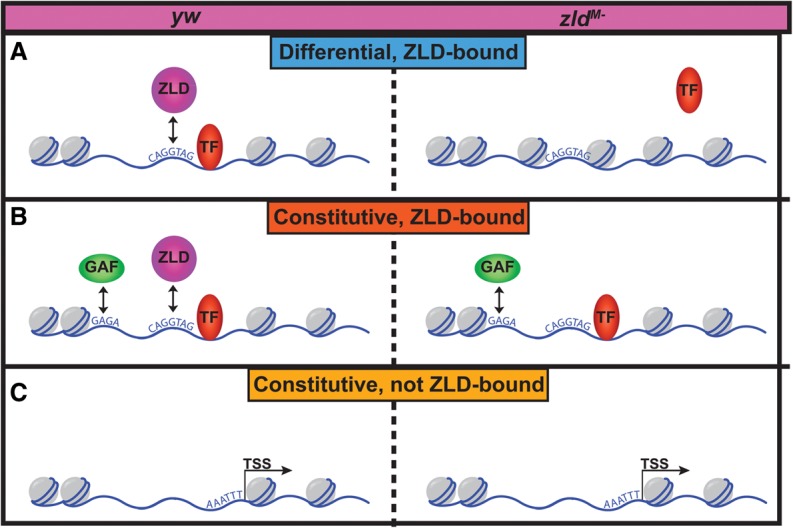
Model for the role of Zld in defining chromatin accessibility at the MZT. (A) Zld determines chromatin accessibility at early embryonic enhancers, allowing other TFs access to their binding sites. (B) At a large subset of regions, Zld binds chromatin but is not required for accessibility. GAF facilitates open chromatin at regions that remain accessible in zldM- embryos, allowing transcription factors to bind to these loci. (C) Promoter regions are enriched among those regions that do not require Zld for accessibility.
The mechanism by which Zld establishes or maintains chromatin accessibility remains unknown. Unlike the pioneer factor FOXA1, which can bind open chromatin by binding through a winged-helix domain (Cirillo et al. 1998, 2002), the Zld DNA-binding domain does not resemble that of a linker histone. Instead, Zld binds DNA through a cluster of four zinc fingers in the C terminus (Struffi et al. 2011; Hamm et al. 2015). In addition, Zld is a large protein with no recognizable enzymatic domains that activates transcription through a low-complexity protein domain (Hamm et al. 2015). Thus, Zld likely facilitates open chromatin through interactions with cofactors, and it is possible that recruitment of different cofactors to distinct Zld-bound loci could partially explain the differential requirement on Zld for chromatin accessibility in the early embryo.
Zld binding but not Zld-mediated chromatin accessibility is a defining feature of HOT regions
HOT regions, loci that are bound by a large number of different transcription factors, have been identified in multiple organisms, including worms, flies, and humans (Moorman et al. 2006; The ENCODE Project Consortium 2007; Li et al. 2008; MacArthur et al. 2009; Gerstein et al. 2010; Roy et al. 2010; Negre et al. 2011). Unexpectedly, these HOT regions are not strongly enriched for the DNA-sequence motifs bound by the transcription factors that define them (Moorman et al. 2006; MacArthur et al. 2009; Gerstein et al. 2010; Roy et al. 2010). Instead, HOT regions are associated with open chromatin, suggesting that chromatin accessibility along with sequence motif enrichment drives the high transcription factor occupancy (Roy et al. 2010). In Drosophila, HOT regions are enriched for developmental enhancers that contain the canonical Zld-binding site, CAGGTAG, as well as for in vivo Zld binding (Li et al. 2008; MacArthur et al. 2009; Harrison et al. 2011; Nien et al. 2011; Kvon et al. 2012). By analyzing the 5000 regions with the highest FAIRE signal, we demonstrate that high transcription factor occupancy is correlated with Zld-bound regions of accessible chromatin and not with open chromatin more generally (Fig. 6A). Furthermore, this association was not specific for those regions that require Zld for accessibility. Thus, HOT regions overlap with Zld-bound regions of open chromatin regardless of whether these loci require Zld for accessibility. Our data suggest that while Zld-mediated chromatin accessibility may facilitate gene expression, it is not this function of Zld alone that defines HOT regions.
