Abstract
The mammary gland is a dynamic organ that undergoes extensive morphogenesis during the different stages of embryonic development, puberty, estrus, pregnancy, lactation and involution. Systemic and local cues underlie this constant tissue remodeling and act by eliciting an intricate pattern of responses in the mammary epithelial and stromal cells. Decades of studies utilizing methods such as transplantation and lineage tracing have identified a complex hierarchy of mammary stem cells, progenitors and differentiated epithelial cells that fuel mammary epithelial development. Importantly, these studies have extended our understanding of the molecular crosstalk between cell types, and signaling pathways maintaining normal homeostasis that often are deregulated during tumorigenesis. While several questions remain, this research has many implications for breast cancer. Fundamental among these are the identification of the cells of origin for the multiple subtypes of breast cancer and the understanding of tumor heterogeneity. A deeper understanding of these critical questions will unveil novel breast cancer drug targets and treatment paradigms. In this review, we provide a current overview of normal mammary development and tumorigenesis from a stem cell perspective.
Introduction
The mammary gland distinguishes itself from other organs since much of its development occurs after birth, allowing for adult developmental studies. Postnatal development of the mammary gland comprises stages of ductal morphogenesis, alveologenesis, lactation and involution, and is regulated by a complex interplay of systemic hormones (notably estrogen, progesterone and prolactin) and local growth factors. The observation that the mammary gland exhibits plasticity through multiple cycles of pregnancy, lactation and involution accompanied by dynamic changes in proliferation, differentiation, cell death and tissue remodeling suggested that there exists a renewable stem or progenitor cell population underlying these processes. It was not until the development of the cleared mammary fat pad technique by DeOme and colleagues in 1959 that it was possible to determine the ability of specific cells to effectively self-renew and differentiate to reconstitute the gland upon transplantation into an epithelium-free fat pad (Deome et al. 1959). This technique was originally employed to investigate whether hyperplastic alveolar nodules were the precursors of mammary tumors. Subsequent studies by Charles Daniel adapted the assay to probe for stem cells and revealed that any portion of the mammary ductal tree could regenerate the entire mammary gland, suggesting that mammary stem cells were distributed throughout the ductal network (Daniel et al. 1975). The transplanted cells responded appropriately to the hormonal environment and were able to functionally differentiate into milk-producing structures. Furthermore, serial transplantation studies using small intact pieces of mammary tissue revealed that the transplanted cells had a finite lifespan and eventually exhibited senescence in contrast to the unlimited division potential of the precancerous lesions (Daniel 1975). Additional advances involved the morphological characterization of putative mammary stem cells by Smith and Medina based on their pale nuclear and cytoplasmic staining properties (Smith & Medina 1988).
It was not until hematopoietic stem cell-based experimental approaches were applied to the mammary gland that significant progress occurred in characterizing specific stem and progenitor cell populations within the mouse and human mammary glands. These studies involved methods for dissociation of mammary tissue followed by fluorescence activated cell sorting (FACS) of cells labeled with specific antibodies against cell surface antigens, allowing for the functional analysis of particular cell populations using both in vitro colony formation and in vivo limiting dilution transplantation assays (Stingl et al. 2001; Welm et al. 2002). Although technical differences existed in these studies, e.g. the sites of transplantation in the cleared mammary fat pad for mouse versus the kidney capsule for human (Eirew et al. 2008), they collectively illustrated the similarity of the mouse and human luminal stem cell hierarchy (Shehata et al. 2012). Mammosphere assays were developed as a surrogate in vitro stem cell assay for the mammary epithelium (Dontu et al. 2003), modeled after in vitro neural stem cell based-assays where stem cells were resistant to anoikis and proliferated under suspension conditions. The assumption upon which many of these studies were based was that cells dissociated from their tissue context would retain cell autonomous properties similar to those observed in the intact tissue. The holy grail of these studies was the eventual demonstration of the ability of a single cell to reconstitute the entire functional mammary gland following transplantation (Shackleton et al. 2006) as had been previously predicted (Kordon & Smith 1998).
