Abstract
Background
Interleukin-18 (IL-18) is a pleiotropic cytokine centrally involved in the cytokine cascade with complex immunomodulatory functions in innate and acquired immunity. Circulating IL-18 concentrations are associated with type 2 diabetes, cardiovascular events and diverse inflammatory and autoimmune disorders.
Methods and Results
To identify causal variants affecting circulating IL-18 concentrations, we applied various omics and molecular biology approaches. By GWAS, we confirmed association of IL-18 levels with a SNP in the untranslated exon 2 of the inflammasome component NLRC4 (NLR family, CARD domain containing 4) gene on chromosome 2 (rs385076, P=2.4×10−45). Subsequent molecular analyses by gene expression analysis and reporter gene assays indicated an effect of rs385076 on NLRC4 expression and differential isoform usage by modulating binding of the transcription factor PU.1.
Conclusions
Our study provides evidence for the functional causality of SNP rs385076 within the NLRC4 gene in relation to IL-18 activation.
Keywords: gene expression, transcription factors, gene regulation, genetic variation, Interleukin 18, Inflammasome, PU.1, NLRC4
Introduction
Interleukin-18 (IL-18), a member of the interleukin-1 family, is a pleiotropic cytokine centrally involved in the cytokine cascade with complex immune-modulatory functions in innate and acquired immunity1,2. Circulating IL-18 levels are associated with diverse inflammatory and autoimmune disorders3, type 2 diabetes 4 and cardiovascular events5,6. The mechanisms through which IL-18 predisposes to these disorders are unknown. Several studies have linked genetic variations in the IL-18 system to circulating biomarker concentrations, disease susceptibility, and progression. Common sequence variants in the four genes that constitute the IL-18 system including IL18 (MIM 600953), IL18R1 (IL-18 receptor 1, MIM 604494), IL18RAP (IL-18 receptor accessory protein, MIM 604509), and the gene coding for the soluble decoy receptor IL18BP (MIM 604113), have been comprehensively genotyped and related to circulating IL-18 levels and cardiovascular events7,8. Genome-wide findings confirm associations at the IL18-BCO2 gene locus on chromosome 11 with circulating IL-18 levels9.
In a recent study Matteini et al.10 identified additional genetic variants within an 8.8 Mb region of chromosome 2 spanning the genes SRD5A2 (MIM 607306), MEMO1 (MIM 611786), DPY30 (MIM 612032), SPAST (MIM 604277), SLC30A6 (MIM 611148), and NLRC4 (MIM 606831) as being associated with circulating IL-18 levels. The NLRC4 gene (NLR family CARD domain-containing protein 4) was discussed as the most likely inflammation- and thus IL-18-related gene within this region. NLRC4 is a central component of the inflammasome, a multimeric protein complex initiating immune responses11. The NLRC4 inflammasome facilitates caspase-1-dependent inflammatory cytokine processing and pyroptosis11,12. In addition to sensing microbial pathogens, the function of the NLRC4 inflammasome is the activation of caspase-1 and subsequent proteolytic maturation of pro-IL-18 and pro-IL1β into their active forms13,14. Based on the involvement of NLRC4 in IL-18 maturation, it can be hypothesized that NLRC4 variants are most likely to influence circulating IL-18 levels via inflammasome action. Furthermore, associations of variants within the NLRC4 locus with serum IL-18 levels were recently confirmed in an IL-18 GWAS in ACS patients15. In particular, the authors identified one SNP, rs385076 which was located in a region of the NLRC4 locus with predicted regulatory relevance. However, both studies10,15 do not provide molecular data supporting a possible causal association of genetic variants within NLRC4 and circulating IL-18 levels.
The aims of the present study were (i) to validate genetic determinants of IL-18 by a GWA study, and (ii) to characterize identified loci on a molecular level by cloning the most promising candidate SNP and investigating transcriptomic and epigenetic (i.e. methylation) mechanisms in relation to IL-18.
Materials and Methods
Genome-wide association study (GWAS)
We assembled three cohorts for the discovery GWAS, comprising 9,562 individuals of European ancestery, the Gutenberg Health Study (GHS), the Framigham Heart Study (FHS) and the Cooperative Health Research in the Region of Augsburg (KORA F4) Study. For replication, three additional cohorts were included totaling 3,348 individuals (AtheroGene, Monitoring of Trends and Determinants in Cardiovascular Diseases (MONICA/KORA S1/S2/S3), PRIME study). Study protocols were approved by the local ethics committees, and all participants provided written informed consent. Details on the GWAS discovery and replication cohorts are outlined in the Supplementary Material and Supplemental Table 1.
Genotyping and Imputation
Single nucleotide polymorphisms (SNPs) were genotyped on the Affymetrix Whole-Genome Human SNP Array 6.0 in GHS and KORA F4 and the Affymetrix Human Mapping 500K Array Set and 50K Human Gene Focused Panel in FHS. SNPs were imputed based on the 1000 Genome Phase 1dataset (version 3 (build 37)). Details of genotyping platform, quality controls, and imputation for each cohort are specified in Supplemental Table 2.
In AtheroGene, MONICA/KORA S1/S2/S3, and PRIME genotyping of rs385076 and rs11606049 (or the respective proxy SNP rs5744222 in MONICA/KORA S1/S2/S3) polymorphisms was performed using 5′ nuclease assay (TaqMan assay, Applied Biosystems, Darmstadt) for replication.
Laboratory analyses
Measurement of circulating IL-18 levels
In all studies except MONICA/KORA S1/S2/S3, circulating IL-18 levels were measured using a commercially available ELISA (MBL Co, Ltd. Nagoya). The detection limit was 128 pg/mL. Intra-assay CV was 6.9%; inter-assay CV was 13%. In MONICA/KORA S1/S2/S3 serum levels of IL-18 were measured by bead-based multiple assay as described before 4 using an antibody pair and recombinant IL-18 protein from MBL (MBL Co, Ltd. Nagoya). The intra- and interassay coefficients of variation (CV) were <10.0 and <25.0%, respectively. Measurements were performed from deep-frozen samples (−80°C).
Cloning of NLRC4 rs385076 luciferase reporter gene constructs
To investigate the influence of the SNP rs385076 (T/C) on binding of the transcription factor PU.1, the 908 bp genomic region 32489378 to 32490286 on the antisense strand of the human chromosome 2 comprising the SNP rs385076 (T/C) and the putative PU.1 binding region within untranslated exon 2 of NLRC4 was amplified by PCR using genomic DNA from carriers of either the C allele or the T allele of rs385076 (T/C). The PCR products were cloned into the KpnI-digested firefly luciferase reporter vector pGL4.10[luc2] (Promega GmbH, Mannheim, Germany). SNP alleles were confirmed by sequencing.
