Kramer et al. discovered that circularization of the Drosophila laccase2 gene is regulated by both intronic repeats and trans-acting splicing factors. Unlike the previously characterized Muscleblind (Mbl) circular RNA, which requires the Mbl protein for its biogenesis, Laccase2 circular RNA levels are not controlled by Mbl or the Laccase2 gene product but rather by multiple hnRNP and SR proteins acting in a combinatorial manner.
Keywords: Laccase2, circRNA, pre-mRNA splicing, noncoding RNA, repetitive element, PlexA, hnRNP
Abstract
Thousands of eukaryotic protein-coding genes are noncanonically spliced to produce circular RNAs. Bioinformatics has indicated that long introns generally flank exons that circularize in Drosophila, but the underlying mechanisms by which these circular RNAs are generated are largely unknown. Here, using extensive mutagenesis of expression plasmids and RNAi screening, we reveal that circularization of the Drosophila laccase2 gene is regulated by both intronic repeats and trans-acting splicing factors. Analogous to what has been observed in humans and mice, base-pairing between highly complementary transposable elements facilitates backsplicing. Long flanking repeats (∼400 nucleotides [nt]) promote circularization cotranscriptionally, whereas pre-mRNAs containing minimal repeats (<40 nt) generate circular RNAs predominately after 3′ end processing. Unlike the previously characterized Muscleblind (Mbl) circular RNA, which requires the Mbl protein for its biogenesis, we found that Laccase2 circular RNA levels are not controlled by Mbl or the Laccase2 gene product but rather by multiple hnRNP (heterogeneous nuclear ribonucleoprotein) and SR (serine–arginine) proteins acting in a combinatorial manner. hnRNP and SR proteins also regulate the expression of other Drosophila circular RNAs, including Plexin A (PlexA), suggesting a common strategy for regulating backsplicing. Furthermore, the laccase2 flanking introns support efficient circularization of diverse exons in Drosophila and human cells, providing a new tool for exploring the functional consequences of circular RNA expression across eukaryotes.
It was long assumed that eukaryotic pre-mRNAs are always canonically spliced to generate a linear mRNA that is subsequently translated to produce a protein. However, it is now becoming increasingly clear that many genes can be noncanonically spliced to produce circular RNAs with covalently linked ends (for review, see Wilusz and Sharp 2013; Jeck and Sharpless 2014; Lasda and Parker 2014; Chen and Yang 2015; Ebbesen et al. 2015; Wilusz 2015). These transcripts are almost exclusively derived from exons, accumulate in the cytoplasm, and are thought to be products of alternative splicing events known as “backsplicing.” In contrast to canonical splicing, which joins the exons in a linear order (joining exon 1 to exon 2 to exon 3, etc.), backsplicing joins a splice donor to an upstream splice acceptor (e.g., joining the 3′ end of exon 2 to the 5′ end of exon 2). A handful of RNAs generated in this manner were identified in the 1990s (Nigro et al. 1991; Cocquerelle et al. 1993; Zaphiropoulos 1997), and recent deep sequencing studies have expanded this observation to thousands of circular RNAs expressed across eukaryotes, including humans (Salzman et al. 2012; Jeck et al. 2013; Memczak et al. 2013; Guo et al. 2014; Conn et al. 2015; Kelly et al. 2015), mice (Memczak et al. 2013; Rybak-Wolf et al. 2015; You et al. 2015), Caenorhabditis elegans (Memczak et al. 2013; Ivanov et al. 2015), Drosophila (Salzman et al. 2013; Ashwal-Fluss et al. 2014; Westholm et al. 2014), Schizosaccharomyces pombe (Wang et al. 2014), and plants (Wang et al. 2014; Ye et al. 2015). Perhaps surprisingly, for some genes, the abundance of the circular RNA exceeds that of the associated linear mRNA by a factor of 10 (Salzman et al. 2012; Jeck et al. 2013), suggesting that the major function of some protein-coding genes may be to generate circular RNAs.
Most exons in eukaryotic genomes have splicing signals at both ends and theoretically can circularize. However, only certain exons are observed in circular RNAs, and these backsplicing events often occur in a tissue-specific manner (Salzman et al. 2013; Guo et al. 2014; Westholm et al. 2014). This suggests that circular RNA biogenesis is tightly regulated. As splicing generally occurs cotranscriptionally (for review, see Brugiolo et al. 2013), most introns, along with their upstream splice acceptors (which are needed for backsplicing), are rapidly removed. Therefore, for circular RNAs to be produced, canonical splicing likely must occur more slowly around these exons (Ashwal-Fluss et al. 2014), and/or exon skipping events may be coupled to circular RNA biogenesis (Zaphiropoulos 1997; Surono et al. 1999; Barrett et al. 2015; Kelly et al. 2015). In the latter, the circular RNA is derived from an exon-containing lariat, allowing a pre-mRNA to yield both a linear mRNA and a circular RNA comprised of the skipped exons.
There is little known about the splicing factors that regulate these events. In some cases, the Muscleblind (Mbl) and Quaking proteins appear to facilitate backsplicing by bridging between two introns and causing the splice sites from the intervening exons to be brought into close proximity (Ashwal-Fluss et al. 2014; Conn et al. 2015). For example, circular RNA production from the Drosophila mbl gene is triggered when the Mbl splicing factor binds to its own introns (Ashwal-Fluss et al. 2014). However, in humans, mice, and C. elegans, the predominant determinants of whether a pre-mRNA is subjected to backsplicing are intronic repetitive elements, such as sequences derived from transposons (for review, see Wilusz 2015). Almost 90% of human circular RNAs have complementary Alu elements in their flanking introns (Ivanov et al. 2015), and, analogous to the protein-bridging mechanism, base-pairing between complementary sequences allows the intervening splice sites to be brought close together (Dubin et al. 1995; Jeck et al. 2013; Liang and Wilusz 2014; Zhang et al. 2014; Ivanov et al. 2015; Li et al. 2015). Interestingly, repeats <40 nucleotides (nt) can drive circular RNA production in human cells (Liang and Wilusz 2014), but it is clear that more than simple thermodynamics regulates circularization. For example, base-pairing interactions can be disrupted by ADAR (adenosine deaminase acting on RNA), which converts adenosines in double-stranded regions to inosines (Ivanov et al. 2015; Rybak-Wolf et al. 2015). In addition, most mammalian pre-mRNAs contain multiple intronic repeats, allowing distinct circular (or linear) RNAs to be produced depending on which repeats base-pair to one another (Zhang et al. 2014). Therefore, other factors likely help dictate splicing outcomes by regulating these exon circularization events.
Despite key regulatory roles for intronic repeats in multiple eukaryotes, it has been suggested that circular RNA biogenesis in Drosophila melanogaster is not driven by base-pairing interactions (Westholm et al. 2014). Instead, a positive correlation between the length of the flanking introns and circular RNA abundance was identified in Drosophila (Westholm et al. 2014). However, the effect of modulating intron lengths on backsplicing has not yet been directly addressed. It is also completely unknown how Drosophila circular RNAs besides Mbl, of which there are >2500 annotated circular RNAs derived from other genomic loci (Salzman et al. 2013; Ashwal-Fluss et al. 2014; Westholm et al. 2014), are generated or post-transcriptionally regulated. Therefore, it is still unclear whether circular RNA biogenesis strategies are conserved across eukaryotes or whether species such as Drosophila use unique mechanisms to determine which exons should be backspliced.
Once produced, circular RNAs are stable transcripts that are naturally resistant to degradation by exonucleases. Two circular RNAs (ciRS7/CDR1as and Sry) modulate the activity of specific microRNAs (Hansen et al. 2013; Memczak et al. 2013), but most other RNA circles (in species other than Drosophila) (Westholm et al. 2014) contain few microRNA-binding sites and likely function differently (Guo et al. 2014). For example, it has been proposed that many circular RNAs may regulate neuronal functions (Westholm et al. 2014; Rybak-Wolf et al. 2015; You et al. 2015), and artificial circular RNAs containing an IRES (internal ribosome entry site) can be translated (Chen and Sarnow 1995; Wang and Wang 2015). However, the lack of efficient methods for modulating circular RNA levels or ectopically expressing circular RNAs (for review, see Petkovic and Muller 2015) has limited our ability to define functions for these transcripts.
