Abstract
The strain dynamics of methicillin-resistant Staphylococcus aureus (MRSA) isolates from people and the household dog were investigated. The isolates were identified in the context of a randomized controlled trial that tested household-wide decolonization of people. Genotypic comparison of MRSA isolates obtained from two household members, the dog, and home surfaces over a three-month period failed to implicate the pet or the home environment in recurrent colonization of the household members. However, it did implicate the pet’s bed in exposure of the dog prior to the dog’s infection. Whole genome sequencing was performed to differentiate the isolates. This report also describes introduction of diverse strains of MRSA into the household within six weeks of cessation of harmonized decolonization treatment of people and treatment for infection in the dog. These findings suggest that community sources outside the home may be important for recurrent MRSA colonization or infection.
Keywords: MRSA, canine, SSTI, SSI, antimicrobial resistance
Introduction
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA), including USA300 strains, plays a major part in the epidemic of MRSA in people in the United States (Klein et al. 2009). Of particular concern is the propensity for cases or their household members to develop recurrent colonization or episodes of MRSA skin or soft tissue infection (SSTI) (Lautenbach et al. 2010; Fritz et al. 2012). Interrupting human-to-human transmission has been tested using household-wide decolonization treatment approaches, but with mixed success (Fritz et al. 2012), suggesting that other sources inside or outside the home may influence recurrent colonization. Within the home, surfaces and companion animals have been identified as potential reservoirs of MRSA, highlighting the importance of a one health approach to investigate drivers of household transmission of MRSA (Davis et al. 2012b).
Previous reports have strongly implicated companion animals in potential maintenance or recurrence of human MRSA colonization or infection (Faires et al. 2009; Bramble et al. 2011; Ferreira et al. 2011), with multiple case reports documenting that treatment of pets was required to clear human MRSA (Manian. 2003; van Duijkeren et al. 2004; van Duijkeren et al. 2005; Sing et al. 2008). Hence, as part of a randomized controlled trial that tested household-wide decolonization treatment of people, patients were enrolled with CA-MRSA skin or soft tissue infection (SSTI). In addition to these index cases, their household members and pet(s) also were enrolled. MRSA was isolated from a SSTI in a patient (a dog owner), and also from a subsequent surgical site infection (SSI) in the patient’s dog. MRSA colonization was observed in the people up to three-months later. This report describes the strain dynamics of these isolates using multiple genetic typing methods.
Methods
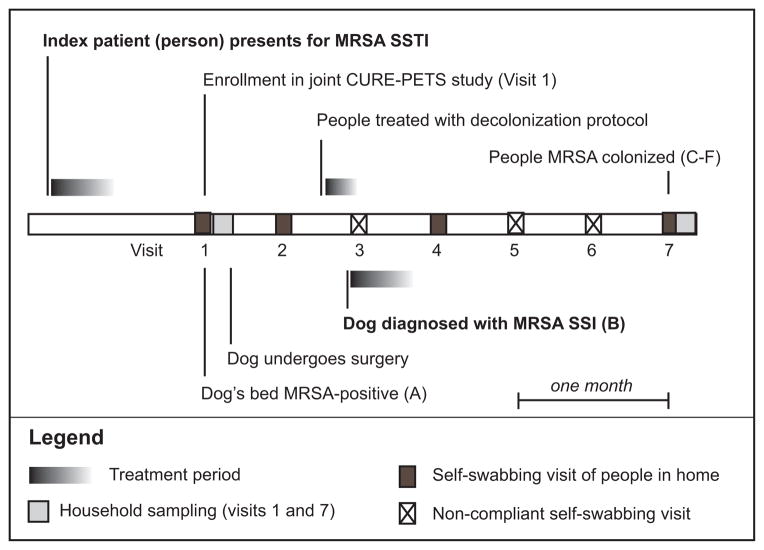
The household, which consisted of the index patient and a white female household member in her 30s, enrolled in the Epidemiology and Prevention of MRSA Transmission in the Community trial (NCT00966446) in July 2012 (Cluzet et al. 2015a; Cluzet et al. 2015b). Four weeks after the index patient’s MRSA SSTI diagnosis, the two people in the household were randomized at the baseline study visit to receive one week of twice-daily nasal mupirocin treatment and two body washes with 4% chlorhexidine gluconate (Hibiclens®, Mölnlycke Health Care, Norcross, Georgia). Mupirocin and Hibiclens® were received one week prior to development of SSI in the dog. This treatment involved daily reminders via text message during the week of treatment and a daily log of treatment adherence. The household members reported full compliance with the protocol. At the baseline visit and at subsequent biweekly intervals, household members also provided Eswabs™ (Copan Diagnostics, Murrieta, CA) obtained from nares, axillae/groin, and (for the index patient) the healed lesion site to test for S. aureus (MSSA and MRSA) by culture. Household members provided swabs at baseline (visit one), two weeks later (visit two), eight weeks later (visit four), and 12 weeks later (visit seven) (Figure 1).
