Abstract
Notch receptors direct the differentiation of T helper (TH) cell subsets, but their influence on regulatory T (Treg) cell responses is obscure. We here report that lineage-specific deletion of components of the Notch pathway enhanced Treg cell-mediated suppression of TH1 responses, and protected against their TH1 skewing and apoptosis. Expression in Treg cells of gain of function transgene encoding Notch1 intracellular domain resulted in lymphoproliferation, exacerbated TH1 responses and autoimmunity. Cell-intrinsic canonical Notch signaling impaired Treg cell fitness, promoted the acquisition by Treg cells of a TH1 cell-like phenotype, whereas Rictor-dependent non-canonical Notch signaling activated the AKT-Foxo1 axis and impaired Foxp3 epigenetic stability. These findings establish a critical role for Notch signaling in controlling peripheral Treg cell functions.
Notch signaling serves pleiotropic roles in the immune system by influencing multiple lineage decisions of developing lymphoid and myeloid cells 1, 2. In mammals, the Notch family is composed by 4 Notch receptors (Notch1–4) and 5 ligands (Delta-like1, 3, and 4 and Jagged1 and 2). After ligand-receptor interaction, the intracellular domain of the Notch receptor is cleaved, traffics to the nucleus and forms complexes with the DNA binding factor RBPJ and the transcriptional co-activators MAML1-3, promoting expression of target genes. In addition to this canonical pathway, cleaved intracellular domains of Notch receptors engage non-canonical signaling components, including the metabolic checkpoint kinase complex mTORC2 and its associated adaptor Rictor 3, 4. Notch intracellular domain also interacts with components of the NF-κB, TGF-β and the hypoxia response pathways 5, 6, 7.
Notch signaling is activated at various stages of commitment and development of T cell lineages, such as commitment to the T cell versus the B cell lineage, αβ versus γδ T cell differentiation and CD4 T versus CD8 single-positive T cell differentiation 1, 2, and during T cell-mediated immune responses, such as peripheral cytotoxic and helper T (TH) cell differentiation and function 8. Pathogen-associated molecular patterns are known to promote expression of Notch ligand at the surface of antigen presenting cells. Activation of naive CD8+ T cells requires binding of Delta-like1 on antigen presenting cells by Notch1 or Notch2 leading to expression of Eomes, Gzmb, Ifng and Pfr1 9, 10. In naïve CD4+ T cells, Delta-like1 and 4 activate Notch signaling and Tbx21 transcription, encoding the TH1 transcriptional regulator T-bet 11, 12. During TH2 differentiation, activation of Notch1 and 2 by Jagged1 and Jagged2 favor the expression of Gata3 and Il4 13, 14, 15, 16.
Notch1 signaling has been described to be important in the differentiation of TH17 and TH9 subsets of helper T cells by promoting Rorc and Il9 expression, respectively 5, 17, 18. The role of Notch signaling in the regulatory T (Treg) cell compartment remain controversial. In vitro, Jagged ligands and Notch 1 and Notch3 signaling seem to promote Treg cell differentiation and survival 3, 19, 20, 21. In contrast, several in vivo studies have demonstrated that blockade of the Notch pathway, in particular Notch1 and Notch2, promotes tolerance in murine models of graft versus host disease, in association with the expansion of Treg cells 22, 23. Studies have shown tolerogenic functions for antibodies to Notch1 in a humanized mouse model of vasculitis and in a murine model of aplastic anemia 24, 25. In this study, we have employed Treg cell lineage-specific genetic and functional approaches to identify a key role for the Notch pathway in destabilizing Treg cells, promoting their apoptosis and inhibiting their function in the context of inflammation.
Results
Notch negatively regulates Treg cell functions and homeostasis
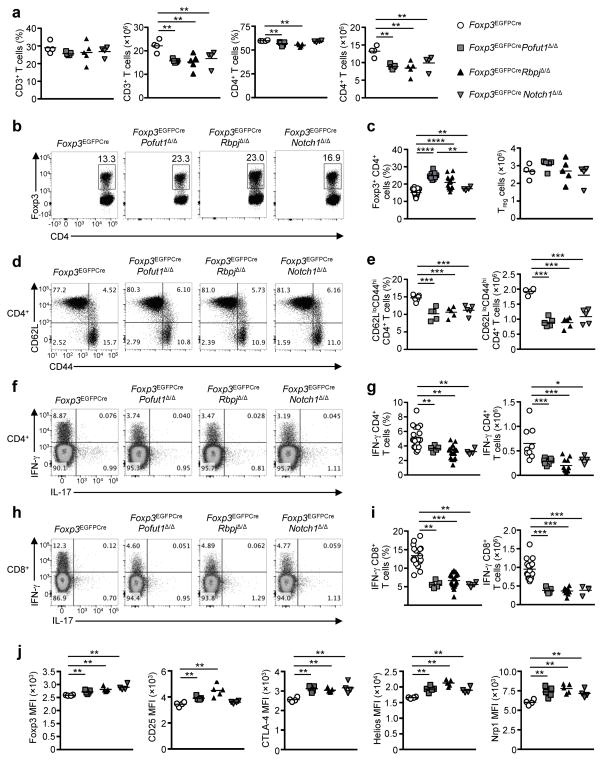
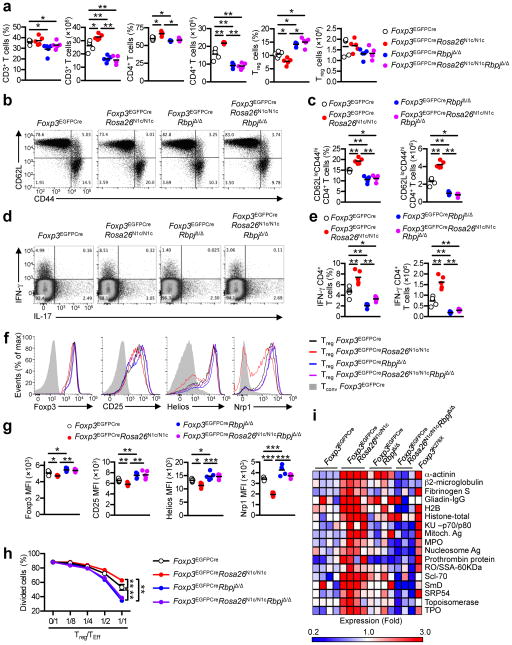
To elucidate the role of the Notch pathway in peripheral tolerance, we examined the functional consequences of interrupting Notch receptor signaling in a Treg cell-specific manner. To this end, we derived mice with a bacterial artificial chromosome (BAC) expressing an enhanced green fluorescent protein fused with the Cre recombinase under the control of Foxp3 promoter together with loxP-flanked Pofut1, encoding the enzyme protein O-fucosyltransferase 1 (called Foxp3EGFPCrePofut1Δ/Δ here; Supplementary Fig. 1a, b) 26. The latter mediates fucosylation of Notch receptors, which is essential for receptor ligand interaction; its deficiency abrogates Notch signaling 26. Treg cell-specific Pofut1 deficiency resulted in a decrease in peripheral CD3+ T cells and CD4+ T cell numbers by about 25% compared to Foxp3EGFPCre mice (Fig. 1a). It also resulted in a reciprocal increase in Treg cell frequency, with decreased CD4+CD62LloCD44hi T effector memory and a relative increase in CD62LhiCD44lo naïve T cells as compared to Foxp3EGFPCre mice (Fig. 1b–e). Expression of IFN-γ in splenic CD4+ T cells was markedly decreased in Foxp3EGFPCrePofut1Δ/Δ as compared to Foxp3EGFPCre mice, whereas expression of IL-17 was unaffected (Fig. 1f, g). Similar results were obtained for the IFN-γ production by CD8+ T cells (Fig. 1h, i). Expression of several Treg cell markers, including Foxp3, CD25, CTLA-4, Helios and neuropilin 1 (Nrp1) was increased in Pofut1-deficient compared to Foxp3EGFPCre Treg cells (Fig. 1j). We examined the role of the canonical Notch signaling in Treg cells by lineage-specific deletion of loxP-flanked Rbpj (Foxp3EGFPCreRbpjΔ/Δ; Supplementary Fig. 1a, b) 27. The key phenotypes of Foxp3EGFPCrePofut1Δ/Δ mice were recapitulated in Foxp3EGFPCreRbpjΔ/Δ mice (Fig. 1a–j), indicating that the canonical pathway is the primary mediator of Notch signaling in Treg cells. Of the four Notch receptors, Notch1 was the most highly expressed in Treg cells, followed by Notch 2, whereas Notch3 and Notch4 expression was negligible (Supplementary Fig. 1c, d). The phenotype of mice with Notch1- deficient Treg cells, achieved by lineage-specific deletion of loxP-flanked Notch1 (Foxp3EGFPCreNotch1Δ/Δ; Supplementary Fig. 1a, b), approximated those of mice with Pofut- or RBPJ-deficient Treg cells, indicating that Notch1 was the main receptor through which Notch signaling was triggered in Treg cells (Fig. 1a–j) 28.
