Abstract
Background
Statin therapy influences not only low-density lipoprotein-cholesterol (LDL-C) levels, but also LDL-related biomarkers including non-high-density lipoprotein cholesterol (non-HDL-C), apolipoprotein B (apo B), total number of LDL particles (LDL-P), and mean LDL particle size (LDL-size). Recent studies have identified many genetic loci influencing circulating lipid levels and statin-induced LDL-C reduction. However, it is unknown how these genetic variants influence statin-induced change in LDL subfractions and non-HDL-C.
Methods and Results
One hundred and sixty candidate SNPs for effects on circulating lipid levels or statin-induced LDL-C lowering were tested for association with response of LDL subfractions and non-HDL-C to rosuvastatin or placebo over 1 year among 7,046 participants from the JUPITER trial. Of the 51 SNPs associated with statin response for one or more of the LDL subfractions, or non-HDL-C, 20 SNPs could be clustered according to effects predominantly on LDL-size, predominantly on LDL particle number, and on apo B but not LDL-C or non-HDL-C.
Conclusions
These differential associations point to pathways of LDL response to statin therapy and possibly to mechanisms of statin dependent CVD risk reduction.
Keywords: genetics, genetics, association studies, cholesterol, lipoprotein, JUPITER, nuclear magnetic resonance spectroscopy, rosuvastatin
Introduction
Statin-mediated inhibition of hydroxy-3-methylglutaryl-Co-A reductase, an enzyme catalyzing the rate-limiting step in cholesterol synthesis, lowers circulating low-density lipoprotein (LDL) cholesterol primarily by inducing uptake of LDL particles from plasma into peripheral tissues1. As a consequence, statins also affect alternative measures related to LDL particles including levels of non-high-density lipoprotein cholesterol (non-HDL-C), levels of apolipoprotein B (apo B), the total number of LDL particles (LDL-P), and the mean LDL particle size (LDL-size)2. The responses to statin of these alternative LDL measures are somewhat but not entirely correlated with each other and with LDL-C, and are discordant in up to a quarter of healthy individuals3, in the sense that statin-lowered LDL-C is not always accompanied by lower LDL particle number, or apo B concentration. These alternative measures of LDL response to statin therapy thus are not necessarily reflected in LDL-C reduction and may have clinical relevance. Among agents targeting LDL-C lowering for prevention of incident CVD, statin therapy appears to be especially beneficial. For example, statins may cause as much as 50% or more lowering of LDL-C, and throughout this range, there is approximately a 10% lowering of CVD risk per a 10% reduction in LDL-C4. By contrast LDL-C lowering by inhibitors of Niemann-Pick protein5 (i.e. ezetemibe, via inhibition of intestinal cholesterol absorption) or CETP 6, 7(cholesteryl ester transfer protein, via inhibition of the exchange of cholesterol and triglyceride in HDL particles and these lipids in LDL and VLDL particles), both are only somewhat less potent at lowering LDL-C than statins and are accompanied by smaller reductions in CVD risk. These findings are consistent with the hypothesis that the benefit of statin therapy is not solely induced by LDL-C lowering per se, a notion also consistent with the observed modest differences in the magnitude of residual vascular risk with statin therapy depending upon which LDL-related measure is evaluated8. Understanding the detailed consequences of statins’ effects on LDL metabolism may reveal the biological basis for the dramatic risk reduction observed in many statin trials.
To date, few if any clinical variables have been identified that distinguish among statin-induced changes in LDL-related measures. Meanwhile, recent work has demonstrated substantive genetic influence not only on circulating lipid levels9-11 but also on statin-induced changes in LDL-C12. The primary aim of the present study was to examine if candidate genetic loci associated with circulating lipids, that represent several different biological pathways, could explain differential response of LDL-C compared to response of other LDL-related lipoprotein fractions and non-HDL-C to statin treatment. We thus sought in the context of a randomized placebo-controlled trial to address whether a panel of 160 SNPs known from the literature to influence basal levels of conventional lipid fractions, and for comparison LDL-C change with statin therapy, might be associated with statin-induced changes in non-HDL-C, apo B, LDL-P, and LDL-size and further whether there may be differential associations among these alternative LDL measures. These markers were selected for analysis on the basis that the LDL fraction is the target of statin therapy and therefore statin effects on these measures would be larger and more likely to be detectable than for other lipid fractions such as HDL, which are also influenced by statin therapy but to a lesser degree13. As shown here, in a study of 7,046 men and women of European ancestry allocated to rosuvastatin 20 mg or placebo over a 12 month period, there were three groups of SNPs that were predominantly associated with statin-response in one or a subset of lipid measures but not with the others. The groups of genes implied by the selective SNP effects appear to influence substantially different aspects of lipid metabolism and catabolism, reflecting the complexity of cholesterol transport and biosynthesis and highlighting the basis of complex statin effects that are not revealed by analysis of LDL-C response alone.
Methods
Study Population
The study population for this analysis was derived from the Justification for Use of statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER trial: NCT00239681). JUPITER is an international, randomized, placebo-controlled evaluation of rosuvastatin (20 mg/day) conducted among men and women free of cardiovascular disease with moderate-to-low LDL cholesterol (LDL-C) levels (<130 mg/dL) and elevated C-reactive protein (CRP) levels (≥2 mg/L) at baseline14. Blood samples were obtained at time of randomization and after 12 months of treatment with either placebo or rosuvastatin. Participants included in this analysis consented to procedures used in JUPITER for the genetic testing of samples. All procedures and protocols in this study were approved by the Partners Human Research Committee (Institutional Review Board for Partners Healthcare on behalf of Brigham and Women's Hospital).