Here, we focused on the Drosophila laccase2 gene, as it produces an abundant circular RNA in vitro and in vivo. We provide evidence that intronic repeats collaborate with trans-acting splicing factors to regulate circularization in flies. Mechanistically, we found that miniature introns (<150 nt) containing the splice sites and inverted repeats were sufficient to support Laccase2 circular RNA production. The intronic repeats must base-pair to one another for circularization to occur, as has been observed in other eukaryotes. Furthermore, we found that the strength of these base-pairing interactions dictates whether backsplicing occurs co- or post-transcriptionally: Long flanking repeats appear to allow cotranscriptional processing. Screening a panel of genes, we found that multiple hnRNP (heterogeneous nuclear ribonucleoprotein) and SR (serine–arginine) family proteins regulate Laccase2 circular RNA levels in a combinatorial manner. Comparisons with the mbl locus suggest that the circularization mechanisms are distinct, as the Laccase2 circular RNA was not regulated by the Mbl or Laccase2 gene products. We then identified additional circular RNAs that are regulated by unique combinations of hnRNP and SR proteins, suggesting that combinatorial control may be a common regulatory strategy that modulates circular RNA levels. This led us to test whether this biogenesis mechanism is active in human cells, and we found that the laccase2 introns can indeed robustly generate circular RNAs. It is thus now possible to efficiently generate “designer” circular RNAs in cells with minimal linear RNA production. In total, our results reveal new insights into how trans-acting factors and intronic repeats collaborate to regulate circular RNA biogenesis across eukaryotes as well as provide new tools for exploring the functions of circular RNAs.
Results
The Drosophila laccase2 gene produces an abundant circular RNA
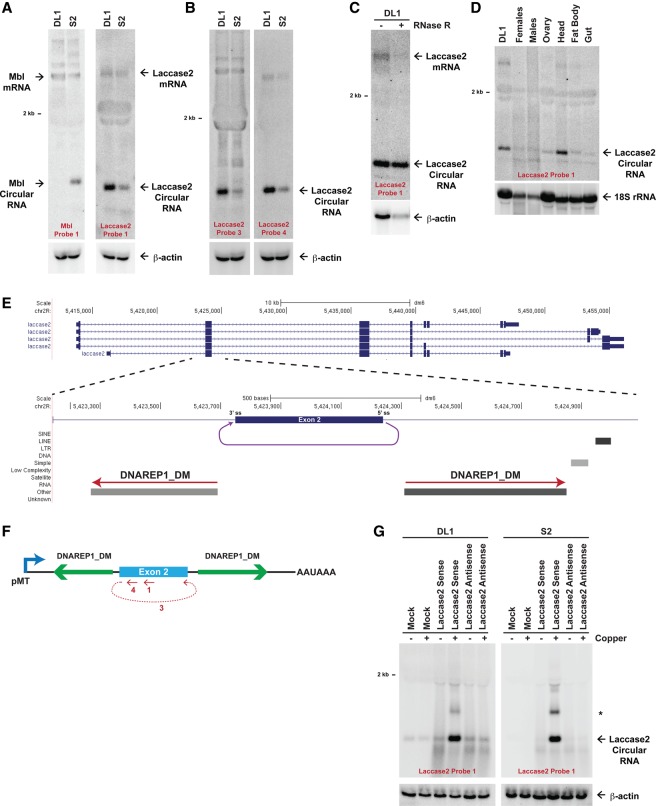
To characterize the mechanisms by which circular RNAs are generated in flies, we first determined whether Drosophila S2 and DL1 cell culture lines express a panel of previously identified circular RNAs (Ashwal-Fluss et al. 2014; Westholm et al. 2014). Northern blots detected the Mbl circular RNA (652 nt) only in S2 cells (Fig. 1A, left), whereas the Plexin A (PlexA) circular RNA (1437 nt) (Supplemental Fig. 1A) and a 490-nt transcript derived from the laccase2 gene were detected in S2 and DL1 cells (Fig. 1A, right). Notably, unlike the PlexA locus, the 490-nt Laccase2 transcript was more abundant (approximately fivefold and ∼2.5-fold in DL1 and S2 cells, respectively) than the linear Laccase2 mRNA, which encodes an enzyme implicated in cuticle formation and pigmentation (Futahashi et al. 2011; Jacobs et al. 2015). Using RT–PCR (Supplemental Fig. 1B) and additional Northern blot probes, including one (probe 3) that spans the backspliced junction (Fig. 1B; Supplemental Fig. 1C), we confirmed that the transcript is a circular RNA that is generated when the 5′ splice site at the end of exon 2 is joined to the 3′ splice site at the beginning of exon 2 (Fig. 1E). Furthermore, the Laccase2 circular RNA was resistant to digestion by the 3′–5′ exonuclease RNase R, unlike the linear Laccase2 or β-actin mRNAs (Fig. 1C). Upon examining Laccase2 expression in adult flies, we determined that the circular RNA is predominately expressed in D. melanogaster heads (Fig. 1D), which is consistent with the known enrichment of circular RNAs in neural tissues (Ashwal-Fluss et al. 2014; Westholm et al. 2014).
Figure 1.
The D. melanogaster laccase2 gene generates a circular RNA. (A) Twenty micrograms of total RNA from DL1 and S2 cells was subjected to Northern blot analysis and probed for Mbl and Laccase2 expression. β-Actin was used as a loading control. (B) The Laccase2 circular RNA was detected with multiple oligonucleotide probes, including one complementary to the backspliced junction (probe 3). (C) The Laccase2 circular RNA is resistant to RNase R digestion. (D) Eleven micrograms of total RNA from adult D. melanogaster tissues was probed for Laccase2 expression. 18S ribosomal RNA was used as a loading control. (E) Exon/intron structure of the D. melanogaster laccase2 locus, highlighting a 1945-nt region that includes exon 2. A circular RNA is formed when the 5′ splice site at the end of exon 2 is joined to the 3′ splice site at the beginning of exon 2 (purple). Repetitive elements in the designated orientations are shown. (F) The 1945-nt region of the Laccase2 pre-mRNA was cloned downstream from the metallothionein promoter to generate the Laccase2 sense plasmid. The regions targeted by Northern oligonucleotide probes are denoted in red. (G) Plasmids containing the laccase2 region in the sense or antisense orientation were transfected into DL1 (left) or S2 (right) cells, and Northern blots were performed. Endogenous Laccase2 circular RNA expression was observed in the mock-treated samples. (*) Concatenated and/or intertwined circular RNA. β-Actin was used as a loading control.
Base-pairing between complementary intronic repeats is sufficient for Laccase2 circular RNA production
Although most introns in Drosophila are <150 nt in length (Lim and Burge 2001), circular RNAs are usually generated from exons that are flanked by long introns (Westholm et al. 2014). Indeed, >9-kb introns are present upstream of and downstream from exon 2 of the Drosophila laccase2 gene (Fig. 1E). Within these intronic sequences, we identified a pair of inverted DNAREP1_DM family transposons that are located very close to the circularizing exon (Fig. 1E). These repeats are highly complementary to each other (75% identical over a 106-nt region and 69% identical over a separate, nonoverlapping 272-nt region) (Supplemental Fig. 2A) but are not conserved in most Drosophila species, including Drosophila yakuba, which encode the laccase2 gene. As Northern blots failed to detect the Laccase2 circular RNA in the heads of D. yakuba (Supplemental Fig. 2B), we hypothesized that base-pairing between the intronic repeats promotes Laccase2 circular RNA production in D. melanogaster.
To test this model, we first asked whether relatively short introns are in fact sufficient to support Laccase2 circularization. A 1945-nt region of the D. melanogaster Laccase2 pre-mRNA, spanning from 607 nt upstream of exon 2 to 848 nt downstream from exon 2, was cloned in the sense or antisense orientation downstream from the metallothionein promoter (pMT) (Fig. 1F). Following transient transfection of DL1 or S2 cells with the plasmids, CuSO4 was added to induce pMT transcription, and total RNA was isolated 14 h later. Only after CuSO4 induction was circular RNA expression from the Laccase2 sense plasmid observed by Northern blots (approximately ninefold and >100-fold over endogenous levels in DL1 and S2 cells, respectively) (Fig. 1G). The transcript was further detected using multiple oligonucleotide probes, including one that spans the spliced junction (Supplemental Fig. 2C), confirming that the RNA generated from our plasmid is backspliced. As the 490-nt circular RNA was the predominant transcript that accumulates in cells (Fig. 1G), we conclude that RNAs from these plasmids backsplice at a high efficiency. In addition to the 490-nt transcript, we also observed some concatenated and/or intertwined (∼1 kb in length) circular RNAs (marked by asterisk in Fig. 1G).
