Abstract
A conserved hallmark of eukaryotic chromatin architecture is the distinctive array of well-positioned nucleosomes downstream from transcription start sites (TSS). Recent studies indicate that trans-acting factors establish this stereotypical array. Here, we present the first genome-wide in vitro and in vivo nucleosome maps for the ciliate Tetrahymena thermophila. In contrast with previous studies in yeast, we find that the stereotypical nucleosome array is preserved in the in vitro reconstituted map, which is governed only by the DNA sequence preferences of nucleosomes. Remarkably, this average in vitro pattern arises from the presence of subsets of nucleosomes, rather than the whole array, in individual Tetrahymena genes. Variation in GC content contributes to the positioning of these sequence-directed nucleosomes and affects codon usage and amino acid composition in genes. Given that the AT-rich Tetrahymena genome is intrinsically unfavorable for nucleosome formation, we propose that these “seed” nucleosomes—together with trans-acting factors—may facilitate the establishment of nucleosome arrays within genes in vivo, while minimizing changes to the underlying coding sequences.
Nucleosomes are the fundamental packaging unit of eukaryotic chromatin. Each nucleosome consists of ∼147 bp of DNA wrapped around a histone octamer (Luger et al. 1997). The organization of nucleosomes across the genome plays an important regulatory role as it lowers the physical accessibility of DNA to cellular factors and may directly impact DNA-based transactions, such as transcription (Piña et al. 1990; Lam et al. 2008). In light of this, it is crucial to understand how nucleosomes are organized across the genome.
Genome-wide nucleosome maps in major model organisms have revealed strikingly similar nucleosome patterns near gene starts, where a nucleosome-depleted region upstream of the TSS is followed by a stereotypical array of nucleosomes inside the gene (Yuan et al. 2005; Lee et al. 2007; Mavrich et al. 2008b; Lantermann et al. 2010; Chang et al. 2012; Chen et al. 2013b; Zhang et al. 2014). Recent studies have used Saccharomyces cerevisiae as a model to understand nucleosome positioning mechanisms underlying the stereotypical nucleosome pattern near eukaryotic TSSs. In principle, nucleosome organization can be guided both by the intrinsic DNA sequence preferences of histone octamers and by the action of trans-acting factors (Kaplan et al. 2009; Zhang et al. 2009, 2011b; Hughes et al. 2012; Struhl and Segal 2013). These two mechanisms, which are distinct but not mutually exclusive, have been studied by comparing nucleosome positions across the genome in vivo and in vitro. The in vitro nucleosome maps were generated by reconstituting nucleosomes on yeast genomic DNA, in the presence or absence of trans-acting factors, represented by cell extracts or ATP-dependent chromatin remodelers (Kaplan et al. 2009; Zhang et al. 2009, 2011b). Such experiments revealed that trans-acting factors, rather than the DNA sequence preferences of nucleosomes (Zhang et al. 2009, 2011b; Gkikopoulos et al. 2011; Hughes et al. 2012; Yen et al. 2012), mainly underlie the characteristic nucleosome array downstream from TSSs. This stands in contrast to the regulatory nucleosome-depleted regions upstream of TSSs, which were found to be intrinsically unfavorable to nucleosome formation (Kaplan et al. 2009; Zhang et al. 2009). These findings have since been considered the consensus in the field (Struhl and Segal 2013).
Here, we dissect the respective contributions of nucleosome sequence preferences and trans-acting factors to nucleosome organization in the somatic macronuclear genome of the ciliate Tetrahymena thermophila, an established model for chromatin biology. The Tetrahymena genome is GC-poor (22% GC), second only to Plasmodium falciparum (Gardner et al. 2002). It exhibits an unconventional structural organization, with ∼225 unique chromosomes, each amplified to ∼45n (Eisen et al. 2006). In order to characterize nucleosome organization and dissect their underlying positioning mechanisms in Tetrahymena, we performed genome-wide MNase-seq nucleosome mapping on log-phase and starved cells, as well as on histones assembled on sheared naked Tetrahymena DNA in vitro (Supplemental Figs. S1, S2). These data together represent, to our knowledge, the first global analysis of chromatin structure in a ciliate.
Results
Genome-wide nucleosome maps of the Tetrahymena thermophila macronuclear genome
We established comprehensive maps of nucleosome organization in the Tetrahymena macronuclear genome through paired-end MNase-seq across two different nutritional conditions in vivo. In addition, we performed paired-end MNase-seq on reconstituted Tetrahymena chromatin in vitro, obtained by assembling histone octamers on sheared naked genomic DNA, in the absence of any other trans-acting factors (see Methods and Supplemental Fig. S1). All data sets are summarized in Supplemental Table S1. Direct comparisons of the in vivo and in vitro data sets allow the inference of distinct nucleosome positioning mechanisms acting on the genome. For all analyses, we examined nucleosome positioning, rather than nucleosome occupancy, by assessing the distribution of nucleosome dyads across the genome. This was inferred from the midpoints of individual MNase-seq read pairs (see Methods).
First, we verified that MNase-seq data sets exhibit high coverage of the Tetrahymena nucleosome landscape by subsampling read pairs at varying proportions and subsequently calling nucleosomes using DANPOS (Chen et al. 2013a). The detected number of nucleosomes approached saturation well before full sampling of each MNase-seq data set, indicating that Tetrahymena nucleosomes are well-sampled (Supplemental Fig. S3). We measured the nucleosome repeat length as 199 bp (Supplemental Fig. S4), agreeing well with previous estimates obtained from gel analysis of MNase-treated macronuclear chromatin (Supplemental Fig. S2A; Gorovsky et al. 1978). The average nucleosome linker length remained constant in both log-phase and starved nutritional conditions, consistent with previous studies showing that this property remains largely invariant between diverse developmental and nutritional states (Zhang et al. 2011a). Other reports have suggested that nucleosome spacing may vary between nutritional conditions (Chang et al. 2012) and cell types (Valouev et al. 2011), though such changes are generally subtle.
We annotated ∼200,000 well-positioned nucleosomes in the Tetrahymena genome in vivo (202,028 in log-phase cells; 191,472 in starved cells). Given that the nucleosome repeat length is ∼200 bp in Tetrahymena, and the portion of the genome that we analyzed is 55.8 Mb (we only analyzed complete chromosomes in our study), we estimate that ∼72% of the genome is occupied by well-positioned nucleosomes. Using the same bioinformatic pipeline, we call 52,277 well-positioned nucleosomes in the 12.1 Mb S. cerevisiae genome in vivo, using a previously published MNase-seq data set (Kaplan et al. 2009). Given that the nucleosome repeat length is ∼165 bp in yeast, we calculate that ∼71% of the yeast genome is occupied by well-positioned nucleosomes, similar to Tetrahymena. We then validated our MNase-seq data sets by analyzing nucleosome positions at the 5′ nontranscribed spacer (NTS) of the Tetrahymena ribosomal DNA (rDNA) locus. Well-positioned nucleosomes flank both origins of replication within the 5′ NTS in vivo (Supplemental Fig. S5), closely corroborating independent studies that mapped nucleosomes at this locus through Southern analysis (Palen and Cech 1984). We observe similar patterns of nucleosome positioning in both log-phase and starved chromatin, consistent with previous reports (Palen and Cech 1984). Interestingly, our data suggest the presence of a small MNase-protected fragment at the proximal origin of replication in log-phase chromatin, possibly due to the presence of bound replication machinery. However, it is susceptible to increased MNase digestion at elevated temperatures (Supplemental Fig. S5). The 5′ NTS of the rDNA locus exhibits lower nucleosome positioning and occupancy in vitro than in vivo, suggesting that its distinctive chromatin organization arises from trans-acting factors, possibly associated with the replication machinery.