Genotyping and Imputation
Study participants were genotyped using the Omni 1M Quad platform and GenomeStudio v 1.6.2 (both Illumina, San Diego, CA, USA) by the manufacturer. A total of 1,006,348 single nucleotide polymorphisms (SNPs) passed quality control standards as previously described12. In short, only a small subset of markers had poor characteristics regarding clustering metrics for ABrMean (intensity), cluster separation, Hardy-Weinberg Equilibrium, or call frequency. These markers were visually inspected and either flagged, removed, or re-clustered manually. Detailed experimental data on individual genotypes and plots for manually clustered variants are available in Chasman et al12. SNPs were retained in the final data set if the updated clusters met quality standards and the genotyping was successful in at least 98.5% of the samples. A multidimensional scaling (MDS) procedure in PLINK15 was used to verify self-reported European ancestry; 37 participants were excluded due to discordance between self-reported ancestry and assignment of European ancestry by MDS. A total of 33 individuals from 31 family groups were excluded in the JUPITER data to eliminate 1st degree relatives as judged by PLINK15. EIGENSTRAT was used to estimate sub-European stratification and calculate principal components16. Approximately 6.8 million SNPs from the pilot data of the 1000 Genomes Project were imputed using MaCH v.1.0.1617, 18. All SNPs were imputed from a genotyping panel with SNPs that either met HWE p>10−6 or for the case of APOE (rs7412), included manual review of genotyping clusters12. Imputation quality scores for all candidate SNPs were above 0.7 (MaCH R2>0.7); please see Supplemental Table 1 for imputation quality scores of the individual SNPs.
Biomarker measurement
LDL-C, non-HDL-C, apolipoprotein B100 (apo B), HDL cholesterol (HDL-C) and total cholesterol (TC) were measured in a core laboratory facility at the time of randomization and after 12 months of randomized allocation to placebo- or rosuvastatin-treatment as previously described12. Non-HDL-C was calculated by subtracting HDL-C from TC measurements and captures the amount of cholesterol carried by apo B particles. LDL-P and LDL-size were determined by proton nuclear magnetic resonance (NMR) spectroscopy analysis (NMR LipoProfile3, LipoScience, Raleigh, NC, USA)19 and capture the total number of LDL particles and the mean size of all LDL particles, respectively.
SNP selection
A total of 154 candidate SNPs from the published GWAS of circulating plasma lipids from the Global Lipids Genetics Consortium (GLGC)9, 10 were selected for analysis (see Supplemental Table 2a). When multiple SNPs mapped to the same locus and were in LD, the SNP with the most significant p-value (in association with any lipid) was chosen as the locus representative (index SNP). All index SNPs that mapped to the same locus but were nevertheless not in LD were included. A proxy (rs8035382) was used for the original index SNP (rs292982) at one locus FRMD5 (R2=1; 1000 Genomes) where the original index was neither genotyped nor imputed. Three SNPs were neither genotyped nor imputed and did not have any proxies available; therefore these SNPs were excluded from analysis (rs2412710 CAPN3, rs1047891 CPS1, rs13238203 TYW1B).
In addition, 6 candidate SNPs (rs17111584, PCSK9; rs2199936, ABCG2; rs10455872, LPA; rs12317268, SLCO1B1; rs11672123, LDLR; rs7412 APOE) for statin-induced LDL-C reduction in JUPITER12 were included in the analysis (see Supplemental Table 2b). Four loci (APOE, LDLR, LPA, and PCKS9) contain two SNPs each, however the variants are independent of one another (R2<0.03 between the two SNPs at each of the four loci). A listing of all candidate SNPs can be found in Supplemental Table 2c.
Analysis
Primary outcomes examined in this study were absolute change and percentage change in LDL-C, non-HDL-C, apo B, LDL-P, and LDL-size after 12 months of rosuvastatin or placebo therapy. Absolute change was calculated as the difference between the 12-month and baseline value; and percentage change was calculated by dividing the absolute change by the baseline value such that, for example, greater reduction in LDL-C was reflected in negative values with greater magnitude. The percentage change calculation implicitly accounts for baseline measures. Analysis of each outcome was stratified by statin-allocation arm and restricted to the 7,046 participants with both baseline and 12-month lipoproteins measures, compliance with study medication based on pill counts, and the absence of self-reported non-trial statin use. To assess the effect of the recruitment criteria in JUPITER (LDL-C < 130mg/dL) on known lipid associations, additional analyses were performed among a combined total of 7,046 participants (allocated to either rosuvastatin or placebo) for association of candidate SNPs from GLGC and baseline measures of LDL-C, non-HDL-C, apo B, LDL-P and mean LDL-size. Spearman correlation coefficients were calculated between all baseline and change measures in up to 3,534 statin-allocated participants.
