Abstract
An anti-inflammatory phenotype with pronounced immunosuppression develops during sepsis, during which time neutrophils and monocytes/macrophages limit their Toll-like receptor 4 responses to bacterial lipopolysaccharide (LPS/endotoxin). We previously reported that during this endotoxin-tolerant state, distinct signaling pathways differentially repress transcription and translation of proinflammatory cytokines such as TNFα and IL-6. Sustained endotoxin tolerance contributes to sepsis mortality. While transcription repression requires chromatin modifications, a translational repressor complex of Argonaute 2 (Ago2) and RNA-binding motif protein 4 (RBM4), which bind the 3′-UTR of TNFα and IL-6 mRNA, limits protein synthesis. Here, we show that Dcp1 supports the assembly of the Ago2 and RBM4 repressor complex into cytoplasmic processing bodies (p-bodies) in endotoxin-tolerant THP-1 human monocytes following stimulation with LPS, resulting in translational repression and limiting protein synthesis. Importantly, this translocation process is reversed by Dcp1 knockdown, which restores TNFα and IL-6 protein levels. We also find this translational repression mechanism in primary macrophages of septic mice. Because p-body formation is a critical step in mRNA translation repression, we conclude that Dcp1 is a major component of the translational repression machinery of endotoxin tolerance and may contribute to sepsis outcome.
Key Words: Severe systemic inflammation, Sepsis, Endotoxin tolerance, MicroRNA, Toll-like receptor signaling, Dcp1, Translation repression
Introduction
Sepsis is initiated via Toll-like receptor (TLR)-mediated activation of mitogen-activated protein kinase (MAPK) and NF-κB pathways in circulating monocytes and neutrophils as well as tissue macrophages [1, 2, 3, 4, 5]. These cells respond to the initial TLR activation by rapidly increasing the production of proinflammatory cytokines such as TNFα, IL-6 and IL-1β, which in turn activate the innate and adaptive immune systems to defend against the invading microbes [5, 6, 7]. As often seen during animal and human sepsis, this early-phase systemic response is replaced by an anti-inflammatory and immunosuppressive state, which contributes to high sepsis mortality rates [5, 8]. This response is reflected by endotoxin ‘adaptation’ or tolerance of septic monocytes/macrophages and neutrophils to TLR4-dependent expression of proinflammatory TNFα, IL-6 and IL-1β cytokines upon ex vivo stimulation with bacterial endotoxin [2, 9, 10]. Endotoxin tolerance can be modeled in cultures of primary monocytes and human monocytic cell lines, such as THP-1 cells, by prolonged stimulation and subsequent restimulation with the Gram-negative bacterial lipopolysaccharide (LPS) [2, 11, 12]. This late-phase response increases the risk of recurring infections and elevates mortality, the hallmark of late/chronic sepsis in animals and humans [3, 4, 5].
We reported that proinflammatory cytokine transcription is repressed by an epigenetic complex that includes RelB, histone code and DNA modifiers in a THP-1 sepsis cell model of endotoxin tolerance and in human sepsis leukocytes [9, 13, 14, 15]. We further found that dissembling this chromatin complex reversed transcription repression, but had no effect on restoring protein synthesis [9, 11, 14]. We then discovered that translation and transcription are differentially regulated during sepsis immunosuppression and endotoxin tolerance [11, 16]. We later found that TLR4-dependent induction of microRNA-146a (miR-146a) contributed to translational repression by promoting interactions between the RNA-binding proteins Argonaute 2 (Ago2) and RNA-binding motif protein 4 (RBM4), a known translation repressor complex [16, 17, 18]. Importantly, combined knockdown of RelB and miR-146a can restore repressed transcription and translation of TNFα, IL-6 and IL-1β [16, 17]. Restored proinflammatory gene expressions improve the immune response and resolve immunosuppression, a phenotype that correlates with better sepsis survival [3, 4, 5].
We [16, 17, 19] and others [20, 21] have reported that miRNA-mediated translational repression is a dynamic and multistep process that involves assembly of the target mRNA with ribonucleoproteins, such as Ago2, to form an miRNA-mediated translation repressor complex (or miRISC). miRISC enters discrete cytoplasmic foci known as processing bodies (p-bodies or stress granules), which contain decapping enzymes and exonucleases involved in mRNA catabolism [22, 23]. Thus, p-bodies are a critical component of mRNA translational repression [20, 24].
In this study, we further defined the mechanism of translational repression of the proinflammatory cytokines during endotoxin tolerance and sepsis immunosuppression. Our data suggest that the RNA-binding protein and decapping enzyme cofactor Dcp1 supports assembly of the translational repressor complex (miRISC) at the mRNAs of TNFα and IL-6, and formation of p-bodies in endotoxin-tolerant THP-1 cells. This process is specific to the proinflammatory genes, as it does not target anti-inflammatory/immunosuppressive genes like IL-10. Reversing this process ex vivo restores the proinflammatory cytokine production and thus the immune response. This mechanism also occurs in tolerant macrophages from septic mice.
Materials and Methods
Cells
The THP-1 human monocytic cell line, obtained from the American Type Culture Collection (Manassas, Va., USA), was maintained in RPMI-1640 medium (Invitrogen, Carlsbad, Calif., USA) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mML-glutamine (all from Hyclone Laboratories, Logan, Utah, USA), and 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, Ga., USA) at 37°C and 5% CO2 atmosphere. Cells were made tolerant by overnight incubation with 1 μg/ml of LPS from Gram-negative bacteria (Escherichia coli serotype 0111:B4; catalog No. L3024; Sigma-Aldrich, St. Louis, Mo., USA) as described previously [25], where LPS dose responses and generation of tolerance in THP-1 cells were defined and optimized. The LPS preparation used in these experiments was free of contaminating proteins and RNAs that activate cells via a non-TLR4-dependent mechanism.
Induction of Endotoxin Tolerance
Endotoxin tolerance is characterized by decreased cytokine production after a challenge dose of LPS, as compared with those levels produced after a primary dose of LPS [25]. The temporal effect of LPS on the establishment of endotoxin tolerance in THP-1 cells was reported previously [25]. Briefly, endotoxin tolerance can be induced in these cells by a primary dose of LPS as low as 10 ng/ml, but there is little change in kinetics with up to 1 μg/ml of primary dose. The shift from the TLR4 initiation state of proinflammatory cytokine transcription to endotoxin tolerance is rapid and can be detected within 6–10 h; it is fully established by 16 h and sustained for up to 96 h after the primary dose [25]. Similar kinetics in LPS tolerant monocytes and macrophages occur in infectious-induced sepsis in mice. The gold standard to test that LPS tolerance has occurred is to stimulate with a second dose of 1 μg/ml between 8 and 24 h after the primary dose, as in this study. LPS tolerance after the second dose provides a gold standard for being certain that the cell phenotype has shifted from responsive to tolerant. This promonocyte sepsis cell model simulates innate immune responses of phagocytes in both mice and human models of severe stress from sepsis.
Animal Experiments
Male BALB/c mice 8 weeks old (Harlan Sprague Dawley, Indianapolis, Ind., USA) were housed in a pathogen-free, temperature-controlled room and were acclimated to the new environment for at least 3 days before surgery. These experiments were conducted in accordance with the National Institutes of Health Guidelines and were approved by the East Tennessee State University Animal Care and Use Committee.