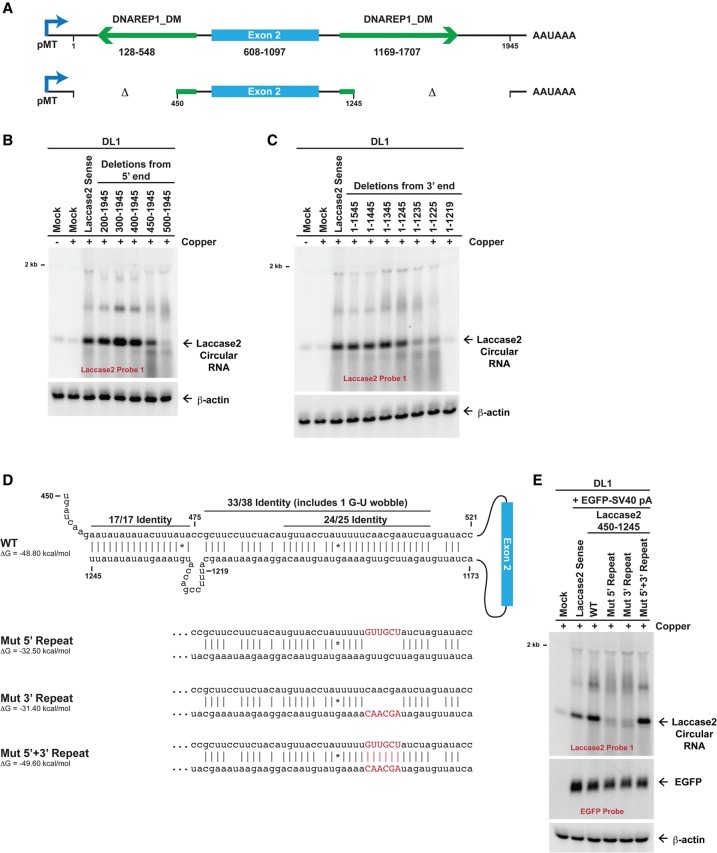
Having shown that the intronic regions immediately flanking exon 2 are sufficient for Laccase2 circular RNA production, we next identified the minimal sequence elements required (Fig. 2A). By progressively deleting nucleotides from the 5′ (Fig. 2B) or 3′ (Fig. 2C) ends, we determined that most of the upstream and downstream DNAREP1_DM repeats are dispensable for circularization. However, completely deleting either of the repeats largely eliminated circular RNA production from the plasmids. Further analysis revealed that ∼100 nt of each repeat (beginning at nucleotide 450 for the upstream repeat and ending around nucleotide 1245 for the downstream repeat) is sufficient for efficient circularization (Fig. 2A–C). As these minimal repeat regions are highly complementary to one another over an ∼55-nt region (Fig. 2D; Supplemental Fig. 2A), we tested the effect of disrupting base-pairing between the two introns. Circularization was not observed when six of the base pairs were disrupted (Mut 5′ repeat and Mut 3′ repeat) (Fig. 2D,E). When base-pairing was re-established by introduction of mutations in both repeats (Mut 5′ + 3′ repeat), efficient Laccase2 circular RNA production was again observed (Fig. 2E). As cotransfection of an EGFP expression plasmid confirmed equal transfection efficiency across samples (Fig. 2E), we conclude that base-pairing between the intronic DNAREP1_DM repeats is necessary for Laccase2 circularization. In addition, these results parallel our prior findings in human cells that short complementary regions (∼40 nt) are sufficient to promote backsplicing and exon circularization (Liang and Wilusz 2014).
Figure 2.
Base-pairing between intronic repeats facilitates Laccase2 circular RNA production. (A) Numbering scheme for the Laccase2 sense expression plasmid. The minimal sufficient introns that support circularization (450–1245 plasmid) are shown at the bottom. (B,C) Laccase2 expression plasmids containing deletions at their 5′ ends (B) or 3′ ends (C) were transfected into DL1 cells, and CuSO4 was added for 14 h where indicated. Northern blots were subsequently performed. (D) Mutations in the repeats (denoted in red) were introduced into the Laccase2 nucleotide 450–1245 expression plasmid. mFold was used to calculate hairpin stabilities, assuming a 7-nt linker (AGAAUUA) between the repeats. (E) An EGFP expression plasmid and Laccase2 expression plasmids containing wild-type (WT) or mutant repeats were transfected into DL1 cells. CuSO4 was then added for 14 h, and Northern blots were performed.
Multiple hnRNP and SR proteins regulate Laccase2 circular RNA levels
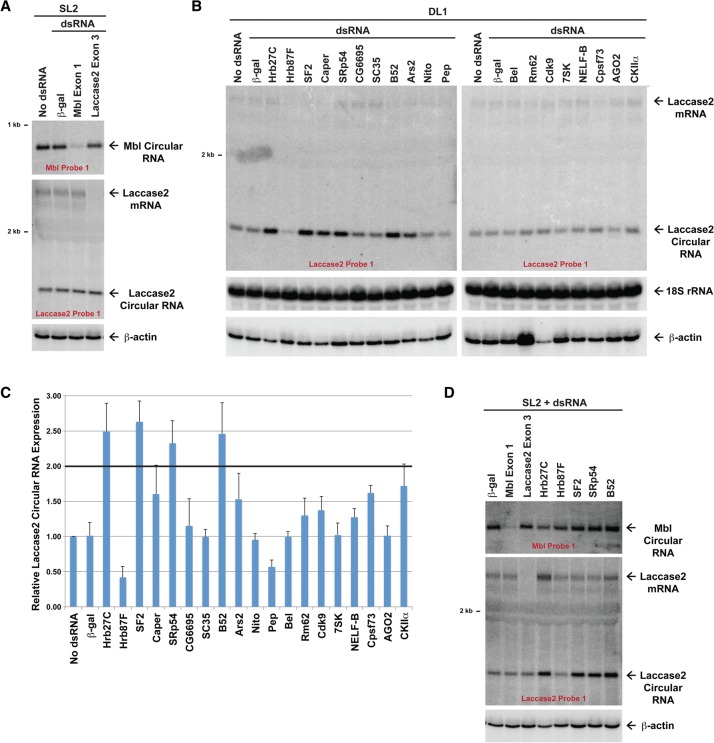
Recent work has begun to identify trans-acting factors that regulate exon circularization, including the RNA-editing enzyme ADAR1 (Ivanov et al. 2015; Rybak-Wolf et al. 2015), the RNA-binding protein Quaking (Conn et al. 2015), and the splicing factor Mbl (Ashwal-Fluss et al. 2014). Production of the fly Mbl circular RNA is tightly controlled in cis by the Mbl protein via an elegant feedback loop, and overexpression of this splicing factor results in increased Mbl circular RNA production (Ashwal-Fluss et al. 2014). However, it is unclear whether other circular RNAs are controlled via Mbl or analogous cis-acting feedback loops. We first validated that knockdown of Mbl using dsRNA targeting the first exon of Mbl (which is not included in the Mbl circular RNA) significantly reduces Mbl circular RNA levels in SL2 cells (Fig. 3A, top). In contrast, expression of the Laccase2 mRNA and Laccase2 circular RNA was unaffected by Mbl knockdown in either SL2 (Fig. 3A, bottom) or DL1 cells (Supplemental Fig. 3A). This suggests that Laccase2 circularization is regulated in a manner distinct from the mbl locus. Indeed, no direct feedback mechanism exists at the laccase2 locus, as knockdown of the Laccase2 mRNA (using dsRNA that targets exon 3) had no effect on Laccase2 circular RNA levels (Fig. 3A, bottom).
Figure 3.
Multiple hnRNPs and SR proteins regulate Laccase2 circular RNA expression. (A) Drosophila SL2 cells were treated with the indicated dsRNAs for 4 d, and Northern blots were performed to analyze expression of the endogenous mbl and laccase2 loci. Knockdown of the linear Mbl transcript caused depletion of the Mbl circular RNA but had no effect on Laccase2 circular RNA levels. (B) Northern blots were used to examine Laccase2 expression in DL1 cells that had been treated with 4 μg of the indicated dsRNAs for 3 d. Representative blots are shown. (C) Laccase2 circular RNA levels were quantified using ImageQuant from three independent experiments and were normalized to the “no dsRNA” samples. Data are shown as mean ± SD. (D) Northern blots were used to examine Mbl expression in SL2 cells that had been treated with the indicated dsRNAs for 4 d.
To identify regulators of Laccase2 circularization, we screened a panel of proteins with well-established roles in transcriptional elongation (e.g., 7SK and NELF-B), RNA splicing (e.g., hnRNP and SR proteins), and other RNA processing events (e.g., helicases or 3′ end cleavage factors). Each protein was individually knocked down in DL1 cells for 3 d using dsRNAs, and expression of the endogenous Laccase2 circular RNA was measured using Northern blots (Fig. 3B,C). Although only minor changes were observed when most proteins were knocked down, depletion of three SR proteins (SF2 [SRSF1], SRp54 [SRSF11], and B52 [SRSF6]) each caused Laccase2 circular RNA levels to reproducibly increase by greater than twofold (t-test, P < 0.01). This indicates that these factors may act to repress the Laccase2 circular RNA. The hnRNP protein Hrb27C likewise acts to inhibit Laccase2 circular RNA accumulation, whereas Hrb87F acts to enhance circularization (t-test, P < 0.01). Quantitative PCR (qPCR) was used to validate that each dsRNA depleted its target gene by >80% (Supplemental Fig. 3B), and we confirmed that an independent, nonoverlapping dsRNA against each factor caused similar effects on Laccase2 circular RNA levels (Supplemental Fig. 3A). In stark contrast, the levels of the Mbl circular RNA were largely unaffected by modulating the expression of these hnRNP and SR proteins (Fig. 3D, top; Supplemental Fig. 3C). This indicates that distinct sets of factors regulate circularization from the mbl and laccase2 loci.