Figure 1. Timeline for the household, demonstrating when isolates were collected from the dog’s bed, the dog, the index patient (dog owner), and the household member.
The household concurrently enrolled in the Pets and Environmental Transmission of Staphylococci (PETS) study, which involved sampling of the dog and the home environment at visits one and seven. Electrostatic cloths (Swiffer™, Proctor & Gamble) were used as previously described for surface sampling (Davis et al. 2012a). Samples were preserved in sterile, sealed stomacher bags until culture. Sites sampled were 1) the top of the refrigerator, 2) the handle of the refrigerator, 3) the top of the television, 4) the television remote, 5) a kitchen towel, 6) the bathroom faucet handle, 7) the index patient’s pillow, and 8) the dusty surface of the headboard of the bed. Sterile BBL™ culturettes (BD, Franklin Lakes, NJ) were used to collect samples from the dog’s nares, mouth, inguinal skin, and perineum. Electrostatic cloths were used to collect a sample from the “petting zone” on the dorsum of the dog and from the dog’s bed.
Human Eswab™ (Copan Diagnostics, Murrieta CA) samples were streaked onto CHROMagar MRSA agar plates (BD, Sparks MD) and incubated at 37°C for 24–48 hours. Environmental cloths and animal swabs and cloths were cultured for methicillin-susceptible and methicillin-resistant coagulase-positive staphylococci using parallel broth enrichment protocols. Presumptive staphylococcal isolates were identified on Columbia CNA blood agar (BD, Sparks MD) and individual isolates were sub-cultured to Baird Parker agar (BD, Sparks MD) and incubated at 37°C for 48hours as previously described (Davis et al. 2012a). Isolates were frozen at −80°C in Microbank™ tubes (Pro-Lab Diagnostics, Canada) until further testing.
The staphylococcal species was confirmed using a multiplex PCR assay that amplifies species-specific segments of the nuclease gene (nuc) (Hirotaki et al. 2011). MRSA isolates were confirmed to carry mecA by presence of a universal mecA/C sequence and absence of the mecC gene using sequential PCR, with ATCC43300 as mecA-positive and LGA251 as mecC-positive controls (Garcia-Alvarez et al. 2011). Isolates were tested for antimicrobial susceptibility to amikacin (AMK), cefoxitin (FOX), chloramphenicol (C), ciprofloxacin (CIP), clindamicin (CLI), erythromycin (ERY), gentamicin, linezolid (LZD), quinupristin-dalfopristin (SYN), tetracycline (TE), and trimethoprim-sulfamethoxazole (SXT) prior to cryopreservation using Kirby-Bauer disc diffusion testing according to CLSI standards (CLSI. 2013). Mupirocin and vancomycin susceptibilities were evaluated via E-test® analysis (Biomerieux, Marcy l’Etoile, France).