Tetrahymena exhibits stereotypical nucleosome patterns near TSSs in vivo
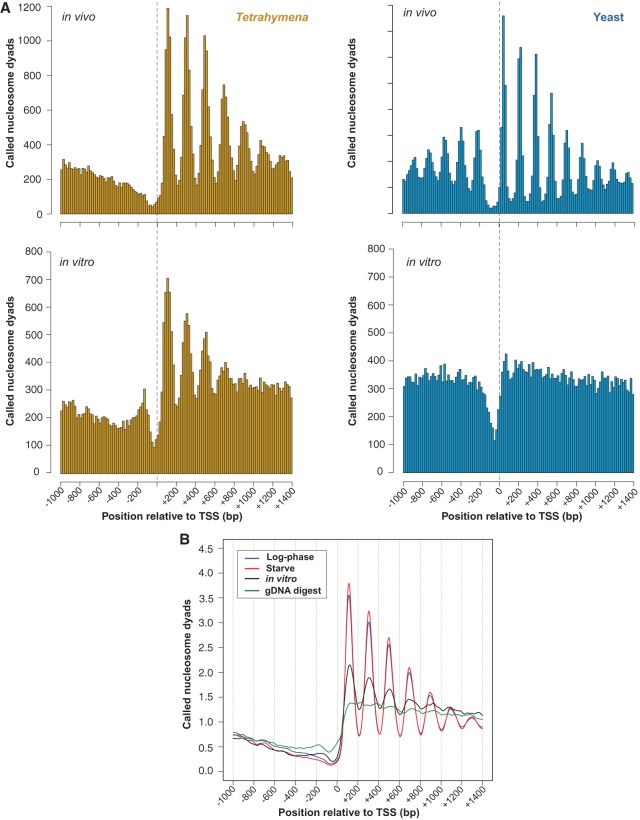
Eukaryotic nucleosome organization is most distinct near the 5′ ends of genes, where regularly spaced nucleosomes lie downstream from a nucleosome-depleted region (Yuan et al. 2005; Lee et al. 2007; Mavrich et al. 2008b; Lantermann et al. 2010; Chang et al. 2012; Chen et al. 2013b). We find that this pattern is conserved in both Tetrahymena and yeast (Fig. 1), though the regularly spaced nucleosome array lies further downstream from the TSS in Tetrahymena than yeast. The pattern in Tetrahymena is maintained between different nutritional conditions in vivo (Figs. 1B, 2A). In contrast to yeast, we do not observe nucleosome arrays upstream of Tetrahymena TSSs, the most apparent being the absence of a well-positioned −1 nucleosome. We also observe that the nucleosome-depleted region (NDR) is narrower in yeast than Tetrahymena. Notably, genes are arranged in a more compact manner in yeast, as evidenced by its significantly shorter inter-genic regions (Supplemental Fig. S6). Thus, the narrow NDR width and upstream nucleosome pattern observed in yeast might be in part explained by the presence of nearby coding sequences that exhibit well-positioned nucleosomes.
Figure 1.
In vivo-like nucleosome organization without trans-acting factors. (A) Histograms of nucleosome positions relative to the TSS were computed from yeast and Tetrahymena MNase-seq data using the same bioinformatic pipeline. A phased distribution of nucleosome positions downstream from the TSS is observed in chromatin from log-phase Tetrahymena and yeast grown in rich media. Surprisingly, an in vivo-like pattern of nucleosome positioning is observed in vitro for Tetrahymena but not yeast. (B) Averaged nucleosome dyad counts around the TSS reveal an in vivo-like distribution of called nucleosomes within in vitro data. MNase-digested naked DNA does not resemble in vivo data (green curve), thus ruling out potential sequence biases associated with MNase preferences.
Figure 2.
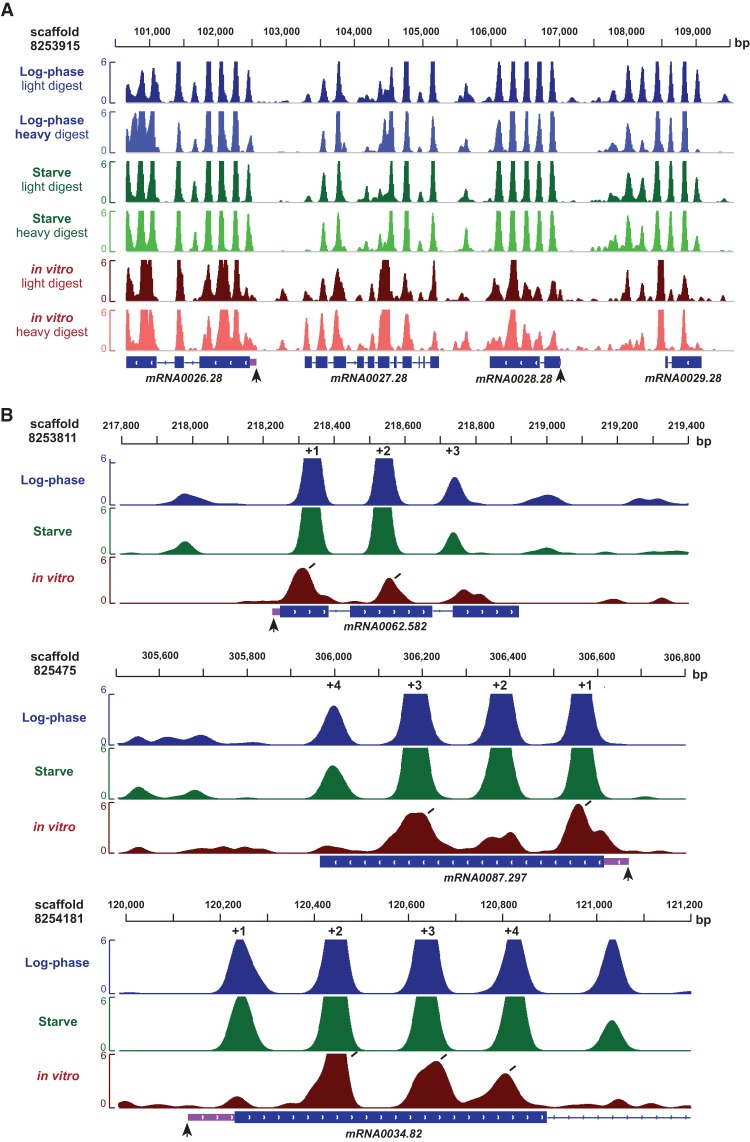
Nucleosome organization in the Tetrahymena genome. (A) Stereotypical nucleosome organization near the 5′ end of genes in a large genomic region. Vertical black arrows represent the TSS, while light purple boxes represent 5′ UTRs. (B) The in vitro nucleosome organization at individual genes resembles in vivo patterns. Diagonal black lines indicate the presence of in vitro nucleosomes at in vivo-like locations. Most genes exhibit a subset of nucleosomes at standard positions in vitro.