Polymicrobial sepsis was induced by cecal ligation and puncture (CLP) as described previously [26]. Briefly, mice were anesthetized via inhalation with 2.5% isoflurane (Abbott Laboratories, Abbott Park, Ill., USA). A midline abdominal incision was made and the cecum was exteriorized, ligated distal to the ileocecal valve, and then punctured twice. A small amount of feces was extruded into the abdominal cavity. The abdominal wall and skin were sutured in layers with 3–0 silk. Sham-operated mice were treated identically except that the cecum was neither ligated nor punctured. Mice received, intraperitoneally, 1 ml of lactated Ringer's solution plus 5% dextrose for fluid resuscitation. To create the late sepsis phenotype, mice were subcutaneously administered antibiotic (imipenem; 25 mg/kg body weight) or saline and 1 ml of lactated Ringer's solution plus 5% dextrose or an equivalent volume of 0.9% saline. The presence of early sepsis was confirmed by transient systemic bacteremia and elevated cytokine levels in the first 5 days after CLP. Late/chronic sepsis (after day 5) was confirmed by enhanced peritoneal bacterial overgrowth and reduced circulating proinflammatory cytokines.
Isolation and Culture of Peritoneal Macrophages
Peritoneal cells were harvested from septic mice that were moribund at days 10–28 (i.e. during late sepsis) and from healthy-appearing sham mice. Mice were sacrificed by an overdose of isoflurane, and the abdominal cavity was opened and flushed with 5 ml of cold phosphate-buffered saline (PBS). The peritoneal exudate was then collected and centrifuged at 1,500 rpm for 5 min. Cells were washed and resuspended in complete RPMI-1640 medium, and then cultured on Petri dishes. Nonadherent cells were removed by washing with cold PBS. Adherent cells (macrophages) were incubated in RPMI-1640 medium, allowed to ‘rest’ overnight, and then stimulated with 100 ng/ml of LPS for the indicated times. Because sham mice have few macrophages, they were intraperitoneally injected 48 h before cell harvest with 2 ml of 3% sterile inflammatory stimulus thioglycollate (Remel, Lenexa, Kans., USA) to elicit peritoneal cells.
Function of Peritoneal Macrophages Assessed ex vivo
To determine the immune function of peritoneal macrophages, harvested peritoneal lavage cells (see above) were washed with sterile PBS, resuspended in RPMI-1640 medium supplemented with 10% FBS, and then cultured for 2 h in a 24-well plastic plate. Nonadherent cells were removed by washing with PBS. Adherent cells (macrophages) were incubated in complete RPMI 1640 medium and stimulated with 100 ng/ml of the TLR4 agonist LPS (serotype 0111:B4, Sigma-Aldrich) for 24 h. Supernatants were collected and stored at -20°C for future cytokine analysis.
Protein Extraction
Whole cell lysates were obtained by lysing cells on ice for 30 min with 1× RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholic acid and 1 mM EDTA (Millipore, Temecula, Calif., USA) plus 1× protease inhibitor cocktail. Cell lysates were cleared by centrifugation for 5 min at 4°C and 14,000 rpm.
Cytoplasmic and nuclear proteins were extracted using the NE-PER nuclear and cytoplasmic protein extraction kit (Thermo Fisher Scientific, Waltham, Mass., USA) per the manufacturer's instructions. Briefly, cells were washed in PBS and resuspended in CER1 lysis buffer with protease inhibitor cocktail and incubated on ice for 1 min. CER2 buffer was added and the incubation continued for 5 min. Supernatants (cytoplasmic proteins) were recovered by centrifugation for 5 min at 4°C and 14,000 rpm. The nuclear pellets were resuspended in NER lysis buffer with protease inhibitor cocktail and incubated for 40 min on ice with occasional vortexing. The nuclear proteins were recovered by centrifugation for 10 min at 4°C and 14,000 rpm. Protein concentrations were determined by the Bradford assay (Bio-Rad, Hercules, Calif., USA) and aliquots were kept at -20°C.
Immunoprecipitation
Dcp1, GW182, Ago2 or RBM4 protein complexes in whole cell lysates or cytoplasmic/nuclear extracts were immunoprecipitated as described previously [18]. Briefly, cytoplasmic or nuclear extracts were precleared by incubation with preblocked protein G-agarose beads for 1 h at 4°C. Beads were preblocked by incubation for 1 h with 100 μg/ml of bovine serum albumin (BSA). Preblocked beads were washed with buffer C [250 mM sucrose, 10 mM Tris-HCl (pH 7.5), 25 mM KCl, 5 mM MgCl2, 2 mM DTT, 30 U/ml RNase inhibitor, and 1× protease inhibitor cocktail]. Protein extract was then centrifuged at 2,000 rpm for 5 min and supernatant (900 μl) was added to 100 μl of preblocked protein G-agarose beads that were coated with 10 μl of antibody against human Ago2 (clone No. 4G8; Wako, Richmond, Va., USA), Dcp1 (Genway, San Diego, Calif., USA), RBM4 (Abcam, Cambridge, Mass., USA), GW182 or IgG isotype control (Santa Cruz Biotechnology, Santa Cruz, Calif., USA). After overnight rotation at 4°C, the beads were centrifuged and washed three times with buffer C. Aliquots of bound protein complexes were used for protein analysis by Western blotting, and the remainder was used to isolate the bound mRNA and miRNA. The RNA was then analyzed by PCR.
Western Blot Analysis
Equal amounts of protein extracts or immunoprecipitated protein complexes were mixed with 5× Laemmli sample buffer, separated by SDS-10% polyacrylamide gel (Bio-Rad) and subsequently transferred to nitrocellulose membranes (Thermo Fisher Scientific). After blocking with 5% milk in Tris-buffered saline/Tween-20 for 1h at room temperature, membranes were probed overnight at 4°C with the appropriate primary antibody. After washing, blots were incubated with the appropriate HRP-conjugated secondary antibody (Life Technologies, Grand Island, N.Y., USA) for 2 h at room temperature. Proteins were detected with the enhanced chemiluminescence detection system (Thermo Fisher Scientific), the bands were visualized using the ChemiDoc XRS System (Bio-Rad), and the images were captured with Image Lab Software v.3.0 (Bio-Rad). Densitometry analysis of protein bands was performed using the same software.