Combinatorial control of circular RNA levels by hnRNPs and SR proteins
In their well-characterized roles, hnRNPs and SR proteins direct pre-mRNA splicing patterns through site-specific binding to target RNAs (for review, see Caceres and Kornblihtt 2002; Busch and Hertel 2012). Each exon (along with its flanking introns) contains a unique set of hnRNP- and SR protein-binding sites, each of which aids or blocks spliceosome assembly. Therefore, there is combinatorial control over pre-mRNA splicing patterns, which helps ensure that only the desired RNA isoforms are ultimately produced (for review, see Smith and Valcarcel 2000). We were thus interested in whether Laccase2 circular RNA levels were similarly controlled in a combinatorial manner (Fig. 4A). Simultaneous depletion of Hrb27C with SF2, SRp54, or B52 resulted in additive increases in Laccase2 circular RNA expression (Fig. 4A, lanes 10–12), suggesting that each of these factors plays a nonredundant role. In contrast, simultaneous depletion of SRp54 and B52 did not result in any further increase in Laccase2 circle levels (Fig. 4A, lane 15) compared with depletion of either factor alone (Fig. 4A, lanes 8,9).
Figure 4.
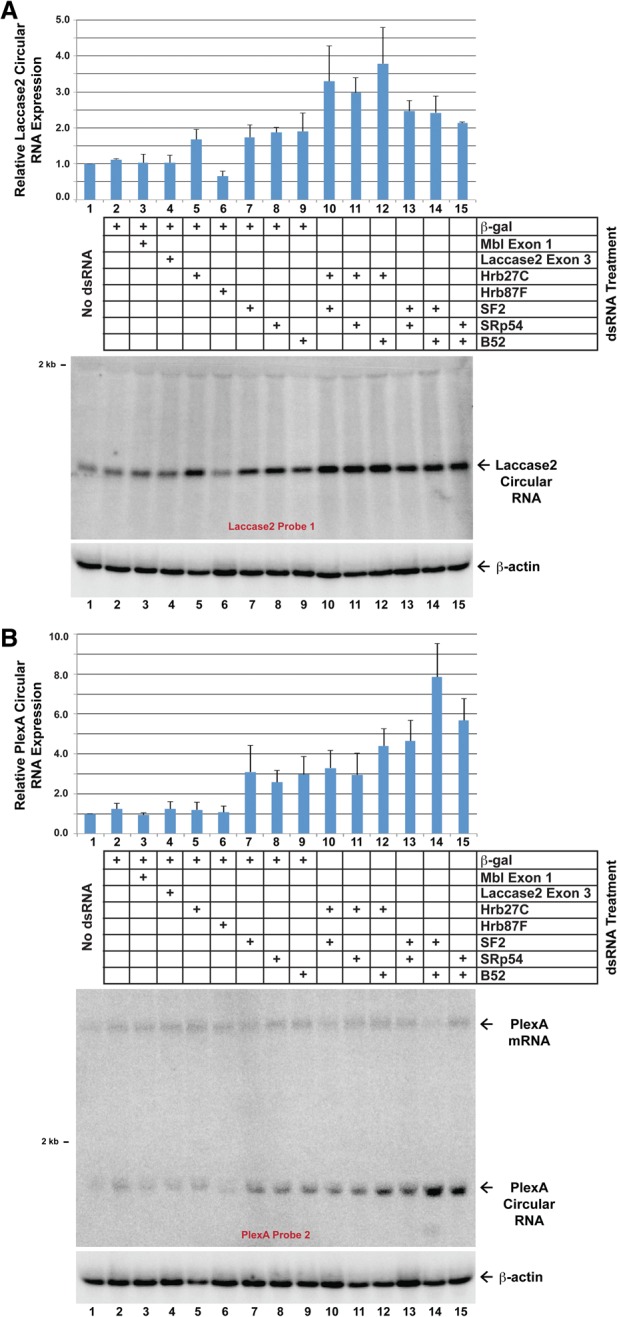
Combinatorial control of circular RNA levels by hnRNPs and SR proteins. DL1 cells were treated with the indicated pairs of dsRNAs (2 μg of each) for 3 d. Northern blots were then used to examine the expression of the endogenous Laccase2 (A) and PlexA (B) circular RNAs. Representative blots are shown. Circular RNA levels were quantified using ImageQuant from three independent experiments and were normalized to the “no dsRNA” samples. Data are shown as mean ± SD.
Since we demonstrated that the biogenesis of the Laccase2 circular RNA was dependent on base-pairing between the introns (Fig. 2), we set out to test whether other circular RNAs flanked by such intronic structures are regulated by similar proteins. Complementary DNAREP1_DM repeats flank the PlexA circular RNA, which is expressed in DL1 and S2 cells (Supplemental Fig. 1A). Whereas depletion of Hrb27C or Hrb87F did not alter PlexA circular RNA levels (Fig. 4B, lanes 5–6), knockdown of SF2, SRp54, or B52 each caused PlexA circular RNA levels to significantly increase (Fig. 4B, lanes 7–9). PlexA expression is also regulated in a combinatorial manner, as simultaneous depletion of B52 with SF2 or SRp54 resulted in additive increases in PlexA circular RNA levels (Fig. 4B, lanes 14–15). This suggests that backsplicing and circular RNA levels may generally be regulated in a combinatorial manner via the combined activities of intronic repeats, hnRNPs, and SR proteins.
Using qPCR, we found six other circular RNAs that are regulated by Hrb27C, Hrb87F, SF2, SRp54, and/or B52 (Supplemental Fig. 4). The Uex and Dbp80 (exon 5/6) circular RNAs appear to be regulated similarly to the Laccase2 circular RNA, whereas other circular RNAs are only regulated by a subset of these factors. This is consistent with each gene containing a unique set of SR protein- and hnRNP-binding sites that dictate splicing outcomes. Interestingly, none of the nine circular RNAs examined were significantly regulated by Mbl (Supplemental Fig. 4), which suggests that the mbl locus may be regulated by a unique mechanism.
The flanking introns likely dictate whether circular RNA production occurs co- or post-transcriptionally
Introns are generally removed as a gene is being transcribed (for review, see Brugiolo et al. 2013), but the introns that flank exons that circularize appear to be spliced more slowly (Ashwal-Fluss et al. 2014). Nevertheless, conflicting data have been reported on whether backsplicing occurs co- or post-transcriptionally. In Drosophila, circular RNAs can be detected in the chromatin-bound nascent RNA fraction, suggesting that they are generated cotranscriptionally (Ashwal-Fluss et al. 2014). However, human expression plasmids lacking a downstream 3′ end processing signal [e.g. poly(A) signal] failed to generate the ZKSCAN1 circular RNA, suggesting that circularization may at least in part occur post-transcriptionally (Liang and Wilusz 2014).
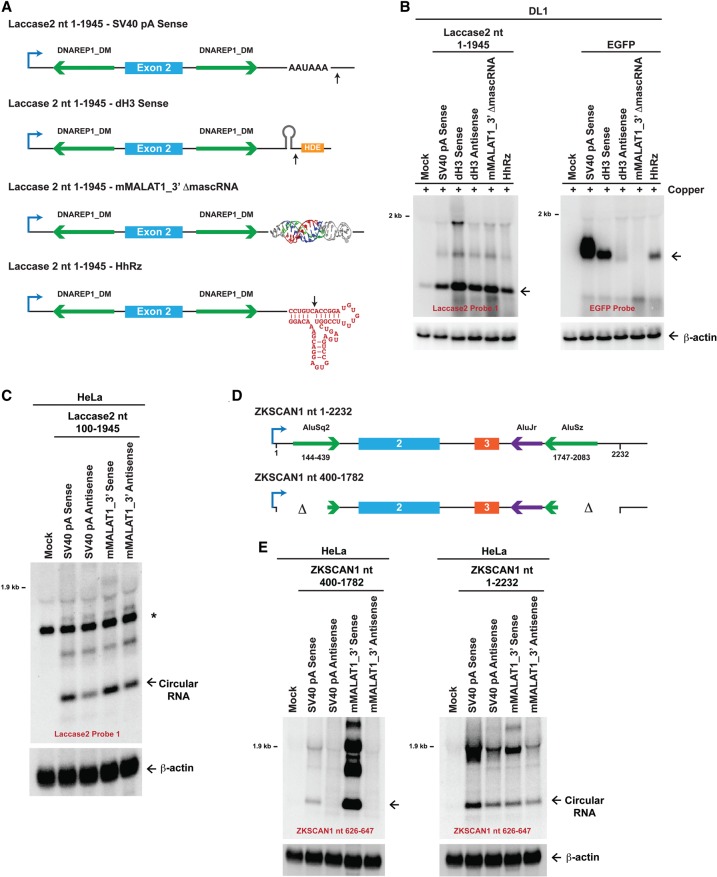
To clarify the mechanism of circular RNA biogenesis, we took advantage of our Drosophila expression plasmids and replaced the downstream SV40 poly(A) signal with other sequences that are uniquely regulated (Fig. 5A). In particular, we inserted (1) the histone 3′ end processing signals (Drosophila histone H3 [dH3]) in the sense orientation, which direct cleavage downstream from a stem–loop structure (for review, see Marzluff et al. 2008); (2) the histone 3′ end processing signals (dH3) in the antisense orientation, which are not cleaved; (3) the MALAT1 triple helix without the downstream tRNA-like structure (denoted mMALAT1_3′ ΔmascRNA), which fails to be cleaved (Wilusz et al. 2012; Brown et al. 2014); or (4) the self-cleaving hammerhead ribozyme (HhRz) (Fig. 5A; Haseloff and Gerlach 1988). When each of these sequences was placed downstream from the EGFP ORF, we confirmed that only the SV40 poly(A), dH3 sense, and HhRz sequences supported EGFP mRNA 3′ end formation and stability (Fig. 5B, right). No EGFP mRNA was observed with the dH3 antisense or mMALAT1_3′ ΔmascRNA sequences. This indicates that there are likely no functional cryptic poly(A) signals or other 3′ processing signals present in our plasmids.