The open chromatin landscape and transcriptional profile of the embryo are likely defined by the functions of both GAGA factor and Zld
Our FAIRE data showed that more than 400 regions are bound by Zld and require Zld for chromatin accessibility. However, at least three times as many regions are bound by Zld but remain open even in its absence. Our data predict GAF functions at many of these constitutively open chromatin regions to maintain chromatin accessibility, even in the absence of Zld (Fig. 7). Along with the CAGGTAG element, GAF-binding motifs are enriched in HOT regions (Kvon et al. 2012; Slattery et al. 2014). Like Zld, GAF is maternally deposited into embryos. Furthermore, GAF is known to facilitate nuclease-hypersensitive regions and interact with members of the NURF ATP-dependent chromatin-remodeling complex (Lu et al. 1993; Tsukiyama and Wu 1995).
Our data show that at early expressed genes there is a correlation between regions that require Zld for chromatin accessibility (differential, Zld-bound) and Zld-dependent gene expression (cycles 10–11) (Fig. 3C). However, this association is not found for genes expressed during cycle 14 (Fig. 3D). Instead, our data suggest that at loci associated with this later gene expression, GAF is functioning together with Zld to regulate chromatin accessibility and gene expression. Maternally deposited GAF is required for robust transcription and nuclear divisions during the MZT (Bhat et al. 1996; Lagha et al. 2013). GAF is thought to mediate transcription, at least in part, through a role in the establishment of poised polymerase (Shopland et al. 1995; Bhat et al. 1996; Lee et al. 2008). The fact that poised polymerase is not established until cycle 13 (Chen et al. 2013; Blythe and Wieschaus 2015) supports our model that GAF is required specifically for gene expression at cycles 13–14. Thus, we suggest that Zld-dependent early embryonic enhancers may be unique in that they rely only on Zld for chromatin accessibility. Although there are likely additional factors involved, our data demonstrate that later in development, Zld and GAF likely function together to define the chromatin landscape of the early embryo.
Pioneer factors as drivers of zygotic genome activation
Pioneer factors are a specialized class of transcription factors that bind nucleosomal DNA and initiate chromatin remodeling, allowing the recruitment of additional transcription factors (Iwafuchi-Doi and Zaret 2014). Zld binding is strongly driven by DNA sequence, much more so than the binding of other transcription actors (Harrison et al. 2011). This observation combined with our FAIRE data and analyses demonstrates that Zld exhibits many of the characteristics of a pioneer factor as defined by Iwafuchi-Doi and Zaret (2014): (1) engaging chromatin prior to gene activity, (2) establishing or maintaining chromatin accessibility to facilitate transcription factor binding, and (3) playing a primary role in cell reprogramming. Additional properties have been shown for classical pioneer factors, including remaining bound to the mitotic chromosomes (i.e., bookmarking) and binding to nucleosomal DNA (Cirillo et al. 2002; Sekiya et al. 2009; Kadauke et al. 2012; Caravaca et al. 2013). It will be important to determine whether Zld shares these characteristics with other pioneer factors.
Pioneer factors, such as FOXA1, can bind to closed chromatin and subsequently increase accessibility of the target site (Cirillo et al. 2002). However the chromatin of the early embryo may provide a unique environment with little compacted chromatin. Heterochromatin formation is not observed until the 14th nuclear cycle (Foe and Alberts 1983). Chromatin-bound H3 levels increase through the MZT, and histone modifications indicative of silent genes, such as H3K27 trimethylation, are not evident until there is widespread activation of the zygotic genome (Li et al. 2014). Thus, while Zld binds to genes prior to zygotic genome activation, this activity may not require binding to compacted chromatin. It may be that Zld is distinctive in the timing of its expression rather than in its chromatin-binding properties and that the sequence-driven binding of Zld is a property of the open chromatin and rapid nuclear divisions that characterize the earliest stages of embryonic development.
Despite the fact that we have demonstrated a critical role for Zld in determining chromatin accessibility at hundreds of genomic regions, our data show that this role is limited to specific regions associated with the earliest-expressed embryonic genes. Other factors, such as GAF, likely work redundantly with Zld to define chromatin accessibility during the MZT. The coordinated function of multiple factors in determining chromatin structure and genome activation is not without precedent. It has recently been demonstrated that homologs of the core pluripotency factors, Nanog, Pou5f3 (previously known as Pou5f1 and Oct4), and Sox19B (a member of the SoxB1 family), act analogously to Zld during the zebrafish MZT to drive genome activation (Lee et al. 2013; Leichsenring et al. 2013). Furthermore, Pou5f3 and Sox2 are known to be pioneer factors instrumental in reprogramming differentiated cells to a pluripotent state (Soufi et al. 2012, 2015). Together, these data suggest that chromatin remodeling in the early embryo requires the function of multiple factors, and this activity facilitates the transition from the specified germ cells to the pluripotent cells of the early embryo.