Aggregate analysis reveals a stereotypical nucleosome array in vitro in Tetrahymena but not yeast
We then compared the in vitro organization of nucleosomes around TSSs between Tetrahymena and yeast. Unlike the in vivo data, surprisingly we find that the in vitro nucleosome patterns were markedly different between Tetrahymena and yeast (Fig. 1). Reconstituted Tetrahymena nucleosomes preferentially occupied positions that closely resemble the in vivo pattern (Fig. 1). We also observed that in vitro nucleosome peaks were less distinct and slightly shifted downstream relative to their matching in vivo peaks. No such in vitro organization was observed in yeast.
Following this, we performed several controls to validate this unusual observation. In order to rule out the possibility that the observed nucleosome organization in vitro arose from overamplification during PCR, we removed duplicate reads from each MNase-seq data set and analyzed the distribution of nucleosome dyads around TSSs. The distinct organization of in vitro nucleosome dyads persisted even when duplicate reads were removed from MNase-seq data sets, ruling out this possibility (Supplemental Fig. S7). Our finding is also robust over a wide range of parameters used for nucleosome calling (Supplemental Fig. S8), even when using highly stringent filtering criteria. Sequencing of MNase-digested naked Tetrahymena DNA did not show such a pattern (Fig. 1B), confirming that the nucleosome pattern observed in vitro does not result from biases in MNase cleavage. We then asked whether our result is robust to variation in different aspects of the chromatin reconstitution procedure. To this end, we repeated the in vitro reconstitution experiment at different reaction volumes and at a different histone:DNA ratio (7:10, instead of the original 4:10) (Supplemental Fig. S2A). We also digested in vitro reconstituted chromatin with two different concentrations of MNase (Supplemental Fig. S2C) to assess whether the observed nucleosome organization was sensitive to the extent of digestion. The characteristic in vitro pattern is maintained across all experimental conditions tested (Supplemental Fig. S9), confirming the reproducibility of this result.
Using these data, we found that nucleosome positioning is more similar between in vivo and in vitro data sets near TSSs, compared to all other nucleosomes in the genome (Supplemental Fig. S10), reinforcing the notion that endogenous DNA sequences play an especially important role in organizing chromatin within Tetrahymena genes.
Distinct modes of nucleosome organization underlie similar in vitro and in vivo aggregate patterns
It is important to realize that aggregate analysis of genomic data can be misleading. Specifically, the fact that we observe a well-positioned nucleosome array after averaging over many genes does not necessarily imply that such an array exists in individual genes. We thus asked whether the unexpected similarity between average nucleosome patterns in vivo and in vitro also holds at the level of individual genes in Tetrahymena. To address this, we systematically measured the prevalence of positioned +1, +2, and +3 nucleosomes across the genome by analyzing nucleosome patterns in individual genes (Fig. 3). These nucleosomes are henceforth termed as “standard nucleosomes.” We note that a gene is designated as having a +1 nucleosome (for example) if there is a well-positioned nucleosome within 35 bp of the aggregate +1 position. If a gene is annotated as “lacking” a +1 nucleosome, it does not imply that there is no nucleosome occupancy at the +1 position. There could be (1) a well-positioned nucleosome 35–147 bp away, or (2) a poorly positioned nucleosome nearby, both of which could contribute to nucleosome occupancy in this region.
Figure 3.
Distinct nucleosome patterns underlie similar aggregate patterns in Tetrahymena. (A) Individual genes were annotated as either possessing or lacking standard nucleosomes at the +1, +2, and +3 positions. For example, the pattern represented by a nucleosome only at the +1 position denotes genes with a +1, but not a +2 or +3 nucleosome. Standard nucleosomes in individual genes were annotated if they lie ≤35 bp from the aggregate position. The aggregate positions are +113, +306, +498, for log-phase (in vivo); +114, +307, +497 for starve; and +122, +310, +505 for in vitro, as defined from the peak positions in Figure 1B. The dominant pattern for in vivo data sets is the full nucleosome array, while a distribution of patterns is observed for in vitro data sets. (B) Standard nucleosome positions are preferred in vitro for Tetrahymena but not yeast. For each gene, the number of nucleosomes in standard and “nonstandard” positions was calculated. Then, the ratio of the total number of standard nucleosomes to nonstandard nucleosomes was calculated across all genes. Standard nucleosomes are defined as in A. Nonstandard nucleosomes lie ≤35 bp from the midpoint between the aggregate +1/+2 and +2/+3 positions, respectively. These are calculated as (113 + 306)/2 = 210 and (306 + 498)/2 = 402 for log-phase; (114 + 307)/2 = 211 and (307 + 497)/2 = 402 for starve; (122 + 310)/2 = 216 and (310 + 505)/2 = 408 for in vitro. As expected, the standard/nonstandard nucleosome ratio is high in vivo for both organisms. However, this ratio is threefold as high in vitro for Tetrahymena compared to yeast.
Strikingly, we find that most genes possess subsets of standard nucleosomes in vitro, rather than completely recapitulating the average pattern. The majority of genes (61.8%) exhibit at least one standard nucleosome in vitro, similar to that in vivo (78.2%). In contrast, only 37.5% of genes had at least two standard nucleosomes in vitro, compared to approximately twice as many genes in vivo (66.1%). Only a minority of genes (11.2%) had nucleosomes at all three standard positions in vitro, while this was much more extensively observed in vivo (47.9%). Additionally, analysis of the genome-wide distribution of distances between individual nucleosomes did not show evidence of regular nucleosome arrays in vitro (Supplemental Fig. S4). Thus, unexpectedly, the average in vivo-like pattern that we observe in vitro is mainly explained by nucleosomes occupying various subsets of standard positions within individual genes, rather than all positions near the TSS. This is clearly observed in profiles of nucleosome organization within individual genes (Fig. 2B).