Fig. 1. Interruption of Notch signaling in Treg cells results in a super-regulatory phenotype.
(a) Frequencies and numbers of CD3 and CD4 T cells from the spleen of 8 weeks old Foxp3EGFPCre, Foxp3EGFPCrePofutΔ/Δ, Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreNotch1Δ/Δ mice. (b) Flow cytometric analyses of CD4 and Foxp3 markers on CD3+ T cells are shown. (c) Frequencies and numbers of Treg cells for each group of panel (a). (d) Flow cytometric analysis of CD62L and CD44 markers on CD4+ T cells are shown. (e) Frequencies and numbers of memory CD4+ T cells for each group. (f) Flow cytometric analyses of IFN-γ and IL-17 on CD4+ T cells are shown. (g) Frequencies and numbers of IFN-γ producing CD4+ T cells for each group. (h) Flow cytometric analyses of IFN-γ and IL-17 expression in CD8+ T cells. (i) Frequencies and numbers of IFN-γ producing CD8+ T cells shown in panel (H). (j) Expression of Foxp3, CD25, CTLA4, Helios and Nrp1 markers were evaluated of splenic Treg cells and expressed as mean fluorescence intensity (MFI). Results are representative of at least 3 experiments per panel. * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001 by one way ANOVA with post test analysis.
We also assessed the effect of loss of function mutations in genes encoding members of the Notch pathway on the generation of thymus-derived Treg cells. We observed similar frequencies and numbers of Foxp3+ Treg cells among the mature CD4 single positive (CD4SP) thymocyte compartment in Foxp3EGFPCre, Foxp3EGFPCrePofut1Δ/Δ Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreNotch1Δ/Δ mice (Supplementary Fig. 2a, b) we employed Foxp3YFPCre mice, which express a fusion of yellow fluorescent protein (YFP) and Cre recombinase in Treg cells under control of the endogenous Foxp3 locus 29. We found that the in vitro differentiation of naive CD4+ T cells from Foxp3YFPCreRbpjΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ into induced Treg (iTreg) cells was similar to that of Foxp3YFPCre control cells (Supplementary Fig. 2c, d). These results indicate normal thymic development and peripheral differentiation of Treg cell populations.
To further elucidate the cell-intrinsic impact of loss of function Notch mutations on Treg cells, we took advantage of random X chromosome inactivation in females to analyze both central thymic and peripheral splenic Treg cells in heterozygous Foxp3YFPCre/+RbpjΔ/Δ female mice. Compared to heterozygote Foxp3YFPcre/+ littermate control female mice, in which approximately 50% of Treg cells within the thymus and spleen are YFP+, a higher proportion of YFP+ Treg cells was observed in the periphery of Foxp3YFPCre/+RbpjΔ/Δ females (Supplementary Fig. 2e, f). YFP+ Treg cells had higher expression of several Treg cell markers, including CD25, Helios and Nrp1 compared to YFP− Treg cells (Supplementary Fig. 2g, h). Overall, loss of function Notch signaling mutations exerted a positive, cell intrinsic effect on Treg cell fitness and function both at steady state and in the context of inflammation.
Treg cell-specific loss of Notch function protects mice from GVHD
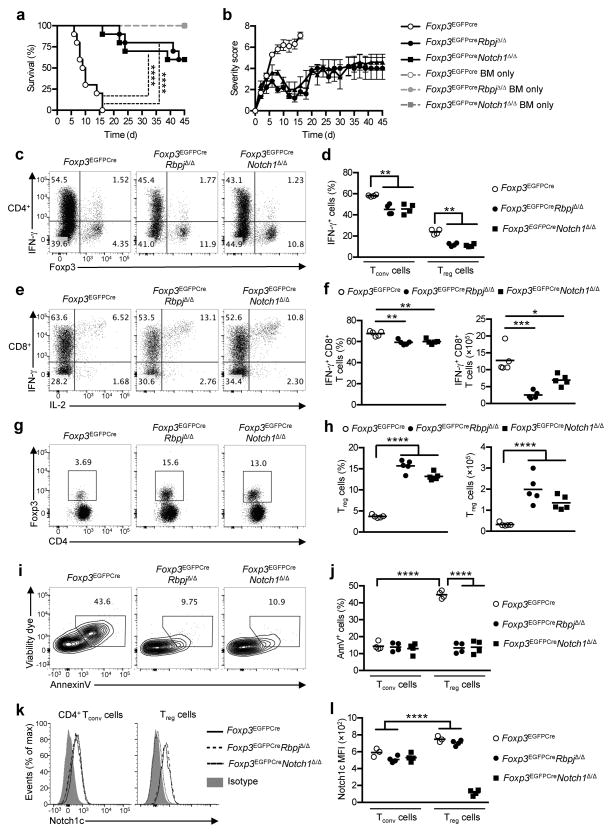
We examined the suppressive capacities of Notch signaling-deficient Treg cells in the context of a strong inflammatory response, using the model of major histocompatibility complex (MHC) class I&II disparate graft versus host disease (GVHD). Adoptive transfer of T cell-depleted bone marrow from Foxp3EGFPCre, Foxp3EGFPCreRbpjΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ C57Bl/6 mice into BALB/c mice lead to recovery from the lethal irradiation (Fig. 2a). Co-transfer of total spleen cells from Foxp3EGFPCre mice induced lethal GVHD, associated with expansion of donor-derived (H-2Kb+) IFN-γ-producing CD4+ and CD8+ T cells (Fig. 2a–f), while co-transfer of Foxp3EGFPCreRbpjΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ total spleen cells resulted in protection from lethal GVHD in recipient mice, with attenuated GVHD severity scores and decreased IFN-γ production by donor T cells (Fig. 2a–f). Moreover, transfer of total spleen cells from Foxp3EGFPCreRBPJΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ mice lead to increased frequency and number of donor Treg cells, associated with decreased apoptosis, as assessed by Annexin V (AnnV) staining, and decreased IFN-γ production (Fig. 2c–d, g–j). In the context of exaggerated inflammation induced by GVHD, donor Foxp3EGFPCre Treg cells exhibited higher expression of the cleaved intracellular domain of Notch1 (N1c), as compared to their CD4+Foxp3− Tconv cell counterparts (Fig. 2h–i) Together, these results reveal a direct role for cell-intrinsic Notch signaling in destabilizing Treg cells in the context of inflammation by promoting their apoptosis and IFN-γ production.
Fig. 2. Treg cell-specific loss of function Notch signaling mutations protect mice from lethal graft versus host disease.
(a) Survival and (b) severity score of lethally irradiated (8.5/9Gy) BALB/c mice infused with C57Bl/6 Foxp3EGFPCre, Foxp3EGFPCreRbpjΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ T cell-depleted bone marrow, either alone (BM only) or together with spleen cells of the respective genotypes. For T cell subpopulation analyses were carried out on spleen cells at day 5 post adoptive transfer. The BM only groups were not included in panel b. (c) Flow cytometric analyses of IFN-γ and Foxp3 markers on CD4+ T cells are shown. (d) Frequencies of IFN-γ producing CD4+ Tconv (Foxp3−) and Treg (Foxp3+) cells for each group. (e) Flow cytometric analyses of IFN-γ and IL-2 markers on CD8+ T cells. (f) Frequencies and numbers of IFN-γ producing CD8+ T cells for each group. (g) Flow cytometric analyses of Foxp3 marker on CD4+ T cells are shown. (h) Frequencies and numbers Treg cells for each group. (i) Viability dye and Annexin V (AnnV) staining of Treg cells are shown (j) Frequencies of apoptotic (AnnV+) Tconv and Treg cells for each group. (k) Overlay of representative N1c expression on Tconv and Treg cells are shown. (l) MFI of N1c expression in Tconv and Treg cells for each group. Results are representative of at least 2 experiments per panel. * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001 by log-rank test, one way ANOVA and two way ANOVA with post test analysis.