To decrease the influence of extreme outliers on the change outcomes, all measures were transformed using inverse-quantile normalization9, 20, 21, which was carried out in the statin- and placebo-allocated arms (preserving the ranks within each allocation group). We chose to transform our data by inverse-quantile normalization due to the long right-tail we observed for statin-induced absolute change in many of the examined traits; log-transformation was not possible because the distributions of absolute and percentage change include negative values. P-values for association were obtained from linear regression of transformed outcome measures while estimates of the genetic effects were obtained from linear regression of untransformed outcome measures, both encoding genetic information with a standard additive genetic model assuming proportionality between the number of inherited copies of the minor (i.e. coded) allele and mean LDL phenotype. Thus, negative regression coefficients implied greater reduction of LDL-C, non-HDL-C, apo B, and LDL-P levels or greater reduction in the mean size of LDL particles with each additional inherited copies of the minor allele. In interaction analysis where statin- and placebo-allocated arms are combined, the values/ranking of the inverse-quantile normalized traits created within the groups are no longer valid for between group comparisons, i.e. interactions. Therefore we reverted back to the untransformed outcomes for interaction analysis. Interaction analysis was performed by introducing a standard multiplicative drug-by-SNP interaction term in the linear regression models of statin response. All regression models were adjusted for age, sex, region and 10 principal components calculated from EIGENSTRAT.
In total, 160 candidate SNPs (for full list see Supplemental Table 2c) were evaluated for association with absolute and percentage change in LDL-C, non-HDL-C, apo B, LDL-P and LDL-size. Variants associated with any of the change measures among statin-allocated participants at a nominal significance level (p<0.05) were carried forward for further analysis if they were either not associated with any of the change measures among placebo-allocated participants (p≥0.05) or had drug-by-SNP interaction (p<0.1); excluding SNPs for associations with change measures among the placebo-allocated participants and excluding SNPs lacking evidence for SNP-by-drug interactions directly assures the SNPs included are better candidates for statin-response. We used complete linkage hierarchical clustering with a standard Euclidean distance metric to cluster SNP associations with change in LDL-C, non-HDL-C, apo B, LDL-P and LDL-size among statin-allocated participants with binary encoding, specifying 1 for significant association (p<0.05) and 0 otherwise; absolute change and percentage change were clustered separately. In the “complete linkage” method the distance between clusters is defined as the maximum distance between any of the individual SNP/biomarker pairs (one in each cluster). To delineate clusters in our association results, we used the NbClust package in R22, 23 which determines credible clustering structures through consensus among 30 clustering criteria. Variance explained (R2) was calculated from a regression of the inverse-quantile normalized residualized trait on each SNP.
To examine the biological connectivity and functional relationships among the genes within clusters based on SNP associations with LDL-size and LDL-P, we used GRAIL (Gene Relationships Among Implicated Loci)24, which is based on text mining of PubMed abstracts. As recommended, to emphasize relationships that might suggest biological pathways rather than associations derived from GWAS findings, GRAIL was run with a database derived from PubMed abstracts published before 2009, pre-dating most GWAS findings.
Gene-set enrichment analysis (GSEA) using MAGENTA25 and R22 were performed to test for enrichment of genes in the mean LDL particle size (n=11 genes, tagged by 8 SNPs), and LDL particle number (n=11 genes, tagged by 8 SNPs) gene clusters and used 1722 predefined genesets from Gene Ontology, BioCARTA, INGENUITY, KEGG, PANTHER, and REACTOME. Genes analyzed in GSEA were derived from the gene name annotations of each SNP from the published GLGC papers; if a SNP was annotated with multiple genesd9, 10, 12, all listed genes were used in GSEA. Multiple testing was addressed by permutation with 10,000 replicates.
Results
As shown in Supplemental Table 3, clinical characteristics of the 7,046 JUPITER participants of European ancestry who consented to genetic research, had successful LDL-related biomarkers measured at baseline and 12 months, and were compliant with study medications were indistinguishable between those allocated to rosuvastatin (N=3,534) and those allocated to placebo (N=3,512). At one year, rosuvastatin reduced LDL-C by 54 mg/dL (−52%), non-HDL-C by 59 mg/dL (−45%), apo B by 43 mg/dL (−37%), LDL-P by 528 nmol/L (−37%) and LDL-size by 0.4 nm (−1.7%). Among statin-allocated participants, we observed high correlation between absolute (and percentage) change in LDL-C, non-HDL-C and apo B, and moderate correlations between change in these three measure and change in LDL-P, but little to no correlation between change in LDL-size and the other LDL-related measures or non-HDL-C (please refer to Supplemental Table 4 for pairwise correlations between all analyzed measures).
Candidate SNPs were selected from genome-wide significant associations9, 10 with one or more of LDL-C (N=58), HDL-C (N=74), triglycerides (N=43), total cholesterol (N=75), or statin response of LDL-C (N=6)12 for a total of 160 unique SNPs. Of the candidates from analysis of circulating lipid levels, 64 SNPs were nominally significantly associated (p<0.05) with baseline levels of either LDL-C, non-HDL-C, apo B, LDL-P, or LDL-size (Supplemental Table 5) among the 7,046 participants with baseline lipoprotein measures representing 132 associations out of the 800[=160 SNPs × 5 traits] total associations (Supplementary Table 5, which also shows associations meeting Bonferroni and false discovery rate [FDR] thresholds). Of the 96 SNPs evaluated that did not associate with any of these LDL-related measures, 10 had been described in prior literature as being primarily associated with triglycerides, 36 with HDL-C, 4 with both triglycerides and HDL-C. Of the remaining SNPs, 31 had previously been described in the literature as being associated with LDL-C (alone or in addition to HDL-C and/or triglycerides). However, not all of the candidate variants were expected to be significantly associated with baseline LDL-related measures due to a smaller size of the JUPITER sample compared to the discovery sample (7,046 vs. 200,000 participants9, 10), the primary association of some candidates with non-LDL-C lipid measures, and possibly the enrollment criteria in JUPITER that required baseline LDL-C<130mg/dL. Of the six candidate SNPs derived from previous analysis of LDL-C response to statin only one (in LDLR) was also associated with LDL-C at baseline in JUPITER.