Analogous to the elegant genetic studies performed in the Drosophila eye, where cell autonomous and non-cell autonomous interactions can be carefully analyzed in chimeras, the mammary gland provides a unique mammalian modeling platform for phenotypic evaluation of genetic alterations in vivo. Taking advantage of mouse genetics and the availability of wildtype and knockout cells, a series of seminal experiments performed by Cathrin Brisken and colleagues have shown that progesterone receptor (PR)-positive cells can rescue PR-null cells to facilitate alveolar development, and similarly, that estrogen receptor alpha (ERα)-positive cells can rescue ERα-negative cells to facilitate ductal morphogenesis, both mediated by paracrine mechanisms (Brisken et al. 1998; Mallepell et al. 2006). These studies were based upon earlier observations that ERα/PR+ cells did not proliferate in mature ducts (Clarke et al. 1997; Russo et al. 1999; Seagroves et al, 2000). Intriguingly, the analysis of an enriched population of basally-located mammary stem cells (MaSCs) exhibited a lack of ERα and PR expression (Asselin-Labat et al. 2006), yet recent studies have illustrated the importance of steroid hormones for mammary stem cell function (Asselin-Labat et al. 2010; Joshi et al. 2010). Transplantation of a single ERα-negative MaSC should, therefore, a priori not be able to give rise to a mammary outgrowth unless it were able to undergo asymmetric division and ultimately give rise to an ERα-positive luminal cell, which indeed studies have now shown.
The mammary fat pad transplantation assay has proven essential for the assessment of stem cell capacity relating to gene-specific loss- or gain-of-function studies and the assessment of self-renewal and differentiation phenotypes. Importantly, lineage tracing has unveiled distinct differences in transplantation versus in situ developmental potential of MaSCs, where the lineage commitment and developmental competence of basal cells is strongly influenced by luminal cells, highlighting the importance of considering whether single cell types or interacting populations should be utilized in the transplantation assay (Van Keymeulen et al. 2011; De Visser et al. 2012; van Amerongen et al. 2012). Indeed, this is reminiscent of the scenario in the intestine where a single Lgr5+ stem cell is able to give rise to organoids ex vivo, presumably through early asymmetric divisions, but forms organoids more efficiently when cultured together with a niche supporting Paneth cell (Sato et al. 2011). While paracrine communication between luminal and basal epithelial compartments is essential, stem cells are also highly dependent upon their direct interactions with the microenvironment (Inman et al. 2015). Additionally, the transplantation of tissue fragments versus dissociated single cells should also be considered, where unlike the transplantation of intact pieces of mammary tissue, single cells injected into the mammary fat pad require the ability to adhere and survive prior to engraftment, properties that are dependent on cell surface integrins. Such factors have been raised in recent studies that distinguish RANKL and Wnt4 functions on MaSC dynamics in vivo (Rajaram et al. 2015), discussed later in this review.
A major impetus for the advancement of mammary gland stem cell biology was the first description of breast cancer stem cells primarily isolated from patient derived xenografts, using similar approaches that had been applied earlier by Dick and colleagues in acute myeloid leukemia (Bonnet & Dick 1997; Al-Hajj et al. 2003). In addition to a relatively small number of investigators initially focused on understanding the roles of stem cells in mammary gland development, numerous investigators have now entered the field, leading to an explosion of studies aimed at understanding the cell of origin of different breast cancer subtypes as well as the mechanisms responsible for therapeutic resistance and metastasis. Thus, the application of the stem cell paradigm to solid cancers and its potential importance in both etiology and treatment has led to a better appreciation of the potential mechanisms responsible for both inter- and intratumoral heterogeneity in breast cancer (Rosen & Jordan 2009). In the following review, we will discuss several of these concepts in detail by focusing primarily on the extensive work performed in mouse models of mammary gland development and tumorigenesis, while directing the reader to comprehensive reviews on studies concerning stem cells in the human breast (Petersen & Polyak 2010; Visvader & Stingl 2014). Importantly, we will address how studies on stem cells during normal murine mammary development have helped inform our understanding of breast cancer.