We next validated the robustness of this observation. It may be possible that a proportion of standard nucleosomes remained undetected because they were digested away during MNase treatment of reconstituted chromatin. To address this, we computed the number of genes that possess 1, 2, and 3 standard nucleosomes in reconstituted chromatin preparations that were treated with different concentrations of MNase. The frequency of genes containing standard nucleosomes is qualitatively and quantitatively similar between different MNase treatments, indicating that our results are not sensitive to the extent of MNase digestion (Supplemental Figs. S11, S12). We next annotated “fragile” and “resistant” nucleosomes in the Tetrahymena genome, based on their susceptibility to MNase digestion (see Methods). Coding regions are enriched in resistant nucleosomes in vivo and in vitro, while inter-genic regions exhibit the reverse trend (Supplemental Fig. S13). Importantly, the +1, +2, and +3 standard nucleosome positions also lie within coding regions (Supplemental Fig. S14), suggesting that they are resistant to MNase digestion. We then directly compared the number of resistant and fragile nucleosomes that occupy these standard positions and found that there are 4.5-fold as many resistant nucleosomes at these positions in vitro. This ratio increases further in vivo to 10.5-fold in log-phase cells and 52.5-fold in starved cells. Taken together, it is unlikely that the unusual nucleosome organization in vitro arises from subsets of nucleosomes being inadvertently digested away due to MNase sensitivity.
Because the bioinformatic pipeline used in this study includes a filtering step to remove poorly positioned nucleosomes (see Methods), it may be possible that subsets of standard nucleosomes were being discarded during this step. We tested this by omitting the filtering step. Yet, there was no appreciable increase in the number of genes with multiple nucleosomes in standard positions (Supplemental Fig. S15).
We also addressed whether our in vitro data are sensitive to the histone concentration used for reconstitution. To this end, we measured the prevalence of standard nucleosomes in chromatin reconstituted at a histone:DNA ratio of 7:10. Again, our results remained essentially unchanged (Supplemental Figs. S12, S15), confirming that variation in histone concentration does not alter the observed in vitro nucleosome organization. This may be attributed to the nature of the genomic DNA used in our reconstitution experiments. Since the DNA was sheared to a size of 0.85–2 kb, nucleosome long-range steric effects—normally accentuated by a high histone:DNA ratio—would be limited in this template. To sum, our results are highly consistent across all the reconstitution experiments performed (Supplemental Table S2) and are thus robust to experimental variation. We therefore conclude that two contrasting scenarios underlie the similar aggregate patterns observed in vivo and in vitro: A large proportion of individual genes possess the full array comprising +1, +2, and +3 standard nucleosomes in vivo but exhibit only subsets of this array in vitro.
Nonstandard nucleosome positions are intrinsically disfavored in Tetrahymena but not yeast
Given the relatively low frequencies of individual nucleosome patterns at standard positions in vitro in Tetrahymena (Fig. 3A), we asked whether these are sufficient to account for the notable difference between yeast and Tetrahymena aggregate nucleosome patterns. However, we find that individual in vitro nucleosome patterns at standard positions are only slightly less frequent in yeast than Tetrahymena (Supplemental Table S2), suggesting that these may not be sufficient to explain the difference in the aggregate patterns. Next, we calculated the ratio of the number of standard to nonstandard nucleosomes in yeast and Tetrahymena (Fig. 3B). We find that, while in yeast in vitro, there is no enrichment for nucleosomes in standard positions, in Tetrahymena, standard nucleosomes are enriched approximately threefold. This finding is again consistent across all Tetrahymena reconstitution experiments performed (Supplemental Fig. S16). Taken together, we conclude that, while both yeast and Tetrahymena aggregate patterns consist of a mix of different individual nucleosome organizations, only the Tetrahymena genome encodes a clear preference for standard over nonstandard nucleosome positions. This, in turn, explains its distinct in vitro aggregate nucleosome pattern.
GC-rich sequences underlie DNA-guided nucleosomes in Tetrahymena genes
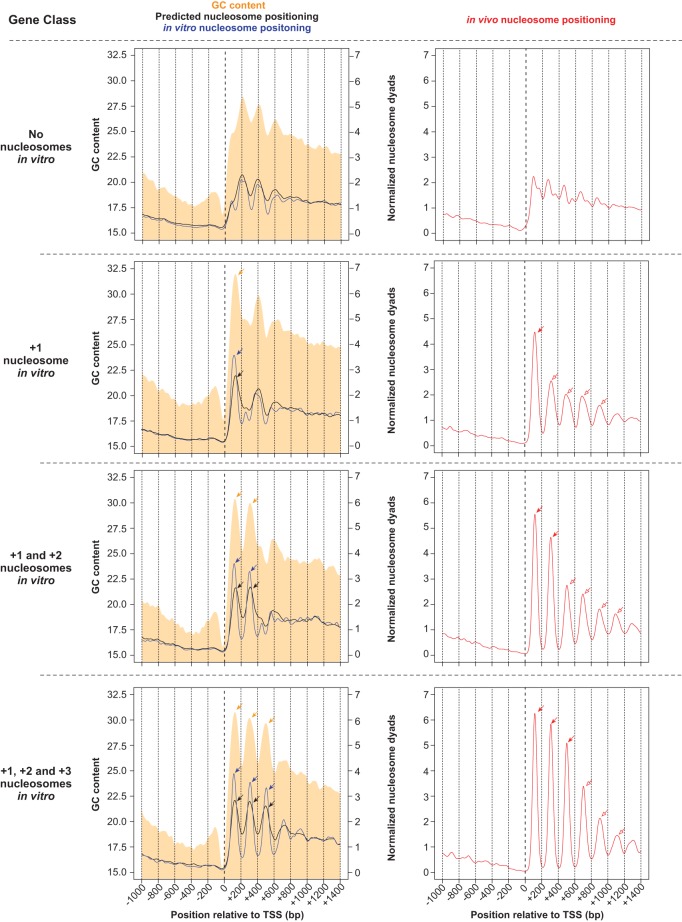
Since in vitro nucleosomes were reconstituted in the absence of trans-acting factors, we asked what DNA sequence preferences of nucleosomes underlie their stereotypical distribution near TSSs in vitro. GC content has previously been identified as a major component of such sequence preferences (Tillo and Hughes 2009). In particular, AT-rich sequences, such as poly(dA:dT) tracts, are refractory to nucleosome formation (Nelson et al. 1987; Suter et al. 2000; Field et al. 2008; Segal and Widom 2009). Similar to other eukaryotes, we observe a decrease in GC content at TSSs, coinciding with nucleosome-depleted regions in vitro and in vivo (Fig. 4). Due to the low histone:DNA concentration used for reconstitution (4:10), the size of sheared DNA used in reconstitution (0.85–2 kb), and our observation that subsets of standard nucleosome positions are occupied in vitro, we conjectured that local sequence features specifically located downstream from the TSS could underlie nucleosome organization in vitro, rather than previously suggested statistical concentration-based nucleosome positioning effects (Kornberg and Stryer 1988; Mavrich et al. 2008a). We thus examined the nucleotide composition of individual genes whose in vitro nucleosome maps show in vivo-like nucleosome organization. Notably, these genes exhibit oscillations in GC content downstream from the TSS, with an average amplitude of 3%–4% GC and a period of ∼200 bp, coincident with standard nucleosome positions (Fig. 4). The data collectively suggest that GC content oscillations within Tetrahymena coding sequences may contribute to regularly spaced nucleosomes in vitro and in vivo.