Figure 5.
The flanking introns dictate whether 3′ end processing is required for exon circularization. (A) Schematics of Drosophila Laccase2 expression plasmids. The complete SV40 poly(A) (pA) signals, which include the AAUAAA sequence, were replaced with dH3 3′ end processing signals, the MALAT1 triple helix, or the HhRz. Whereas the dH3 sense sequence is cleaved by CPSF73, and HhRz self-cleaves, the mMALAT1_3′ ΔmascRNA sequence (nucleotides 6581–6690 of mouse MALAT1) lacks the tRNA-like structure and is unable to be cleaved by RNase P (Wilusz et al. 2012). (B) Laccase2 (left) or EGFP (right) expression plasmids ending in the designated sequences were transfected into DL1 cells. CuSO4 was added, and Northern blots were then performed. (C) HeLa cells were transfected with mammalian expression plasmids containing laccase2 nucleotides 100–1945 followed by differing 3′-terminal sequences. Unlike in A, the mMALAT1_3′ region (nucleotides 6581–6754 of mouse MALAT1) was inserted, which includes the tRNA-like structure. When present in the sense orientation, the mMALAT1_3′ region is recognized and cleaved by RNase P (Wilusz et al. 2012). (*) A nonspecific band that is also present in mock-treated cells. (D) Schematics of human ZKSCAN1 expression plasmids (Liang and Wilusz 2014). (E) Plasmids containing ZKSCAN1 nucleotides 400–1782 (left) or ZKSCAN1 nucleotides 1–2232 (right) followed by differing 3′-terminal sequences were transfected into HeLa cells, and Northern blots were performed.
When we then analyzed how these various 3′-terminal sequences affected processing of the Laccase2 pre-mRNA (Fig. 5A), we surprisingly observed that all of the plasmids generated high levels of Laccase2 circular RNA (Fig. 5B, left). Therefore, 3′ end processing is not required for circularization in Drosophila cells, which is distinct from what was observed with human ZKSCAN1 expression plasmids (Liang and Wilusz 2014). We thus asked whether these conflicting results were due to species-specific regulation or differences in the transcripts tested. The fly Laccase2 pre-mRNA was inserted into a mammalian expression plasmid upstream of (1) the SV40 poly(A) signal or (2) the MALAT1 triple helix along with the downstream tRNA-like structure (which is cleaved by RNase P) (Fig. 5C). When either of these 3′-terminal sequences is placed in the antisense orientation, it is not cleaved (Wilusz et al. 2012). As was observed in DL1 cells (Fig. 5B), significant amounts of the Laccase2 circular RNA were generated in HeLa cells regardless of whether 3′ end processing occurred (Fig. 5C). While the antisense plasmids did generate less circular RNA, these results are consistent with exon circularization occurring cotranscriptionally in human cells. Furthermore, it is clear that the Drosophila laccase2 introns can support exon circularization in human cells (Fig. 5C; below), and thus the circular RNA biogenesis machinery recognizes similar features in both human and fly cells.
Why, then, is 3′ end processing required for circular RNA production in certain contexts? One key difference between the Laccase2 transcript and the previously examined ZKSCAN1 transcript is that the Laccase2 transcript contains ∼500 nt of inverted intronic repeats (Fig. 2A), while the ZKSCAN1 nucleotide 400–1782 transcript has only ∼40 nt of repeats (Fig. 5D,E, left; Liang and Wilusz 2014). We thus hypothesized that extending the length of the ZKSCAN1 flanking introns may alter the timing of biogenesis. Indeed, including an additional ∼260 nt of intronic repeats allowed the ZKSCAN1 nucleotide 1–2232 transcript to produce circular RNAs independently of a 3′ end processing signal (Fig. 5E, right). Therefore, the length of the flanking intronic repeats dictates the timing of circularization, with long repeats (∼300 nt or longer) likely promoting cotranscriptional backsplicing.
The laccase2 intronic repeats support circularization of many long exons in fly cells
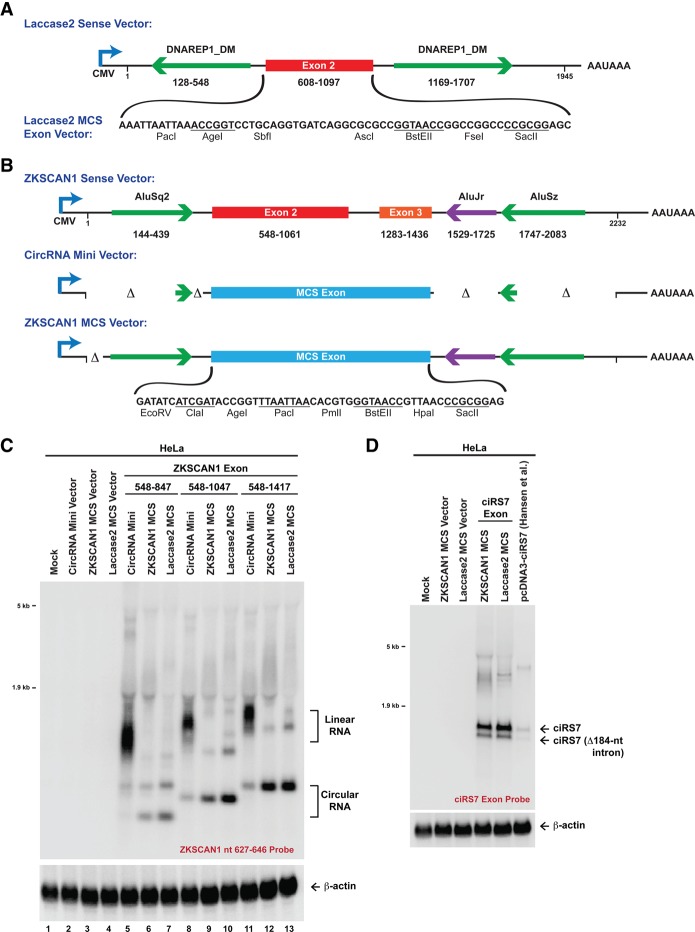
It is still largely unclear how much of a role exonic sequences play in circular RNA formation. Therefore, we tested whether the laccase2 exon 2 sequence impacts Laccase2 circular RNA production. First, we tested length requirements. For these studies, we inserted an artificial 57-nt exon composed of a multiple cloning site (MCS) between the laccase2 introns (Fig. 6A). This “Laccase2 MCS exon” vector failed to generate a circular RNA when transfected into DL1 cells, but reinsertion of exon 2 of laccase2 (nucleotides 608–1097) between the KpnI and XmaI sites restored efficient circular RNA production (Fig. 6B,C). Inserting smaller regions of the laccase2 exon revealed that only sequences ≥300 nt circularized efficiently (e.g., Fig. 6B [nucleotides 608–907], C [nucleotides 808–1097]). This is consistent with previous computational analyses (Jeck et al. 2013; Zhang et al. 2014) as well as experiments in humans (Liang and Wilusz 2014) and S. pombe (Barrett et al. 2015) that revealed a positive correlation between exon length and circularization efficiency.
Figure 6.