Treg cell-specific gain of Notch function disrupts peripheral tolerance
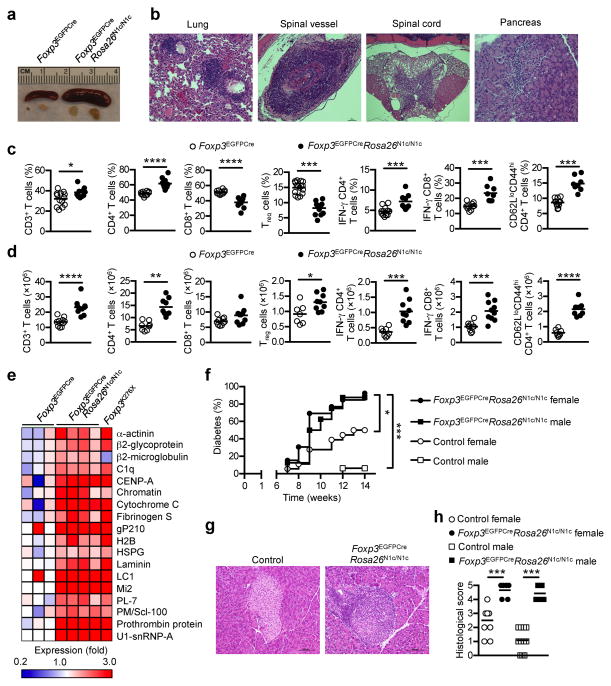
To investigate the role of Notch1 signaling in Treg cells, we generated Foxp3EGFPCreRosa26N1/N1c mice, which constitutively express N1c in their Treg cells (Supplementary Fig. 1e) 30. We found increased expression of N1c in Foxp3EGFPCreRosa26N1/N1c Treg cells, associated with heightened expression of several Notch signaling target genes, including Hes1, Hey1, Heyl and Dtx1 (Supplementary Fig. 1f, g). In contrast to the mutations that resulted in loss of Notch function, constitutive expression of N1c in Treg cells resulted in an autoimmune lymphoproliferative disease, whose manifestations included large vessel vasculitis and lymphocytic end organ infiltration in the Foxp3EGFPCreRosa26N1/N1c mice (Fig. 3a, b). The CD4+CD62LloCD44hi T effector memory cell pool was expanded and the numbers of CD4+ and CD8+ T cells expressing IFN-γ were increased by 50%, while the frequency of Treg cells was decreased by 45% (Fig. 3c, d). There was also dysregulation of the B cell compartment with significant increase of several circulating autoantibodies (to 18 out of 98 screened endogenous antigens) in Foxp3EGFPCreRosa26N1/N1c mice as compared to Foxp3EGFPCre mice (Fig. 3e). Overexpression of N1c in Treg cells dramatically worsened the incidence and severity of type I diabetes in the genetically susceptible NOD mice, particularly in normally resistant NOD males (Fig. 3f–h). Whereas N1c expression in T cell precursors precipitates the occurrence of T cell acute lymphocytic leukemia, no evidence of leukemia was found in Foxp3EGFPCreRosa26N1c/N1c mice up to 6 months of age (data not shown) 4.
Fig. 3. Exacerbated Notch signaling in Treg cells results in immune dysregulation and autoimmunity.
(a) A representative picture of spleens and peripheral lymph nodes from 6 months old Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c mice. (b) Representative pictures of H&E staining of lung, spinal vessels, spinal cord and pancreas from 6 month old Foxp3EGFPCreRosa26N1c/N1c mice. Frequencies (c) and numbers (d) of CD3, CD4, CD8 T cells, Treg cells, IFN-γ producing CD4 and CD8 T cells and memory CD62LloCD44hi CD4 T cells from the spleen of 8 weeks old Foxp3EGFPCre (white circles) and Foxp3EGFPCreRosa26N1c/N1c (black circles) mice. (e) Heatmap summarizing the expression of circulating autoantibodies significantly increased in 8 weeks old Foxp3EGFPCreRosa26N1c/N1c compared to Foxp3EGFPCre mice (serum from Foxp3K276X was used as a positive control). (f) Diabetes incidence of littermate control and Foxp3EGFPCreRosa26N1c/N1c female (n=18 and n=13 respectively) and male (n=16 and n=8 respectively) mice on NOD background. (g, h) Representative pictures of H&E staining of pancreas and histological score of 15 weeks old control females (white circles) or males (white squares) and Foxp3EGFPCreRosa26N1c/N1c females (Black circles) or males (black squares) on NOD background. * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001 by unpaired two-tailed Student’s t-test and log-rank test.
Analysis of Foxp3EGFPCreRosa26N1c/N1c mice revealed an age-dependent increase in the frequency of Treg cells that do not express the Foxp3 BAC-driven EGFP-Cre transgene (data not shown). Accumulation of EGFP− Treg cells was observed during thymic development and reached up to 30% of the peripheral Treg cell pool at 2 month of age (Supplementary Fig. 3a, b). EGFP− Treg cells were observed at very low frequencies in Foxp3EGFPCre, Pofut1, RBPJ or Notch1 mice (data not shown). Whereas the GFP+ Foxp3EGFPCreRosa26N1c/N1c Treg cells expressed high amounts of N1c, consistent with the expression of the transgene from the Rosa26 locus, N1c expression in GFP− Foxp3EGFPCreRosa26N1c/N1c Treg cells was similar to that of control Foxp3EGFPcre Treg cells (Supplementary Fig. 3c), suggesting that the GFP− Treg cells never expressed the Foxp3EGFPcre BAC gene and that transgenic expression of N1c induced a profound competitive disadvantage in Treg cells. To overcome this 'escape' phenotype, we used Foxp3YFPCre mice that expressed a YFP-Cre fusion protein under control of the endogenous Foxp3 locus 29. Unlike Foxp3EGFPCreRosa26N1c/N1c mice, 95% of Foxp3YFPCreRosa26N1c/N1c Treg cells expressed the Foxp3-driven Cre recombinase (Supplementary Fig. 3d, e). Analysis of heterozygous Foxp3YFPCre/WTRosa26N1c/N1c females revealed marked skewing of X-chromosome utilization by Treg cells in the periphery in favor of expressing the Foxp3WT allele (Supplementary Fig. 3f, g). YFP+ Foxp3YFPCre/WTRosa26N1c/N1c Treg cells expressed significantly lower amounts of Foxp3 compared to their YFP− Treg cell counterparts (Supplementary Fig. 3h, i). Double mutant Foxp3YFPCreRosa26N1c/N1c male mice developed a more aggressive and accelerated autoimmune lymphoproliferative disease than Foxp3EGFPCreRosa26N1c/N1c males, as revealed by tissue histology and immunological analyses (Supplementary Fig. 4). Altogether, these data indicate a profoundly deleterious impact of N1c expression on Treg cell fitness.
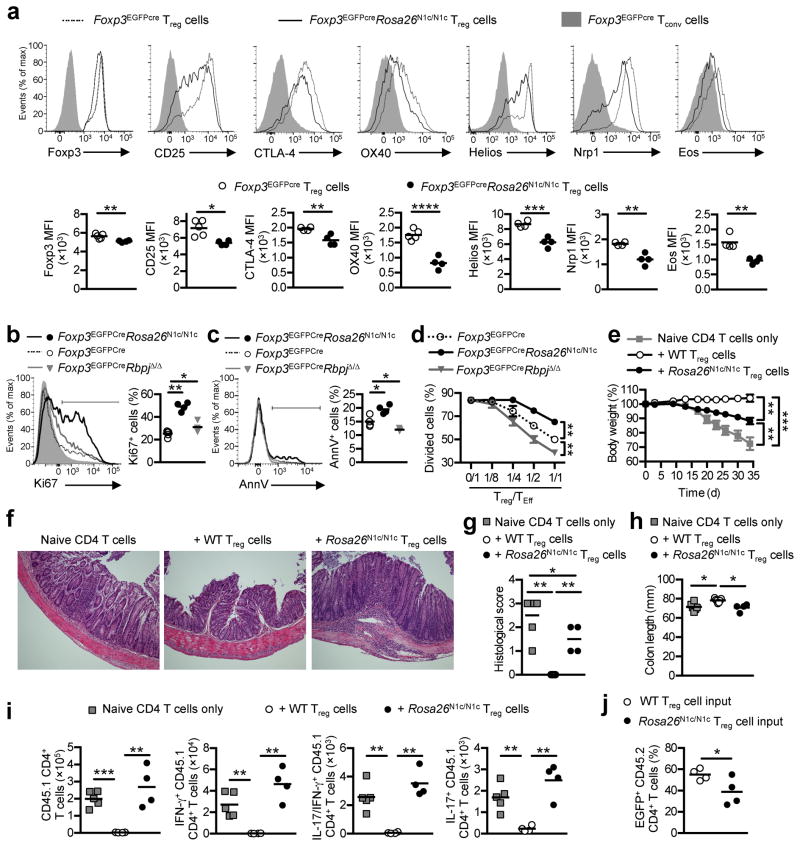
Flow cytometric analysis of peripheral Treg cells in the Foxp3EGFPCreRosa26N1c/N1c and Foxp3YFPCreRosa26N1c/N1c mice revealed decreased expression of Treg cell markers, including Foxp3, CD25, CTLA-4, OX40, Helios, Nrp1 and Eos (Fig. 4a and Supplementary Fig. 5a, b). Foxp3EGFPCreRosa26N1c/N1c Treg cells exhibited increased Ki67 expression, consistent with heightened cellular proliferation, and increased apoptosis, as detected by AnnV staining, as compared to Foxp3EGFPCre and Foxp3EGFPCreRbpjΔ/Δ Treg cells (Fig. 4b, c). In vitro, the suppressive capacity of Foxp3EGFPCreRosa26N1c/N1c Treg cells was decreased, whereas that of Foxp3EGFPCreRbpjΔ/Δ Treg cells was enhanced compared to Foxp3EGFPCre Treg cells (Fig. 4d). The generation of thymic Treg cells in Foxp3EGFPCreRosa26N1c/N1c mice proceeded unaffected as similar frequencies and numbers of Foxp3+ cells among CD4 single positive thymocytes were found in Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c mice (Supplementary Fig. 5c–d). In addition, the demethylation status of the conserved non-coding sequence 2 (CNS2) CpG elements in the Foxp3 promoter was similar in thymic Treg cells of Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c mice. (Supplementary Fig. 5e–f). In vitro, the differentiation of naive CD4+ Foxp3YFPCreRosa26N1c/N1c T cells into induced Treg (iTreg) cells following stimulation with anti-CD3+CD28 mAb and TGF-β was decreased compared to those of Foxp3YFPCre mice (Supplementary Fig. 5g, h), suggesting an inhibitory function of N1c in both thymus and periphery-derived Treg cells. Next, we investigated the effect of N1c on the ability of adoptively transferred Treg cells to suppress colitis in Rag1-deficient mice. Naive CD45RbhiCD4+ Tconv cells were isolated from CD45.1+ Foxp3EGFPCre donor mice and transferred alone or in combination with Treg cells from CD45.2+ Foxp3EGFPcre or CD45.2+ Foxp3EGFPcreRosa26N1c/N1c mice into T cell–deficient Rag1−/− host mice. We found that such co-transfer of Treg cells from CD45.2+ Foxp3EGFPcreRosa26N1c/N1c mice failed to suppress the colitis induced by the transfer of naive CD45.1+ CD45RbhiCD4+ Tconv cells: the recipient Rag1−/− mice exhibited substantial weight loss and tissue inflammation, as well as shorter colons and excess generation of IFN-γ+ and IL-17+ CD45.1+ effector T cells (Fig. 4e–i). Analysis of the CD45.2 cell compartment in recipient mice at d35 showed increased EGFP loss in Foxp3EGFPCreRosa26N1c/N1c (60%) as compared to control Foxp3EGFPCre cells (45%) (Fig. 4j), indicative of the heightened instability of N1c-expressing Treg cells.