All 160 SNPs were evaluated by regression for association with absolute and fractional response to statin for the five LDL-related lipoprotein measures (Supplemental Tables 6a-d). SNPs with effects on statin response were selected for further examination based on the following criteria for a total of 51 SNPs: nominally significant association with 12-month change in absolute or fractional lipoprotein response among the 3,534 study participants allocated to rosuvastatin (Supplemental Tables 6a-d) and either no significant association with absolute or fractional response among the 3,512 study participants allocated to placebo (Supplemental Tables 6c and 6d), or at least marginally significant interaction effect (p<0.1) for allocation to rosuvastatin vs. placebo in a complementary analysis (see Methods and Supplemental Table 6e).
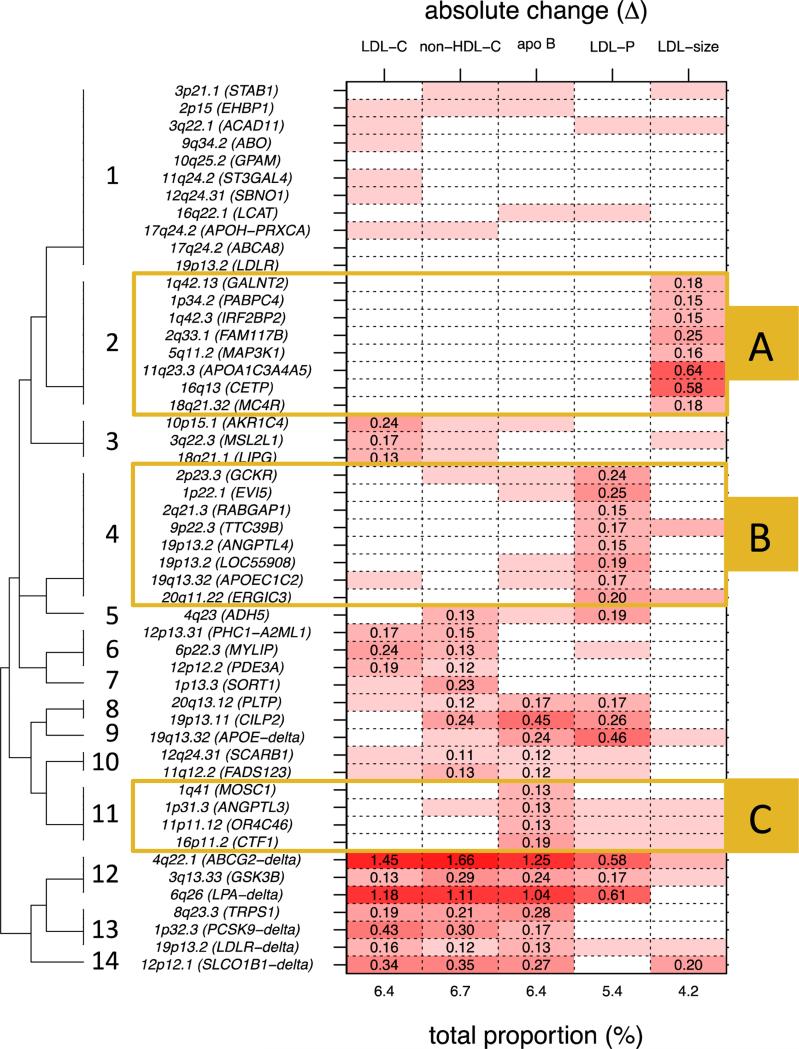
Complete linkage hierarchical clustering of the 51 SNPs selected for association with absolute change in non-HDL-C and LDL-related subfractions indicated the existence of 14 clusters (see Figure 1, see Methods). An identical clustering procedure performed on statin-induced percentage change in non-HDL-C and LDL-related subfractions resulted in the same number of optimal clusters (N=14 clusters; see Methods and Supplemental Figure 1). As expected, these clusters were similar but not completely identical to those identified in the absolute change results. This discrepancy is likely due to the presence of SNPs that are highly influential on circulating levels and therefore have a stronger effect on percentage change compared to absolute change. Therefore we only focused follow-up analyses on the clusters identified by the absolute change analysis.
Figure 1.
Dendrogram of 14 clusters from complete linkage hierarchical clustering and variance explained in rosuvastatin-induced absolute change for LDL cholesterol (LDL-C), non-HDL cholesterol (non-HDL-C), apolipoprotein B (apo B), LDL particle number (LDL-P), and LDL particle size (LDL-size) by candidate variants (p<0.05). A) cluster of SNPs associated with change in LDL-size, B) cluster of SNPs associated with change in LDL-P, and C) cluster of SNPs associated with change in apo B.