Cloning of PU.1 overexpression plasmid
For PU.1 overexpression, the PU.1 coding sequence was amplified by PCR using macrophage genomic DNA (Supplemental Table 3) and cloned into EcoRV- and SalI -digested pVITRO2-MCS (InvivoGen, Toulouse, France). The insert was confirmed by sequencing.
Transfection of cells and measurement of luciferase activity
HEK293A cells were transfected with 0.5 μg/mL pGL4.10[luc2] containing either the rs385076 C or T allele and 0 μg/mL , 0.125 μg/mL, 0.25 μg/mL or 0.5 μg/mL pVITRO2-PU.1 using 2 μL/mL Lipofectamine2000 (Life Technologies). After 24 h, Bright-Glo Reagent (Promega) was added and cells were immediately frozen at −80 °C for 30 min. Cells were then thawed to room temperature, shaken at 14,000 rpm and luciferase activity was detected via luminescence reader.
Assessment of NLRC4 isoforms expression
Out of four NLRC4 isoforms, only isoforms 2 and 4 (ENST00000402280 and ENST00000404025) contain the 5’UTR exon 2. Therefore, the association between expression of NLRC4 isoforms 2 and 4 and isoforms 1 and 3 (ENST00000360906 and ENST00000342905) with rs385076 genotype was determined. Expression of total NLRC4 transcript expression, NLRC4 isoforms 2 and 4, NLRC4 isoforms 1 and 3 and GAPDH as reference gene was measured in monocytes from 1,444 GHS subjects via real-time qPCR using a 7900 TaqMan system (Applied Biosystems). Briefly, RNA was purified using TRIzol (Life Technologies, Darmstadt, Germany) and reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Darmstadt, Germany) according to manufacturers’ protocols. Real-time PCR was performed in a 10μl reaction volume using 5 μlL ABsolute qPCR SYBR Green Mix (Life Technologies, Darmstadt, Germany), 0.2 μl ROX Reference Dye (Jena Bioscience, Oxford, England), 0.25 ng cDNA and 150 – 500 nM primers (Supplemental Table 3). After 15 min activation at 95°C, 40 PCR cycles were run with 15 sec denaturation at 95 °C, 30 sec annealing at 60/62°C and 30 sec extension at 72°C. Primer specificity was verified by melting curve analysis. NLRC4 mRNA expression was normalized to GAPDH as an endogenous control.
Measurement of DNA methylation
For assessement of DNA methylation sites (CpG sites) in relation to IL-18 levels, genome-wide methylation was performed using the Illumina HumanMethylation450 BeadChip in a subgroup of the KORA F4 cohort (n=1,814) (for details see Supplementary Material).
Statistical analyses
Statistical analysis of genotyping data
IL-18 levels showed a skewed distribution and were natural log-transformed for analyses. For genetic association analysis, age- and sex-adjusted linear regression models were applied in all discovery and replication cohorts separately, using an additive genetic model. In GHS, genetic relatedness was considered by identifying and removing outliers based on multidimensional scaling of genetic data prior to GWAS. In FHS, linear mixed effect models were performed to account for relatedness among family members using the function lmekin from the R kinship package. KORA is a population representative subsample of the population in the study region. Cryptic relatedness in the study sample has been investigated based on genome-wide genotypic data. The degree of relatedness between samples is negligible. Therefore, no further adjustments have been made in this analysis. For meta-analysis, individual estimates of allelic effects from GHS, FHS and KORA F4 were combined after excluding genotyped and imputed SNPs not meeting the quality control filters. An inverse-variance weighted fixed-effects approach was applied as implemented in METAL16. The genome-wide significance level for the GWAS was set to P<5×10−8.
Additive effects of lead SNPs from different loci were tested in all discovery cohorts. SNPs were combined by adding the T allele dosages of rs385076 and rs11606049 for each individual resulting in a number of protective alleles between 0 and 4. The estimated percentage of explained variance for log-transformed ILe-18 by SNPs was calculated by substracting the explained variance of sex and age on loge-transformed IL-18 from the multivariable model. For replication in AtheroGene, MONICA/KORA S1/S2/S3 and PRIME associations were calculated by applying the inverse-variance based fixed-effects meta-analysis for the selected SNPs.
Conditional analysis within genome-wide significant loci
To determine if the signals at each locus were independently associated with IL-18 levels, a conditional analysis was carried out in the GHS dataset using the variant with the lowest p-value (with a region of ± 500kb) as a lead SNP using SNPTEST v2.5 (www.mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html), built-in functions.
Expression quantitative trait loci (eQTL) analysis
Associations between selected SNPs and gene expression of transcripts in cis (± 500kb) were assessed for all samples with available gene expression data in GHS (n=1,133)17, and KORA F4 (n=699)18. In both studies, gene expression analysis was performed using the Illumina Human HT-12 v3 BeadChip. Technical variables were used for adjustment as previously described18. In both studies, a linear model was used with log2-transformed gene expression as dependent variable. For SNP-gene expression analyses data were adjusted for age and sex. PheGenI (www.ncbi.nlm.nih.gov/gap/phegeni) and the Geuvadis Data Browser (www.ebi.ac.uk/Tools/geuvadis-das/) were used to compare significant eQTLs from our study to publicly available data from large scale studies.
Expression values of NLRC4 and its isoforms measured by qPCR were normalized for GAPDH Ct values prior to association analysis and are represented as deltaCt values (deltaCt_transcript = Ct_transcript – Ct_GAPDH). A linear mixed model with the plate as random variable, transcript levels as dependent variable and age and sex as covariates was used to calculate effects of rs385076 allele dosage on NLRC4 isoform expression. Reported effect sizes refer to the beta from the linear regression, which represents the change of deltaCt per rs385076 T allele.
Estimating the proportion of variance in IL-18 levels
The proportion of explained variance of log-transformed IL-18 levels attributable to genetic variants was calculated by substracting the coefficient of variation estimated from a linear model with adjustments for sex and age from the multivariate model, which additionally included the allele dosage as independent variable.
Association with cardiovascular mortality
Hazard ratios for rs385076 and rs11606049 related to cardiovascular mortality were estimated using Cox proportional hazards regression in the AtheroGene study. Analyses were adjusted for age and sex and an additive genetic model was used.