Fig. 4. Notch signaling compromises Treg cell phenotype and suppressive functions.
(a) Expression of Foxp3, CD25, CTLA4, OX40, Helios, Nrp1 and Eos markers were evaluated in splenic Treg cells of Foxp3EGFPCre (Dashes lines and white circles) and Foxp3EGFPCreRosa26N1c/N1c (Solid lines and black circles) mice. Gray plains represent expression of those markers in Tconv of Foxp3EGFPCre mice. Cell turn-over was assessed in Treg cells of Foxp3EGFPCre (Dashes lines and white circles), Foxp3EGFPCreRosa26N1c/N1c (Solid black lines and black circles) and Foxp3EGFPCreRbpjΔ/Δ (Solid gray lines and Gray triangles) by Ki67 staining (b) for active cell cycle phases and by AnnV staining (c) for apoptosis. (d) In vitro suppression of responder CD4+ T cell proliferation (TEff) was assessed by evaluation of proliferation dye dilution upon anti- CD3/C28 stimulation in the presence of various number of Treg cells from each genotype. In vivo suppressive capacity of Treg cells from Foxp3EGFPCre (white circles), Foxp3EGFPCreRosa26N1c/N1c (black circles) was assessed in the CD4 T cell transfer-induced colitis model. (e) Changes in body weight over time are shown (n=4–7). One representative experiment of 3 is shown. (f–h) Disease severity was evaluated by H&E staining of colon sections and end point colon length. (i) Absolute number of total, IFN-γ and IL-17-producing CD45.1+CD4+ T cells from the naïve T cell input was quantified within the entire colon. (j) Stability of Treg cell lineage was shown by percent of EGFP expression of the CD45.2 T cell compartment. * p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001 by unpaired two-tailed Student’s t-test, One way ANOVA with post test analysis and two way ANOVA.
To delineate the intrinsic impact of N1c on the functional phenotype of Treg cells, as opposed to a cell-extrinsic effect mediated by systemic inflammation, we analyzed heterozygous Foxp3YFPCre/WTRosa26N1c/N1c female mice for lymphoproliferation, immune cell activation and Treg cell phenotype. In contrast to homozygous Foxp3YFPCre/YFPCreRosa26N1c/N1c females, Foxp3YFPCre/WTRosa26N1c/N1c females exhibited no signs of lymphoproliferation or TH1 skewing of CD4+ Tconv cells at steady state (Supplementary Fig. 6a–d). However, YFP+ Treg cells from Foxp3YFPCre/WTRosa26N1c/N1c females showed decreased expression of Treg cell markers such as Foxp3, CD25, CTLA4, OX40, Helios and Nrp1, as compared to YFP− Treg cells from the same mice (Supplementary Fig. 6e, f). These results indicate that N1c overexpression destabilizes the phenotype and impairs the function of Treg cells by a cell-intrinsic mechanism, leading to immune cell activation, lymphoproliferation and autoimmunity.
The canonical Notch pathway mediates Treg cell destabilization
To determine the overall contribution of the canonical Notch pathway to the inflammation and autoimmunity observed in Foxp3EGFPCreRosa26N1c/N1c mice, we concurrently deleted Rbpj in the Treg cells of these mice. Most major phenotypes of Foxp3EGFPCreRosa26N1c/N1c mice, including lymphoproliferation, T cell activation, TH1 and TC1 skewing, autoantibody production and alterations in Treg cell markers and suppressor function were reversed in Foxp3EGFPCreRosa26Nc1/N1cRbpjΔ/Δ mice (Fig. 5a–i). Thus, activation of canonical Notch signaling has a predominant role in the immune dysregulation observed in the Foxp3EGFPCreRosa26N1c/N1c mice.
Fig. 5. Treg cell failure and disease manifestations in Foxp3EGFPCreRosa26N1c/N1c mice proceed by a canonical pathway dependent mechanism.
(a) Frequencies and numbers of CD3, CD4 and Treg cells from the spleen of 8 weeks old Foxp3EGFPcre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (b) Flow cytometric analysis of CD62L and CD44 markers on CD4+ T cells. (c) Scatter plots represent frequencies and numbers of memory CD4+ T cells for each group. (d) Flow cytometric analysis of IFN-γ and IL-17 secretion by CD4+ T cells are shown. (e) Scatter plots represent frequencies and numbers of IFN-γ producing CD4+ T cells for each group. (f, g) Expression of Foxp3, CD25, Helios, Nrp1 markers were evaluated in splenic Treg cells of each group. n=4–5 per group. (h) In vitro suppression of responder CD4+ T cell proliferation (TEff) was assessed by evaluation of proliferation dye dilution upon anti-CD3/C28 stimulation in the presence of various number of Treg cells from each genotype. (I) Heatmap summarizing the expression of significantly modulated circulating autoantibodies in 8 weeks old Foxp3EGFPCre, Foxp3EGFPcreRosa26N1c/N1c and Foxp3EGFPcreRBPJΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice (serum from Foxp3K276X was used as positive control). One representative experiment of 2 or 3 is shown for panels (a-h). * p<0.05 and ** p<0.01 by One way ANOVA with post test analysis and two way ANOVA.
Notch activates RBPJ-dependent and independent transcription in Treg cells
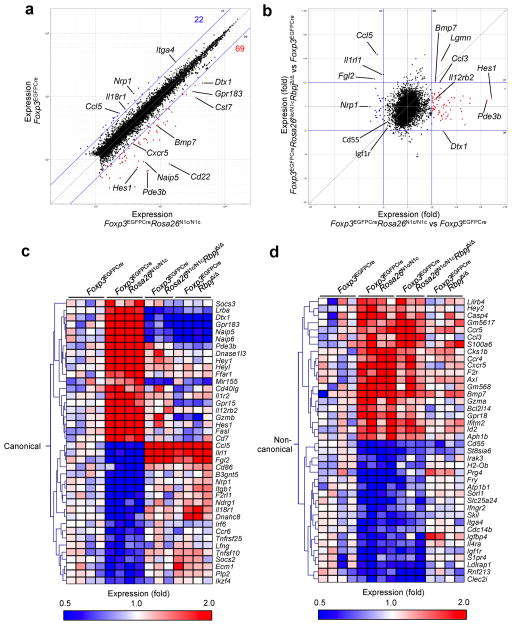
Treg cells have a transcriptional signature reflective of their regulatory function 31, 32, 33. We compared the transcriptional profiles of splenic Treg cells isolated from Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. We compared the transcriptional profiles of splenic Treg cells isolated from Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. Comparison of the transcriptional profiles of Foxp3EGFPCreRosa26N1c/N1c Treg cells and Foxp3EGFPCre Treg cells revealed a limited set of genes whose transcription was affected by N1c expression (mean expression value, >120; false-discovery rate, <0.1; coefficient variation, <0.5; Fig. 6a). Plotting of the transcriptional profiles as the difference in expression in Foxp3EGFPCreRosa26N1c/N1c Treg cells versus Foxp3EGFPCre Treg cells against the difference in expression Foxp3EGFPCreRosa26N1c/N1c RbpjΔ/Δ Treg cells versus Foxp3EGFPCre Treg cells revealed both RBPJ-dependent alterations in gene expression and RBPJ-independent alterations in gene expression (Fig. 6b). Hierarchal clustering analysis of genes whose expression was significantly altered by N1c expression revealed a subgroup of genes that were either upregulated (Dtx1, Ifng, Gzmb, Pde3b) or down-regulated (Nrp1, Socs2, Il1rl1, Ikzf2, Ikzf4) in an RBPJ-dependent manner (Fig. 6c). A second subgroup included genes that were modulated by N1c in an RBPJ-independent manner, consistent with their regulation by non-canonical pathway(s) (Fig. 6d). This subgroup included genes downstream of Foxo1 signaling, such as Bmp7, Gzma, Cd55 and others 34, 35, 36. These results indicated a profound impact of Notch signaling on the Treg cell transcriptome by both canonical and non-canonical pathways.