Among these 14 clusters identified in the absolute change results, there were three large clusters that were predominantly associated with a single fraction (clusters 2, 4 and 11 in Figure 1; to be referred to as Clusters A, B and C, respectively in Figure 1), one large cluster that was not associated with any specific LDL-related measure nor appeared to follow a discernable pattern (cluster 1; which was not followed up in subsequent analyses), and 10 smaller clusters containing 3 SNPs or less (clusters 3, 5-10 and 12-14; also not followed up in subsequent analyses). The first trait-specific cluster (cluster A) included 8 SNPs - rs964184 (APOA1-A5 cluster), rs3764261 (CETP), rs11694172 (FAM117B), rs4846914 (GALNT2), rs514230 (IRF2BP2), rs9686661 (MAP3K1), rs12967135 (MC4R), and rs4660293 (PABPC4) - that were associated only with statin-induced response in LDL-size. The second trait-specifc cluster (Cluster B) included 8 SNPs - rs4420638 (APOE-C1-C2), rs7255436 (ANGPTL4), rs2277862 (ERGIC3), rs7515577 (EVI5), rs1260326 (GCKR), rs737337 (LOC55908-DOCK6), rs6759321 (RABGAP1), and rs643531 (TTC39B) – that were associated almost exclusively with statin-induced response in LDL-P. The third trait-specifc cluster (cluster C) included 4 SNPs - rs2131925 (ANGPTL3), rs11649653 (CTF1), rs2807834 (MOSC1), and rs11246602 (OR4C46) – that were associated with statin-induced response primarily in apo B and had virtually no association with statin-induced reductions in LDL-C or non-HDL-C. One branch of the top level split in the hierarchical clustering eventually leads to several small clusters (clusters 12-14) and captures associations that were very strong for statin response of LDL-C and also for the related subfractions of non-HDL-C, apoB, and LDL-P, but not for LDL-size. These loci were rs2199936 (ABCG2-delta), rs10455872 (LPA-delta), rs17111584 (PCSK9-delta), rs11672123 (LDLR-delta) and rs12317268 (SLCO1B1-delta). Of note, two SNPs not previously identified in association with statin-response of LDL-C are also present in this branch of combined clusters, rs6805251 (GSK3B) and rs2293889 (TRPS1).
Bioinformatics tools were used to assess the correspondence between known biological pathways related to LDL and SNP clusters A and B, the clusters reflecting selective associations with LDL-size and LDL-P, respectively. This analysis was not done for cluster C due to the small number of SNPs in the cluster (n=4). For cluster A that associated with change in LDL-size only, two genes tagged by the 8 SNPs had significant functional connections based on text-mining of PubMed abstracts before 2009 (pre-dating the main lipoprotein GWAS results) using GRAIL (CETP (rs3764261, p=9.7e-3), and APOA5 (rs964184, p=9.0e-3). In addition, gene set enrichment analysis (GSEA) of the SNPs in cluster A using 1,724 predetermined gene-sets showed enrichment in phosphatidylcholine binding pathway (ppermutation=0.03) that is related to vesicle transport; no other pathways reached statistical significance after multiple testing correction. However both marginally enriched pathways (ppermutation<0.1), “high-density lipoprotein particle” and “cholesterol binding”, are related to lipid metabolism; a list of pathways are available in Supplemental Table 7. For cluster B that related predominantly to LDL-P, none of the 11 genes tagged by the 8 index SNPs were functionally connected in GRAIL analysis – the most significant result was for rs4420638 at the APOE-C1-C2 locus (p=0.06). GSEA using 1,724 predetermined gene-sets identified one marginally enriched pathway, chylomicron remnant clearance (ppermutation=0.09).
Discussion
Previous genetic analysis of statin response has primarily focused on associations between LDL-C and genetic variants or SNPs either in known pathways of statin action26 or arising from genome-wide analysis of LDL-C lowering12, the latter limited in statistical power by the relatively small size of suitable cohorts with genome-wide genetic data. Recently, however, genome-wide association studies of LDL-C and other lipid fractions including as many as 200,000 samples have dramatically increased the number of credible candidate variants for statin response analysis. This advance is complemented by high throughput NMR-based assays of lipoprotein sub-fractions that provide higher resolution lipoprotein profiles than can be inferred from standard plasma lipid measures alone. We examined the effects of 160 candidate SNPs on rosuvastatin response of LDL-C, non-HDL-C, apo B and two NMR-based LDL subfraction measures over 1 year of follow-up. This investigation highlighted clearly delineated subsets of SNPs implicating clusters of genes with selective effects on LDL properties or LDL-related measures distinct from LDL-C. One of the two larger clusters with effects on LDL properties was selective for effects on LDL-size while the other was selective for LDL-particle number. SNPs in both had essentially no effect on the rosuvastatin-induced change in LDL-C.
Several of the genes implicated for selective association with LDL-size (cluster A) may be understood in terms of known pathways regulating triglyceride levels, lipids which constitute much of the volume of LDL particles. CETP encodes cholesteryl ester transfer protein that exchanges triglycerides and cholesteryl esters between HDL or LDL and VLDL27, 28. The apoA5 protein encoded in the APOA1-APOA5 cluster is also a strong determinant of circulating triglyceride levels29. The candidate SNPs at these two genes conferred among the strongest total effects on LDL-size at baseline but exert relatively little effect on LDL-C, likely by affecting baseline triglyceride content. The exact mechanisms underlying these associations are unknown, although it may be relevant that for both loci, the alleles associated with greater LDL particle size at baseline are also associated with a smaller change in size with statin allocation (Supplemental Tables 5, 6a and 6b). Although the MC4R gene was initially identified for association with HDL-C by the GLGC9, 10, this locus was also associated with triglycerides albeit not at genome-wide significance; and the effect at MC4R may be related to its predominant role in regulating adiposity30, itself highly correlated with triglyceride levels. The mechanistic relationships of the other candidate SNPs in this cluster and the change in LDL-size with statin treatment are less clear, but both PABPC4 and GALNT2 are involved in protein expression and may act through regulation of protein, rather than lipid components of LDL particles. None of the remaining SNPs (IRF2BP2, FAM117B, MAP3K1) was significant for association with LDL-C at baseline (p>0.05) in JUPITER and only IRF2BP2 was associated with circulating LDL-C at genome-wide significance in previous analyses9, 10.