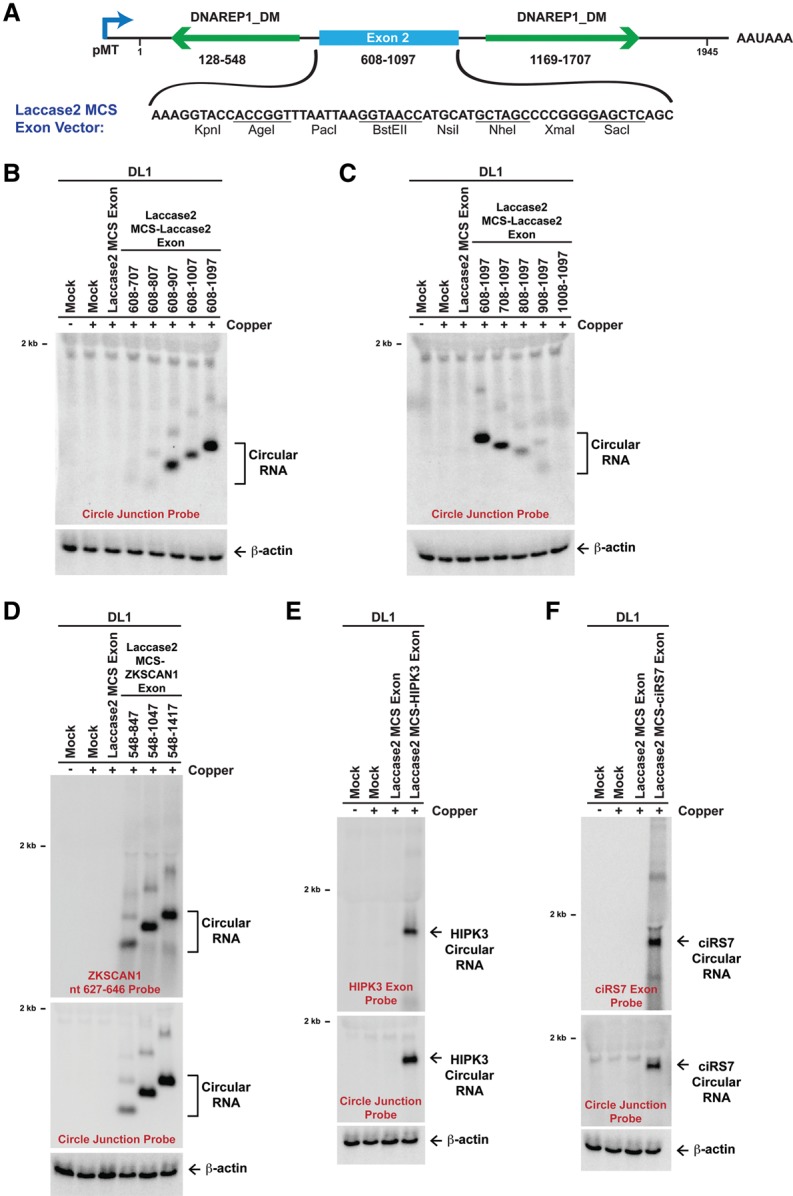
The laccase2 introns facilitate circularization of diverse exons in fly cells. (A) To facilitate the identification of exon sequences that can be circularized in Drosophila, exon 2 of the Laccase2 sense expression plasmid was replaced with an artificial 57-nt exon composed of restriction enzyme sites. (B,C) Segments of laccase2 exon 2 (numbering scheme as in A) were inserted between the KpnI and XmaI sites. DL1 cells were then transfected, CuSO4 was added, and Northern blots were performed. To avoid detection of the endogenous Laccase2 circular RNA, a probe complementary to the MCS backspliced junction was used. (D) The Laccase2 MCS vector was able to circularize segments of human ZKSCAN1 exons 2 and 3 (numbering scheme from Liang and Wilusz 2014). The ZKSCAN1 nucleotide 627–646 probe (top) detects linear and circular RNAs derived from the plasmid, whereas the circle junction probe (bottom) detects only properly backspliced RNAs. (E,F) Exon 2 of human HIPK3 (E) and the ciRS7 (CDR1as) exon (F) were likewise circularized in DL1 cells when placed between the laccase2 introns.
Next, we tested whether other exons ≥300 nt could be circularized with the laccase2 flanking introns. Exons of different lengths from human ZKSCAN1 (300, 500, and 650 nt) (Fig. 6D), human HIPK3 (1098 nt) (Fig. 6E), and human ciRS7 (CDR1as; 1485 nt) (Fig. 6F), all of which naturally circularize from their endogenous loci, were inserted into the Laccase2 MCS exon vector. Upon addition of CuSO4, all of the exons efficiently circularized in both DL1 (Fig. 6D–F) and S2 (Supplemental Fig. 5A) cells. Furthermore, the laccase2 introns also promoted circularization of an exon consisting of the GFP ORF (Supplemental Fig. 5B), indicating that the flanking introns play a dominant role in circular RNA formation and can drive the intervening exons to be backspliced. Importantly, minimal linear RNA was generated in all cases (Fig. 6D–F), although some concatenated and/or intertwined circular RNAs were detected with the ZKSCAN1 transcripts (Fig. 6D).
The laccase2 intronic repeats facilitate efficient circular RNA production in mammalian cells
As the laccase2 DNAREP1_DM repeats can also promote efficient exon circularization in human cells (Fig. 5C) while generating only a minimal amount of linear RNA (Fig. 7A; Supplemental Fig. 6A), we reasoned that the laccase2 introns could be used to improve mammalian circular RNA expression methods. Current plasmids, such as our previously described “CircRNA Mini Vector” (Fig. 7B; Liang and Wilusz 2014), have limited utility, as they generate significantly greater amounts of linear RNA than circular RNA (Fig. 7C). In addition, the CircRNA Mini Vector (which contains ∼40-nt intronic repeats from the human ZKSCAN1 gene) is unable to circularize long exons, such as human HIPK3 exon 2 (1098 nt) (Supplemental Fig. 6B). We thus generated two new mammalian expression plasmids: (1) the Laccase2 MCS exon vector, in which the laccase2 exon was replaced with a multicloning site (Fig. 7A), and (2) the ZKSCAN1 MCS vector, which contains the full ZKSCAN1 intronic repeats (Fig. 7B).
Figure 7.
Plasmids for efficient circular RNA expression in mammalian cells. (A) To test the ability of the laccase2 exon to circularize in mammalian cells, the 1945-nt region of the Laccase2 pre-mRNA was cloned into pcDNA3.1(+). (Bottom) In the Laccase2 MCS exon vector, exon 2 was replaced with a 63-nt artificial exon. (B) These plasmids were then compared with analogous pcDNA3.1(+) expression plasmids that are based on the human ZKSCAN1 gene. The CircRNA Mini Vector has short Alu repeats flanking the MCS exon, whereas the ZKSCAN1 MCS vector includes intronic sequences from nucleotides 100–2232 of the previously described ZKSCAN1 sense expression plasmid (Liang and Wilusz 2014). (C) Segments of human ZKSCAN1 exons 2 and 3 (numbering scheme from Liang and Wilusz 2014) were inserted into the multicloning sites of the designated plasmids. Plasmids were then transfected into HeLa cells, and Northern blots were performed. The presence of long flanking repeats (in either the ZKSCAN1 MCS vector or the Laccase2 MCS vector) greatly improved circularization efficiency. (D) The ZKSCAN1 and laccase2 introns generated significantly more ciRS7 circular RNA than the previously described ciRS7 expression plasmid (Hansen et al. 2013).
To determine the efficiencies at which each of these plasmids generates circular RNAs in human cells, ZKSCAN1 exons of different lengths (300, 500, and 650 nt) (Fig. 7C) or the HIPK3 exon (Supplemental Fig. 6B) were inserted into each vector. The plasmids were then transfected into HeLa cells and analyzed by Northern blots (Fig. 7C; Supplemental Fig. 6B–F). As observed previously (Liang and Wilusz 2014), the CircRNA Mini Vector backbone predominately generated linear transcripts (Fig. 7C, lanes 5,8,11). In contrast, the ZKSCAN1 MCS-based vector (Fig. 7C, lanes 6,9,12) and Laccase2 MCS-based vector (Fig. 7C, lanes 7,10,13) produced significantly greater amounts of the ZKSCAN1 and HIPK3 circular RNAs, with a concomitant decrease in linear RNA production (Fig. 7C; Supplemental Fig. 6B). As expected, the circular RNAs were resistant to degradation by RNase R (Supplemental Fig. 6D) and localized to the cytoplasm (Supplemental Fig. 6E).
To further examine whether these vectors are able to generate translatable circular RNAs, we inserted portions of GFP into the ZKSCAN1 MCS vector (Supplemental Fig. 7A). Exons containing a portion of the GFP ORF or the full-length ORF (split across the backsplicing junction) were efficiently circularized in HeLa cells (Supplemental Fig. 7B). Upon inserting the encephalomyocarditis virus (EMCV) IRES upstream of the start codon, GFP protein production was observed (Supplemental Fig. 7C). This strongly suggests that circular RNAs can be translated in vivo, as has been suggested previously (Wang and Wang 2015).
Finally, we tested whether perfect intronic repeats promote greater circular RNA production than imperfect repeats. We took advantage of a previously described ciRS7 circular expression plasmid (Hansen et al. 2013) in which >800 nt of perfectly complementary sequences had been inserted into the introns flanking the ciRS7 exon. After cloning the ciRS7 exon into our vectors, we compared the circularization efficiency obtained from the Hansen et al. (2013), ZKSCAN1 MCS, and Laccase2 MCS vectors (Fig. 7D). With all three vectors, we observed that an optional intron within the ciRS7 exon was spliced out ∼25% of the time. Nevertheless, the overall ciRS7 expression levels were ∼10-fold greater with the imperfect repeat-based vectors (Fig. 7D). In total, these results indicate that long (∼300- to 500-nt) imperfect repeats from the human ZKSCAN1 and fly laccase2 introns allow robust circular RNA production from mammalian expression plasmids. As these plasmids generate circular RNAs at a high efficiency, they should be of great utility in deciphering the functions of circular RNAs across eukaryotes.