Fig. 6. Impact of Notch signaling on the Treg cell transcriptome.
(a) Microarray analysis of gene expression (mean) of Foxp3EGFPCreRosa26N1c/N1c (n = 4) (X axis) versus Foxp3EGFPCre Treg cells (n=4). Numbers in plots indicate total genes downregulated (red) or upregulated (blue) in N1c overexpressing Treg cells relative to their expression in Foxp3EGFPCre Treg cells (cutoff of a two fold change). (b) Comparison of changes in gene expression induced by Notch1 signaling (Foxp3EGFPCreRosa26N1c/N1c vs Foxp3EGFPCre; horizontal axis) and those induced by Notch gain of function in absence of canonical pathway (Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ vs Foxp3EGFPCre; vertical axis) within Treg cells. Blue lines mark a fold change of 2. (c, d) Genes whose expressions are significantly modulated (p<0.05 by one way ANOVA) were segregated in 2 clusters based on the pattern of modulation (canonical and non-canonical) and representative heat maps for each cluster are shown.
We further analyzed the binding of RBPJ at two Foxp3-regulated genes that were affected by N1c expression. These included Pde3b, whose expression is normally suppressed by Foxp3, but was upregulated by N1c expression in Treg cells, and Ikzf2, whose expression is normally upregulated by Foxp3, but was decreased by N1c. Chromatin-immunoprecipitation analysis indicated that Foxp3 binding to the Pde3b and Ikzf2 promoters was decreased, and that of RBPJ was increased, in Foxp3EGFPCreRosa26N1c/N1c as compared to Foxp3EGFPCre Treg cells (Supplementary Fig. 7a, b). Analysis of epigenetic histone-methylation markings at the Pde3b promoter revealed more H3K4me3 and less H3K27me2 in Foxp3EGFPCreRosa26N1c/N1c Treg cells than in Foxp3EGFPCre Treg cells (Supplementary Fig. 7a), a pattern associated with decreased Foxp3-dependent suppression of gene expression37. Reciprocally, H3K27me2 was increased at the Ikzf2 promoter (Supplementary Fig. 7b). These results indicate that canonical Notch signaling directly targeted a subset of Foxp3-regulated genes, antagonized Foxp3 binding and altered Foxp3-induced epigenetic markings at those loci.
The Notch canonical pathway mediates TH1 reprograming of Treg cells
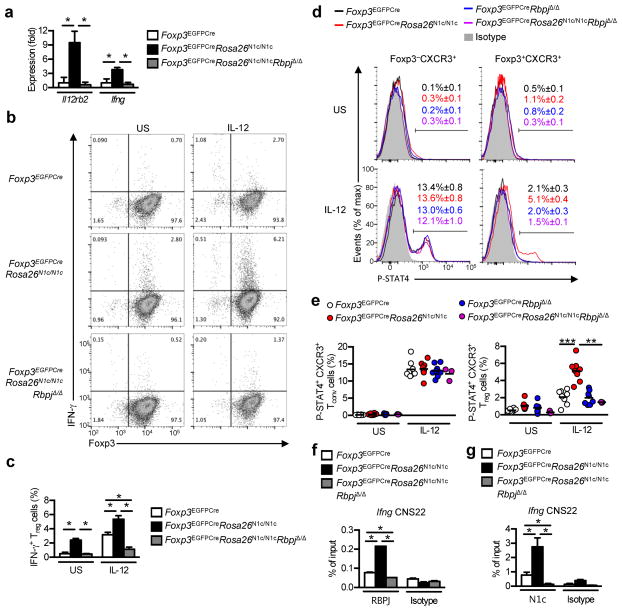
Aborted TH1 programming provides a mechanism by which Treg cells restrain TH1 responses 38. Treg cells respond to IFN-γ treatment by upregulating Tbx21 expression in a STAT1-dependent manner 38. T-bet expression in Treg cells induces a partial Th1 program, including expression of CXCR3, but fails to upregulate expression of IL-12Rβ2, necessary to complete STAT4-dependent TH1 differentiation 39. N1c over-expression in Foxp3EGFPCreRosa26N1c/N1c Treg cells upregulated the transcription of several genes associated with TH1 cells, including Il12rb2 and Ifng as well as a number of genes downstream of IL-12-STAT4 signaling, including Ffar1, Id2 and Nkg7 (Fig. 7a and Supplementary Table 1). Treatment of Foxp3EGFPCreRosa26N1c/N1c Treg cells with IL-12 lead to expression of IFN-γ, which was lost in Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ Treg cells (Fig. 7b, c). Treatment with IL-12 resulted in the phosphorylation of STAT4 in a subset of N1c-expressing Treg cells in an RBPJ-dependent manner (Fig. 7d, e). Foxp3EGFPCreRosa26N1c/N1c Treg cells exhibited increased RBPJ and N1c binding to the Ifng CNS22 element, which mediates transcriptional activation of Ifng by Notch signaling in cooperation with T-bet (Fig. 7f, g) 40. IL-12-induced STAT4 phosphorylation was only observed in YFP+, but not YFP− Treg cells from heterozygous Foxp3YFPcre/+Rosa26N1c/N1c females (Supplementary Fig. 6g, h), indicating that expression of Il12rb2 in Foxp3YFPcre/WTRosa26N1c/N1c Treg cells was due to intrinsic N1c-overexpressing and not to systemic inflammation. These results indicated that N1c promoted TH1 programming of Treg cells in a cell-intrinsic and canonical pathway-dependent manner.
Fig. 7. N1c mediates TH1 reprogramming of Treg cells.
(a) qPCR analysis of Il12rb2 and Ifng transcripts in Treg cells isolated from Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice (n=4–6 per group). Results represent mean fold change + S.E.M. compared to mean of Foxp3EGFPCre Treg cells. Representative flow cytometry dot plots (b) and histograms (c) of IFN-γ production by sorted Treg cells after ex-vivo IL-12 stimulation (n=3 per group). Representative histograms (d) and dot plots (e) of P-STAT4 on gated Foxp3−CXCR3+ and CD4+Foxp3+CXCR3+ cells after CD4 enrichment and IL-12 stimulation (n=3–8 per group). ChIP graphs represent quantitative PCR analysis of the ratio of enriched Ifng CNS22 binding site immuneprecipitated with anti-RBPJ (f) and anti-N1c (g) to the input DNA on Treg cells isolated from Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice (n=3 per group). * p<0.05, ** p<0.01 and *** p<0.001 by unpaired two-tailed Student’s t-test and One Way ANOVA with post test analysis.