For the most part, the associations with the cluster of determinants for rosuvastatin response of LDL particle number (LDL-P; cluster B) are not explained by a simple model of cholesterol and triglyceride regulation. The major exception to this is the APOE-C1-C2 SNP, rs4420638, which is in linkage disequilibrium with APOE4 (rs429358; R2=0.7). A well-studied variant in plasma lipid metabolism, APOE4 influences LDL-receptor binding31, but has not previously shown evidence of association with statin-induced response in LDL-C12, 32. Of the remaining genes in this cluster, ANGTPL4 and GCKR have been highlighted for roles in regulation of triglyceride levels, but it is not obvious why SNPs in these genes are specifically associated with statin response of LDL-P and not, for example, mean LDL particle size. GCKR's regulation is mediated through effects on glucokinase and therefore glucose metabolism33, 34, and these effects are manifest in association with LDL-P at baseline (Supplemental Table 5). ANGTPL4 appears to modulate the triglyceride hydrolyzing activity of lipoprotein lipase35, 36 and was also associated with LDL-P at baseline. Half of the candidate genes in this cluster are also associated with baseline LDL-P, which suggests the genes that mediate the number of circulating LDL particles may also play a role in change of LDL particle number on statin therapy. However, SNPs associated with baseline LDL-P are not particularly enriched in the statin response LDL-P cluster compared to the other three clusters, indicating a complex mechanism may influence statin-induced LDL-P response.
While the gene cluster identified as cluster C (containing candidate variants from the MOSC1, ANGPTL3, CTF1 and OR4C46 genes) was most significantly associated with statin-induced changes in apo B, there was much less specificity of this cluster for change in apo B compared to the LDL-size and LDL-P clusters (clusters A and B). In part, the lack of specificity may be due to mechanisms influencing change in apo B that were not adequately captured by selecting candidate SNPs from analyses of circulating lipoproteins. We did not observe an association between the APOB SNP and statin-induced change in any of the LDL-related measures analyzed (Supplemental Table 6a), and only observed associations with baseline apo B and LDL-P that were marginally significant (p=0.085 and p=0.054; Supplemental Table 4). It is possible the enrollment criteria based on low LDL-C levels affected the distribution of baseline apo B and thus affected the power to detect variants associated with higher levels of apo B; there have not been any prior associations of the APOB SNP and statin-induced reduction in LDL-related measures or subfractions.
Several strengths and limitations should be considered when interpreting our results. The chief strength of the study is the unique nature of the data representing a large population-based sample with measures of lipoprotein particle concentration by NMR at baseline and after 1 year of follow up after randomized allocation to rosuvastatin or placebo. The study also benefited from the large scale of recent the genome-wide genetic analysis of conventional plasma lipid measures among up to 200,000 individuals10 identifying 62 loci beyond the 95 that had been identified previously9, all combined with 6 loci with prior evidence for effects on statin response directly12, 26. The large number of candidates poses a risk for associations due to chance. However, this risk is offset by the strong prior evidence for roles of the candidate SNPs in lipoprotein metabolism or statin response. Moreover, our statistical criteria included verified interaction with randomized allocation to placebo. Our a priori selection of common candidate variants does not address the possibility of rare genetic variants with effects on statin-induced changes in LDL-related subfractions or non-HDL-C. Targeted sequencing of candidate genes or whole exome sequencing would be ideal methods to implement in future investigations. We also acknowledge our lack of replication as a limitation. Ideally, we would replicate our analysis in an independent sample; however an adequately powered sample with genotypes and the biomarkers examined in our study is currently not available. Finally, it is possible that the ascertainment in JUPITER could influence association with genetics and potentially limit generalizability; although any limitations on the population variance, as for example the LDL < 130 mg/dL study entry criterion, would be expected to diminish rather than accentuate the strength of association. In addition, at baseline, the strongest GLGC SNPs remain associated with lipid fractions in the JUPITER sample (Supplemental Table 5). The recently published GIST consortium paper32, which did not represent trials or studies ascertained on lipoprotein level and did not include JUPITER in discovery, observed highly comparable loci for statin-induced change in LDL-C that were identified in JUPITER12. Thus, we believe that the other genetic associations we report with statin response are likely to be generalizable.
Focusing on LDL-related biomarker alternatives to LDL-C, the genetic associations reported here highlight pathways for statin response of LDL particle number, LDL particle size, and apo B that differ at least in part from pathways for statin response of LDL-C. The clinical literature evaluating these alternative LDL-related measures in outcomes-driven statin trials has been limited to comparing LDL-C with apo B or non-HDL-C (the amount of cholesterol carried by apo B particles)37-42. Meta-analysis of eligible studies suggests, for example, that achieved levels of apo B and non-HDL-C may more accurately reflect residual CV risk on statin therapy than achieved LDL-C levels8, 43, 44. These studies also emphasize the possibility that variation in the residual risk assessed by on-statin LDL-C levels may reflect discordance between statin responses of LDL-C and the alternative LDL-related measures45. It remains to be seen whether statin modification of the pathways identified by the current genetic analysis for LDL particle number and size may influence CV risk in ways that are distinguishable from statin effects on LDL-C alone.