RT-PCR
Semi-qPCR was performed to detect the presence of the TNFα, IL-6 and IL-10 mRNA in the immunoprecipitated protein complexes. Briefly, DCP1 or IgG protein complexes (see above) were treated at 37°C with DNase for 15 min and subsequently with proteinase K for 15 min. The protein-bound RNA was then purified with TRIzol reagent (Invitrogen). The RNA was reverse-transcribed and then amplified by 25 cycles. The PCR primers (Integrated DNA Technologies, Coralville, Iowa, USA) were designed to amplify ∼250-bp fragments within the 3′-UTR of the target mRNA and were as follows: TNFα forward 5′-ggagccagctccctctattt-3′, reverse 5′-ggctacatgggaacagccta-3′; IL-6 forward 5′-gtccactgggcacagaactt-3′, reverse 5′-caaactgcatagccactttcc-3′; IL-10 forward 5′-tggtgaaaccccgtctctac-3′, reverse 5′-ctggagtacaggggcatgat-3′. The 18S RNA was also amplified as an input control using RNA extracted from a portion of the whole cell lysate before the immunoprecipitation (see above). The PCR primers were: 18S forward 5′-cagccacccgagattgagca-3′ and reverse 5′-tagtagcgacgggcggtgtg-3′. Real-time PCR was performed as described previously [11] to measure TNFα and IL-6 mRNA levels after Dcp1 knockdown.
siRNA-Mediated Gene Silencing
Cells were seeded at 0.5 × 106 cells/ml 1 day prior to transfection. Cells were transfected by electroporation using the GenePulser MXcell system (Bio-Rad) and HiPerFect transfection reagent per the manufacturer's instructions (Qiagen, Valencia, Calif., USA). For Dcp1 and/or RelB knockdown, endotoxin-tolerant THP-1 cells were transfected with pools of Dcp1- and/or RelB-specific siRNAs or scrambled (control) siRNAs at 0.5 μM final concentration (Santa Cruz Biotechnology). For miR-146a knockdown, cells were transfected with negative control or miR-146a-specific antisense 2′-O-Me oligonucleotides (final concentration 100 nM, antagomiRs; Ambion, Austin, Tex., USA).
DNA Constructs
Full-length, Flag-tagged human Ago2 and GFP-tagged human RBM4 cDNAs cloned in pReceiver-M43 and pEZ-M11 expression plasmids, respectively, and full-length Flag- or GFP-tagged human Dcp1 cloned in pEZ-M11 plasmid were generated by GeneCopoeia (Rockville, Md., USA). For transfection, endotoxin-tolerant THP-1 cells, with or without miR-146a knockdown, were cotransfected with Dcp-1 plus Ago2 or RBM4 expression plasmids using Attractene transfection reagent (Qiagen). Endotoxin-responsive (normal) THP-1 cells were similarly cotransfected and served as a control. After 36 h, cells were stimulated with 1 μg/ml of LPS for 0–3 h and then harvested for immunofluorescence staining.
Immunofluorescence
Colocalization of Dcp-1, Ago2 and RBM4 was evaluated by immunofluorescence staining using antibodies against Flag and GFP tags. Briefly, cytospin cell preparations were made. Cells were fixed in 4% paraformaldehyde for 10 min, permeabilized in PBS with 0.2% Triton X-100 and 0.5% BSA for 10 min, and blocked with 2.5% goat serum in PBS with 0.2% Triton X-100 and 0.5% BSA. Cells were incubated overnight at 4°C with human anti-Flag (1:2,000; Sigma) or anti-GFP (1:400; Life Technologies) antibody. After washing, fluorescence visualization of the Flag and GFP tags was achieved using Alexa Fluor 568-conjugated goat anti-mouse IgG (1:1,000) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:1,000; Life Technologies). Slides were examined on a Leica TCS SP8 confocal laser scanning microscope (Leica, Buffalo Grove, Ill., USA) and images were acquired and analyzed using Adobe Photoshop version 9.0.
Cytokine Assays
Levels of TNFα and IL-6 proteins in the culture supernatants were determined by an enzyme-linked immunosorbent assay (ELISA) using commercially available kits according to the manufacturer's instructions (eBioscience, San Diego, Calif., USA). Samples were assayed in triplicate.
Statistical Analysis
Data were analyzed with Microsoft Excel 3.0. Differences between two groups were analyzed by an unpaired Student t test. One-way ANOVA with Tukey's multiple comparison tests was used to analyze data with more than two groups/treatments. All values are expressed as means (± SD). Statistical significance was set at p ≤ 0.05.
Results
p-Bodies Enriched with TNFα and IL-6 mRNAs Are Assembled in Endotoxin-Tolerant THP-1 Cells
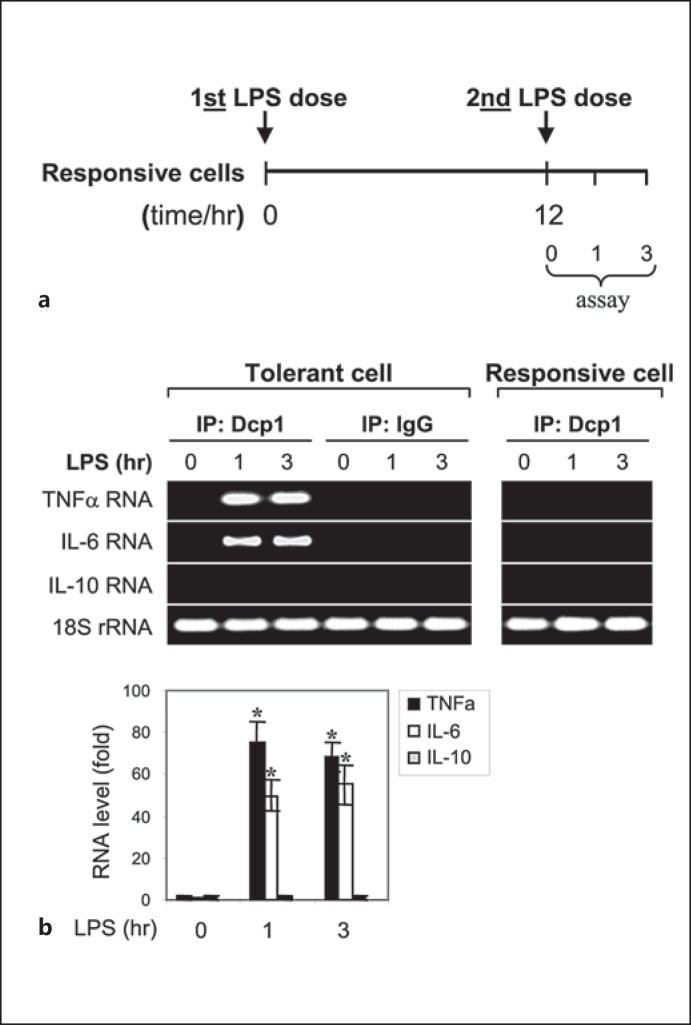
p-Bodies are the primary sites for miRNA-mediated translational repression [27, 28], and the decapping enzyme cofactor Dcp1 is a critical component and a marker of p-body formation [24, 29]. We investigated whether the p-bodies play a role in the translational repression of the proinflammatory cytokines TNFα and IL-6 in endotoxin-tolerant THP-1 cells. Because the transcription of these cytokines is also repressed by the transcription factor RelB, the levels of which are increased in endotoxin-tolerant THP-1 cells [30], we reactivated the cytokine gene transcription first in order to optimally investigate their translational repression, as reported [11]. To do this, THP-1 cells were made tolerant by an overnight incubation with LPS and then RelB was knocked down for 24 h, which restores mRNAs of TNFα and IL-6 after stimulation with LPS, but protein synthesis remains repressed [16, 17]. Next, the tolerant THP-1 cells with RelB knockdown, along with responsive/normal THP-1 cells (as a control), were stimulated with LPS for 0–3 h to activate the cytokine gene transcription. Whole protein extracts were prepared and immunoprecipitated with anti-Dcp1 specific or IgG isotype control antibody. RNA was extracted from the immunoprecipitated Dcp1 protein complexes and analyzed by RT-PCR for enrichment of TNFα and IL-6 mRNAs. Because anti-inflammatory/immuosuppressive cytokine genes such as IL-10 are not repressed in endotoxin-tolerant cells [2, 31], we also measured IL-10 mRNA as a control. As shown in figure 1, both TNFα and IL-6 mRNAs were markedly enriched in the Dcp1 protein complex in endotoxin-tolerant but not responsive THP-1 cells, and this was only observed after LPS stimulation. We did not detect the IL-10 mRNA in the Dcp1 protein complex. These results support the theory that p-bodies form in endotoxin-tolerant cells and specifically target the proinflammatory cytokine mRNAs for translation repression.