Notch regulates Foxp3 epigenetic stability via Rictor
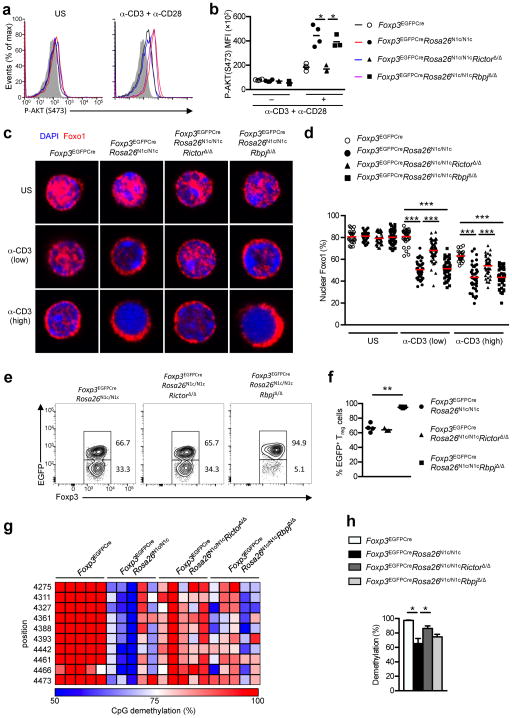
Notch signaling activates the mammalian target of Rapamycin (mTOR) kinase complex 2 (mTORC2) and its downstream kinase AKT independent of RBPJ3, 4, 41. AKT phosphorylates the transcription factor Foxo1, resulting in its retention in the cytosol and its ubiquitinaiton and degradation42, 43. In turn, Foxo1 negatively regulates Th1 differentiation of Treg cells by suppressing Ifng transcription, as well as other type 1 genes 35, 44. Because N1c overexpression increased the transcription of Foxo1- suppressed genes in Foxp3EGFPCreRosa26N1c/N1c Treg cells, we examined mTORC2-Akt activation in these cells. Anti-CD3+CD28 mAb treatment resulted in increased phosphorylation of S473 in AKT, which is a target of mTORC2, in Foxp3EGFPCreRosa26N1c/N1c Treg cells, but not in control Foxp3EGFPCre Treg cells. Anti-CD3 mAb treatment did not induce the phosphorylation of T308 in AKT, which is a target of phospho-inositide 3-kinase (PI3K) (Fig. 8a, b and data not shown). AKT S473 phosphorylation was detected in Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ Treg cells as well, suggesting it was independent of RPBJ (Fig. 8a, b). AKT S473 phosphorylation was not observed in Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ Treg cells, bearing a Foxp3-driven deletion of Rictor, an essential component of the mTORC2 complex following anti- CD3+CD28 mAb stimulation (Supplementary Fig. 1a, b). In contrast, AKT S473 phosphorylation proceeded unaffected in stimulated CD4+ Tconv cells of all genotypes (Fig. 8a, b). There was increased Foxo1 translocation to the cytosol upon CD3 activation in Foxp3EGFPCreRosa26N1c/N1c Treg cells, which was partially reversed by Rictor but not by RBPJ deficiency in these cells (Fig. 8c, d).
Fig. 8. Rictor-dependent non-canonical signaling dysregulates AKT-Foxo1 axis in Foxp3EGFPCreRosa26N1c/N1c Treg cells.
(a) Flow cytometric analysis and (b) scatter plot analysis of MFI of unstimulated and anti-CD3/anti-CD28 stimulated Treg cells from Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (c) N1c augments anti-CD3 mAb-induced translocation of nuclear Foxo1 to the cytosol in a Rictor dependent manner. Unstimulated and anti-CD3 mAb stimulated Treg cells were stained for nuclear DNA (DAPI) and Foxo1 and examined by confocal microscopy for Foxo1 distribution in the nucleus vs. cytosol. (d) Quantitation of percent Foxo1 nuclear expression. Each point represents one cell. (e) Flow cytometric analyses of Foxp3 and EGFP expression in peripheral Treg cells from Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (f) Fractions of EGFP+ cells among Foxp3+ Treg cells from panel (e). (g) Methylation status of individual CpG motifs within the TSDR of CNS2 in Foxp3. Individual CpG motifs are numbered with reference to the transcription initiation site of Foxp3. (h) Global methylation status of the TSDR of CNS2 in Foxp3. * p<0.05, ** p<0.01 and *** p<0.001 by unpaired two-tailed Student’s t-test and One Way ANOVA with post test analysis.
Analysis of Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ mice revealed that the frequencies and absolute numbers of splenic Treg and IFN-γ+CD4+ Tconv cells were decreased and the lymphoproliferative disease was reduced as compared to Foxp3EGFPCreRosa26N1c/N1c mice, (Supplementary Fig. 8a,b). However, in the absence of N1c overexpression, Rictor deficiency did not impact the frequencies or the total numbers of IFN-γ+ CD4+ T cells, memory CD4+ T cells or Treg cells in Foxp3EGFPCreRictorΔ/Δ as compared to Foxp3EGFPCre mice (Supplementary Fig. 8c, d). Importantly, the escape of Foxp3EGFPCre Rosa26N1c/N1c Treg cells away from expressing the Foxp3EGFPCre BAC transgene was ameliorated by concurrent RBPJ but not Rictor deficiency, indicative of the failure of Rictor deficiency to correct the impaired fitness of N1c–expressing Treg cells (Fig. 8e, f). However, expression of key Treg cell markers was improved in Foxp3EGFPCre Rosa26N1c/N1cRictorΔ/Δ as compared to Foxp3EGFPCreRosa26N1c/N1c Treg cells (Supplementary Fig. 8e, f).
Epigenetic demethylation of the Foxp3 CNS2 region has been linked to sustained Foxp3 expression and overall Treg cell stability 45, 46. The demethylation of the Foxp3 CNS2 region was decreased in Foxp3EGFPCreRosa26N1c/N1c as compared to Foxp3EGFPCre Treg cells, which was largely reversed by concurrent Rictor but not RBPJ deficiency (Fig. 8g, h). These findings indicated that N1c destabilized Treg cells in part by altering their Foxp3 CNS2 epigenetic demethylation signature in a Rictor-dependent manner.
Discussion
In this study, we provide evidence that cell-intrinsic Notch signaling regulates the Treg cell compartment in the periphery and controls Treg cell programming towards a TH1 phenotype, as well as TH1 cell responses. Blockade of different steps of the Notch signaling pathway in Treg cells, by means of lineage-specific targeted gene inactivation of Pofut1, Rbpj or Notch1, resulted in increased Treg cell frequency, decreased CD4+ T cell compartment size, and decreased IFN-γ production by CD4 and CD8 cells. Treg cell-specific deletion of Notch signaling components protected against full MHC-mismatched GVHD in association with decreased Treg cell apoptosis, Treg cell programing towards a TH1 phenotype and alloreactive TH1 and TC1 responses. Reciprocally, over-expression of N1c in Treg cells resulted in lymphoproliferation, increased Treg cell apoptosis, TH1 programing of Treg cells, dysregulated TH1 and TC1 responses and autoimmunity. N1c expression down-regulated the expression of several critical components of the Treg cell transcriptome and weakened the Treg cell epigenetic imprint. Thus, homeostatic Notch signaling in Treg cells defines the size and function of the Treg cell compartment and controls the magnitude of TH1 and TC1 cell responses in vivo.
The effects of Notch signaling on Treg cells were predominantly exerted by the canonical pathway. Interruption of the canonical Notch pathway by means of Treg cell-specific Rbpj deletion rescued the key manifestations associated with enforced N1c expression in Treg cells, including lymphoproliferation and autoimmunity. Several Treg cell phenotypic, transcriptional and functional alterations induced by enforced N1c expression were reversed upon RBPJ deletion, indicating their dependence on canonical Notch signaling. RBPJ bound to a subset of Foxp3 target gene, such as Pde3b and Ikzf2, antagonized the binding of Foxp3 and reversed the Foxp3-associated histone methylation profile at those loci, indicative of a direct effect of the canonical pathway on at least a subset of Foxp3-regulated genes. Additional regulation may also be exerted by downstream gene targets of Notch signaling, such as Hes1 and Dtx1, which may engage in secondary transcriptional circuits that contribute to gene expression changes induced by Notch in Treg cells.
In contrast to the above, a number of Treg cell transcripts appeared to respond to Notch signaling in a canonical pathway-independent manner. Several of these have been previously associated with Foxo1 dependent regulation, consistent with their induction by the mTOR-AKT-Foxo1 pathway. N1c-driven impairment of epigenetic demethylation of Foxp3 CNS2 was also independent of the canonical pathway, suggesting a putative role for non-canonical signaling in Notch-mediated Treg cell destabilization. This conclusion was reinforced by the demonstration that N1c-induced increase in CpG methylation of Foxp3 CNS2 was largely reversed in N1c overexpressing but Rictor deficient Treg cells
These studies highlight the important role played by canonical Notch signaling in Treg cells in enabling TH1 and TC1 responses. In the course of controlling a TH1 response, Treg cells normally undergo an aborted program of TH1 differentiation that limits their capacity to express IL-12Rβ2 38. Activation of the canonical Notch signaling bypassed this blockade, leading to STAT4 activation and IFN-γ production in response to IL-12 in Treg cells. RBPJ and N1c associated with CNS22 in the Ifng locus in N1c-overexpressing Treg cells, consistent with previous data showing direct activation of Ifng expression by Notch signaling in synergy with T-bet 40. N1c also enabled more effective activation of mTORC2-AKT by TCR engagement and Foxo1 cytosolic sequestration, a mechanism previously implicated (by means of CD4+ T cell- or Treg cell-specific Foxo1 deletion) in promoting TH1 differentiation of Treg cells 34, 35, 44. The two mechanisms, canonical and non-canonical, may thus act cooperatively to induce TH1 skewing.