Discussion
Recent deep sequencing studies have identified thousands of circular RNAs that are generated from eukaryotic protein-coding genes. However, we are only beginning to understand the mechanisms by which the pre-mRNA splicing machinery selects certain exons to circularize. In the present study, we demonstrated that intronic repeats and trans-acting hnRNPs and SR proteins combinatorially regulate circularization of the Drosophila laccase2 gene. Base-pairing between transposable elements in the flanking introns facilitates circularization, and the strength of these interactions likely dictates whether backsplicing occurs co- or post-transcriptionally. This mechanism is distinct from the one that regulates Drosophila Mbl circular RNA production (Ashwal-Fluss et al. 2014) but is similar to that used to generate many circles in humans, mice, and C. elegans (Dubin et al. 1995; Jeck et al. 2013; Liang and Wilusz 2014; Zhang et al. 2014; Ivanov et al. 2015; Li et al. 2015). This suggests that base-pairing between intronic repeats may be a major mechanism promoting exon circularization across eukaryotes. Moreover, we found that the laccase2 exon is dispensable, allowing the laccase2 introns to be used to efficiently generate “designer” circular RNAs from plasmids in diverse organisms. Altogether, our results suggest that circular RNA biogenesis strategies are conserved across eukaryotes and provide new tools for exploring the functions of circular RNAs.
Inverted repeat sequences facilitate circularization in Drosophila and other eukaryotes
Over 2500 circular RNAs have so far been described in Drosophila, many of which are expressed in fly heads and increase with aging (Ashwal-Fluss et al. 2014; Westholm et al. 2014). The pre-mRNA splicing machinery plays a key role in their biogenesis, as exons that circularize are almost always flanked by canonical 5′ and 3′ splice sites. Nevertheless, our current understanding of why only certain exons are circularized in Drosophila is rather limited. A recent computational analysis found that the introns flanking exons that circularize are longer than average, but no enriched motifs or differences in sequence conservation were noted (Westholm et al. 2014). Instead, there was a positive correlation between circular RNA abundance and intron length (Westholm et al. 2014), which suggested that long flanking introns somehow serve as intrinsic determinants for circularization.
Long introns also generally flank mammalian circular RNAs, but the length of the introns does not reliably determine which mammalian exons are backspliced (Jeck et al. 2013; Salzman et al. 2013; Zhang et al. 2014). Instead, complementary sequences, such as Alu elements in humans, are statistically enriched in the introns flanking exons that circularize, and backsplicing is triggered when these sequences base-pair to one another (Jeck et al. 2013; Liang and Wilusz 2014; Zhang et al. 2014). In fact, one can accurately predict many circular RNAs in humans, mice, and C. elegans by searching for pairs of complementary regions in the flanking introns (Ivanov et al. 2015). As no similar enrichment of complementary regions was found in the 500 nt that flank Drosophila circular RNAs, it was suggested that circularization mechanisms are distinct in Drosophila compared with other eukaryotes (Westholm et al. 2014).
However, there was little experimental evidence for this assertion. Indeed, our results on the laccase2 locus indicate that base-pairing between complementary intronic sequences efficiently promotes RNA circularization in flies. As the DNAREP1_DM repeats closely flank exon 2 of the laccase2 gene (Fig. 1E), we propose a model in which the repeats base-pair to one another, bringing the intervening splice sites into close proximity and facilitating catalysis. The Laccase2 circular RNA then accumulates as one of the most abundant circular RNAs in Drosophila (fifth most abundant across >100 Drosophila RNA sequencing libraries) (Westholm et al. 2014). At the endogenous laccase2 gene locus, the long introns that flank this exon likely slow the overall speed of cotranscriptional splicing, thereby allowing the backsplicing reaction to effectively compete with canonical splicing. Indeed, we found that the strength of the base-pairing interactions between the flanking introns dictates how quickly backsplicing can occur (Fig. 5). When very stable interactions are present, it is possible that exon definition is improved, allowing the rapid and cotranscriptional generation of a circular RNA. Nevertheless, further studies are still required to clarify the exact role that long flanking introns may play in regulating circularization.
Upon examining the introns that flank other abundant Drosophila circular RNAs, we identified other examples in which complementary regions >60 nt in length flank circularizing exons, including CaMKI, CG11155, CG2052, Parp, and PlexA (which are among the top 25 most abundant Drosophila circular RNAs) (Westholm et al. 2014). Interestingly, the Semaphorin-2b (CG33960) circular RNA (39th most abundant circular RNA) is flanked by introns containing short (CA)n simple repeats that are complementary to each other over a <30-nt region (Supplemental Fig. 8A). Upon cloning a 980-nt region of the Semaphorin-2b pre-mRNA downstream from the pMT, we observed circular RNA production from the plasmid in DL1 cells (Supplemental Fig. 8B,C). Removal of either of the (CA)n simple repeats, however, strongly reduced circularization (Supplemental Fig. 8D). This suggests that diverse inverted repeat sequences, including short simple repeats, may play a general role in facilitating circularization in Drosophila.
Trans-acting splicing factors act to combinatorially regulate circular RNA levels
Complementary repeats, however, are not observed at all Drosophila loci that generate circular RNAs. Furthermore, many exons that do not circularize are flanked by complementary repeats (Westholm et al. 2014), so there must be other mechanisms that regulate circularization. This has been most notably demonstrated at the Drosophila mbl locus, which requires the Mbl splicing factor for its circularization (Ashwal-Fluss et al. 2014). When Mbl protein is in excess, an intricate feedback mechanism is induced: The Mbl protein decreases the production of its own mRNA by binding its pre-mRNA. This blocks canonical splicing and promotes the biogenesis of the Mbl circular RNA, which further functions as a sponge that binds and sequesters the excess Mbl protein. However, this Mbl-driven mechanism appears to be specific for the mbl locus, as we found that knockdown of the Mbl linear mRNA had no effect on Laccase2, PlexA, or a panel of other circular RNAs (Figs. 3, 4; Supplemental Fig. 4). Knockdown of the Laccase2 linear mRNA likewise did not affect Laccase2 circular RNA levels, indicating that the laccase2 locus is not subjected to a similar direct cis-acting feedback mechanism. Instead, we found that other splicing factors, including hnRNPs and SR proteins, regulate Laccase2 RNA levels (Figs. 3, 4).
At the laccase2 locus, we propose that hnRNPs (e.g., Hrb27C and Hrb87F) and SR proteins (e.g., SF2 [SRSF1], SRp54 [SRSF11], and B52 [SRSF6]) add an additional layer of control on top of the DNAREP1_DM intronic repeats. Base-pairing between the intronic repeats promotes circularization, but protein binding likely helps ensure that the appropriate ratio of linear to circular Laccase2 RNA is produced. Depletion of any one of these splicing factors alters Laccase2 circle levels, and additive effects were observed when multiple factors were depleted (Fig. 4). This suggests combinatorial control, with each protein playing a nonredundant role. Furthermore, Laccase2 circular RNA production does not appear to be linked to exon skipping, and thus these proteins may specifically modulate spliceosome assembly, the speed of splicing, and/or the stability of the mature circular RNA. Notably, it does not seem that Hrb27F, SF2, SRp54, or B52 affects Laccase2 circular RNA stability, as depletion of these factors did not cause the expression of a plasmid-derived Laccase2 circular RNA (Laccase2 MCS–Laccase2 608–1097) (Fig. 6B) to increase (Supplemental Fig. 9). We thus instead propose that these hnRNPs and SR proteins regulate Laccase2 circular RNA biogenesis (e.g., by binding to the flanking introns or exons), but further studies are required to understand exactly how the intronic repeats and trans-acting factors collaboratively dictate the splicing outcome. Nevertheless, we found that the same SR proteins that regulate the laccase2 locus also regulate the PlexA circular RNA (Fig. 4B) but not the Mbl circular RNA (Fig. 3D). Since the laccase2 and PlexA exons are both flanked by inverted repeats, we hypothesize that intronic repeats may generally provide the opportunity for circularization to occur. This is then further regulated by trans-acting factors that combinatorially fine-tune the amount of each circular RNA that the cell ultimately produces.
Improved methods for expressing circular RNAs in cells
Catalogs of circular RNAs expressed in various species and cell types have been reported (Glazar et al. 2014), but the functions for nearly all of these transcripts, including Laccase2, are currently unknown. This is due in part to the current lack of methods for efficiently generating circular RNAs in cells. For example, the circular RNA expression plasmids that have been described (Hansen et al. 2013; Ashwal-Fluss et al. 2014; Liang and Wilusz 2014; Zhang et al. 2014; Starke et al. 2015; Wang and Wang 2015) all generally produce circular transcripts at a low efficiency (often 20% or less). These plasmids instead generate abundant amounts of linear RNA, which limits their utility for defining circular RNA functions. Here, using the Drosophila laccase2 and human ZKSCAN1 introns, we largely overcame this hurdle and generated circular RNAs (ranging in size from 300 to 1500 nt) at a high efficiency in human and fly cells (Figs. 6, 7). We verified that these transcripts accumulate in the cytoplasm (Supplemental Fig. 6E), are resistant to RNase R treatment (Supplemental Fig. 6D), and are likely translated when an IRES is present (Supplemental Fig. 7). Furthermore, easy-to-use restriction sites are present in the plasmids, allowing any desired sequence to be queried. Beyond allowing ectopic expression of circular RNAs, these plasmids can be designed to sponge microRNAs or proteins as well as identify novel IRES sequences.