Mammary stem cells in development
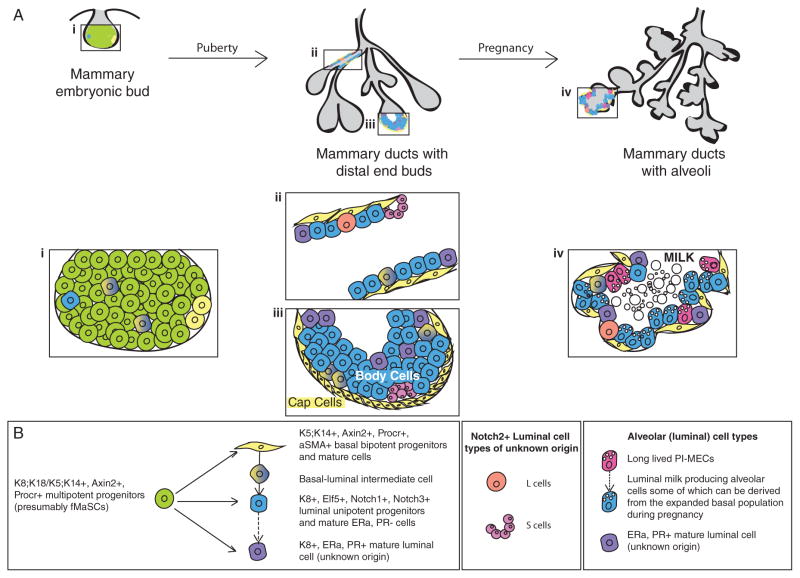
The murine mammary gland anlage is established from the ventral ectoderm as a result of inductive influences from the underlying mesenchyme. This occurs during embryogenesis at E12.5, at which point the rudiment is stably committed to the mammary lineage. The mammary rudiment continues to invade into the fat pad precursor until a primitive branched structure exists at E18.5 (Veltmaat et al. 2003). This is accompanied by a concomitant expansion in the mammary stem cell population termed fetal mammary stem cells (fMaSCs), which constitutes the earliest mammary stem cell population (Spike et al. 2012). fMaSCs can be enriched using the CD24hiCD49fhi cell surface protein marker profile and may presumably be the same pool of bipotent mammary stem cells (K14+/K8+) observed in multiple lineage tracing studies (Fig 1) (Van Keymeulen et al. 2011; Rios et al. 2014). Interestingly, the gene expression signature of fMaSCs differed significantly from the adult CD29hi24+ mammary stem cells (aMaSCs), yet had the capacity to give rise to fully functional, morphologically normal mammary outgrowths upon transplantation. The discordance between the fMaSC and aMaSC gene signature could be partially attributed to the fact that fMaSCs are not yet restricted into basal and luminal lineages. This study also unveiled several reciprocal gene expression patterns in the embryonic bud and the fetal stromal environment further elucidating the paracrine interactions thought to occur early during mammary development. Furthermore, the enrichment of stem cells as measured by stem cell frequency upon limiting dilution transplantation was about 4-fold higher using fMaSC markers as compared to aMaSCs (*Frequency = 1/14 compared to 1/50 in the aMaSC population in this study) (Spike et al. 2012).
Figure 1.