Results
GWA study of circulating IL-18 levels
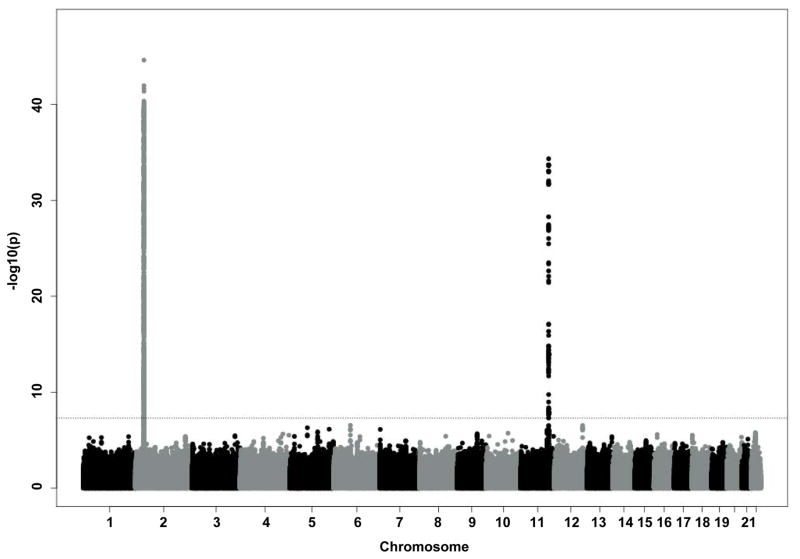
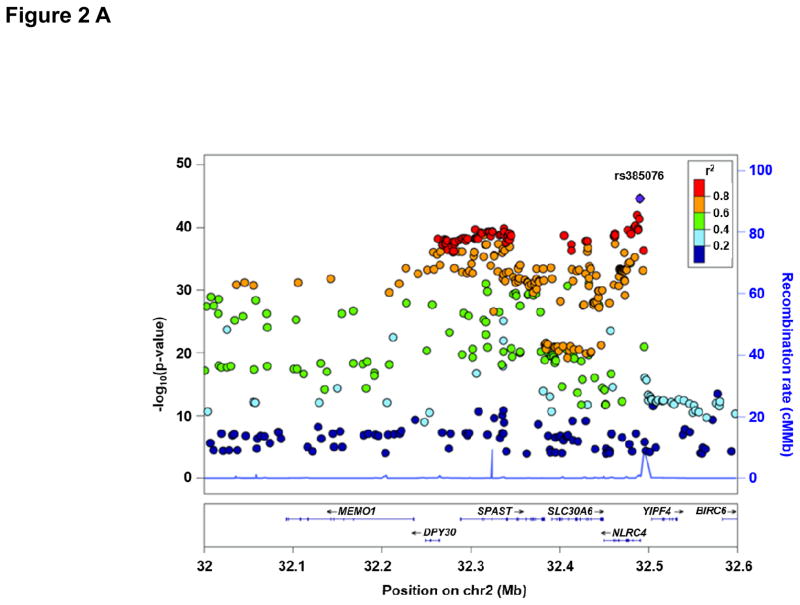
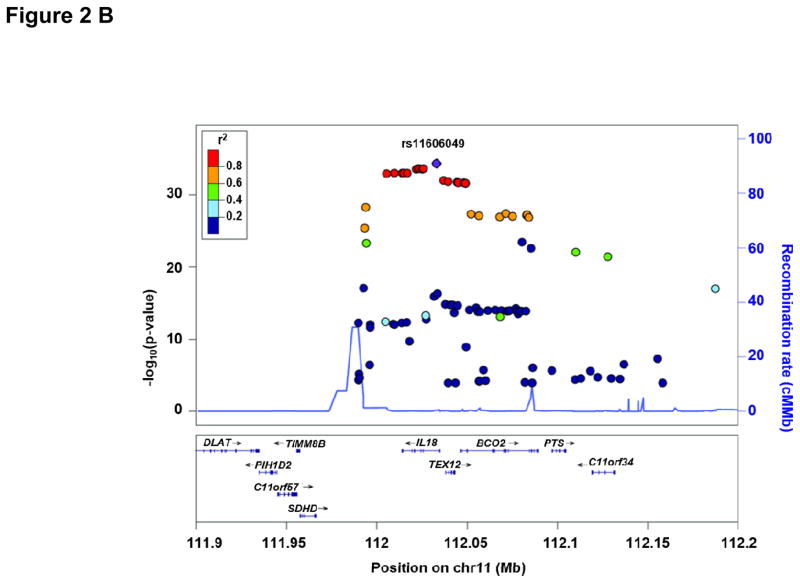
Characteristics of all discovery and replication cohorts participating in the GWA study are presented in Supplemental Table 1. Figure 1 depicts the Manhattan Plot of our initial GWAS approach demonstrating two regions of genome-wide significance for circulating IL-18 levels. Regional plots of these two regions including functional annotation are presented in Figure 2. Summary GWAS results are available in the Supplemental Table 4. The strongest evidence of association was located within the NLRC4 gene on chromosome 2 (lead SNP: rs385076, Pmeta = 2.4×10−45). On chromosome 11, the strongest signal was detected within the IL18 gene (lead SNP: rs11606049, Pmeta = 4.6×10−35). Carriers of the respective T allele of both SNPs had lower IL-18 levels. Successful replication of these two SNPs (or the respective proxy SNP rs5744222 in MONICA/KORA S1/S2/S3) was carried out in three independent cohorts (AtheroGene (n=1,165), PRIME (n=440), MONICA/KORA S1/S2/S3 (n=1,743)). T allele frequencies ranged between 35.4% and 39.7% for rs385076 and between 19.9% and 26% for rs11606049 respectively (Table 1). The associations of rs385076 and rs11606049 explained between 2.05% and 2.55% (rs385076) and 1.51% and 3.32% (rs11606049) of the inter-individual variability of IL-18 levels among studies (Table 2). Combination of the lead SNP allele dosages from both loci led to an explained variance between 3.15% (FHS) and 5.49% (GHS) (Table 2). After conditional analysis on the most significant SNP on chromosome 2 (rs385076) and chromsome 11 (rs11606049), no additional SNPs remained significant at the discovery p-value threshold of <5×10−8 (Supplemental Figure 1 A, B).
Figure 1.
Results from the GWAS meta-analysis of IL-18 concentrations in GHS I, GHS II, FHS and KORA F4. The Manhattan plot shows the associations between allele dosages of 1000- Genomes imputed variants and loge-transformed IL-18 concentrations for all autosomes. The horizontal line indicates the genome-wide significance level (P<5×10−8).
Figure 2.
Regional plot for the NLRC4 locus (A) and the IL18 locus (B). P-values of SNPs from loge IL-18 meta-analysis were plotted as –log10 values against their physical position on chromsome 2 (NLRC4) and chromosome 11 (IL18), respectively (NCBI build 37). (A) The lead SNP rs385076 on chromsome 2 is represented by a blue diamond and lies within the 5’ region of NLRC4. (B) The lead SNP rs11606049 on chromsome 11 is represented by a blue diamond and lies within an intronic region of the IL18 gene. The color code for the pairwise linkage disequilibirum structure is based on 1,000-Genomes 2012 EUR.
Table 1.