Figure 4.
Standard in vitro nucleosomes coincide with GC content oscillations and are associated with increased nucleosome positioning in vivo. Tetrahymena genes were classified according to the number of standard in vitro nucleosomes downstream from their TSS. Nucleosome positioning data are obtained from in vitro (blue line) and in vivo (red line) experiments, as well as from predictions of a thermodynamic model formulated by Kaplan et al. (2009) (black line). Log-phase MNase-seq data were used as the in vivo sample. GC content is represented as a filled orange curve. Different gene classes are separated by horizontal dotted lines. The nucleosome-depleted region upstream of standard nucleosomes coincides with GC-poor DNA. Pronounced peaks in GC content (orange arrows) exhibit a ∼200-bp periodicity, and coincide with nucleosome positions in vitro (blue arrows). This is consistent with GC-rich DNA being intrinsically favorable for nucleosome formation. Genes with no standard nucleosomes in vitro (top row) exhibit an indistinct nucleosome pattern in vivo (right panel). On the other hand, genes with a +1 nucleosome in vitro (blue arrow within left panel) exhibit increased nucleosome positioning in vivo, not only at the +1 position (red filled arrow), but also around this region (red open arrows). A model based on nucleosome sequence preferences successfully predicts in vitro nucleosome positions (black arrows), which in turn overlap with in vivo nucleosomes (red filled arrows). However, the model fails to predict in vivo nucleosomes in surrounding regions (red open arrows), suggesting that such nucleosomes are instead positioned by trans-acting factors. These trends are also observed in other gene classes, with varying numbers of nucleosomes in vitro. DNA sequences favorable for nucleosome formation may thus function as nucleation sites that aid trans-acting factors in positioning nucleosomes in flanking regions in vivo.
Next, we asked whether species-specific variation in the DNA affinity of histone octamers (Allan et al. 2013) underlies the in vitro pattern observed uniquely in Tetrahymena. We addressed this by comparing Tetrahymena nucleosome sequence preferences to those previously measured by in vitro reconstitution of chicken nucleosomes on yeast DNA. We find in vitro that the average nucleosome occupancies of nucleotide 5-mers correlate well between Tetrahymena and yeast (Spearman ρ = 0.93) (Supplemental Fig. S17). We also observe ∼10-bp periodic dinucleotide patterns within Tetrahymena nucleosomes (Supplemental Fig. S18), similar to previous analyses of yeast and human nucleosomes (Kaplan et al. 2009; Gaffney et al. 2012). Finally, we used a previously published thermodynamic model (Kaplan et al. 2009), trained on the same yeast data set, to predict nucleosome positioning in Tetrahymena. We find that the genome-wide distribution of nucleosome dyads is similar between the in vitro data set and predictions from the model (Spearman ρ = 0.69). These data together argue that the observed differences in nucleosome organization in vitro between Tetrahymena and yeast likely arise from distinct DNA sequence features encoded within each genome (Fig. 5), rather than species-specific DNA sequence preferences of Tetrahymena and yeast histone octamers. However, we cannot entirely rule out contributions from the latter possibility to the establishment of in vivo-like nucleosome patterns in vitro.
Figure 5.
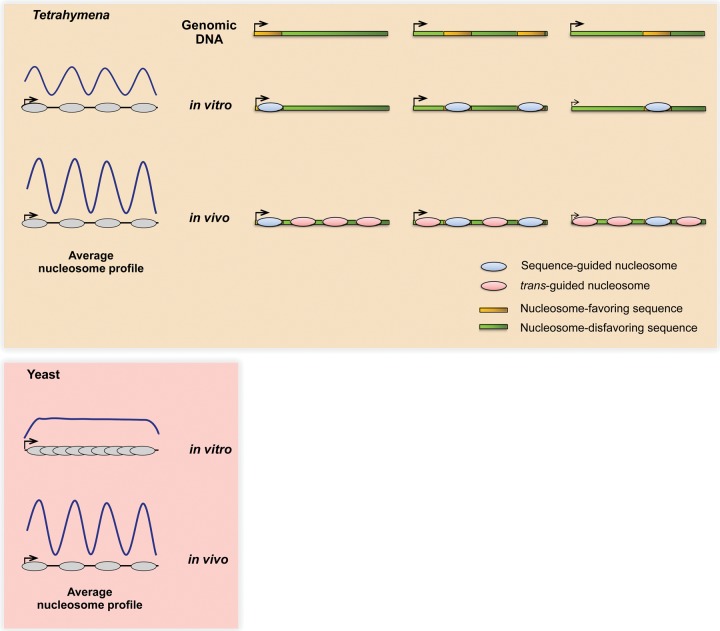
Contrasting mechanisms may underlie conserved nucleosome patterns in vivo between Tetrahymena and yeast. The Tetrahymena genome is GC-poor and is generally unfavorable for nucleosome formation. The majority of Tetrahymena genes encode nucleosome-favoring sequences at subsets of standard positions downstream from TSSs, which might in turn facilitate nucleosome positioning in and around these regions in vivo. On the other hand, yeast genes generally show no such DNA-guided specificity near TSSs, instead relying mainly on trans-acting factors to generate the distinctive nucleosome organization in vivo. As a result, the average in vitro and in vivo nucleosome patterns appear similar in Tetrahymena but not yeast.
Sites containing DNA-guided nucleosomes exhibit greater in vivo positioning in their vicinity
We then addressed the in vivo consequences of encoding only a subset of standard nucleosomes in the Tetrahymena genome. Curiously, genes with a DNA-guided nucleosome at a standard position in vitro exhibited more distinct in vivo nucleosome positioning, at and around this location (Fig. 4). For example, genes with a +1 nucleosome in vitro were not only significantly more likely to possess a +1 nucleosome (P < 2.2 × 10−16) but also a +2 (P < 2.2 × 10−16) and +3 nucleosome in vivo (P < 2.2 × 10−16, all with Fisher's exact test). Similarly, those with a +2 nucleosome in vitro were more likely to exhibit +1 (P = 6.27 × 10−15), +2 (P < 2.2 × 10−16), and +3 nucleosomes in vivo (P < 2.2 × 10−16, all with Fisher's exact test). Conversely, genes without any standard nucleosomes in vitro lacked the regular pattern in vivo (Fig. 4). These results may suggest that DNA-guided nucleosomes—observed at standard positions in vitro—act as nucleation sites to position adjacent nucleosomes in vivo, possibly through packing effects or the action of chromatin remodelers. We then annotated DNA-guided and trans factor-guided nucleosomes throughout the genome by comparing the in vivo and in vitro nucleosome maps (see Methods). DNA-guided nucleosomes are identified from their presence both in vivo and in vitro, while trans factor-guided nucleosomes are identified as being present in vivo but not in vitro. We find that DNA-guided nucleosome positions are more robust to prolonged MNase digestion (Supplemental Fig. S19A). Such nucleosomes also exhibit smaller changes in translational positions between different environmental conditions and are more strongly positioned in vivo than trans factor-guided nucleosomes (Supplemental Fig. S19A). These properties of DNA-guided nucleosomes hold true, not only at standard positions near TSSs, but across the entire genome (Supplemental Fig. S19B).