While the primary focus of this study was to examine differential genetic effects on statin-induced changes in LDL-related subfractions and non-HDL-C, statin therapy is also known to beneficially influence levels of other lipoproteins such as HDL-C13. To fully explore alternate the pathways influencing differential CVD risk reduction by statin therapy, compared to Niemann-Pick or CETP inhibitors, future investigations would benefit from examination of genetic influences on statin-induced changes in HDL-C and other lipid biomarkers.
In conclusion, when examining LDL-related biomarker alternatives to LDL-C, we found that the genetic pathways for statin response of LDL particle number, LDL particle size, and apo B only partially overlapped with pathways for statin response of LDL-C. These differences in LDL-related statin responses may provide potential therapeutic targets that could be exploited to reduce residual CV risk for individuals on statin therapy.
Supplementary Material
Acknowledgments
We thank Jean MacFadyen, Lynda Rose, the JUPITER study participants, and the >1000 physicians worldwide for their personal time, effort, and commitment to JUPITER.
Funding Sources: This work was supported by research funds from AstraZeneca to Drs Ridker and Chasman. The JUPITER trial was funded by AstraZeneca. Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number R01HL117861 to Dr Mora and T32 HL007575 to Dr. Ridker. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Clinical Trial Registration - http://clinicaltrials.gov; Unique Identifier: NCT00239681
Conflict of Interest Disclosures: Drs Barratt, Ding and Nyberg are employees of AstraZeneca. Dr Ridker reports being listed as a coinventor on patents related to the use of inflammatory biomarkers in cardiovascular disease, which are held by the Brigham and Women's Hospital and have been licensed by Siemens and AstraZeneca. Dr Ridker receives funding from the National Heart, Lung, and Blood Institute, the National Cancer Institute, AstraZeneca, Sanofi-Aventis, and Novartis. Dr. Mora has received research support from AstraZeneca, Atherotech Diagnostics, and NHLBI, served as a consultant to Quest Diagnostics, Lilly, Sanofi-Genzyme, Pfizer, and Cerenis Therapeutics. The other authors report no conflicts.
References
- 1.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 2.Mora S, Glynn RJ, Boekholdt SM, Nordestgaard BG, Kastelein JJ, Ridker PM. On-treatment non-high-density lipoprotein cholesterol, apolipoprotein B, triglycerides, and lipid ratios in relation to residual vascular risk after treatment with potent statin therapy: JUPITER (Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin). J Am Coll Cardiol. 2012;59:1521–1528. doi: 10.1016/j.jacc.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mora S, Buring JE, Ridker PM. Discordance of low-density lipoprotein (LDL) cholesterol with alternative LDL-related measures and future coronary events. Circulation. 2014;129:553–561. doi: 10.1161/CIRCULATIONAHA.113.005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr., Clark LT, Hunninghake DB, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 5.Kastelein JJ, Akdim F, Stroes ES, Zwinderman AH, Bots ML, Stalenhoef AF, et al. Simvastatin with or without ezetimibe in familial hypercholesterolemia. N Engl J Med. 2008;358:1431–1443. doi: 10.1056/NEJMoa0800742. [DOI] [PubMed] [Google Scholar]
- 6.Bots ML, Visseren FL, Evans GW, Riley WA, Revkin JH, Tegeler CH, et al. Torcetrapib and carotid intima-media thickness in mixed dyslipidaemia (RADIANCE 2 study): a randomised, double-blind trial. Lancet. 2007;370:153–160. doi: 10.1016/S0140-6736(07)61088-5. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 8.Boekholdt SM, Arsenault BJ, Mora S, Pedersen TR, LaRosa JC, Nestel PJ, et al. Association of LDL cholesterol, non-HDL cholesterol, and apolipoprotein B levels with risk of cardiovascular events among patients treated with statins: a meta-analysis. JAMA. 2012;307:1302–1309. doi: 10.1001/jama.2012.366. [DOI] [PubMed] [Google Scholar]
- 9.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen AK, Stark K, Musameh MD, Nelson CP, Romisch-Margl W, Kremer W, et al. Genetic associations with lipoprotein subfractions provide information on their biological nature. Hum Mol Genet. 2012;21:1433–1443. doi: 10.1093/hmg/ddr580. [DOI] [PubMed] [Google Scholar]
- 12.Chasman DI, Giulianini F, Macfadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic Determinants of Statin-Induced Low-Density Lipoprotein Cholesterol Reduction: The Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin (JUPITER) Trial. Circ Cardiovasc Genet. 2012;5:257–264. doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- 13.Barter PJ, Brandrup-Wognsen G, Palmer MK, Nicholls SJ. Effect of statins on HDL-C: a complex process unrelated to changes in LDL-C: analysis of the VOYAGER Database. J Lipid Res. 2010;51:1546–1553. doi: 10.1194/jlr.P002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM, Jr., Kastelein JJ, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359:2195–2207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mora S, Otvos JD, Rifai N, Rosenson RS, Buring JE, Ridker PM. Lipoprotein particle profiles by nuclear magnetic resonance compared with standard lipids and apolipoproteins in predicting incident cardiovascular disease in women. Circulation. 2009;119:931–939. doi: 10.1161/CIRCULATIONAHA.108.816181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens M. A unified framework for association analysis with multiple related phenotypes. PLoS One. 2013;8:e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team RC. R: A language and environment for statistical computing. 2014 [Google Scholar]