Effects of lead SNPs on circulating IL-18 levels in discovery and replication
| Meta-analysis | AtheroGene | PRIME | MONICA/KORA S1/S2/S3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP (coding/non coding allele) | Chr | Gene | Beta | SE | P | AF† | Beta | SE | P | AF† | Beta | SE | P | AF† | Beta | SE | P | AF† |
| rs385076 (T/C) | 2 | NLRC4 | −0.093 | 0.007 | 2.4×10−45 | 35.6 | −0.107 | 0.017 | 9.0×10−10 | 39.7 | −0.098 | 0.032 | 0.002 | 35.4 | −0.107 | 0.026 | 4.0×10−5 | 36.5 |
| rs11606049 (T/C) | 11 | IL18 | −0.089 | 0.007 | 4.6×10−35 | 24.5 | −0.060 | 0.020 | 0.0027 | 26.0 | −0.109 | 0.029 | 0.004 | 19.9 | - | - | - | |
| rs5744222 (A/C)* | −0,085 | 0,007 | 9.2×10−32 | 75.1 | - | - | - | - | - | - | - | −0.110 | 0.030 | 3.0×10−4 | ||||
Beta refers to the effect estimate and SE to the standard error from the linear regression model after adjustment for sex and age. For each SNP, the chromosome (Chr) and nearest gene (Gene) is shown. Discovery meta-analysis in GHS I and II, FHS and KORA F4. AtheroGene, PRIME and MONICA/KORA S1/S2/S3 were used as independent replication cohorts.
proxy SNP for rs11606049 with R² > 0.9 according to SNAP19. † Allele frequency (AF) of the coding allele in %.
Table 2.
Variance of circulating IL-18 levels explained by SNPs.
| Variant (Allele) | Gene | Explained variance (R²) | |||
|---|---|---|---|---|---|
| GHS I | GHS II | KORA F4 | FHS | ||
| Sample size | 2,743 | 1,073 | 1,802 | 2,940 | |
| rs385076 (T) | NLRC4 | 2.05% | 2.22% | 2.55% | 2.19% |
| rs11606049 (T) | IL18 | 1.51% | 3.32% | 1.89% | 1.03% |
| rs385076 (T) + rs11606049 (T) | NLRC4 +IL18 | 3.68% | 5.49% | 4.50% | 3.15% |
SNPs were combined by adding the allele dosages of rs385076 T and rs11606049 T for each individual resulting in a number of protective alleles between 0 and 4. The R² is the estimated percentage of explained variance for loge-transformed IL-18 by SNPs. It is calculated by substracting the explained variance of sex and age on log-transformed IL-18 from the multivariate model.
Our meta-analysis data confirmed previous results of strong associations of circulating IL-18 levels with loci on chromosome 2 and 119,10. For the IL18 locus on chromosome 11, previous reports have already investigated the functional consequences of variants within this locus7,9,20. However, to our knowledge, for the locus on chromosome 2, no functional data in relation to circulating IL-18 levels have been presented. Therefore, we performed in silico and molecular characterization of the NLRC4 locus.
In silico functional analysis of the NLRC4 locus
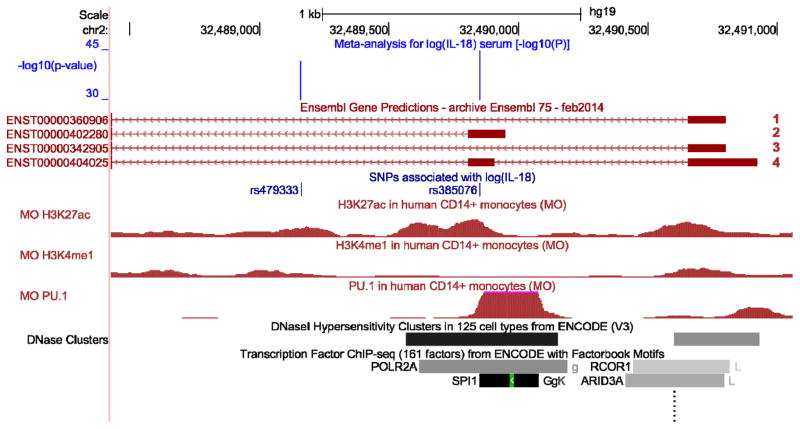
Regulatory and epigenetic features of the NLRC4 locus from the Encyclopedia of DNA Elements (ENCODE)21 and a data set by Pham et al.22 were explored using UCSC Genome Browser23 and are displayed in Figure 3. The NLRC4 gene encompasses 4 distinct isoforms and the SNP rs385076 lies within the 5′UTR of the NLRC4 cDNA, in the untranslated exon 2 included in isoforms 2 and 4. This region directly falls with in a region enriched for the H3K27ac histone acetylation mark (often found near active regulatory elements) in human CD14+ monocytes and several additional cell lines22. Furthermore, the region surrounding the SNP rs385076 lies within a DNase hypersensitivity cluster in multiple cell lines as indicated by ChIP-seq data. DNase hypersensitivity is characteristic for open chromatin regions, which are accessible for DNA binding proteins. ChIP-seq data in monocytes22 showed that strong binding of the transcription factor PU.1 occurs around rs385076. Together, these in silico data point towards regulatory region(s) within the NLRC4 5'UTR and suggest that different alleles of rs385076 might lead to different NLRC4 expression levels.
Figure 3.
In silico functional analyses for rs385076 within the NLRC4 gene region. Co-localization of IL-18 associated variants and regulatory and epigenetic features were investigated in the UCSC Genome Browser (genome-euro.ucsc.edu) using genome release hg19. Only SNPs with P-values <5×10−8 for the association with circulating IL-18 levels are shown (green track). Tracks showing histone modifications and PU.1 binding sites were uploaded into the UCSC Genome Browser from an external study22 (red tracks). The PU.1 track represents binding sites of the transcription factor PU.1 in monocytes from a CHIP-seq experiment. The H3K27ac track shows histone modification by acetylation from CHIP-seq experiments in monocytes, which can be considered as a mark for active enhancers26. The H3K4me1 mark shows regions undergoing histone methylation, which are likely to reflect cell-type specific regulation of a gene27. Enhancer regions are highlighted by DNaseI hypersensitivity clusters from the ENCODE project21. In the bottom track, CHIP-seq results for transcription factors (TF) from the ENCODE project are shown, indicating TF binding regions. rs385076 falls into the 5‘UTR exon of NLRC4 and a binding region of PU.1. DNase hypersensitivity clusters around rs385076 suggest a good DNA accessibility in monocytes and histone modification data highlights the region as an enhancer.
NLRC4 expression may be affected in several ways: i) a direct effect on NLRC4 mRNA expression, ii) effects on differential mRNA isoform usage, iii) an effect on NLRC4 protein level and iv) by modulated binding of transcription factor PU.1. These possibilities were investigated at the molecular level.