Notch signaling in Treg cells played a particularly deleterious role in the context of severe inflammation in the full MHC-mismatched GVHD. Whereas control (Foxp3EGFPCre) Treg cells were susceptible to apoptosis and TH1 programming, Notch1 or RBPJ-deficient Treg cells were resistant to both processes, leading to the accumulation of Treg cells, decreased skewing towards the TH1 phenotype and increased survival. Notch1 inactivation in T cells protects against GVHD22, and our studies indicated that Treg cells were the key cellular effectors of this effect. In the context of overwhelming TH1 immune responses, such as experimental T. gondii infection, Treg cells undergo TH1 programming and apoptosis, recapitulating the fate of Treg cells in GVHD 47. The exaggerated TH1 response in T. gondii-infected mice was associated with decreased IL- 2 production, which contributed to Treg cell apoptosis. In addition, interference with Notch signaling in Treg cells attenuated the TH1 and TC1 response and increased IL-2 production, consistent with a role for IL-2 in enhanced survival of Notch-deficient Treg cells in GVHD.
The inhibitory effects of Notch signaling on Treg cell function offer mechanistic insights into how blockade of Notch receptors may induce tolerance, as has been reported in some experimental mouse models of graft versus host disease. Intervention strategies targeting the Notch pathway may thus offer innovative therapeutic approaches in transplant and autoimmune diseases.
Online Methods
Mice
Foxp3EGFPCre, Foxp3YFPCre, Notch1fl/fl, Rosa26N1c/N1c, Rag1−/−, CD45.1, Foxp3EGFP and BALB/c mice were obtained from the Jackson Laboratory 28, 30, 48. Rictorfl/fl mice were obtained from the Mutant Mouse Regional Resource Center. Pofut1fl/fl and Rbpjfl/fl were kind gifts of Pamela Stanley and Tasuku Honjo, respectively, and were generated as described 26, 27. All Foxp3 mutant mouse strains and their respective crosses were backcrossed 8–10 generations on C57BL/6 or NOD background where specified. Excepted when it was specified, 8 weeks old mice were used in this study. The mice were housed under specific pathogen-free conditions and used according to the guidelines of the institutional Animal Research Committees at the Boston Children’s Hospital.
Real-time PCR
Total RNA was isolated from sorted cells with RNeasy kit (Qiagen). Reverse transcription was performed with the SuperScript III RT-PCR system (Invitrogen) and quantitative real-time reverse transcription (RT)-PCR with Taqman® Fast Universal PCR master mix, internal house keeping gene mouse (GAPD VIC-MGB dye) and specific target gene primers (FAM Dye) (Applied Biosystems) on Step-One- Plus machine. Relative expression was normalized to GAPD for Notch1-4 receptor and calculated as fold change compared to wild-type CD4+GFP− conventional T cells for Pofut1, Notch1, RBPJ and Rictor regarding the regulatory T cell-specific deficiency and fold change normalized to wild-type Treg cells for IFNg and IL12rb2.
Flow cytometry
Annexin V and antibodies against CD4, CD8, CD16/CD32, CD90.2, CTLA4, ICOS, H-2Kd (biolegend), CD3Σ, CD25, CD44, CD45.1, CD45.2, CD45Rb, CD62L, Eos, Foxp3, Helios, IFN-γ, IL-17A, IL-2, Ki67, OX40, CXCR3, H-2Kb (eBioscience), N1c, P-STAT4 (BD biosciences), P-AKT (S473) (Cell signaling) and Nrp1 (R&D system) were used. Cell suspensions were incubated for 10 min with CD16/CD32 then stained for surface markers for 20 min on PBS/1%FCS. Foxp3, Helios, Eos, CTLA4, Ki67 staining was performed by using eBioscience Fixation/Permeabilization kit. For cytokine detection, cell suspensions were pre-incubated with 50 ng/mL PMA, 500 ng/mL ionomycin and 10 μg/mL brefeldin A for 4h in complete medium before CD16/32 blocking followed by surface staining, permeabilization and intracellular Foxp3, IFN-γ, IL-2 and IL-17 staining. For phospho-AKT experiments, spleen cells were stimulated for 30 min with soluble anti-CD3 (1μg/mL) and anti-CD28 (5μg/mL). In some experiments, spleen cells were pre-treated with Rapamycin (250nM, Tocris) for 0, 1 or 24h. After stimulation, cells are fixed with PBS/2%PFA for 20min, permeabilized in 90% methanol for 30 min on ice and stained for CD4, Foxp3 and PS473-AKT. For phospho-STAT4 experiments, splenic CD4+ T cells were isolated and stimulated for 30min with 25ng/mL of recombinant mouse IL-12 (Biolegend). Cells were fixed with PBS/2%PFA for 20min, permeabilized in 90% methanol for 30 min on ice and stained for CXCR3, CD4, Foxp3 and PY693-STAT4. For ex-vivo Treg cell stimulation, isolated Treg cells were cultured for 3 days with IL-2 at 200U/mL ± IL-12 at 25 ng/mL (Biolegend) and PMA/ionomycin/BrefeldinA the last 4 hours before staining for IFN-γ and Foxp3. All flow cytometry acquisitions were performed on a Fortessa cytometer using DIVA software (BD Biosystems) and analyzed using FlowJo (Tree Star).
Graft versus host disease
Eight weeks old Balb/c mice were lethally irradiated (8.5–9 Gy) 4 hours prior reconstitution with 5.106 T cell depleted bone marrow cells alone or in presence of 107 spleen cells from C57BL/6 Foxp3EGFPCre, Foxp3EGFPCreRbpjΔ/Δ or Foxp3EGFPCreNotch1Δ/Δ mice. T cell deplete bone marrow was prepared using CD90.2 microbeads (Miltenyi Biotech). Clinical GVHD score was evaluated every 2 to 3 days by assessment of five clinical parameters as followed: Weight loss (<10%, grade 0; >10% to <20%, grade 1; >20%, grade 2), posture (Normal, grade 0; Hunching noted only at rest, grade 1; Severe hunching and/or impairs movement, grade 2), activity (Normal, grade 0; Mild to moderately decreased, grade 1; Stationary unless stimulated, grade 2), fur texture (Normal, grade 0; Mild to moderate ruffling, grade 1; Severe ruffling/poor grooming, grade 2) and skin integrity (Normal, grade 0; Scaling of paws/tail, grade1; Obvious areas of denuded skin, grade 2). In selected experiments, mice were sacrificed 5 days post transplantation and donor H-2Kd−H-2Kb+ T cells were evaluated for apoptosis, cytokine production and Foxp3 expression.
Adoptive transfer induced Colitis
Naïve (CD4+CD45RBhighGFP−) and Treg (CD4+GFP+) cells are respectively isolated from the spleen of CD45.1 and CD45.2 Foxp3EGFPCre or Foxp3EGFPCreRosa26N1c/N1c mice. Colitis was induced in RAG1-deficient males by i.p. injection of 5.105 CD45.1 naive ± 2.105 Treg cells. Mice were weighed and monitored for signs of disease twice weekly.
Suppression assays
CD4+ T cells were isolated using a CD4 negative isolation kit (Miltenyi), then labeled with CellTrace™ Violet Cell Proliferation dye (Life technologies) according to the manufacturer’s instructions and used as responder cells. Treg (CD4+GFP+) cells were isolated on FACSAria and used as suppressor cells. Responders were used at a fixed concentration of 105 cells per well and stimulated for 3 days with 2 μg/mL of soluble anti-CD3 antibody and 5 μg/mL of soluble anti-CD28 antibody in the presence of 4.105 feeder Rag1−/− spleen cells in 96-well, round-bottom plates in triplicate.
Gene-expression profiling
Spleen Treg (CD3+CD4+GFP+) cells were double-sorted from 6 week old male Foxp3EGFPCre, Foxp3EGFPcreRbpjΔ/Δ, Foxp3EGFPcreRosa26N1c/N1c and Foxp3EGFPcreRosa26N1c/N1cRbpjΔ/Δ mice (n=3–4 per group). Cells were collected directly into TRIzol. RNA was purified and used for probe synthesis for hybridization to Affymetrix Mouse Gene M1.0 ST microarrays. Raw data were background-corrected and normalized with the RMA algorithm in the GenePattern software package 49.
ChIP assays
Chromatin immunoprecipitation on purified Treg cells was performed with Agarose ChIP Kit (Pierce) and anti-RBPJ (Cell signaling), anti-Notch1 (biolegend), anti- Foxp3 (MBL), anti-Histone H3 trimethylated on Lysine 4 (H3K4me3) (abcam), anti-dimethylated on Lysine 27 (H3K27me2) (Millipore) and respective isotype control antibodies. Purified DNA was subjected to real-time PCR with primers flanking RBPJ binding site at Ifng CNS-22 or with primers flanking Foxp3 binding sites on Pde3b and Ikzf2 as previously described 40 37.
Histology
Pancreatic inflammation was scored based on the degree of inflammatory cellular infiltrations present in the islets of Langerhans: 0, no inflammation; 1, mild inflammatory infiltrates; 2, periinsulitis; 3, moderate and diffuse or severe and focal insulitis; 4, severe and diffuse insulitis; 5, severe insulitis and destruction of the tissue. For colitis experiments, colon sections were stained by H&E and scored as followed: 0, no inflammation; 1, mild, scattered infiltrates; 2, moderate infiltrates without loss of epithelium integrity; 3, moderate and diffuse or severe inflammation; 4, Severe inflammation associated with loss of the epithelial barrier integrity.