Schematic illustration of the mammary epithelial cell hierarchy in a developmental context. A) This panel indicates 3 critical stages in the ontogeny of the mammary epithelium. (i) The mammary gland begins development as an invaginated bud in the embryo. Upon the onset of puberty, (iii) terminal end buds (TEBs) drive the rapid expansion of (ii) ductal structures through the fat in a proliferation dependent manner. TEBs (iii) comprise an outer layer of cap cells and multilayered body cells that are thought to largely give rise to basal and luminal cells respectively. During pregnancy, (iv) the epithelium proliferates and expands to form grape-like clusters of alveoli that secrete milk during lactation. Magnified snapshots of the (i) embryonic bud, (ii) ductal, (iii) TEB and (iv) alveolar structures indicate the complex and rapidly dynamic cellular compositions during different stages of mammary development. B) The cells observed spatially in their epithelial context in A) are displayed based on their position in the epithelial stem cell hierarchy as gleaned from lineage tracing data. Multipotent progenitors in the embryonic bud differentiate to generate all mature mammary lineages. In the adult, ducts are comprised of basal and luminal lineages. Lineage tracing data have identified cells in the basal compartment with bipotential capacity i.e. ability to give rise to both basal and luminal lineages. More recently, novel Notch2+ luminal populations were described whose origin and function remain ambiguous. Finally, studies in multiparous mice have identified long-lived progenitors termed Parity-Induced Mammary Epithelial Cells (PI-MECs) that are capable of initiating multiple rounds of alveologenesis during repeated pregnancies. Dotted lines indicate hypothesized derivative cell lineages in the epithelial hierarchy.
The rudimentary branched mammary structure at birth remains seemingly dormant until the initiation of puberty at about 3 weeks of age. The spike in estrogen levels that accompanies puberty manifests in a burst of proliferation and resultant bulbous structures termed Terminal End Buds (TEBs) at the distal ends of the mammary ducts. The TEBs are thought to drive the growth and arborization of the ducts throughout the mammary fat pad as a consequence of resident cells undergoing several rounds of cell division (Hinck & Silberstein 2005). The TEB is composed of a single outer layer of cap cells and underlying multilayered body cells that are the precursors to the adult mature K5+/K14+ basal layer and K8+/K18+/K19+ luminal layer, respectively. Interestingly, early studies also demonstrated that the rapidly dividing epithelial end bud structures in rats are the targets of dimethyl benzanthracene (DMBA)-mediated mammary carcinogenesis, emphasizing the importance of a deeper understanding of this developmental stage (Russo & Russo 1980). A small proportion of cap cells have also been observed to migrate into the inner body cell layers and have long been hypothesized to be a potential pool of bipotent stem cells i.e. cells capable of giving rise to both basal and luminal cells (Williams & Daniel 1983). Based on the relatively undifferentiated ultrastructure of the cap cells in the TEB, these cells are a putative population of enriched mammary stem cells. Indeed, a stem cell associated s-SHIP promoter-driven GFP reporter allowed specific isolation of the cap cell population that could efficiently repopulate fully functional mammary glands in a limiting dilution transplantation assay (*Frequency = 1/71 in s-SHIP+ aMaSCs compared to 1/333 in the s-SHIP− aMaSC population in this study. However, in this experiment, the comparative frequency to the total aMaSC population was not determined) (Bai & Rohrschneider 2010). This population of cells was recently demonstrated to possess high canonical Wnt signaling activity, a hallmark of stem cells in multiple organs (Roarty et al. 2015). Intriguingly, a protein in the Par family of polarity regulators called Par3L was identified in cap cells in the TEB (and at luminal tight junctions) and found to be crucial for the maintenance of stem cells (Huo & Macara 2014). Whether these polarity proteins have a secondary role in orienting the different planes of cell division observed in cap cells (Regan et al. 2013), similar to the role of the Par complex in Drosophila neuroblasts remains unknown.