Association of rs385076 and NLRC4 gene expression
To determine the association of NLRC4 mRNA expression and the SNP rs385076, the expression data sets of the GHS (monocytes) and KORA F4 (whole blood) studies were used. In monocytes, our data demonstrated an impact of rs385076 on NLRC4 expression levels with a decrease of NLRC4 mRNA in carriers of the T allele (beta=−0.037; P=3.95×10−3). In contrast, in our whole blood dataset, no association between rs385076 and NLRC4 expression was observed (beta=−0.085; P=0.14). We further compared our results to publicly available eQTL data of lymphoblastoid cell lines from 270 individuals24 using PheGenI. The lymphoblastoid data were in line with our monocytic data, as decreased NLRC4 mRNA expression for rs479333 C allele carriers (a proxy SNP of rs385076, R2=0.83) was described (beta= −0.133; P=3.4×10−8). In RNA-seq data from lymphoblastoid cell lines of 373 European individuals from the 1000 Genomes project25, which is publicly available at the Geuvadis Data Browser, rs385076 T allele was strongly associated with NLRC4 gene expression (beta=−0.373; P=9.1×10−14) . A summary of rs385076 and rs479333 eQTLs is provided in Supplemental Table 5.
Association of rs385076 and NLRC4 isoform expression
Expression of the NLRC4 gene including all isoforms as well as two distinct groups of NLRC4 mRNA isoforms were determined in monocytes of 1,487 GHS participants by qPCR. The group NLRC42,4 (encompassing isoforms 2 and 4) includes the rs385076 containing exon 2, whereas the group NLRC41,3 (encompassing isoforms 1 and 3) does not include the rs385076-exon 2 as depicted in Figure 3.
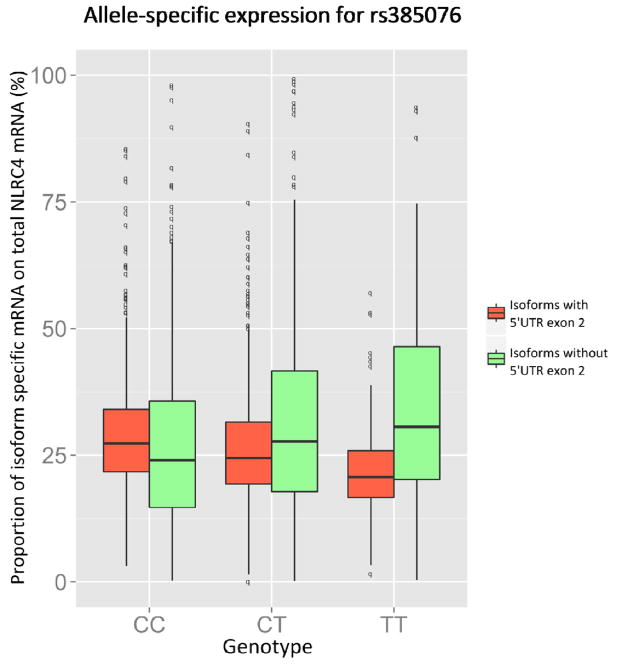
Consistent with our monocytic gene expression data, overall NLRC4 expression was decreased in carriers of the rs385076 T allele (beta=0.134; SE=0.05; P=6.9×10−3). Interestingly, expression of NLRC42,4 (beta= 0.305; SE=0.047; P=2.0×10−10) and NLRC41,3 (beta= −0.103; SE=0.03; P=7.4×10−4) isoforms were associated with the T allele in opposite directions. Comparing the proportion of NLRC42,4/NLRC41,3 isoforms on overall NLRC4 gene expression, we observed a shift towards usage of isoforms 1 and 3 in carriers of the rs385076 T allele as depicted in Figure 4. In relation to increased IL-18 concentrations, NLRC41,3 was significantly lower (beta=0.027; SE=0.014; P=0.049), whereas overall NLRC4 (beta=0.014; SE=0.008; P=0.092) and NLRC42,4 expression (beta=0.009; SE=0.008; P=0.254) showed no significant change (Table 3).
Figure 4.
Allele-specific NLRC4 isoform expression in relation to the rs385076 genotype. Box plots show the distribution of different isoform usage, which is defined as proportion of isoform expression on overall NLRC4 gene expression, for each allele of rs385076. All transcripts were quantified by qPCR and normalized for Ct values of the housekeeping gene GAPDH. Isoforms including 5’UTR exon (isoforms 2 and 4) are marked red, others (isoforms 1 and 3) are marked green. We can observe a switch in isoform usage with a higher number of T alleles leading to a higher proportion of transcripts, which do not include 5’UTR exon.
Table 3.
Allele-specific NLRC4 isoform expression in relation to the rs385076 genotype.
| Association between mRNA (deltaCt) and rs385076 T alleles | Association between mRNA (deltaCt) and log(IL-18) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isoforms | Description | delta Ct | Beta | SE | P | N | Beta | SE | P | N |
| 2, 4 | Including exon 2 | 5.1±1.4 | 0.305 | 0.047 | 2.0×10−10 | 1,175 | 0.009 | 0.008 | 0.254 | 1,459 |
| 1, 3 | Excluding exon 2 | 5.2±0.8 | −0.103 | 0.030 | 7.4×10−4 | 1,171 | 0.027 | 0.014 | 0.049 | 1,438 |
| 1, 2, 3, 4 | Total NLRC4 mRNA | 3.2±1.3 | 0.134 | 0.050 | 6.9×10−3 | 1,180 | 0.014 | 0.008 | 0.092 | 1,467 |
Expression levels of mRNA are represented as deltaCt values (deltaCt(transcript) = Ct(transcript) – Ct(GAPDH)), which are normalized for the housekeeping gene GAPDH. Beta estimates were calculated in a linear mixed model and refer to the change in deltaCt for each T allele of rs385076 and the change in log(IL-18), respectively. Therefore, a negative beta implies an increase in gene expression. N refers to the number of individuals with non-missing values used for a test.