Fig. 1.
The mRNAs of proinflammatory cytokines TNFα and IL-6 but not the anti-inflammatory IL-10 associates with Dcp1 protein in endotoxin-tolerant but not endotoxin-responsive THP-1 cells after LPS stimulation. a THP-1 cells were made tolerant through the pretreatment of responsive (normal) cells overnight with 1 μg/ml of LPS, followed by RelB knockdown for 24 h, which restores mRNAs of TNFα, IL-6 and IL-β, but protein synthesis remains repressed (see first paragraph of Results). b The tolerant THP-1 cells with RelB knockdown, along with responsive (normal) THP-1 cells (as a control), were stimulated (2nd LPS dose) with 1 μg/ml of LPS for the indicated times. Whole cell lysates were prepared and immunoprecipitated with an anti-Dcp1 or IgG isotype (control) antibody. RNA was then extracted from the immunoprecipitated protein complexes and analyzed by RT-PCR to detect the presence of TNFα, IL-6 or IL-10 RNA in the protein complexes using primers that amplify the 3′-UTR sequences. RNA was also extracted from a portion of the cell lysate before the immunoprecipitation and was used to measure the level of the 18S ribosomal RNA as an internal (input) control. The quantitation of RNA levels is shown below. Values were normalized to the 18S rRNA and are presented as the fold change relative to the IgG-immunoprecipitated samples (set at 1-fold). Data are expressed as the mean ± SD from three experiments. * p < 0.05, compared with 0 h.
The Translational Repressor Protein Complex of Ago2 and RBM4 Is Bound to Dcp1 after LPS Stimulation in Endotoxin-Tolerant Cells
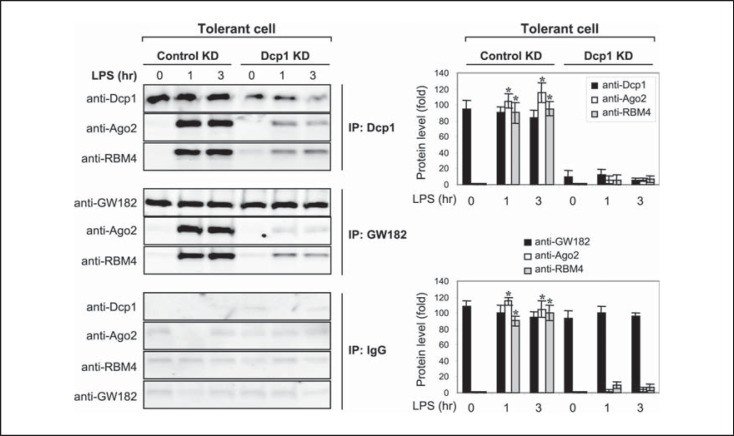
Ago2 protein acts as a platform for assembling a translational repressor complex (or miRISC), which contains regulatory RNA-binding proteins and the mRNAs that are the target of translation repression [20, 21, 24]. We have previously shown that Ago2 assembles a translational repressor complex with RBM4, and that this complex was enriched with TNFα and IL-6 mRNAs after LPS stimulation in endotoxin-tolerant THP-1 cells [16, 17]. Because TNFα and IL-6 mRNAs were present in the immunoprecipitated Dcp1 protein, as part of p-bodies (fig. 1), we examined whether the Ago2-RBM4 repressor complex is deposited into p-bodies by measuring their association with Dcp1. To do this, we stimulated tolerant THP-1 cells with LPS for 0–3 h. Whole protein extracts were isolated and immunoprecipitated with anti-Dcp1 antibody, followed by immunoblotting with anti-Ago2 or anti-RBM4 antibody. As shown in figure 2 (upper panel), both Ago2 and RBM4 were found as a complex with Dcp1 after LPS stimulation, suggesting that the Ago2-RBM4 translational repressor complex is assembled into p-bodies.
Fig. 2.
Dcp1 forms a protein complex with Ago2 and RBM4 after LPS stimulation in endotoxin-tolerant cells. Endotoxin-tolerant THP-1 cells were prepared by pretreatment of responsive (normal) cells overnight with 1 μg/ml of LPS. Tolerant cells were transfected with a pool of Dcp1-specific or scrambled siRNA (control KD). After 24 h, cells were washed and restimulated with 1 μg/ml of LPS for the indicated times. Whole cell lysates were prepared and immunoprecipitated with an anti-Dcp1, GW182 or IgG isotype antibody. The immunoprecipitated protein complexes were resolved by SDS-10% PAGE and then immunoblotted with a Dcp1, Ago2 or RBM4 antibody. The quantitation of protein levels is shown on the right. Values are presented as the fold change relative to the IgG-immunoprecipitated samples (set at 1-fold). Data are expressed as the mean ± SD from three experiments. * p < 0.05, compared with 0 h. KD = Knockdown.
Based on the results from Dcp1 coimmunoprecipitation with Ago2 and RBM4, we hypothesized that Ago2-RBM4 complex should be disrupted in the absence of Dcp1. Tolerant cells were subjected to Dcp1 knockdown for 24 h and then stimulated with LPS for 0–3 h. The immunoprecipitation of whole protein extracts with anti-Dcp1 antibody followed by immunoblotting with Ago2 or RBM4 antibody showed that Ago2 and RBM4 binding was disrupted (fig. 2; upper panel). In addition, and to support specificity of Dcp1, immunoprecipitation with antibody against GW182, another p-body marker [20, 32], gave similar results (fig. 2; middle panel). These results suggest that, as a marker of p-body formation, Dcp1 is required for the assembly of the translational repressor complex in endotoxin-tolerant cells and that this process is initiated by the LPS signal.
Ago2 and RBM4 Binding to Dcp1 and p-Body Formation Are Restricted to the Cytoplasmic Compartment and Prevented by Inhibiting miR-146a
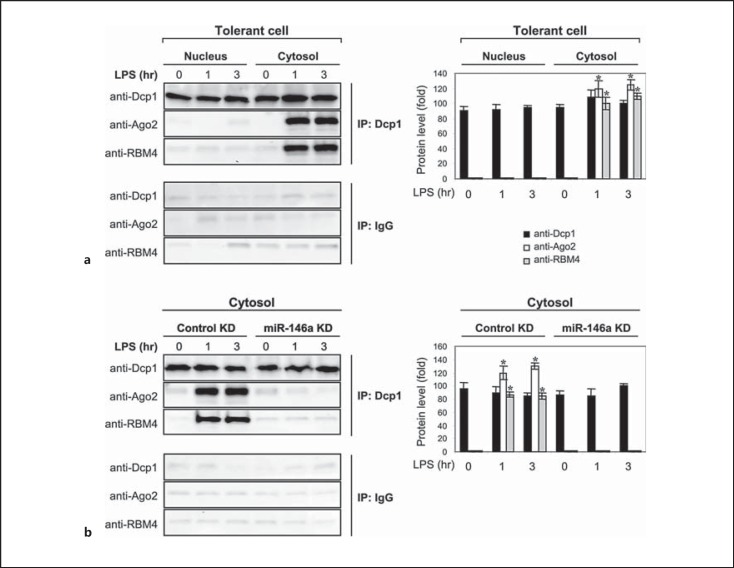
To examine the dynamic of p-body formation, we first investigated Dcp1 binding to Ago2 and RBM4 in the nucleus and cytosol of tolerant THP-1 cells. The nuclear and cytoplasmic proteins were isolated before and after LPS stimulation, immunoprecipitated with anti-Dcp1 antibody and then immunoblotted with Ago2 or RBM4 antibody. Despite the presence of Dcp1 protein in the nucleus and cytosol, the binding of Dcp1 to Ago2 and RBM4 was detected in the cytosol only (fig. 3a). It should be noted that while Ago2 and RBM4 proteins are ubiquitously present in the nucleus and cytosol of tolerant THP-1 cells, RBM4 is present in the cytosol in an unphosphorylated form, which facilitates its binding to Ago2 [17].