- 23.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. Journal of Statistical Software. 2014;61:1–36. URL: http://www.jstatsoft.org/v61/i06/ [Google Scholar]
- 24.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre AV, Consortium D, investigators M, Groop L, Mootha VK, Daly MJ, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010:6, e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangravite LM, Medina MW, Cui J, Pressman S, Smith JD, Rieder MJ, et al. Combined influence of LDLR and HMGCR sequence variation on lipid-lowering response to simvastatin. Arterioscler Thromb Vasc Biol. 2010;30:1485–1492. doi: 10.1161/ATVBAHA.110.203273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuivenhoven JA, Jukema JW, Zwinderman AH, de Knijff P, McPherson R, Bruschke AV, et al. The role of a common variant of the cholesteryl ester transfer protein gene in the progression of coronary atherosclerosis. The Regression Growth Evaluation Statin Study Group. N Engl J Med. 1998;338:86–93. doi: 10.1056/NEJM199801083380203. [DOI] [PubMed] [Google Scholar]
- 28.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993;34:1255–1274. [PubMed] [Google Scholar]
- 29.Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 30.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 32.Postmus I, Trompet S, Deshmukh HA, Barnes MR, Li X, Warren HR, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayward BE, Dunlop N, Intody S, Leek JP, Markham AF, Warner JP, et al. Organization of the human glucokinase regulator gene GCKR. Genomics. 1998;49:137–142. doi: 10.1006/geno.1997.5195. [DOI] [PubMed] [Google Scholar]
- 34.Hayward BE, Fantes JA, Warner JP, Intody S, Leek JP, Markham AF, et al. Co-localization of the ketohexokinase and glucokinase regulator genes to a 500-kb region of chromosome 2p23. Mamm Genome. 1996;7:454–458. doi: 10.1007/s003359900132. [DOI] [PubMed] [Google Scholar]
- 35.Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci U S A. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon JC, Chickering TW, Rosen ED, Dussault B, Qin Y, Soukas A, et al. Peroxisome proliferator-activated receptor gamma target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Mol Cell Biol. 2000;20:5343–5349. doi: 10.1128/mcb.20.14.5343-5349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen TR, Olsson AG, Faergeman O, Kjekshus J, Wedel H, Berg K, et al. Lipoprotein changes and reduction in the incidence of major coronary heart disease events in the Scandinavian Simvastatin Survival Study (4S). Circulation. 1998;97:1453–1460. doi: 10.1161/01.cir.97.15.1453. [DOI] [PubMed] [Google Scholar]
- 38.Simes RJ, Marschner IC, Hunt D, Colquhoun D, Sullivan D, Stewart RA, et al. Relationship between lipid levels and clinical outcomes in the Long-term Intervention with Pravastatin in Ischemic Disease (LIPID) Trial: to what extent is the reduction in coronary events with pravastatin explained by on-study lipid levels? Circulation. 2002;105:1162–1169. doi: 10.1161/hc1002.105136. [DOI] [PubMed] [Google Scholar]
- 39.Gotto AM., Jr Establishing the benefit of statins in low-to-moderate--risk primary prevention: the Air Force/Texas Coronary Atherosclerosis Prevention Study (AFCAPS/TexCAPS). Atheroscler Suppl. 2007;8:3–8. doi: 10.1016/j.atherosclerosissup.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Charlton-Menys V, Betteridge DJ, Colhoun H, Fuller J, France M, Hitman GA, et al. Targets of statin therapy: LDL cholesterol, non-HDL cholesterol, and apolipoprotein B in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS). Clin Chem. 2009;55:473–480. doi: 10.1373/clinchem.2008.111401. [DOI] [PubMed] [Google Scholar]
- 41.Holme I, Cater NB, Faergeman O, Kastelein JJ, Olsson AG, Tikkanen MJ, et al. Lipoprotein predictors of cardiovascular events in statin-treated patients with coronary heart disease. Insights from the Incremental Decrease In End-points Through Aggressive Lipid-lowering Trial (IDEAL). Ann Med. 2008;40:456–464. doi: 10.1080/07853890801964955. [DOI] [PubMed] [Google Scholar]
- 42.Ovbiagele B, Goldstein LB, Amarenco P, Messig M, Sillesen H, Callahan A, 3rd, et al. Prediction of major vascular events after stroke: the stroke prevention by aggressive reduction in cholesterol levels trial. J Stroke Cerebrovasc Dis. 2014;23:778–784. doi: 10.1016/j.jstrokecerebrovasdis.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Gotto AM, Jr., Whitney E, Stein EA, Shapiro DR, Clearfield M, Weis S, et al. Relation between baseline and on-treatment lipid parameters and first acute major coronary events in the Air Force/Texas Coronary Atherosclerosis Prevention Study (AFCAPS/TexCAPS). Circulation. 2000;101:477–484. doi: 10.1161/01.cir.101.5.477. [DOI] [PubMed] [Google Scholar]
- 44.Kastelein JJ, van der Steeg WA, Holme I, Gaffney M, Cater NB, Barter P, et al. Lipids, apolipoproteins, and their ratios in relation to cardiovascular events with statin treatment. Circulation. 2008;117:3002–3009. doi: 10.1161/CIRCULATIONAHA.107.713438. [DOI] [PubMed] [Google Scholar]
- 45.Sniderman AD. Differential response of cholesterol and particle measures of atherogenic lipoproteins to LDL-lowering therapy: implications for clinical practice. J Clin Lipidol. 2008;2:36–42. doi: 10.1016/j.jacl.2007.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.