PU.1 binding and influence on NLRC4 expression in relation to rs385076 alleles
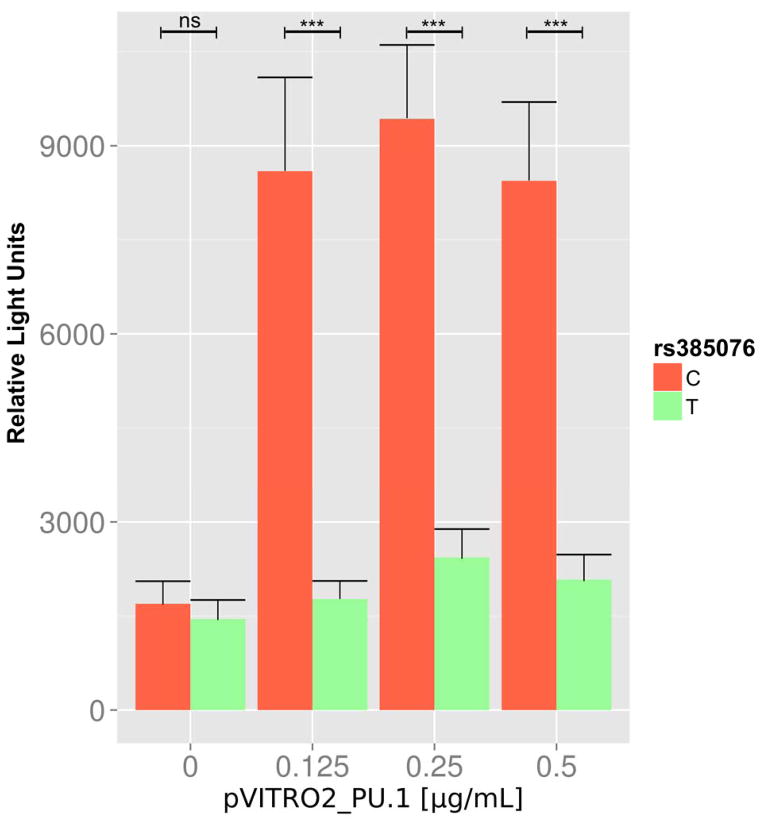
In order to evaluate whether the rs385076 variant influences binding of the transcription factor PU.1 and subsequently NLRC4 expression, a luciferase reporter gene assay in the presence of overexpressed PU.1 levels were performed. Overexpression of PU.1 was confirmed on mRNA and protein levels (Supplemental Figure 2). Without PU.1 overexpression, no significant difference in luciferase activity between the rs385076 C and T alleles was observed. However, increasing PU.1 concentrations significantly raised luciferase activity in the presence of the rs385076 C allele but not the T allele (Figure 5).
Figure 5.
PU.1 overexpression increases NLRC4 gene expression in relation to the rs385076 C allele. HEK293A cells were transfected with pGL4.10[luc2] plasmid, containing either rs385076 C (shown in red) or T allele (shown in green) and the putative PU.1 binding site, as well as the pVITRO2-PU.1 plasmid. Expression of yellow fluorescent protein after transfection of an empty pEYFP-N1 plasmid was used to control for transfection efficiency. PU.1 overexpression significantly increased luciferase activity of the rs385076 C allele but not the T allele in the plasmids. Without PU.1 overexpression, there was no significant difference between C and T alleles. Cells with 0.125 μg/mL (4.24-fold, P=3×10−4), 0.25 μg/mL (3.34-fold, P=3×10−4) and 0.5 μg/mL (3.5-fold, P=5×10−4) of the pVITRO2-PU.1 plasmid showed significant increases of luciferase activity for the C allele compared to the T allele. ns = not significant, *** P <0.001; n=4.
DNA methylation in relation to circulating IL-18 levels and NLRC4 expression
We screened for genome-wide associations between DNA methylation in whole blood and circulating IL-18. As shown in Supplemental Figure 3, six CpG–IL-18 associations at the Bonferroni threshold 1×10−7 were identified. No genome-wide significant CpG sites were found for the chromosome 2 (NLRC4) locus. However, two CpG sites cg07055315 and cg22805603 within the NLRC4 locus showed moderate associations (P=2.25×10−5; P=8.5×10−3, respectively). Genome-wide significance was found for the two CpG sites on chromosome 16 (cg07839457 (P=2.85×10−12); cg16411857 (P=8.67×10−08)), both located within the NLRC5 gene, another member of the NLRC inflammasome protein family.
Clinical relevance of the IL-18 GWAS loci
As circulating IL-18 levels have been associated to cardiovascular disease (MIM 611139) in previous studies5,6,28, a possible link between the SNPs in NLRC4 and IL-18 and cardiovascular outcome was investigated. For this, we tested the association of rs385076 and rs11606049 with cardiovascular death in 2,585 coronary heart disease patients of the AtheroGene cohort (median follow-up 4.9 years, maximum 7.6 years, 159 cardiovascular deaths). The rs385076 T allele, related to lower IL-18 concentration, was associated with a protective effect on cardiovascular mortality with an age- and sex-adjusted hazard ratio of 0.78 (95% confidence interval: 0.62–0.98, p=0.03). However, the IL-18 lowering allele of rs11606049 was not significantly associated with cardiovascular mortality.
Discussion
Our study aimed to confirm and functionally characterize genetic determinants of circulating IL-18 levels by molecular approaches. Our molecular data show that the SNP rs385076 affects NLRC4 expression and differential NLRC4 isoform usage by influencing the binding of the transcription factor PU.1. Our data suggest that the IL-18-lowering T allele of rs385076 associates with cardiovascular events in a protective manner, thereby providing a link for clinical relevance.
Molecular analyses of circulating IL-18 levels
Genetic determinants of circulating IL-18 levels
Our GWA meta-analysis and replication study revealed two loci, on chromsomes 2 and 11, that associated with circulating IL-18 levels at genome-wide significance. Previous GWA studies reported significant associations of several SNPs on both of these chromosomes with circulating IL-18 levels9,10. The IL18-BOC2 locus associated with IL-18 levels in two independent GWAS samples with the strongest association found for SNP rs2115763, located in intron 2 of BCO29. Our data confirmed the strong association of this BCO2 SNP and showed additional SNPs with even stronger association in the IL18 gene region, including our top SNP rs11606049 (P=4.6×10−35). Moreover, our GWAS and conditional studies confirm and extend findings on further genetic determinants of circulating IL-18 identified by Matteini et al.10 and Johannson et al.15 on chromosome 2 and point towards NLRC4 as the “determinant” locus within this region. SNP rs385076 showed the strongest association of all SNPs in our data (beta: -0.093, SE: 0.007, P=2.4×10−45) and is located in the untranslated exon 2 (also designated as 5′UTR) of NLRC4, suggesting a functional effect of this SNP. This assumption was further strengthened by our in silico investigation of the NLRC4 5′untranslated region and our experimental data.
Effect of SNP rs385076 on NLRC4 gene expression and function
NLRC4 encodes a cytosolic protein with a caspase recruitment domain (CARD) found primarily in monocytes and macrophages. Upon activation, NLRC4 assembles into a multiprotein complex, the inflammasome, which in turn leads to activation of caspase-1 and subsequent maturation of IL-1813,14. Our in silico functional analyses of the NLRC4 5′UTR/exon 2 region point towards the presence of a binding region of the transcription factor PU.1 within the exact region, where the top GWAS SNP (rs385076) is located.