At the end of puberty, the TEBs disappear resulting in the mature adult mammary gland, which comprises the epithelial skeleton upon which alveoli-like buds and alveoli form during the estrus cycle and pregnancy respectively. FACS allowed the identification and prospective isolation of aMaSCs based on their CD24+CD29hi/CD49fhi cell surface marker profile (Shackleton et al. 2006; Stingl et al. 2006). This population of aMaSCs is transiently expanded upon progesterone exposure during the diestrus phase of each estrus cycle and efficiently repopulates the mammary gland in limiting dilution transplantation assays (Joshi et al. 2010). Additionally, the top 5% most brightly stained CD49fhi cells are thought to further enrich for aMaSCs (Stingl et al. 2006). Based on the observation that the aMaSC population harbored label-retaining cells, RNA-seq for cell surface markers on the long term label-retaining population revealed CD1d as a prospective marker of quiescent stem cells (dos Santos et al. 2013). When used in combination with the CD24+29hi basal/aMaSCs, CD1d was able to significantly enrich the stem cell frequency as scored by limiting dilution analysis (*Frequency = 1/8 in CD1d+ aMaSCs compared to 1/44 in the total aMaSC population in this study). Recently, Procr, a previously described CD44+ cancer stem cell (CSC)-associated protein (Shipitsin et al. 2007), has been identified as a Wnt pathway target and a novel aMaSC marker that also repopulates the mammary gland at a high frequency upon transplantation (*Frequency = ~1/12 in Procr+ aMaSCs compared to 1/69 in the total aMaSC population in this study). Interestingly, these Procr+ cells have a low keratin profile and some epithelial to mesenchymal transition (EMT)-like properties. The Procr+ stem cells appear to be cycling but do not overlap with the transient cap cell population suggesting that they may comprise another non-quiescent subset of aMaSCs. Also of note is that Procr+ cells largely do not overlap with Axin2+ cells in the mammary gland, though both markers are canonical Wnt pathway targets, suggesting additional layers of complexity (Wang et al. 2014). Whether these Procr+ cells constitute the progesterone-induced amplified aMaSC population remains unclear. Yet it is becoming increasingly apparent that multiple aMaSC populations might exist to carry out specific functions relating to the homeostatic and proliferative demands of the mammary gland.
Finally, alveologenesis and the priming of alveolar cells for milk production and secretion reflect pregnancy-associated changes in the mammary gland. There is an expansion in the MaSC population at mid-pregnancy which strikingly correlates with s-SHIP re-expression specifically in basal alveolar cells (Bai & Rohrschneider 2010). These stem cells are distinct from the aMaSC population based on their gene expression signature and are thought to be poised for alveolar differentiation. Notably, a long-lived population of alveolar Parity Induced–Mammary Epithelial Cells (PI-MECs) is retained through gland remodeling post-pregnancy and is thought to initiate a new round of alveologenesis during subsequent pregnancies (Boulanger et al. 2005). Lineage tracing using an alveolar-specific Whey Acidic Promoter (WAP)-driven Cre demonstrated that PI-MECs contribute to the luminal layer but only to the hormone receptor-negative cells through multiple pregnancies (Chang et al. 2014). This suggests the existence of separate pools of progenitor cells maintaining the basal and/or hormonal receptor-positive cells during pregnancy. Whether the existence of PI-MECs and the pregnancy expanded MaSC populations are coupled or independent remains an open question.
Thus, the mammary gland is thought to house multiple populations of stem cells that fulfill the requirement of self-renewal and differentiation into mature cell lineages. Do these stem cells exist in invariant stable states or do they exhibit plasticity induced by developmental cues, hormones and the microenvironment? The latter possibility has been suggested recently where a large percentage (~60%) of αSMA+ cells display stem cell capacity upon single cell dissociation and assessment by colony formation and limiting dilution transplantation (Prater et al. 2014). Additionally, while some of the stem cell pools, e.g. fMaSCs, s-SHIP+ cells and Procr+ cells (Bai & Rohrschneider 2010; Spike et al. 2012; Wang et al. 2014) demonstrate active cell cycling dynamics, others such as CD1d+ aMaSCs display label retention and are quiescent in vivo (dos Santos et al. 2013). Such a multiplicity of stem cell populations with different cycling dynamics has been observed in other organ systems (Li & Clevers 2010), where a quiescent stem cell pool acts as a long-term reservoir that is co-opted following stress responses e.g. wounding in the epidermis, whereas the proliferative pool is required to maintain normal tissue homeostasis. Whether the quiescent and rapidly cycling stem cell pools in the mammary gland are independently required or if they serve redundant functions remains shrouded in mystery. There is a pressing requirement for improved ex vivo models that recapitulate the entire gamut of mammary epithelial cell types, e.g. the organoid culture models developed for a number of other epithelial tissues (Sato et al. 2009), to better dissect these mammary population subtypes and their crosstalk. Efficient ex vivo models should be designed to help independently confirm the interpretation of differences in outgrowth efficiency observed in the transplantation assay. In its current form, the transplantation assay results in the same readout (i.e. impaired outgrowths) whether there is defective stem cell survival, decreased stem cell frequency, inability to recapitulate the stem cell niche and/or abnormal differentiation into derivative lineages and should therefore be interpreted with caution.