A priori, isolates were compared using PCR methods to identify Panton-Valentine leukocidin (PVL) genes, pulsed-field gel electrophoresis (PFGE), Staphylococcus aureus protein A (spa) typing, and staphylococcal chromosomal cassette (SCCmec) typing (Shopsin et al. 1999; McDougal et al. 2003; Milheirico et al. 2007; Zhang et al. 2008). Isolates then were subjected to pyrosequencing via 454 chemistry using the Genome Sequencer FLX System (Roche, Branford, CT, USA). Single-nucleotide polymorphisms (SNPs) were compared to assess overall relatedness, using a cut-off of ≤60 SNPs for clonal assignment (Tong et al. 2015). Details of these methods and additional results are provided in the online supplement.
Results
The human index patient was a white male in his 30s who presented as an outpatient to the emergency department (ED) after a two-week history of soft tissue swelling of the neck. The patient had a temperature of 37.2°C and reported no co-morbidities present. The attending physician diagnosed a non-draining, indurated abscess on the neck; incised and drained the abscess; administered one dose of intravenous clindamycin in the ED; and prescribed oral, twice daily trimethoprim-sulfamethoxazole and Hibiclens® use for ten days. Culture of pus from the abscess confirmed MRSA. The isolate was discarded by the hospital laboratory before study staff could request it for additional testing.
A 23-month old female spayed 32kg Italian Mastiff presented for outpatient Tibial Tuberosity advancement surgery to repair a ruptured cranial cruciate ligament on the right stifle two days after the study baseline visit (visit one) and four weeks after the index patient’s ED visit. This surgery involved implantation of a 6-prong plate, 12mm cage titanium implant, and cancellous bone graft, as well as additional C-laser treatments. Four weeks later, SSI was diagnosed by the veterinary surgeon. A commercial veterinary diagnostic laboratory identified the infecting organism as MRSA (Isolate B). An additional swab was submitted to study staff. A three-week course of oral, twice daily clindamycin was prescribed and the dog’s bed was laundered.
A MRSA isolate (Isolate A) was cultured from the dog’s bed at visit one, prior to the surgery. No other environmental isolates at either visit were found to be S. aureus. No MRSA isolates were cultured from the index patient and household member at baseline, at visit two, or at visit four. No S. aureus isolates were identified from the dog at visit one. At visit seven, MRSA was isolated from both household members (Isolates C-F). At visit seven, a methicillin-susceptible S. aureus (MSSA) was cultured from the dog’s perineum (Isolate G).
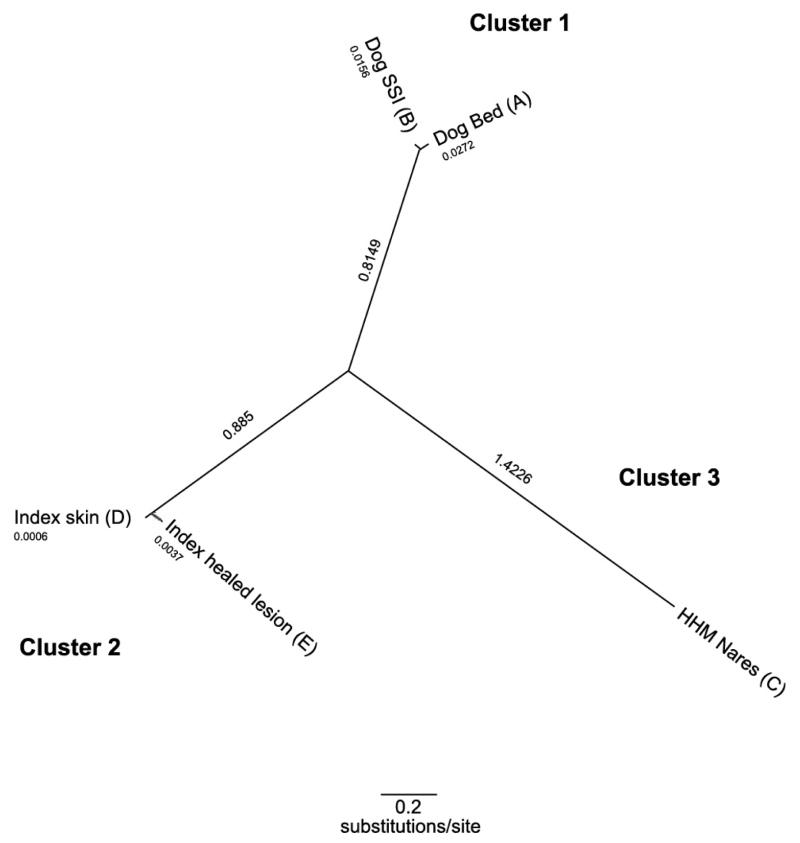