Our molecular data suggested that rs385076 influences the binding of the transcription factor PU.1 to the NLRC4 5′UTR region, with reduced PU.1 binding to the NLRC4 5′UTR region, in the presence of the IL-18-lowering T allele. The most obvious effect of this influence would be an impact on NLRC4 mRNA expression. Indeed, our data demonstrated that carriers of the IL-18-lowering T allele of rs385076 had a decreased NLRC4 mRNA expression in monocytes. Evidence for eQTLs was corroborated in silico in two publicly available studies using lymphoblastoid cell lines24. In monocytes, we observed decreased expression of NLRC4 isoforms containing the 5’UTR exon 2 in carriers of rs385076 T alleles (isoforms 2 and 4) and a switch in isoform usage towards those without exon 2 (isoforms 1 and 3). We detected an association between isoforms without the 5’UTR exon 2 and decreased circulating IL-18, but it still needs to be investigated in more detail, whether an enhanced expression of these isoforms leads to reduced activation of IL-18. Compared to monocytes, our analyses in whole blood showed no significant association between rs385076 and NLRC4 gene expression. Since the inflammasome is mainly active in the innate immune system and is thus more prominent in monocytes, monocyte-specific gene expression might be narrowed by other cell types present in whole blood.
In addition to an influence on PU.1 binding, ENCODE data suggests characteristics for an open chromatin region (hypersensitivity cluster) as well as a binding site for the polymerase 2 (Pol2) around the region of rs385076. Also, phosphorylation of the NLRC4 protein on a specific serin residue also has been described to play a critical role, possibly by driving conformational changes necessary for inflammasome activation29. In our data, SNPs located near this serine residue in the exon 5 also showed a strong association with circulating IL-18 levels (rs408813, P=2.5×10−40 and rs455060, P=5.04×10−34). Epigenetic modifications mirrored by methylation status showed no statistically significant relation of NLRC4 with IL-18 levels in our data. However, methylation analysis identified an additional inflammasome-linked gene, NLRC5, known to play a role in the regulation of IL-18 levels30,31. Thus, further experimental work is required to assess the relation between regulatory elements, inflammasome orchestration and IL-18 levels. Our results of the molecular analyses indicate a decreased NLRC4 gene expression and a switched isoform usage for the rs385076 T allele, putatively modulated by differential PU.1 binding. Since the NLRC4 inflammasome plays a role in IL-18 activation, our observations are consistent with the finding that rs385076 T alleles lead to lower circulating IL-18 concentrations.
Genetic variation and cardiovascular mortality
Since it had previously been shown that circulating IL-18 levels are a marker of cardiovascular mortality in coronary heart disease (CHD)5,32,33, we evaluated the association the SNPs rs385076 (NLRC4) and rs11606049 (IL18) with incident fatal cardiovascular events in 2,585 CHD patients in the AtheroGene study. The IL-18-lowering allele of rs385076 was associated with a lower risk of cardiovascular mortality (OR=0.78, 95% CI: 0.62–0.98, P=0.03). This observation of an association of the genotype with IL-18 concentrations and potentially clinically relevant outcomes could support a causal relationship in disease evolution. Although variations of the IL18 gene had previously been shown to influence circulating IL-18 levels and clinical outcome in patients with coronary artery disease7, our study did not show an association of our lead IL-18 SNP rs11606049 with cardiovascular mortality. Since the inflammasome is a central component to inflammatory activity and immune response, our findings may be applicable beyond the cardiovascular system to a broad range of inflammatory and autoimmune diseases3 and diabetes mellitus4. The detailed work-up of the pathophysiological pathways involved in the genetic and posttranscriptional NLRC4 regulation will show whether causal relations with disease can be identified. Such work may reveal risk indicators and, more importantly, potential therapeutic targets.
Strengths and Limitations
The present study applies a combination of genetic, molecular and methylation approaches to gain insights into the regulation of circulating IL-18 levels in large, well-characterized studies. Nevertheless, it is important to address some limitations. The present data are restricted to individuals of European descent and associations may be different in other ethnicities. The question, whether NLRC4 protein levels associate with NLRC4 gene expression in relation to the SNP rs385076, still remains to be answered. We were not able to show a strong relation of SNP rs385076 with NLRC4 protein levels in peripheral blood mononuclear cells (PBMCs) in 200 individuals from the community-based Gutenberg Health Study (data not shown). This null finding could be due to the low number of samples investigated in rather healthy individuals. Furthermore, the NLRC4 protein was measured in PBMCs, whereas an effect of rs385076 on NLRC4 gene expression was observed in monocytes. Further in-depth experimental studies as well as sequencing approaches are needed to better understand the implications of these newly identified genetic variants in relation to the exact regulation of inflammatory as well as autoimmune pathways. Lastly, DNA methylation data were only available from whole blood. Here, we did not see a significant association of CpG sites with NLRC4. In order to assess whether NLRC4 expression in relation to IL-18 might be influenced by methylation in a cell-type specific manner, additional methylation analyses, e.g in monocytes need to be performed.
In summary, using multiple molecular and genetic approaches we confirmed and extended previous knownledge in relation to circulating IL-18 levels within NLRC4. Genetic variants in NLRC4 affect binding of the transcription factor PU.1, suggesting thereby a mediation of the influence on circulating IL-18 levels. The clinical importance of our results is underlined by an association between rs385076 T allele and a lower risk of cardiovascular mortality. The exact mechanisms in relation to cardiovascular disease and other inflammatory and autoimmune diseases needs to be established and should stimulate further epidemiological and experimental studies.
Supplementary Material
Acknowledgments
Funding Sources: Many funding agencies contributed to this work. None of the funding organizations had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript. This work was supported by the German Center for Cardiovascular Research (DZHK, 81Z1710101) and the Deutsche Herzstiftung (F/21/12) to Tanja Zeller.
Gutenberg Health Study
The Gutenberg Health Study is funded through the government of Rhineland-Palatinate (“Stiftung Rheinland-Pfalz für Innovation”, contract AZ 961-386261/733), the research programs “Wissen schafft Zukunft” and “Center for Translational Vascular Biology (CTVB)” as well as the Center for Thrombosis and Hemostasis (CTH) of the Johannes Gutenberg-University of Mainz, and its contract with Boehringer Ingelheim and PHILIPS Medical Systems, including an unrestricted grant for the Gutenberg Health Study. Specifically, the work described here was supported by the German Heart Foundation (F/21/12).