Sequences underlying DNA-guided nucleosomes bias codon usage and amino acid composition
Given our findings that some standard nucleosome positions are encoded by endogenous DNA sequences within genes, we asked how this feature is reconciled with other sequence constraints, such as the genetic code and the GC-poor nucleotide composition of the genome. Indeed, the GC content oscillations associated with DNA-guided nucleosome positioning in vitro overlap extensively with coding sequences, given the short 5′ UTRs of Tetrahymena genes (Supplemental Fig. S14). We quantified the impact of nucleosome sequence preferences on codon composition by comparing the GC content of each of the three nucleotide positions within codons that are found within DNA-guided nucleosomes versus the corresponding nucleotide positions in codons that are found within trans factor-guided nucleosomes. We find that codons that overlap DNA-guided nucleosomes exhibited significantly higher GC content at all three positions (P < 2.2 × 10−16, Fisher's exact test, for each position, respectively) (Supplemental Table S3) than trans factor-guided nucleosomes. This enrichment in GC-rich codons results in deviations in amino acid composition mainly arising from the first and second codon positions, as well as deviations in synonymous codon usage from the third (wobble) position (Supplemental Tables S4, S5). Indeed, six out of the eight GC-rich codons (100% GC) are elevated in frequency within DNA-guided nucleosomes, while six out of the eight AT-rich codons (0% GC) exhibit lower frequency in these regions. Thus, local variation in GC content—which likely underlies DNA-guided nucleosome patterns in vivo—imposes biases in amino acid composition and codon usage within genes.
Discussion
In this study, we present genome-wide in vivo and in vitro nucleosome maps of the ciliate Tetrahymena thermophila. These maps not only constitute a comprehensive resource for further studies of ciliate chromatin (Coyne et al. 2012) but also provide novel insight into nucleosome positioning mechanisms within genes and allude to their impact on genome evolution. The stereotypical nucleosome array that has been previously observed near transcription start sites in aggregate plots remains somewhat of a mystery. This organization has been observed in diverse eukaryotes (Yuan et al. 2005; Mavrich et al. 2008b; Lantermann et al. 2010; Ponts et al. 2010; Chen et al. 2013b; Zhang et al. 2014), suggesting that it is established by a widely conserved mechanism. However, the functional relationship between the stereotypical nucleosome array and gene transcription is unclear, since even highly expressed genes exhibit this nucleosome organization (Shivaswamy et al. 2008; Lantermann et al. 2010). Paradoxically, such nucleosomes lie within coding regions near the TSS and should thus present a significant barrier to the passage of RNA polymerase II (Teves et al. 2014). Furthermore, recent experiments in yeast demonstrated that in vivo ATP-dependent chromatin remodelers, rather than nucleosome sequence preferences, are mainly responsible for this organization (Zhang et al. 2009; Gkikopoulos et al. 2011). It has also been suggested that the stereotypical nucleosome array downstream from TSSs arises as a byproduct of a conserved process such as transcriptional elongation, which recruits ATP-dependent chromatin remodelers such as Chd1 and Isw1 (Hughes et al. 2012; Struhl and Segal 2013). Our results in Tetrahymena may suggest that the distinct nucleosome organization may, in fact, be more than a byproduct. Our finding that some of the nucleosomes in these stereotypical arrays are guided by the underlying DNA is unexpected per se; yet, even more striking is that they are encoded at specific stereotypical positions amid coding sequences. Because the genetic code is highly constrained, encoding any additional information in parallel can potentially affect both codon and amino acid usage. Indeed, we demonstrate that both codon and amino acid usage are skewed at the positions where DNA-guided nucleosomes are positioned, possibly alluding to their importance.
In previous studies, the stereotypical nucleosome array was mostly studied as a pattern averaged over many genes. This may have been a reasonable mode of analysis, since it reduces measurement noise associated with individual genes. Furthermore, individual genes in previously studied eukaryotes do indeed exhibit the array in vivo, consistent with the average pattern. Unexpectedly, this is not the case in Tetrahymena. Given our surprising observation that the stereotypical nucleosome pattern is present in the averaged pattern in vivo and in vitro, we chose to perform further analysis at the level of individual genes. We found that the ostensibly similar aggregate in vivo and in vitro nucleosome patterns are, in fact, explained by contrasting nucleosome patterns in individual genes. While, on average, the whole array is apparent in vitro, we found that individual genes mostly exhibit only subsets of these stereotypically arranged nucleosomes in vitro. These DNA-guided nucleosomes exhibit less variation in translational positioning between environmental conditions, are flanked by well-positioned nucleosomes in vivo, and are more strongly positioned than nucleosomes guided by trans-acting factors. We propose that these DNA-guided “seed” nucleosomes may contribute to nucleosome array formation within Tetrahymena genes—which are GC-poor and thus intrinsically unfavorable for nucleosome formation—while minimizing the consequences on protein-coding sequences (Fig. 5). Given that well-positioned nucleosomes exhibit different mutation rates from free DNA (Radman-Livaja and Rando 2010; Chen et al. 2012), it is tempting to speculate that seed nucleosomes conserve their own localization. They may thus act as protected nucleation sites to facilitate array formation in flanking regions in vivo, together with the help of trans-acting factors. Indeed, the notion of seed nucleosomes promoting in vivo nucleation of nucleosome arrays has been proposed in the human genome (Valouev et al. 2011) and could be a general mechanism for organizing chromatin within some eukaryotic genomes.
In conclusion, we find that nucleosome sequence preferences and trans-acting factors work together in a previously unreported fashion and extent in Tetrahymena to establish the distinctive nucleosome pattern in genes. These forces may function in concert with epigenetic marks such as DNA methylation, which disfavor nucleosome formation (Huff and Zilberman 2014). The arising evolutionary implications leave open the question of how distinct nucleosome positioning mechanisms operate in the context of numerous other regulatory codes enmeshed within the genome, including the maintenance of transcription factor binding sites (Stergachis et al. 2013), translational efficiency (Fredrick and Ibba 2010), mRNA splicing fidelity (Parmley et al. 2006; Parmley and Hurst 2007), and secondary structure (Shabalina et al. 2006).
Methods
Cell culture
One liter of Tetrahymena thermophila wild-type strain SB210 (Tetrahymena stock center) was grown in 1×SPP at 30°C with shaking at 100 rpm to a log-phase density of ∼35 × 104 cells/mL. The cell density matched that used by a recently published Tetrahymena RNA-seq study (Xiong et al. 2012). To obtain starved samples, the cells were centrifuged at 1100g for 2 min, resuspended in 1.75 volumes of 10 mM Tris pH 7.5, and incubated at 25°C without shaking for 15 h.