The MRSA isolate from the dog bed at visit one, prior to the surgery, and from the dog’s surgical site infection at visit three were 100% identical by PFGE and had an identical spa type (t121, a deletion variant of spa type t008—USA300) (Figure 2). Both isolates were positive for the PVL gene. At visit seven, both household members were positive for MRSA from multiple sites. PFGE indicated that the isolates from the index patient axillae/groin, index patient healed lesion and household member nares were related (Dice coefficient of similarity=0.968) had an identical spa type (t008—USA300), and were positive for the PVL gene. In addition, one MRSA isolate was obtained from the nares of the index patient. This isolate was spa type t002 (USA 100) and was negative for the PVL gene. While the dog infection and three of the human colonizing isolates had the same pulsed-field profile, they had minor differences in spa type (t121 versus t008). Whole genome sequencing and SNP analysis revealed that the dog bed and dog SSI MRSA isolates clustered together, but did not cluster with the subsequent human MRSA isolates (Figure 3).
Figure 2. Genetic and phenotypic characteristics of S. aureus isolates from the household over time.
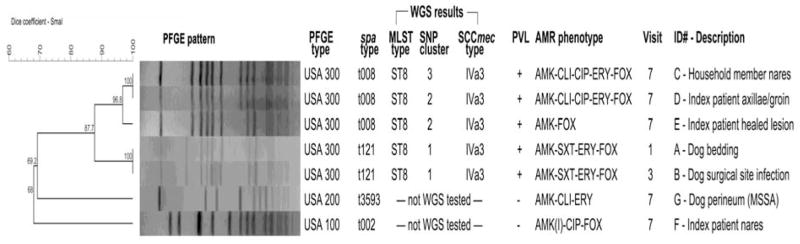
acronyms. AMR: antimicrobial resistance, with (I) indicating intermediate resistance; MLST: multi-locus sequence type; PFGE: pulsed-field gel electrophoresis; PVL: Panton-Valentine leukocidin; spa: staphylococcal protein A; SCCmec: staphylococcal chromosomal cassette mec conferring resistance to β-lactam antimicrobials; SNP: single nucleotide polymorphism; WGS: whole genome sequencing.
Figure 3. Household MRSA USA300 isolate phylogeny inferred with single nucleotide polymorphisms (SNPs).
Discussion
This report is notable in that 1) it describes laboratory-confirmed MRSA infections in a person and his dog in temporal and spatial proximity; 2) it implicates an environmental surface (the pet’s bed) in exposure of the dog prior to onset of the dog’s infection, which to our knowledge is the first report of a potential linkage between MRSA contaminated pet bedding and subsequent pet disease; and 3) it fails to implicate the pet in human MRSA outcomes, which also is distinct in the case literature. This study further demonstrates the critical importance of using multiple typing methods to investigate household transmission. In the absence of spa typing data, we would have concluded that the human and animal strains were related; whole genome sequencing was required to confirm that the dog bed and dog SSI MRSA isolate cluster was distinct from related strains found subsequently in people. In the prior literature, less than half (5 of 11 case reports or case series) were documented using more than one molecular typing technique, typically the same combination (PFGE+spa typing) that we initially employed here (Sing et al. 2008; Rutland et al. 2009; Faires et al. 2009; Ferreira et al. 2011). This example highlights the value of using whole genome sequencing methods for strain comparison to assess the dynamics and epidemiology of household MRSA contamination.