Framingham Heart Study
This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The work was also supported in part by grants from the National Institutes of Health HL076784, AG028321, HL064753 to Dr. Benjamin, and Kalkhof-Rose Foundation for Young Researchers and Deutsche Forschungsgemeinschaft (German Research Foundation) Research Fellowship SCHN 1149/1-1 to Dr. Schnabel. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the NIH NCRR (National Center for Research Resources) Shared Instrumentation grant (1S10RR163736-01A1) and the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
MONICA/KORA
The MONICA/KORA studies were initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFNPlus, project number 01GS0834), the German Research Foundation (DFG: TH 784/2-1) and through additional funds from the University of Ulm. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. IL-18 measurements in KORA F3 were funded by the German Center for Cardiovascular Research (DZHK). This work was further supported by the Ministry of Science and Research of the State of North Rhine-Westphalia (MIWF NRW) and the German Federal Ministry of Health (BMG). Additionally, this study was supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.). AtheroGene The AtheroGene study was supported by a grant of the ‘Stiftung Rheinland-Pfalz für Innovation,’ Ministry for Science and Education (AZ 15202–386261/545), Mainz. This work was further supported by research grants from the Brandenburg Ministry of Economics, Germany, and the European Regional Development Fund (EFRE/ERDF).
Footnotes
Conflict of Interest Disclosures:
None.
References
- 1.Okamura H, Tsutsi H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 2.Dinarello CA, Fantuzzi G. Interleukin-18 and host defense against infection. J Infect Dis. 2003;187 (Suppl 2):S370–384. doi: 10.1086/374751. [DOI] [PubMed] [Google Scholar]
- 3.Muhl H, Pfeilschifter J. Interleukin-18 bioactivity: a novel target for immunopharmacological anti-inflammatory intervention. Eur J Pharmacol. 2004;500:63–71. doi: 10.1016/j.ejphar.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Thorand B, Kolb H, Baumert J, Koenig W, Chambless L, Meisinger C, et al. Elevated levels of interleukin-18 predict the development of type 2 diabetes: results from the MONICA/KORA Augsburg Study, 1984–2002. Diabetes. 2005;54:2932–2938. doi: 10.2337/diabetes.54.10.2932. [DOI] [PubMed] [Google Scholar]
- 5.Blankenberg S, Tiret L, Bickel C, Peetz D, Cambien F, Meyer J, et al. Interleukin-18 is a strong predictor of cardiovascular death in stable and unstable angina. Circulation. 2002;106:24–30. doi: 10.1161/01.cir.0000020546.30940.92. [DOI] [PubMed] [Google Scholar]
- 6.Blankenberg S, Luc G, Ducimetiere P, Arveiler D, Ferrieres J, Amouyel P, et al. Interleukin-18 and the risk of coronary heart disease in European men: the Prospective Epidemiological Study of Myocardial Infarction (PRIME) Circulation. 2003;108:2453–2459. doi: 10.1161/01.CIR.0000099509.76044.A2. [DOI] [PubMed] [Google Scholar]
- 7.Tiret L, Godefroy T, Lubos E, Nicaud V, Tregouet DA, Barbaux S, et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112:643–650. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 8.Grisoni ML, Proust C, Alanne M, Desuremain M, Salomaa V, Kuulasmaa K, et al. Lack of association between polymorphisms of the IL18R1 and IL18RAP genes and cardiovascular risk: the MORGAM Project. BMC Med Genet. 2009;10:44. doi: 10.1186/1471-2350-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He M, Cornelis MC, Kraft P, van Dam RM, Sun Q, Laurie CC, et al. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler Thromb Vasc Biol. 2010;30:885–890. doi: 10.1161/ATVBAHA.109.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matteini AM, Li J, Lange EM, Tanaka T, Lange LA, Tracy RP, et al. Novel gene variants predict serum levels of the cytokines IL-18 and IL-1ra in older adults. Cytokine. 2014;65:10–16. doi: 10.1016/j.cyto.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annu Rev Cell Dev Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 12.Leavy O. Inflammasome: NAIPs: pathogen-sensing proteins. Nat Rev Immunol. 2011;11:644. doi: 10.1038/nri3069. [DOI] [PubMed] [Google Scholar]
- 13.Abdul-Sater AA, Said-Sadier N, Ojcius DM, Yilmaz O, Kelly KA. Inflammasomes bridge signaling between pathogen identification and the immune response. Drugs Today (Barc) 2009;45 (Suppl B):105–112. [PMC free article] [PubMed] [Google Scholar]
- 14.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 15.Johansson A, Eriksson N, Becker RC, Storey RF, Himmelmann A, Hagstrom E, et al. The NLRC4 Inflammasome Is an Important Regulator of Interleukin-18 Levels in Patients with Acute Coronary Syndromes: A Genome-Wide Association Study in the PLATO Trial. Circ Cardiovasc Genet. 2015;8:498–506. doi: 10.1161/CIRCGENETICS.114.000724. [DOI] [PubMed] [Google Scholar]
- 16.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schurmann C, Heim K, Schillert A, Blankenberg S, Carstensen M, Dorr M, et al. Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PLoS One. 2012;7:e50938. doi: 10.1371/journal.pone.0050938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbaux S, Poirier O, Godefroy T, Kleinert H, Blankenberg S, Cambien F, et al. Differential haplotypic expression of the interleukin-18 gene. Eur J Hum Genet. 2007;15:856–863. doi: 10.1038/sj.ejhg.5201842. [DOI] [PubMed] [Google Scholar]
- 21.Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pham TH, Benner C, Lichtinger M, Schwarzfischer L, Hu Y, Andreesen R, et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood. 2012;119:e161–171. doi: 10.1182/blood-2012-01-402453. [DOI] [PubMed] [Google Scholar]
- 23.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian Y, Jia Z, Wang J, Huang Z, Tang J, Zheng Y, et al. Global mapping of H3K4me1 and H3K4me3 reveals the chromatin state-based cell type-specific gene regulation in human Treg cells. PLoS One. 2011;6:e27770. doi: 10.1371/journal.pone.0027770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaptoge S, Seshasai SR, Gao P, Freitag DF, Butterworth AS, Borglykke A, et al. Inflammatory cytokines and risk of coronary heart disease: new prospective study and updated meta-analysis. Eur Heart J. 2014;35:578–589. doi: 10.1093/eurheartj/eht367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012;490:539–542. doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol. 2012;12:813–820. doi: 10.1038/nri3339. [DOI] [PubMed] [Google Scholar]
- 31.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espinola-Klein C, Rupprecht HJ, Bickel C, Lackner K, Schnabel R, Munzel T, et al. Inflammation, atherosclerotic burden and cardiovascular prognosis. Atherosclerosis. 2007;195:e126–134. doi: 10.1016/j.atherosclerosis.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Mallat Z, Corbaz A, Scoazec A, Besnard S, Leseche G, Chvatchko Y, et al. Expression of interleukin-18 in human atherosclerotic plaques and relation to plaque instability. Circulation. 2001;104:1598–1603. doi: 10.1161/hc3901.096721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.