Fig. 3.
The Dcp1 protein binding to Ago2 and RBM4 is detected in the cytoplasm in tolerant cells after LPS stimulation and is disrupted by miR-146a knockdown (KD). Endotoxin-tolerant THP-1 cells were prepared by pretreatment of responsive (normal) cells overnight with 1 μg/ml of LPS. a Tolerant cells were washed and restimulated with 1 μg/ml of LPS for the indicated times. Cytoplasmic and nuclear proteins were isolated and immunoprecipitated with an anti-Dcp1 or IgG isotype antibody. The immunoprecipitated protein complexes were resolved by SDS-10% PAGE and then immunoblotted with a Dcp1, Ago2 or RBM4 antibody. b Tolerant cells were transfected with anti-miR-146a oligonucleotides (antagomiRs) or scrambled anti-miRNA oligonucleotides (control KD). After 24 h, cells were washed and stimulated with 1 μg/ml of LPS for the indicated times. Cytoplasmic proteins were isolated, immunoprecipitated and immunoblotted as in a. The quantitation of protein levels is shown on the right. Values are presented as the fold change relative to the IgG-immunoprecipitated samples (set at 1-fold). Data are expressed as the mean ± SD from three experiments. * p < 0.05, compared with 0 h.
Next, we probed Dcp1 binding to Ago2 and RBM4 in cytoplasmic extracts isolated from tolerant cells lacking miR-146a. We have previously reported that miR-146a levels are increased in tolerant THP-1 cells after LPS stimulation and that its knockdown prevents Ago2 and RBM4 interactions, and restores TNFα and IL-6 protein synthesis [17]. We also showed that miR-146a promotes dephosphorylation of RBM4 and thus its accumulation in the cytosol; unphosphorylated but not phosphorylated RBM4 binds Ago2 [17, 19]. Therefore, we asked if the modification of RBM4 binding activity, induced by miR-146a, also affects its binding to Dcp1. Cytoplasmic proteins from tolerant cells with miR-146a knockdown were immunoprecipitated with anti-Dcp1 antibody and then immunoblotted with Ago2 or RBM4 antibody. As shown in figure 3b, miR-146a knockdown prevented Ago2 and RBM4 binding to Dcp1. Together, these results demonstrate that depositing of the Ago2-RBM4 translational repressor complex into p-bodies occurs in the cytosol and is dependent on the miR-146a-mediated modification of RBM4 binding activity.
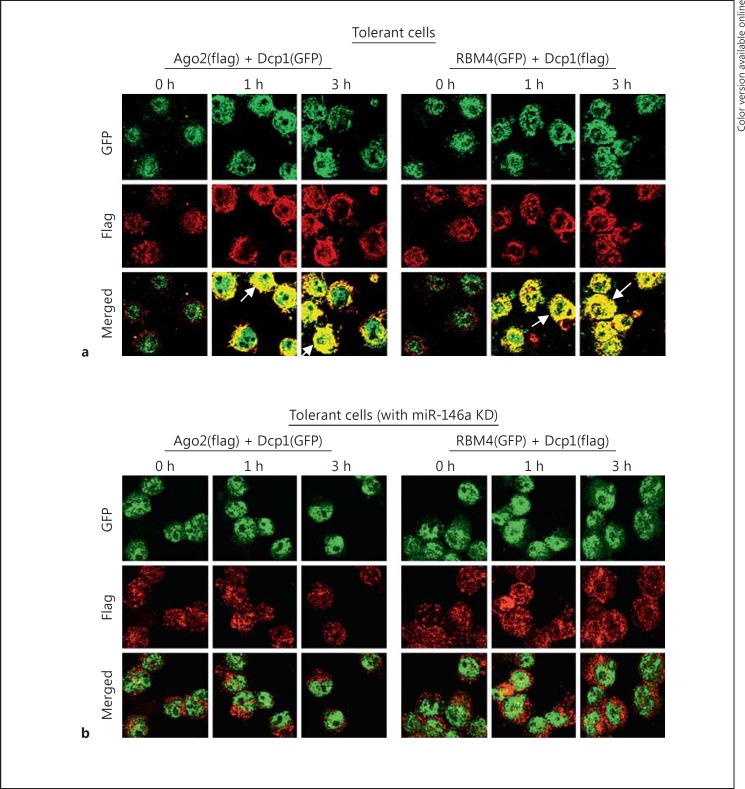
To further investigate the role of Dcp1 in p-body formation, we carried out a protein colocalization study using Flag- or GFP-tagged DNA constructs expressing Ago2, RBM4 or Dcp1. Immunofluorescence analysis showed that both Ago2 and RBM4 colocalized (i.e. formed a complex) with Dcp1 in the cytosol of endotoxin-tolerant THP-1 cells after LPS stimulation, and this colocalization was not observed in endotoxin-responsive cells (fig. 4a and data not shown). In contrast, both Ago2 and RBM4 did not cocolocalize with Dcp1 in endotoxin-tolerant cells after miR-146a knockdown (fig. 4b), despite expression of both proteins in these cells [16, 17]. These results suggest that Dcp1 plays a role in assembling the Ago2-RBM4 translational repressor complex into p-bodies in endotoxin-tolerant cells and that the assembly of this protein complex requires an LPS-mediated signal, which is disrupted by miR-146a.
Fig. 4.
Dcp1 colocalizes with Ago2 and RBM4 in tolerant but not responsive cells after LPS stimulation and miR-146a knockdown (KD) disrupts this colocalization. a Endotoxin-tolerant cells were cotransfected with Flag-tagged Ago2 plus GFP-tagged Dcp1 expression constructs or GFP-tagged RBM4 plus Flag-tagged Dcp1 expression constructs for 36 h and then stimulated with 1 μg/ml of LPS for the indicated times. Arrows indicate colocalized protein complexes (yellow; colors refer to the online version only). b miR-146a was knocked down in endotoxin-tolerant cells for 24 h. Cells were then transfected and stimulated as in a. Cytospin preparations were readied and cells were fixed, permeabilized and costained with anti-Flag (red) and anti-GFP (green) antibodies. Images were captured at 40× magnification. The results are representative of two experiments.