Methods
Fly strains
Embryos lacking maternally supplied Zld were generated according to the method described by Liang et al. (2008). zld294 FRT19A/FM7 females were crossed with y w sn P{mini w+, ovoD1−26}25 FRT19A, hsFLP122/Y males and allowed to lay for 24 h. The resulting offspring were heat-shocked twice for 30 min at 37°C with a 24-h interval between (at ∼24–48 h and 48–72 h after laying) to generate germline clones. In parallel, y w FRT19A females were crossed with y w sn P{mini w+, ovoD1−26}25 FRT19A, hsFLP122/Y males, and the offspring were heat-shocked to serve as paired controls. Since only females with the desired germline clones should produce embryos, non-heat-shocked offspring from each cross were preserved and allowed to mature to check for escapers. Any crosses that produced embryos without heat-shock (escapers) were discarded.
Immunostaining
Embryos depleted for maternal Zld and paired yw controls were dechorionated and fixed in 3 mL fixation buffer (1.3× PBS, 67 mM EGTA at pH 8), 1 mL 37% formaldehyde, and 5 mL heptanes for 25 min. Vitelline membranes were removed with methanol. Embryos were rehydrated and stained using anti-Zld antibodies (Harrison et al. 2011) along with DM1A anti-tubulin (Sigma) as a positive control. Goat anti-rabbit Alexa-488 and goat anti-mouse Alexa-688 were used as secondary antibodies (Life Technologies).
FAIRE
Embryos ranging in age from 2 to 3 h were collected, dechorionated, and crosslinked for 10 min with 3:1 heptane:fix solution at room temperature. Following washing, embryos were flash-frozen in liquid nitrogen and stored at −80°C. FAIRE was performed on ∼0.1 g of embryos, as previously described by McKay and Lieb (2013) with the following exceptions. The homogenized embryos were filtered through Miracloth (EMD Millipore). After resuspension in lysis buffer (10 mM Tris-Cl at pH 8, 100 mM NaCl, 1 mM EDTA, 2% Triton X-100, 1% SDS), the nuclei were subjected to four rounds of bead beating with 0.5-mm glass beads. The chromatin was sonicated three times for 30 sec using a Branson sonifier 250 at a power setting of 1.5 to achieve a size range of 200 bp–1 kb.
Single-embryo RNA preparation
Embryos depleted for maternal Zld and paired yw controls were dechorionated and analyzed under halocarbon oil to determine stage. Individual paired stage 5 embryos were selected, and oil was removed. Embryos were lysed in TRIzol (Life Technologies) supplemented with 150 µg/mL glycogen, and RNA was prepared according to the manufacturer's instructions.
Sequencing and data analysis
High-throughput sequencing libraries were prepared by the University of Wisconsin Biotechnology Center DNA Sequencing Facility (FAIRE) and by Jacqueline Villalta (RNA). Libraries were prepared from both FAIRE-enriched DNA and from sonicated genomic DNA to serve as input material. The libraries were sequenced using the Illumina HiSeq 2000 platform. The total reads for yw were 42,524,780 and for zldM- were 50,462,591. The sequence data were processed essentially as previously described (McKay and Lieb 2013). Briefly, reads were aligned to the genome using Bowtie 2 (Langmead and Salzberg 2012) with the –sensitive and –end-to-end flags. Aligned reads with a MAPQ score less than 10 were removed using SAMtools (Li et al. 2009). Finally, reads were extended to 110 bp. Since replicate data were highly correlated, reads from each replicate were pooled for further analysis, resulting in 23,646,263 reads for yw and 13,371,214 reads for zldM-. This corresponds to 92% of the euchromatic region covered by greater than one read for yw and 87% for zldM-. FAIRE peaks were called with MACS2 (Feng et al. 2012) using a shift size of 125 bp. Differential FAIRE peaks between yw and zldM- were identified with edgeR (Robinson et al. 2010), using the “classic analysis.” The input file consisted of a table of FAIRE read counts for each yw and zldM- replicate intersecting the union set of the top 5000 FAIRE peaks. FAIRE peaks with a P-value lower than 0.05 were called as differential.