Autoantibody arrays
Screening for a broad panel of IgG autoantibodies was performed with autoantibody arrays (University of Texas Southwestern Medical Center, Genomic and Microarray Core Facility) as described 50.
Methylation analysis
The methylation status of Foxp3 Treg-specific demethylation region (TSDR or CNS2) in thymic and splenic Treg cells of 8 weeks old male mice was assessed by bisulfite sequence analysis, as described 51. The TSDR of converted DNA was amplified by methylation-specific primer sequences: Foxp3 CNS2 Forward 5′-TATTTTTTTGGGTTTTGGGATATTA-3′ (forward) and Foxp3 CNS2 Reverse 5′-AACCAACCAACTTCCTACACTATCTAT-3′. The PCR product was subcloned and sequenced 51. Blast analyses were done by comparing the resulting sequences with converted Foxp3 gene sequences.
Confocal microscopy
Treg cells were purified and incubated on pre-coated coverslip (poly-L-lysin 50μg/mL, ± anti-CD3 0.1 or 1 μg/mL for low and high dose respectively) at 37°C for 30 min in RPMI/10%FCS. After fixation with PBS/4% paraformaldehyde, cells were permeabilized with PBS/0.1% saponin and blocked on PBS/4% bovine serum albumin (BSA). Cells were incubated with 1:100 diluted rabbit anti-Foxo1 (C29H4, Cell Signaling) followed by 1:500 diluted Alexa fluor 555-anti rabbit secondary antibody (Life technologies) in PBS/1%BSA. Slides were mounted with gold anti-fade reagent with DAPI (Invitrogen). Images were acquired with a Zeiss LSM700 confocal microscopy and ZEN imaging software. Five to 10 fields were selected randomly and total cells in the field were analyzed for percentage of Foxo1 nuclear localization using ImageJ software. Percentage of nuclear Foxo1 localization was obtained by the formula: 100 × corrected nuclear fluorescence/corrected total cell fluorescence and corrected fluorescence was obtained by the formula: Integrated Density – (Area of selected cell or nucleus × Mean fluorescence of background).
In vitro induced Treg cell generation
Naive CD4+CD62LhiCD44lo T cells were isolated from spleen using FACS Aria (purity>98%, data not shown). Cells are stimulated in vitro with coated anti-CD3 (2μg/mL) and anti-CD28 (5μg/mL) for 5 days in the presence of different concentrations of rhTGF-β1 (0, 1, 2 or 5 ng/mL) 52.
Statistical analysis
Data were analyzed by paired and unpaired two-tailed Student’s t-test, one and two way ANOVA with post test analyses and log-rank test, as indicated. Differences in mean values were considered significant at a p < 0.05.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants 2R01AI065617, 2R01AI085090 and 1R56AI115699-01 (to T.A.C.). We thank Christophe Benoist for critical reading of the manuscript, and Pamela Stanley and Tasuku Honjo for respectively providing us with Pofut1fl/fl and Rbpjfl/fl mice.
Footnotes
Author Contributions
L-M.C. and T.A.C. designed the experiments and evaluated the data; L-M.C., S.W., P.G. and E.S. performed experiments; L-M.C. and E.S. analyzed data and prepared the Figures, T.A.C. conceived of the project and directed the research, L-M.C. and T.A.C. wrote the manuscript.
Accession Codes. Gene Expression Omnibus GSE71343.
References
- 1.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 2.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32(1):14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Perumalsamy LR, Marcel N, Kulkarni S, Radtke F, Sarin A. Distinct spatial and molecular features of notch pathway assembly in regulatory T cells. Sci Signal. 2012;5(234):ra53. doi: 10.1126/scisignal.2002859. [DOI] [PubMed] [Google Scholar]
- 4.Lee K, Nam KT, Cho SH, Gudapati P, Hwang Y, Park DS, et al. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med. 2012;209(4):713–728. doi: 10.1084/jem.20111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elyaman W, Bassil R, Bradshaw EM, Orent W, Lahoud Y, Zhu B, et al. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9- producing T cells. Immunity. 2012;36(4):623–634. doi: 10.1016/j.immuni.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Laboratory investigation; a journal of technical methods and pathology. 2008;88(1):11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- 7.Poellinger L, Lendahl U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Current opinion in genetics & development. 2008;18(5):449–454. doi: 10.1016/j.gde.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Amsen D, Antov A, Flavell RA. The different faces of Notch in T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):116–124. doi: 10.1038/nri2488. [DOI] [PubMed] [Google Scholar]
- 9.Cho OH, Shin HM, Miele L, Golde TE, Fauq A, Minter LM, et al. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182(6):3380–3389. doi: 10.4049/jimmunol.0802598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9(10):1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- 11.Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, et al. Delta1- Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19(4):549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 12.Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6(7):680–688. [PubMed] [Google Scholar]
- 13.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27(1):89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, et al. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202(8):1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117(4):515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 17.Keerthivasan S, Suleiman R, Lawlor R, Roderick J, Bates T, Minter L, et al. Notch signaling regulates mouse and human Th17 differentiation. J Immunol. 2011;187(2):692–701. doi: 10.4049/jimmunol.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol. 2009;182(12):7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbarulo A, Grazioli P, Campese AF, Bellavia D, Di Mario G, Pelullo M, et al. Notch3 and canonical NF-kappaB signaling pathways cooperatively regulate Foxp3 transcription. J Immunol. 2011;186(11):6199–6206. doi: 10.4049/jimmunol.1002136. [DOI] [PubMed] [Google Scholar]
- 20.Ou-Yang HF, Zhang HW, Wu CG, Zhang P, Zhang J, Li JC, et al. Notch signaling regulates the FOXP3 promoter through RBP-J- and Hes1-dependent mechanisms. Molecular and cellular biochemistry. 2009;320(1–2):109–114. doi: 10.1007/s11010-008-9912-4. [DOI] [PubMed] [Google Scholar]
- 21.Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, et al. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 2008;112(5):1813–1821. doi: 10.1182/blood-2008-03-144980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, Shan GT, et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J Clin Invest. 2013;123(4):1590–1604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandy AR, Chung J, Toubai T, Shan GT, Tran IT, Friedman A, et al. T cell-specific notch inhibition blocks graft-versus-host disease by inducing a hyporesponsive program in alloreactive CD4+ and CD8+ T cells. J Immunol. 2013;190(11):5818–5828. doi: 10.4049/jimmunol.1203452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roderick JE, Gonzalez-Perez G, Kuksin CA, Dongre A, Roberts ER, Srinivasan J, et al. Therapeutic targeting of NOTCH signaling ameliorates immune-mediated bone marrow failure of aplastic anemia. J Exp Med. 2013;210(7):1311–1329. doi: 10.1084/jem.20112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piggott K, Deng J, Warrington K, Younge B, Kubo JT, Desai M, et al. Blocking the NOTCH pathway inhibits vascular inflammation in large-vessel vasculitis. Circulation. 2011;123(3):309–318. doi: 10.1161/CIRCULATIONAHA.110.936203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003;100(9):5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269(1):81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18(8):1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 34.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30(3):358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, et al. Novel Foxo1- dependent transcriptional programs control T(reg) cell function. Nature. 2012;491(7425):554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tejera MM, Kim EH, Sullivan JA, Plisch EH, Suresh M. FoxO1 controls effector-to- memory transition and maintenance of functional CD8 T cell memory. J Immunol. 2013;191(1):187–199. doi: 10.4049/jimmunol.1300331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37(3):501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailis W, Yashiro-Ohtani Y, Fang TC, Hatton RD, Weaver CT, Artis D, et al. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 2013;39(1):148–159. doi: 10.1016/j.immuni.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perumalsamy LR, Nagala M, Banerjee P, Sarin A. A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell death and differentiation. 2009;16(6):879–889. doi: 10.1038/cdd.2009.20. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Tindall DJ. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim Biophys Acta. 2011;1813(11):1961–1964. doi: 10.1016/j.bbamcr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278(14):12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 44.Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33(6):890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS biology. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31(5):772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10(9):1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107(13):5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li QZ, Zhou J, Wandstrat AE, Carr-Johnson F, Branch V, Karp DR, et al. Protein array autoantibody profiles for insights into systemic lupus erythematosus and incomplete lupus syndromes. Clin Exp Immunol. 2007;147(1):60–70. doi: 10.1111/j.1365-2249.2006.03251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitt EG, Haribhai D, Williams JB, Aggarwal P, Jia S, Charbonnier LM, et al. IL-10 produced by induced regulatory T cells (iTregs) controls colitis and pathogenic ex-iTregs during immunotherapy. J Immunol. 2012;189(12):5638–5648. doi: 10.4049/jimmunol.1200936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, et al. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity. 2015;42(3):512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.