Dcp1 Knockdown in Endotoxin-Tolerant THP-1 Cells Restores TNFα and IL-6 Protein Synthesis
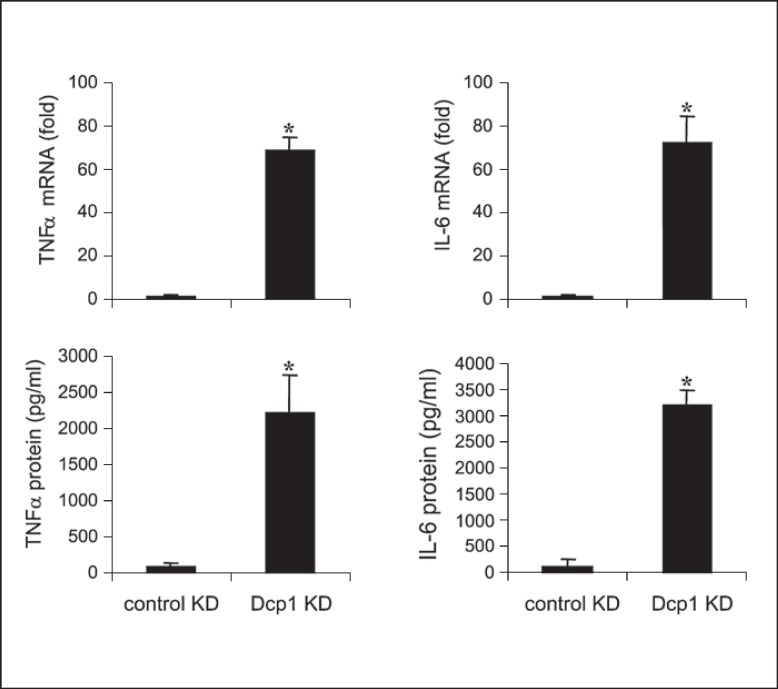
Based on the interactions of Dcp1 with Ago2 and RBM4, we examined the effect of Dcp1 depletion on the proinflammatory cytokine production. Dcp1 was knocked down in tolerant cells, in which RelB was also knocked down (to restore mRNA transcription), and cells were then stimulated with LPS. After 1 h, RNA was isolated, and TNFα and IL-6 mRNA levels were measured by PCR. After 6 h in LPS, culture supernatants were collected and analyzed for TNFα and IL-6 protein production by ELISA. The results showed that both mRNA and protein levels of TNFα and IL-6 were significantly increased due to the Dcp1 knockdown (fig. 5). We observed that mRNAs of proinflammatory genes, including TNFα, rapidly degrade in tolerant THP-1 cells due to the translational repression [16]. These results suggest that Dcp1 binding to the Ago2-RBM4 translational repressor complex and the p-body formation are critical steps in the path leading to translation repression of proinflammatory genes in endotoxin-tolerant cells.
Fig. 5.
Knockdown (KD) of Dcp1 in endotoxin-tolerant THP-1 cells restores the mRNA and protein levels of TNFα and IL-6. Endotoxin-tolerant THP-1 cells were prepared by pretreatment of responsive (normal) cells overnight with 1 μg/ml of LPS. Tolerant cells were transfected with pools of RelB- plus Dcp1-specific or scrambled siRNAs (control KD). After 24 h, cells were washed and restimulated with 1 μg/ml of LPS. After 1 h in LPS, a portion of the cells was harvested and RNA was isolated and used to measure TNFα and IL-6 mRNA levels by real-time RT-PCR. After an additional 5 h, the cells and supernatants were harvested and used to measure TNFα and IL-6 protein levels by ELISA. Data are the mean ± SD from three experiments. * p < 0.05.
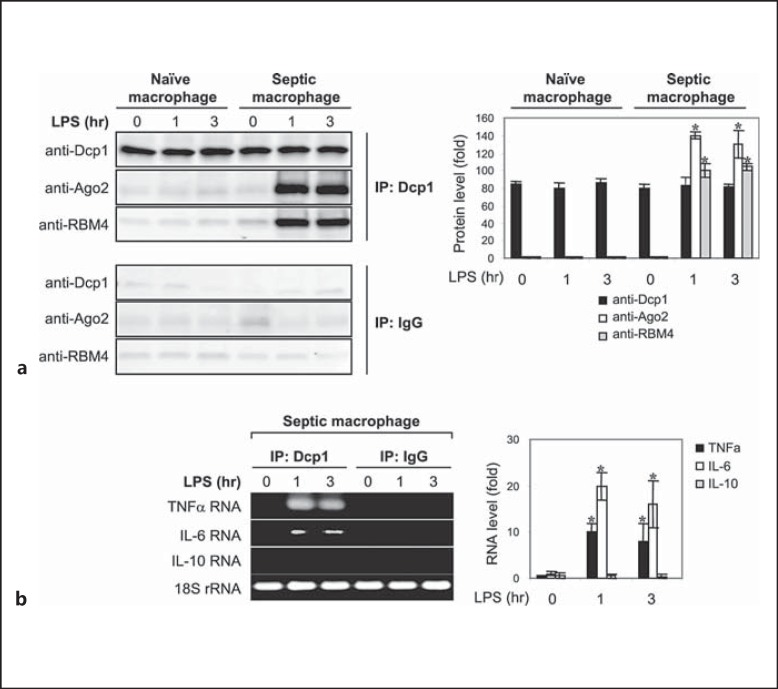
Dcp1 Binds the Ago2-RBM4 Translational Repressor Complex in Septic Macrophages
We investigated the assembly of the translational repressor complex in primary macrophages isolated from septic mice. Septic macrophages, like endotoxin-tolerant THP-1 cells, do not produce proinflammatory cytokines after LPS stimulation due to translational repression [31]. Polymicrobial sepsis was induced by CLP. Peritoneal macrophages were harvested at days 10–28 (i.e. during the late phase of sepsis where proinflammatory cytokine production is repressed) and stimulated with LPS for 0- 3 h. Cytoplasmic proteins were immunoprecipitated with anti-Dcp1 antibody and then immunoblotted with Ago2 or RBM4 antibody. As shown in figure 6a, Dcp1 binding to Ago2 and RBM4 was detected in septic but not naive macrophages after LPS stimulation, similar to what we observed in tolerant THP-1 cells (fig. 2a). In addition, TNFα and IL-6 but not IL-10 mRNAs were enriched in the Dcp1 protein complex (fig. 6b). The level of mRNA enrichment was much less than what we detected in tolerant THP-1 cells (fig. 1). In contrast to THP-1 cells, RelB was not knocked down in these primary macrophages, and therefore they are expected to have low levels of proinflammatory gene transcription. Also, septic macrophages, similar to tolerant THP-1 cells without Dcp1 knockdown (fig. 5), did not produce TNFα and IL-6 cytokines after stimulation with LPS (data not shown), indicating that they remain translationally repressed ex vivo. Together, these results demonstrate that the translational repressor complex that targets the proinflammatory cytokine mRNAs in endotoxin-tolerant THP-1 cells is also assembled in septic macrophages.
Fig. 6.