In summary, our findings provide key insights into how trans-acting factors and intronic repeats regulate circular RNA biogenesis as well as provide new tools for exploring the functions of circular RNAs across eukaryotes. From humans to flies, repetitive elements in introns can act to facilitate backsplicing, but it is still largely unclear why circular RNAs accumulate only in certain tissues. We hypothesize that base-pairing between repeats is only one part of the “splicing code” (Barash et al. 2010), and it is ultimately a combination of cis-acting elements and trans-acting splicing factors, including hnRNPs and SR proteins, that dictates whether canonical splicing or backsplicing occurs. Nevertheless, we have now defined a minimal set of elements that is sufficient for promoting efficient exon circularization, which should facilitate the prediction of circular RNAs as well as enable the functions of many circular RNAs to be revealed. Considering that a surprisingly large number of protein-coding genes generates circular RNAs, these previously overlooked transcripts likely represent key ways that gene functions are expanded and modulated.
Materials and methods
Expression plasmid construction
To generate the laccase2 expression plasmids for transfection into fly cells, the indicated sequences were inserted into a pMK33/pMtHy-based plasmid between the pMT and the SV40 poly(A) signal, as described further in the Supplemental Material. Genomic coordinates for the full-length laccase2 insert are chromosome 2R: 5,423,143–5,425,087 of the D. melanogaster genome (version dm6). For the Laccase2 MCS exon vector depicted in Figure 6A, exonic sequences were inserted between the KpnI and XmaI sites. To generate laccase2 plasmids terminating in various 3′ end sequences in Figure 5A, the SV40 poly(A) signal was replaced with the histone stem–loop (dH3), the mouse MALAT1 triple helix (Wilusz et al. 2012), or the HhRz (Haseloff and Gerlach 1988). To construct a corresponding set of EGFP expression plasmids ending in different 3′ ends, the laccase2 insert was replaced with the EGFP ORF.
To generate laccase2 expression plasmids for transfection into human cells, the indicated sequences were inserted into pcDNA3.1(+) downstream from the CMV promoter, as described further in the Supplemental Material. The CircRNA Mini Vector was previously described (Liang and Wilusz 2014). The ZKSCAN1 MCS vector was constructed from the pcDNA3.1(+) ZKSCAN1 nucleotide 100–2232 plasmid (Liang and Wilusz 2014) by replacing the endogenous ZKSCAN1 exons with an artificial exon. Except where noted, exonic sequences were inserted between the PacI and SacII sites of the Laccase2 MCS exon vector (Fig. 7A) and between the EcoRV and SacII sites of the CircRNA Mini Vector and ZKSCAN1 MCS vector (Fig. 7B). Mammalian expression plasmids ending in different 3′ ends were generated as previously described (Liang and Wilusz 2014). Additional details and the sequences of the inserts for all plasmids are provided in the Supplemental Material.
Transfections, RNAi, and RNA isolation
Drosophila cells (DL1, S2, and SL2 cells) were grown at 25°C in Schneider's Drosophila medium (Life Technologies) supplemented with penicillin–streptomycin, L-glutamine, and 10% fetal bovine serum. Cells (2 × 106) were plated in complete medium, and 2 μg of each expression plasmid was transfected using Effectene (16 μL of enhancer and 8 μL of Effectene reagent; Qiagen). On the following day, a final concentration of 500 μM copper sulfate was added for 14 h (where indicated) to induce transcription from the pMT. Total RNA was then isolated using Trizol (Life Technologies) as per the manufacturer's instructions. Adult wild-type D. melanogaster or D. yakuba were dissected and processed for total RNA using Trizol.
dsRNAs from the DRSC (Drosophila RNAi Screening Center) were generated by in vitro transcription (MEGAscript kit, Life Technologies) of PCR templates containing the T7 promoter sequence on both ends. Primer sequences are provided in Supplemental Table 1. Knockdown experiments were then performed by bathing 3 × 106 cells with 4 μg of dsRNA. DL1 cells were incubated for 3 d, whereas SL2 cells were incubated for 4 d. Total RNA was then isolated using Trizol (Life Technologies).
HeLa cells were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing high glucose (Life Technologies) supplemented with penicillin–streptomycin and 10% fetal bovine serum. One microgram of each expression plasmid was transfected using Lipofectamine 2000 (Life Technologies), and total RNA was isolated after 24 h using Trizol (Life Technologies) as per the manufacturer's instructions. Nuclear and cytoplasmic fractionation was performed as previously described (Wilusz et al. 2008).
Northern blotting
Northern blots using NorthernMax reagents (Life Technologies) and oligonucleotide probes were performed as previously described (Wilusz et al. 2008). All oligonucleotide probe sequences are provided in Supplemental Table 1. Blots were viewed and quantified with the Typhoon 9500 scanner (GE Healthcare). Representative blots are shown. For RNase R treatments, 20 μg of total RNA was treated with 10 U of RNase R (Epicentre) for 10 min at room temperature. mFold was used to calculate hairpin stabilities, assuming a 7-nt linker (AGAAUUA) between the two repeat sequences.
qPCR
For real-time qPCR, 2 μg of total RNA was reverse-transcribed using random hexamers and SuperScript III (Life Technologies). qPCR was then carried out in triplicate using Power SYBR Green PCR Master mix (Life Technologies), and β-actin (Act42a) served as a normalization control. qPCR primer sequences are provided in Supplemental Table 1.
Supplementary Material
Acknowledgments
We thank Jørgen Kjems for providing the pcDNA3-ciRS7 expression plasmid, and Kristen Lynch, Gideon Dreyfuss, Jeff Wilusz, and all members of the Cherry and Wilusz laboratories for discussions and advice. This work was supported by National Institutes of Health grants R00-GM104166 (to J.E.W.) and R01-AI074951 (to S.C.) and start-up funds from the University of Pennsylvania (to J.E.W.). J.E.W. is a Rita Allen Foundation Scholar. S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.270421.115.
References
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56: 55–66. [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. 2010. Deciphering the splicing code. Nature 465: 53–59. [DOI] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J. 2015. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4: e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. 2014. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol 21: 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiolo M, Herzel L, Neugebauer KM. 2013. Counting on co-transcriptional splicing. F1000Prime Rep 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Hertel KJ. 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18: 186–193. [DOI] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. [DOI] [PubMed] [Google Scholar]
- Chen LL, Yang L. 2015. Regulation of circRNA biogenesis. RNA Biol 12: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. 1993. Mis-splicing yields circular RNA molecules. FASEB J 7: 155–160. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H. 1995. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167: 245–248. [DOI] [PubMed] [Google Scholar]
- Ebbesen KK, Kjems J, Hansen TB. 2015. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Tanaka K, Matsuura Y, Tanahashi M, Kikuchi Y, Fukatsu T. 2011. Laccase2 is required for cuticular pigmentation in stinkbugs. Insect Biochem Mol Biol 41: 191–196. [DOI] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N. 2014. circBase: a database for circular RNAs. RNA 20: 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL. 1988. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334: 585–591. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. 2015. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10: 170–177. [DOI] [PubMed] [Google Scholar]
- Jacobs CG, Braak N, Lamers GE, van der Zee M. 2015. Elucidation of the serosal cuticle machinery in the beetle Tribolium by RNA sequencing and functional analysis of Knickkopf1, Retroactive and Laccase2. Insect Biochem Mol Biol 60: 7–12. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Greenman C, Cook PR, Papantonis A. 2015. Exon skipping is correlated with exon circularization. J Mol Biol 427: 2414–2417. [DOI] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2014. Circular RNAs: diversity of form and function. RNA 20: 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264. [DOI] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. 2014. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB. 2001. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci 98: 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. 1991. Scrambled exons. Cell 64: 607–613. [DOI] [PubMed] [Google Scholar]
- Petkovic S, Muller S. 2015. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 43: 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885. [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25: 381–388. [DOI] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. 2015. Exon circularization requires canonical splice signals. Cell Rep 10: 103–111. [DOI] [PubMed] [Google Scholar]
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. 1999. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 8: 493–500. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Z. 2015. Efficient backsplicing produces translatable circular mRNAs. RNA 21: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. 2014. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9: e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. 2014. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9: 1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2015. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sharp PA. 2013. Molecular biology. A circuitous route to noncoding RNA. Science 340: 440–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Chen L, Liu C, Zhu QH, Fan L. 2015. Widespread noncoding circular RNAs in plants. New Phytol 208: 88–95. [DOI] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG. 1997. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 17: 2985–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. 2014. Complementary sequence-mediated exon circularization. Cell 159: 134–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.