Pathways regulating mammary stem cells
The ovarian hormones estrogen and progesterone underlie many of the instructive cues guiding development and tissue homeostasis within the mammary gland (Brisken & O’Malley 2010). Whether during bursts of proliferation in puberty and pregnancy, or simply during fluctuations within the estrous cycle, estrogen and progesterone enlist the assistance of multiple signaling pathways to regulate the coordinated efforts of the epithelial hierarchy during stages of active morphogenesis and maintenance. The mere fact that MaSCs are ERα/PR− implies the requirement of paracrine mechanisms for MaSC regulation by hormones (Asselin-Labat et al. 2006). These paracrine mechanisms have long been appreciated since the discovery of ERα/PR+ cells as the sensor cells necessary to relay proliferative cues to neighboring ERα/PR− cells (Mallepell et al. 2006; Beleut et al. 2010), yet it remains unclear how certain pathways regulate an ever-changing epithelial landscape in the mammary gland.
One pathway touted as an indisputable regulator of self-renewal in stem cells is Wnt signaling (Clevers et al. 2014). Several studies throughout the years have identified multiple Wnt ligands in the mammary gland, characterized by spatiotemporal specific patterns of expression (Gavin & McMahon 1992; Kouros-Mehr & Werb 2006). MaSCs clearly possess active Wnt/β-catenin-dependent signaling, as evidenced by Axin2 (Zeng & Nusse 2010) and 7TCF/Lef reporter activity (Roarty et al. 2015). Axin2+ stem cells additionally express higher levels of the Wnt co-receptors Lrp5 and Lrp6 (Zeng & Nusse 2010), while ablation of Lrp5 disrupts mammary stem cell function and basal cell number (Badders et al. 2009). Intriguingly, lineage tracing studies using an Axin2-driven Cre line reveal stage-specific differences in lineage contributions by Axin2+ stem cells (van Amerongen et al. 2012). Although the precise function of individual Wnt proteins in vivo is still evolving, it is known that some Wnt proteins possess the ability to convey self-renewal signals to MaSCs by Wnt/β-catenin-dependent signaling. The sustained activation of Wnt/β-catenin signaling in vivo by MMTV-dependent control of Wnt1 or active-β-catenin (ΔN-β-catenin) expression can expand stem and progenitor fractions, underpinning the influence of Wnt signaling on primitive epithelial cell populations in vivo (Incassati et al. 2010).