The Dcp1 protein forms a complex with Ago2 and RBM4 in primary macrophages from septic mice and is associated with TNFα and IL-6 but not IL-10 mRNA. Sepsis was induced by CLP. Peritoneal macrophages were harvested from sham and septic mice that were moribund and sacrificed at days 10–28 (i.e. during late sepsis response). Cells were stimulated with 100 ng/ml of LPS for the indicated times. a Whole cell lysates were prepared and immunoprecipitated with anti-Dcp1 or IgG isotype antibody, resolved by SDS-10% PAGE, and then immunoblotted with a Dcp1, Ago2 or RBM4 antibody. The quantitation of protein levels is shown on the right. Values are presented as the fold change relative to the IgG-immunoprecipitated samples (set at 1-fold). Data are expressed as the mean ± SD from three experiments. * p < 0.05, compared with 0 h. b RNA was extracted from the immunoprecipitated protein complexes and then analyzed by RT-PCR for the presence of TNFα, IL-6 or IL-10 RNA. RNA was also extracted from a portion of the cell lysate before the immunoprecipitation and was used to measure the level of the 18S rRNA as an internal (input) control. Cells were pooled from 8 sham mice and 6 late septic mice. The quantitation of RNA levels is shown on the right. Values were normalized to the 18S rRNA and are presented as the fold change relative to the IgG-immunoprecipitated samples (set at 1-fold). Data are expressed as the mean ± SD from three experiments. * p < 0.05.
Discussion
This study further identifies the specific posttranscription pathway that separates translation from transcription regulation of endotoxin tolerance during the acute systemic inflammatory response associated with sepsis. We show that Dcp1, which supports mRNA degradation and protein synthesis disruption in p-bodies, is required for Ago2-RBM4 p-body assembly. Importantly, this pathway is flexible and can be reversed by reducing Dcp1 levels. We also show that miR146a expression, as a feed-forward response to TLR4 stimulation, lies proximal to posttranscriptional regulation of the acute inflammatory response of sepsis. This pathway, if sustained during sepsis, can delay the restoration of homeostasis and increase mortality risk.
Assembly of the translational repressor complex is the first and most critical step in the mechanism of miRNA-mediated translation repression [20, 33]. Ago2 protein functions as a platform for forming the translational repressor complex, since it binds both to miRNAs that are specific to the translationally repressed mRNAs and small RNA-binding proteins [20, 34]. We reported that Ago2 interacts with the RNA-binding protein RBM4 to repress mRNAs of TNFα and IL-6, a gene-specific event. In the case of TNFα, this repressor complex includes three specific miRNAs: miR-221, miR-579 and miR-125b [18]. Direct translational repression occurs in cytoplasmic p-bodies where decapping enzymes and exonucleases induce mRNA catabolism [20, 35, 36]. This study shows that the decapping enzyme cofactor and p-body marker Dcp1 protein mediates the specific translational repression of TNFα and IL-6 mRNAs in endotoxin-tolerant THP-1 cells and septic macrophages by binding to the Ago2-RBM4 protein complex with assembly into p-bodies. Importantly, this process can be prevented by reducing Dcp1.
Translational repression during endotoxin tolerance is tightly controlled. It is gene specific, repressing TNFα and IL-6, but not the IL-10 mRNAs which support endotoxin tolerance, and is known for its strong immunosuppressive effects in acute systemic and local inflammation [2, 37]. In addition, we did not detect the TNFα and IL-6 mRNAs with Dcp1 protein complex in responsive THP-1 cells (fig. 1b) or naive macrophages (data not shown), despite the presence of Dcp1. This study further shows that the translational repressor complex's assembly into p-bodies operates only during endotoxin tolerance and inflammation adaptation. We reported that Ago2 and RBM4 are ubiquitously expressed in endotoxin-responsive and tolerant THP-1 cells [16, 17]. However, they repress TNFα and IL-6 cytoplasmic mRNAs only after LPS stimulation, which activates the MAPK phosphatase MKP-1 leading to accumulation of unphosphorylated RBM4 in the cytosol and thus RBM4 binding to Ago2 protein [17, 19]. Here, we show that Dcp1 protein is needed to disrupt translation by assembling Ago2-RBM4 proteins into p-bodies during endotoxin tolerance and that this process is reversible by removing Dcp1.
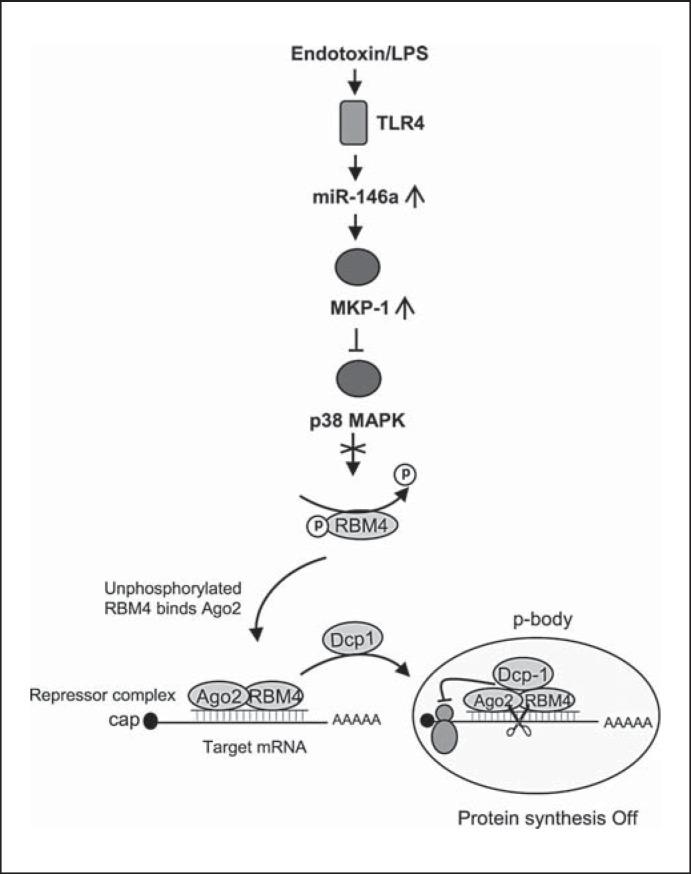
LPS stimulation also increases miR-146a expression in endotoxin-tolerant THP-1 cells [16], during which time miR-146a indirectly deactivates p38 MAPK, which is required for RBM4 phosphorylation [19]. When p38 MAPK is deactivated, unphosphorylated RBM4 accumulates in the cytosol where it binds to Ago2 [17, 19]. Our study shows that this proximal signaling mechanism underlies Dcp1 targeting of Ago2 and RBM4 into p-bodies. Figure 7 summarizes this concept. MiR-146a knockdown prevents generation of endotoxin tolerance at the level of p-body activity by disrupting Dcp1 binding to Ago2 and RBM4 (fig. 3). Thus, a distinct branch of TLR4 signaling, which includes NF-κB transcription activation, also controls mRNA and protein degradation in the p-bodies.
Fig. 7.
A model depicting Dcp1-mediated translational repression in endotoxin-tolerant cells. Based on our present and previous findings, LPS stimulation in endotoxin-tolerant cells elevates miR-146a, which inactivates p38 MAPK. In the absence of activated p38, RBM4 accumulates in the cytosol in an unphosphorylated form, where it binds Ago2 and Dcp1 and forms a translational repressor complex at the target mRNA, which will then be deposited into p-bodies where translation repression occurs. Note RBM4 phosphorylation is disrupted in tolerant cells due to induction of the MAPK phosphatase MKP-1, which is controlled by miR-146a [17, 19]. Not shown is the RelB-induced cytokine gene transcription repression pathway, which is also induced in parallel as described in the first paragraph of Results.