Data for Bcd binding in wild-type and zldM- embryos were from GSE55256 (Xu et al. 2014). GAF binding was from GSE23537 (Negre et al. 2011). Histone modification data were obtained from GSE58935 (Li et al. 2014). Microarray expression data for wild-type and zldM- embryos were from GSE11231 (Liang et al. 2008). Zld-binding sites were from GSE30757 (Harrison et al. 2011). Data for RNA Pol II binding during the MZT were obtained from GSE41703 (Chen et al. 2013).
Peak classification
FAIRE peaks were classified into four functional groups based on their location. Peaks located −500 to +150 bases from a known transcription start site were classified as promoter peaks. Peaks overlapping coding regions were classified as genic or intronic, and the remaining peaks classified as intergenic.
Motif analysis
The DNA sequence in FAIRE peaks was analyzed using SeedSearcher software by Barash (2005). Specifically, we enumerated over all possible k-mers (ranging from length three to eight, while allowing one or two wildcards in specific positions). Each k-mer was then scored using a differential hypergeometric score, comparing the number of motif occurrences within a set of peaks to the overall number of occurrences within a background set of control sequences. Here we compared each group of FAIRE peaks (constitutive or differential, with or without in vivo Zld or GAF binding) to the remaining of the 5000 high-accessibility yw FAIRE peaks. All reported k-mers are highly significant after corrections for multiple hypothesis testing.
Data access
The FAIRE-seq and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65837.
Supplementary Material
Acknowledgments
We thank Michael Eisen for assistance with the single-embryo RNA-seq and helpful discussions. T.K. is a member of the Israeli Center of Excellence (I-CORE) for Gene Regulation in Complex Human Disease (no. 41/11) and the Israeli Center of Excellence (I-CORE) for Chromatin and RNA in Gene Regulation (no. 1796/12). K.N.S. was supported by the National Institutes of Health (NIH) National Research Service award T32 GM007215. This work was supported in part by a Basil O'Connor Starter Scholar Research award (March of Dimes: #5-FY14-29) to M.M.H. and grant 1R01GM111694 from the National Institute of General Medical Sciences to M.M.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.192682.115.
References
- Barash Y. 2005. “Unified models for regulatory mechanisms.” PhD thesis, Hebrew University, Jerusalem. [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. 1996. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development 122: 1113–1124. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. 2015. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160: 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. 2013. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev 27: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A, Biggin MD. 1999. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J 18: 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. 2013. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2: e00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. 1993. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Zaret KS. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell 4: 961–969. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, et al. 2012. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci 109: 21330–21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci 61: 31–70. [DOI] [PubMed] [Google Scholar]
- Foo SM, Sun Y, Lim B, Ziukaite R, O'Brien K, Nien CY, Kirov N, Shvartsman SY, Rushlow CA. 2014. Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr Biol 24: 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–1682. [DOI] [PubMed] [Google Scholar]
- Hamm DC, Bondra ER, Harrison MM. 2015. Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain. J Biol Chem 290: 3508–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7: e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. 2012. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533–538. [DOI] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T, Li X-y, Sabo PJ, Thomas S, Stamatoyannopoulos JA, Biggin MD, Eisen MB. 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet 7: e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. 2012. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev 26: 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. 2013. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153: 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244. [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, Divito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. 2013. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 341: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol 12: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334. [DOI] [PubMed] [Google Scholar]
- Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. 2006. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res 16: 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. 2011. Noncanonical compensation of the zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-Seq. PLoS Biol 9: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wallrath LL, Granok H, Elgin SC. 1993. (CT)n⋅(GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol 13: 2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Lieb JD. 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev Cell 27: 306–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci 103: 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30: 687–696. [DOI] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7: e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Bradley RK. 2012. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res 22: 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18: 735–748. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. 2009. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23: 804–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. 1995. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev 9: 2756–2769. [DOI] [PubMed] [Google Scholar]
- Slattery M, Ma L, Spokony RF, Arthur RK, Kheradpour P, Kundaje A, Negre N, Crofts A, Ptashkin R, Zieba J, et al. 2014. Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. Genome Res 24: 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N, Fellert S, Chung HR, Jackle H, Vorbruggen G. 2006. Mutations of the Drosophila zinc finger–encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Mol Biol Cell 17: 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S. 2011. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development 138: 4291–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133: 1967–1977. [DOI] [PubMed] [Google Scholar]
- Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, et al. 2011. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol 12: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. 1997. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res 25: 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, Small S. 2014. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev 28: 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. 2012. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res 22: 2018–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell 24: 593–602. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. 2014. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.