Purification of macronuclei and MNase digestion in vivo
Macronuclei from log-phase or starved cells were isolated and digested with MNase, essentially as described in Jacob et al. (2004), with minor modifications. The detailed procedure is included in Supplemental Methods.
The MNase-digested in vivo chromatin sample with ∼80% mononucleosomal and ∼20% dinucleosomal DNA (Supplemental Fig. S2A) was labeled “light digest.” This is in accordance with previous recommendations for an adequate level of MNase digestion in nucleosome mapping studies (Zhang and Pugh 2011). Separately, the sample exhibiting almost exclusively mononucleosomal DNA with a significant smear in the subnucleosomal region (Supplemental Fig. S2A) was labeled “heavy digest.” For each nutritional condition (log-phase and starved), we prepared: (A) fixed, lightly digested, and (B) native, heavily digested, each with two biological replicates. We omitted formaldehyde in preparation B, in order to expose the chromatin to increased perturbation. We identified nucleosome positions that were invariant (“resistant”) or labile (“fragile”) between preparations A and B. We describe the annotation of resistant and fragile nucleosomes in the “Nucleosome calling” section below.
Light and heavy digest samples were run on a 2% agarose-TAE gel, and the mononucleosome-sized fragment was excised and purified using a QIAquick gel extraction kit (Qiagen). Illumina libraries were prepared from mononucleosomal DNA according to the manufacturer's instructions and subjected to paired-end sequencing.
Separately, undigested gDNA was sheared on a Covaris LE220 and then directly used for Illumina library preparation, according to the manufacturer's instructions. The sheared gDNA libraries were then subjected to single-read sequencing. The sheared genomic DNA-seq data was used solely for the estimation of chromosome copy number, as described in the “Sequencing data processing pipeline” section below.
Chromatin reconstitution and MNase digestion in vitro
Genomic DNA for reconstitution experiments was prepared from macronuclei of starved Tetrahymena cells through phenol-chloroform extraction, ethanol precipitation, and subsequent RNase treatment. Genomic DNA was then sheared to 850 bp–2 kb using a Covaris LE220. This size range is in accordance with previously published in vitro reconstitution experiments (Valouev et al. 2011). Sheared DNA was end-repaired with DNA polymerase I (NEB), T4 DNA polymerase (NEB), and T4 polynucleotide kinase (NEB), and then subjected to phenol-chloroform extraction and ethanol precipitation before resuspension in nuclease-free water.
Tetrahymena histones were acid-extracted from macronuclei (Wiley et al. 2000), refolded into octamers, and assembled on genomic DNA by salt-gradient dialysis (Luger et al. 1999). Details of all reconstitution experiments—including set A/B designations and MNase digest conditions—are provided in the Supplemental Methods. Unless otherwise noted, the representative in vitro data used for all figures and tables in this study are obtained from set B, light MNase digest (with replicates pooled).
MNase digestion of Tetrahymena gDNA
12.9 μg macronuclear gDNA was made up to 200 μL with TMS, and then digested with 4.74 Kunitz units of MNase (NEB) for 7 min at 25°C. This is depicted in the “++” MNase lane, in Supplemental Figure S2D. The reaction was terminated by adding 112 μL stop buffer (300 mM NaCl, 30 mM Tris pH 8, 75 mM EDTA, 1.5% [w/v] SDS, 1.5 mg/mL Proteinase K), and subsequently purified through phenol-chloroform extraction and ethanol precipitation. MNase-digested gDNA was resuspended in buffer EB (Qiagen) and loaded on a 2% agarose-TAE gel, and the mononucleosome-sized fragment was gel-purified using a QIAquick gel extraction kit (Qiagen). Illumina libraries were then prepared from gDNA according to the manufacturer's instructions and subjected to paired-end sequencing.
Sequencing data processing pipeline
All Illumina sequencing data sets are summarized in Supplemental Table S1. Replicates of respective in vitro and in vivo data sets were pooled for downstream analyses. Raw MNase-seq and genomic DNA-seq reads were quality-trimmed (minimum quality score = 20) and length-filtered (minimum length = 40nt) using Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010) before mapping with Bowtie 2 (Langmead and Salzberg 2012) to the October 2008 build of the Tetrahymena SB210 reference genome (Eisen et al. 2006) using standard settings. Only complete Tetrahymena chromosomes in the genome assembly were included in downstream analyses. For all MNase-seq data sets presented in this study, we calculated the per-bp coverage of nucleosome dyads from the midpoints of 122- to 172-bp fragments (see Supplemental Methods for discussion on fragment size selection). The data were normalized by relative chromosome copy number (as obtained from RPKM of sheared genomic DNA-seq reads for each chromosome) and the whole genome average coverage value. Normalized values were then smoothed with a Gaussian filter of standard deviation = 15. We refer to the resulting values as normalized nucleosome dyads. Data from MNase-digested genomic DNA were processed in the same way as all other MNase-seq data sets.
Nucleosomal dinucleotide frequencies
To strengthen the dinucleotide periodicity signal, we restricted our analysis to MNase-seq read pairs that spanned exactly 147 bp in length. AA/TT/TA/AT dinucleotide frequencies within nucleosomal DNA were calculated as previously described (Kaplan et al. 2009).
Nucleosome calling
Nucleosomes were called from normalized nucleosome dyad data according to Kaplan et al. (2010b), with minor modifications. Details of this procedure are included in Supplemental Methods. Called nucleosomes (peaks) were then filtered according to two previously suggested metrics (Kaplan et al. 2010a): absolute nucleosome positioning and conditional nucleosome positioning. Absolute nucleosome positioning was defined as the number of MNase-seq read centers (normalized by chromosome copy number and the genome-wide average value) that correspond to a particular peak. Conditional nucleosome positioning was defined as the normalized number of read centers that lie within 21 bp of the called nucleosome peak, divided by the normalized number of read centers that lie within 147 bp of the peak. To construct the histograms in Figure 1 and Supplemental Figure S7, ∼35% of originally called nucleosomes were first removed using a stringent filter of minimum absolute positioning and conditional positioning. For all other analyses and figures, nucleosomes with absolute positioning <0.26 and conditional positioning <0.21 were first omitted, resulting in the removal of ∼15% of peaks. These filtering steps removed poorly positioned nucleosomes from the respective data sets.
We annotated sequence-guided and trans-guided nucleosomes by comparing the in vivo and in vitro nucleosome maps. A nucleosome in vivo is classified as “DNA-guided” if it lies within 10 bp of a nucleosome in vitro. On the other hand, a nucleosome in vivo that lies >73 bp from a nucleosome in vitro is classified as “trans factor-guided.”
Separately, we annotated resistant and fragile nucleosomes by comparing the in vivo nucleosome maps of lightly and heavily digested chromatin. A nucleosome in vivo is annotated as “resistant” if there is <10 bp difference in position between the light and heavy digest condition. On the other hand, a nucleosome in vivo is annotated as “fragile” if there is no called nucleosome in the heavy digest condition that is within 73 bp of the corresponding nucleosome in the light digest condition.