In endotoxin tolerance models, downregulated cytokine production at the protein level occurs after LPS interaction with TLR4 as the specific challenge stimulus [10, 38]. However, polymorphonuclear cells from sepsis patients are tolerant to LPS but remain responsive to Staphylococcus aureus stimulation, as demonstrated by increases in IL-1β transcription and protein production [10], suggesting alternative signaling pathways. Moreover, endotoxin-tolerant THP-1 cells are capable of activating cytokine expression upon stimulation with phorbol ester (PMA) [25, 39]. In the current study, LPS tolerance is linked to a repressor pathway initiated downstream of TLR4, and alternative signaling pathways were not tested. In LPS tolerance, it is important to emphasize that transcription and translation control in increased production of proinflammatory cytokines is controlled both at the transcriptional and posttranscriptional levels.
Previous studies reported that p-bodies increase in cells with active translational repression machinery [23, 40] and suggested that inhibiting p-body components can interrupt the miRNA-mediated translational repression [40, 41, 42]. A recent study also reported increases in p-body formation in endotoxin-tolerant THP-1 cells [34]. Reactivation of the proinflammatory cytokine production in endotoxin-tolerant or septic monocyte/macrophages is needed to resolve late sepsis immunosuppression in animals and humans [4, 8, 43, 44], by clearing infection and restoring cellular homeostasis [45, 46]. This study adds Dcp1 as an important component of the TLR4-dependent reprogramming in a sepsis cell model and translates the concept to sepsis in mice, which may inform novel ways to treat late sepsis.
Acknowledgments
The authors thank Rolf Fritz (Department of Biomedical Sciences) for assistance with the confocal microscopy. This work was supported in part by National Institutes of Health Grants R15 GM100322 (to M.E.) and C06RR0306551 (to the College of Medicine).
References
- 1.Bianchi ME. Editorial: A recipe for inflammation. J Leukoc Biol. 2009;86:471–472. doi: 10.1189/jlb.0309121. [DOI] [PubMed] [Google Scholar]
- 2.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 4.McCall CE, Yoza B, Liu T, El Gazzar M. Gene-specific epigenetic regulation in serious infections with systemic inflammation. J Innate Immun. 2010;2:395–405. doi: 10.1159/000314077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remick DG. Pathophysiology of sepsis. Am J Pathol. 2007;170:1435–1444. doi: 10.2353/ajpath.2007.060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghisletti S, Huang W, Jepsen K, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.McCall CE, El Gazzar M, Liu T, Vachharajani V, Yoza B. Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J Leukoc Biol. 2011;90:439–446. doi: 10.1189/jlb.0211075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFα transcription silencing during endotoxin tolerance. J Biol Chem. 2008;283:32198–32208. doi: 10.1074/jbc.M803446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCall CE, Grosso-Wilmoth LM, Larue K, Guzman RN, Cousart SL. Tolerance to endotoxin-induced expression of the interleukin-1 beta gene in blood neutrophils of humans with the sepsis syndrome. J Clin Invest. 1993;91:853–861. doi: 10.1172/JCI116306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Gazzar M, Yoza BK, Hu JY, Cousart SL, McCall CE. Epigenetic silencing of tumor necrosis factor alpha during endotoxin tolerance. J Biol Chem. 2007;282:26857–2664. doi: 10.1074/jbc.M704584200. [DOI] [PubMed] [Google Scholar]
- 12.Fan H, Cook JA. Molecular mechanisms of endotoxin tolerance. J Endotoxin Res. 2004;10:71–84. doi: 10.1179/096805104225003997. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem. 2009;284:27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Gazzar M, Liu T, Yoza BK, McCall CE. Dynamic and selective nucleosome repositioning during endotoxin tolerance. J Biol Chem. 2010;285:1259–1271. doi: 10.1074/jbc.M109.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Gazzar M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-α during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. MicroRNA-146a and RBM4 form a negative feed-forward loop that disrupts cytokine mRNA translation following TLR4 responses in human THP-1 monocytes. Immunol Cell Biol. 2013;91:532–540. doi: 10.1038/icb.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. 2010;285:20940–20951. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. Mitogen-activated protein kinase phosphatase 1 disrupts proinflammatory protein synthesis in endotoxin-adapted monocytes. Clin Vaccine Immunol. 2013;20:1396–1404. doi: 10.1128/CVI.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 21.Hock J, Weinmann L, Ender C, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougherty JD, Reineke LC, Lloyd RE. mRNA decapping enzyme 1a (Dcp1a)-induced translational arrest through protein kinase R (PKR) activation requires the N-terminal enabled vasodilator-stimulated protein homology 1 (EVH1) domain. J Biol Chem. 2014;289:3936–3949. doi: 10.1074/jbc.M113.518191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaRue KE, McCall CE. A labile transcriptional repressor modulates endotoxin tolerance. J Exp Med. 1994;180:2269–2275. doi: 10.1084/jem.180.6.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brudecki L, Ferguson DA, Yin D, Lesage GD, McCall CE, El Gazzar M. Hematopoietic stem-progenitor cells restore immunoreactivity and improve survival in late sepsis. Infect Immun. 2012;80:602–611. doi: 10.1128/IAI.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 28.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoza BK, Hu JY, Cousart SL, Forrest LM, McCall CE. Induction of RelB participates in endotoxin tolerance. J Immunol. 2006;177:4080–4085. doi: 10.4049/jimmunol.177.6.4080. [DOI] [PubMed] [Google Scholar]
- 31.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. Adoptive transfer of CD34+ cells during murine sepsis rebalances macrophage lipopolysaccharide responses. Immunol Cell Biol. 2012;90:925–934. doi: 10.1038/icb.2012.32. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Rivas FV, Wohlschlegel J, Yates JR III, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Pauley KM, Satoh M, Pauley BA, et al. Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol Cell Biol. 2010;88:205–212. doi: 10.1038/icb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 37.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect Immun. 2012;80:2026–2034. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathison JC, Virca GD, Wolfson E, Tobias PS, Glaser K, Ulevitch RJ. Adaptation to bacterial lipopolysaccharide controls lipopolysaccharide-induced tumor necrosis factor production in rabbit macrophages. J Clin Invest. 1990;85:1108–1118. doi: 10.1172/JCI114542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoza BK, Hu JY, Cousart SL, McCall CE. Endotoxin inducible transcription is repressed in endotoxin tolerant cells. Shock. 2000;13:236–243. doi: 10.1097/00024382-200003000-00011. [DOI] [PubMed] [Google Scholar]
- 40.Jakymiw A, Lian S, Eystathioy T, et al. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 41.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 42.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Docke WD, Randow F, Syrbe U, et al. Monocyte deactivation in septic patients: restoration by IFN-γ treatment. Nat Med. 1997;3:678–681. doi: 10.1038/nm0697-678. [DOI] [PubMed] [Google Scholar]
- 44.Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13:260–268. doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyn-Kew K, Standiford TJ. Immunosuppression in sepsis. Curr Pharm Des. 2008;14:1870–1881. doi: 10.2174/138161208784980545. [DOI] [PubMed] [Google Scholar]
- 46.Shubin NJ, Monaghan SF, Ayala A. Anti-inflammatory mechanisms of sepsis. Contrib Microbiol. 2011;17:108–124. doi: 10.1159/000324024. [DOI] [PubMed] [Google Scholar]