While it can be speculated that the MRSA-infected owner contaminated the dog’s bed or another fomite responsible for exposure, and that this led to the dog’s SSI, this route of indirect transmission cannot be confirmed because the clinical isolate from the index patient could not be obtained. Similarly, both dog and owner may have been exposed to a common source outside the home. A third explanation is that the dog was exposed to MRSA through veterinary contact prior to the surgery. However, during 2012 and 2013, the veterinary surgeon performed 300 surgeries, with three staphylococcal surgical site infections (1% SSI rate), including this case. Only one case (this report) was MRSA; the other two dogs were infected with methicillin-resistant S. pseudintermedius, an animal-associated pathogen. While veterinarians and their staff may become MRSA colonized, which could be a risk factor for transmission to their animal patients, Hanselman et al. found MRSA positivity rates of 4.4% among small-animal veterinarians attending a U.S. conference (Hanselman et al. 2006).
The finding of diverse MRSA strains in this household within six weeks of cessation of harmonized decolonization treatment in both people and treatment for infection in the dog is of great concern as it demonstrates rapid re-introduction of MRSA to the household. The scenario described in this report represents one explanation for observed failure of household-wide decolonization protocols to eradicate S. aureus carriage from index cases (Fritz et al. 2012; Cluzet et al. 2015a) and is unique in that it neither implicates the home environment nor the pet dog in re-colonization of the index patient following successful treatment. While future case assessments and research studies should include multiple evaluations of home environmental contamination and companion animal carriage over time to capture the potentially dynamic nature of intra-household transmission, attention to community sources outside the home may be necessary to understand drivers of recurrent colonization or infection.
Supplementary Material
Acknowledgments
Funding
This work was supported by a Commonwealth Universal Research Enhancement Program grant from the Pennsylvania State Department of Health (to E.L.), a grant from the Johns Hopkins Center for a Livable Future (to M.D.), a pilot award from the Morris Animal Foundation (to M.D.), and a grant from the American College of Veterinary Dermatology/American Academy of Veterinary Dermatology (to D.M.). Investigators were supported by a NIAID K24 (AI080942 to E.L.) and a NIEHS T32 grant (ES7141-29 to M.D.).
We thank the participants and study staff, particularly John Ndicu. We thank Alan L. Scott, Anne Jedlicka, and Amanda Dziedzic in the JHSPH Genomic Analysis and Sequencing Core Facility for assistance with genome sequencing and Johanne Ahrenfeldt for helpful discussions about SNP analysis. This work was conducted as part of the CDC Prevention Epicenter.
Footnotes
Data Access
Genome data from this study have been deposited in the Short Reads Archive under accession numbers A: SAMN03755562, B: SAMN03755563, C: SAMN03755566; D: SAMN03755565; and E: SAMN03755564.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bramble M, Morris D, Tolomeo P, Lautenbach E. Potential role of pet animals in household transmission of methicillin-resistant Staphylococcus aureus: a narrative review. Vector Borne Zoonotic Dis. 2011;11:617–620. doi: 10.1089/vbz.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute; 2013. pp. 1–206. [Google Scholar]
- Cluzet VC, Gerber JS, Nachamkin I, Metlay JP, Zaoutis TE, Davis MF, Julian KG, Linkin DR, Coffin SE, Margolis DJ, Hollander JE, Bilker WB, Han X, Mistry RD, Gavin LJ, Tolomeo P, Wise JA, Wheeler MK, Hu B, Fishman NO, Royer D, Lautenbach E. Risk factors for recurrent colonization with methicillin-resistant Staphylococcus aureus in community-dwelling adults and children. Infect Control Hosp Epidemiol. 2015a doi: 10.1017/ice.2015.76. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzet VC, Gerber JS, Nachamkin I, Metlay JP, Zaoutis TE, Davis MF, Julian KG, Royer D, Linkin DR, Coffin SE, Margolis DJ, Hollander JE, Mistry RD, Gavin LJ, Tolomeo P, Wise JA, Wheeler MK, Bilker WB, Han X, Hu B, Fishman NO, Lautenbach E. Duration of colonization and determinants of earlier clearance of colonization with methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2015b;60:1489–1496. doi: 10.1093/cid/civ075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MF, Baron P, Price LB, Williams DL, Jeyaseelan S, Hambleton IR, Diette GB, Breysse PN, McCormack MC. Dry collection and culture methods for recovery of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains from indoor home environments. Appl Environ Microbiol. 2012a;78:2474–2476. doi: 10.1128/AEM.06886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MF, Iverson SA, Baron P, Vasse A, Silbergeld EK, Lautenbach E, Morris DO. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect Dis. 2012b;12:703–716. doi: 10.1016/S1473-3099(12)70156-1. [DOI] [PubMed] [Google Scholar]
- Faires MC, Tater KC, Weese JS. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J Am Vet Med Assoc. 2009;235:540–543. doi: 10.2460/javma.235.5.540. [DOI] [PubMed] [Google Scholar]
- Ferreira JP, Fowler VG, Correa MT, Lyman R, Ruffin F, Anderson KL. Transmission of methicillin-resistant Staphylococcus aureus between human and hamster. J Clin Microbiol. 2011;49:1679–1680. doi: 10.1128/JCM.02469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Epplin EK, Garbutt J, Fraser VJ. Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: a randomized trial. Clin Infect Dis. 2012;54:743–751. doi: 10.1093/cid/cir919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez L, Holden MTG, Lindsay H, Webb CR, Brown DFJ, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan E, Kearns AM, Pichon B, Hill RLR, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotaki S, Sasaki T, Kuwahara-Arai K, Hiramatsu K. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J Clin Microbiol. 2011;49:3627–3631. doi: 10.1128/JCM.00488-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E, Smith DL, Laxminarayan R. Community-associated methicillin-resistant Staphylococcus aureus in outpatients, United States, 1999–2006. Emerging Infectious Diseases. 2009;15:1925–1930. doi: 10.3201/eid1512.081341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenbach E, Tolomeo P, Nachamkin I, Hu B, Zaoutis TE. The impact of household transmission on duration of outpatient colonization with methicillin-resistant Staphylococcus aureus. Epidemiol Infect. 2010;138:683–685. doi: 10.1017/S0950268810000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manian FA. Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clin Infect Dis. 2003;36:e26–8. doi: 10.1086/344772. [DOI] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutland BE, Weese JS, Bolin C, Au J, Malani AN. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerging Infectious Diseases. 2009;15:1328–1330. doi: 10.3201/eid1508.081635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Tuschak C, Hormansdorfer S. Methicillin-resistant Staphylococcus aureus in a family and its pet cat. N Engl J Med. 2008;358:1200–1201. doi: 10.1056/NEJMc0706805. [DOI] [PubMed] [Google Scholar]
- Tong SY, Holden MT, Nickerson EK, Cooper BS, Koser CU, Cori A, Jombart T, Cauchemez S, Fraser C, Wuthiekanun V, Thaipadungpanit J, Hongsuwan M, Day NP, Limmathurotsakul D, Parkhill J, Peacock SJ. Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome Res. 2015;25:111–118. doi: 10.1101/gr.174730.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijkeren E, Wolfhagen MJHM, Heck MEOC, Wannet WJB. Transmission of a Panton-Valentine leucocidin-positive, methicillin-resistant Staphylococcus aureus strain between humans and a dog. J Clin Microbiol. 2005;43:6209–6211. doi: 10.1128/JCM.43.12.6209-6211.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijkeren E, Wolfhagen MJ, Box AT, Heck ME. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2004;10:2235–7. doi: 10.3201/eid1012.040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mcclure J, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton-Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol. 2008;46:1118–1122. doi: 10.1128/JCM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.