Standard nucleosomes were annotated for each gene at the +1, +2, and +3 positions, respectively. A gene was designated as having a standard nucleosome if there was a called nucleosome with absolute positioning ≥0.26 and conditional positioning ≥0.21, that is ≤35 bp from the corresponding aggregate position.
Nucleosome model
To predict nucleosome positioning from DNA sequence, we used the thermodynamic model of Kaplan et al. (2009), which was trained on MNase-seq data measured on chicken histones that were reconstituted onto yeast DNA. We used the same concentration and temperature parameters as used previously. In order to produce a track that is comparable to our smoothed dyad track, we applied a Gaussian filter (standard deviation = 15 bp) to the track of nucleosome start probabilities given by the model and shifted it by 73 bp.
Representative MNase-seq data sets used in figures and tables
Unless otherwise stated, we examined data sets from lightly digested chromatin for all figures and tables, with individual replicates pooled. The in vitro sample corresponds to chromatin from experimental set B (see Supplemental Methods for details). All yeast MNase-seq data used in this work were obtained from a previous study by Kaplan et al. (2009) (GEO accession number GSE13622).
Data access
All sequencing data generated for this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE64061.
Supplementary Material
Acknowledgments
This paper is dedicated to the late Jonathan Widom, whose work inspired L.Y.B. and N.K. We thank Job Dekker, Jason Lieb, Richard Bártfai, Nicole Francis, Galia Debelouchina, Jaspreet Khurana, and Geoff Dann for feedback and comments on the manuscript; Robert Coyne for sharing Tetrahymena gene annotations; David Robinson for advice on MNase-seq read subsampling; Jingmei Wang for general laboratory support; and Jessica Wiggins, Wei Wang, and Donna Storton for assistance with Illumina sequencing. We also thank the three anonymous reviewers for valuable comments. This study was supported by a Princeton Centennial Fellowship to L.Y.B., Human Frontier Science Program Long Term Fellowship LT000706/2012 to N.K., National Institutes of Health (NIH) grant GM59708 to L.F.L., and NSF-1158346 award to R. Coyne.
Author contributions: L.Y.B. conceived the project and performed experimental and bioinformatic analysis for all figures and tables. N.K. provided advice, analysis, and critical interpretations of bioinformatic data. M.M.M. and T.W.M. contributed feedback, core protocols, and equipment for in vitro reconstitution experiments. L.F.L. provided guidance and feedback. L.Y.B. and N.K. prepared the manuscript, which all authors edited.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.188516.114.
References
- Allan J, Fraser RM, Owen-Hughes T, Docherty K, Singh V. 2013. A comparison of in vitro nucleosome positioning mapped with chicken, frog and a variety of yeast core histones. J Mol Biol 425: 4206–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 19: 19.10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GS, Noegel AA, Mavrich TN, Müller R, Tomsho L, Ward E, Felder M, Jiang C, Eichinger L, Glöckner G, et al. 2012. Unusual combinatorial involvement of poly-A/T tracts in organizing genes and chromatin in Dictyostelium. Genome Res 22: 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen Z, Chen H, Su Z, Yang J, Lin F, Shi S, He X. 2012. Nucleosomes suppress spontaneous mutations base-specifically in eukaryotes. Science 335: 1235–1238. [DOI] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. 2013a. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RA-J, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. 2013b. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Stover NA, Miao W. 2012. Whole genome studies of Tetrahymena. Methods Cell Biol 109: 53–81. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Ibba M. 2010. How the sequence of a gene can tune its translation. Cell 141: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. 2012. Controls of nucleosome positioning in the human genome. PLoS Genet 8: e1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA, Glover C, Johmann CA, Keevert JB, Mathis DJ, Samuelson M. 1978. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb Symp Quant Biol 42 Pt (1): 493–503. [DOI] [PubMed] [Google Scholar]
- Huff JT, Zilberman D. 2014. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156: 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. 2012. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Stout AR, Price CM. 2004. Modulation of telomere length dynamics by the subtelomeric region of Tetrahymena telomeres. Mol Biol Cell 15: 3719–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Hughes TR, Lieb JD, Widom J, Segal E. 2010a. Contribution of histone sequence preferences to nucleosome organization: proposed definitions and methodology. Genome Biol 11: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E. 2010b. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17: 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L. 1988. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res 16: 6677–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK. 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Strålfors A, Yuan G-C, Ekwall K, Korber P. 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244. [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008a. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008b. Nucleosome organization in the Drosophila genome. Nature 453: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson HC, Finch JT, Luisi BF, Klug A. 1987. The structure of an oligo(dA)·oligo(dT) tract and its biological implications. Nature 330: 221–226. [DOI] [PubMed] [Google Scholar]
- Palen TE, Cech TR. 1984. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell 36: 933–942. [DOI] [PubMed] [Google Scholar]
- Parmley JL, Hurst LD. 2007. Exonic splicing regulatory elements skew synonymous codon usage near intron-exon boundaries in mammals. Mol Biol Evol 24: 1600–1603. [DOI] [PubMed] [Google Scholar]
- Parmley JL, Chamary JV, Hurst LD. 2006. Evidence for purifying selection against synonymous mutations in mammalian exonic splicing enhancers. Mol Biol Evol 23: 301–309. [DOI] [PubMed] [Google Scholar]
- Piña B, Brüggemeier U, Beato M. 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60: 719–731. [DOI] [PubMed] [Google Scholar]
- Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, Hicks GR, Hardiman G, Lonardi S, Le Roch KG. 2010. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res 20: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. 2010. Nucleosome positioning: How is it established, and why does it matter? Dev Biol 339: 258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. 2009. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov NA. 2006. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res 34: 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, Reynolds A, Raubitschek A, Ziegler S, LeProust EM, Akey JM, et al. 2013. Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342: 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E. 2013. Determinants of nucleosome positioning. Nat Struct Mol Biol 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Schnappauf G, Thoma F. 2000. Poly(dA·dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res 28: 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Weber CM, Henikoff S. 2014. Transcribing through the nucleosome. Trends Biochem Sci 39: 577–586. [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley EA, Mizzen CA, Allis CD. 2000. Isolation and characterization of in vivo modified histones and an activity gel assay for identification of histone acetyltransferases. Methods Cell Biol 62: 379–394. [DOI] [PubMed] [Google Scholar]
- Xiong J, Lu X, Zhou Z, Chang Y, Yuan D, Tian M, Zhou Z, Wang L, Fu C, Orias E, et al. 2012. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS One 7: e30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G-C, Liu Y-J, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Pugh BF. 2011. High-resolution genome-wide mapping of the primary structure of chromatin. Cell 144: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF. 2011a. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 21: 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. 2011b. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, Pauli A, van Hummelen P, Schier AF, Liu XS. 2014. Canonical nucleosome organization at promoters forms during genome activation. Genome Res 24: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.