Initially recognized as a Wnt ligand downstream of progesterone signaling, Wnt4 has gained considerable attention with respect to MaSC regulation. Joshi et al. identified an increase in MaSC number and activity upon initiation of the luteal diestrus phase of the mouse estrous cycle, coinciding with elevated progesterone levels (Joshi et al. 2010). Moreover, Wnt4 and RANKL expression were induced downstream of progesterone in the luminal compartment, implicating both as mediators of progesterone-regulated MaSC control (Asselin-Labat et al. 2010; Joshi et al. 2010). While RANKL has been shown to be an important paracrine mediator in the mouse and human mammary gland (Gonzalez-Suarez et al. 2010; Tanos et al. 2013), studies conflict as to whether it plays a primary or secondary role. Recently, Rajaram et al. discovered the activation of Wnt/β-catenin signaling in diestrus using an Axin2+/LacZ reporter model, with ablation of Wnt4, not RANKL, severely impairing the regenerative capacity of the mammary epithelium together with Wnt/β-catenin activity, based on the serial transplantation of epithelial fragments (Rajaram et al. 2015). On the other hand, RANK signaling reinforced Wnt-responsiveness of mammary stem and progenitor cells through R-Spondin in a separate study (Joshi et al. 2015). Though slightly inconsistent, the above studies collectively provide evidence for an integrated RANKL/Wnt network downstream of progesterone, instrumental for proliferative and self-renewal cues within the mammary epithelium. Additionally, alternative Wnt/β-catenin-independent ligands, Wnt5a and Wnt5b, can negatively regulate Wnt/β-catenin signaling (Roarty et al. 2015), and Werb and colleagues identified an indirect role for the metalloproteinase MMP3 in the regulation of MaSCs by inactivating Wnt5b function (Kessenbrock et al. 2013), thus extending our understanding of Wnt pathway biology as it relates to microenvironmental regulation of MaSC dynamics. Other factors within the microenvironment, such as the tissue inhibitors of metalloproteinases (TIMPs), can also instruct decisions of MaSC fate (Jackson et al. 2015).
In collaboration with Wnt signaling, additional pathways also play an integral part in the establishment of cellular fate in the mammary gland. While the Wnt/β-catenin-dependent pathway oversees MaSC instructive cues, the activity of Notch and Hippo pathways can tweak the ability of a MaSC to commit to a luminal fate. For instance, Pygo2, a histone methylation reader and Wnt/β-catenin co-activator, can direct the balance of self-renewal Wnt signals with luminal-specific Notch signals in mammary epithelial lineage determination within the MaSC (Gu et al. 2013). Absence of Pygo2 shifts this balance toward a luminal-directed fate by favoring Notch activation. Additionally, absence of the Notch effector Cbf-1 can expand the MaSC pool and regenerative potential of the mammary gland, where low levels of Cbf-1 normally quell MaSC functions (Bouras et al. 2008). Given the link between Notch and Aurora A Kinase (AURKA) in mitotic spindle orientation in the TEB (Regan et al. 2013), Wnt and Notch could have a collaborative function in orientation of the spindle pole, thus influencing MaSC fate determination during puberty. With regard to Hippo signaling, the transcriptional co-activator TAZ interacts with components of the SWI/SNF complex to specify basal-specific gene expression, where loss of TAZ coerces basal cells down a luminal fate (Skibinski et al. 2014). Given that TAZ can mediate a subset of Wnt/β-catenin-dependent transcriptional outputs, MaSC control likely requires a combination of Hippo and Wnt signaling, with Notch signaling initiating a luminal path of fate specification for the MaSC in the mouse mammary gland.
Mammary epithelial cell hierarchy
The mammary epithelium is a bilayered structure that comprises the cytokeratin K5+/K14+ basal or myoepithelial cell layer and the K8+/K18+/K19+ luminal cell layer. The basal compartment is thought to house both the aMaSCs and basal cells in a continuum of differentiation. Therefore, caution should be employed when interpreting stem cell phenotypes, as they maybe secondary to a broader impairment of basal cells. In addition, basal cells have contractile abilities that help the pumping of milk during lactation. The luminal cells in the mammary gland consist of ERα and PR-positive as well as -negative cells. A majority of the luminal progenitor cells are a subset of the ERα/PR− population and are thought to contain the alveolar progenitor subset (Fig 1).
Hormonal cues are the harbingers of developmental changes in the mammary gland and orchestrate a series of paracrine interactions between epithelial and stromal cell types to elicit phenotypic changes in the mammary gland. Despite the presence of ERα (Lemmen et al. 1999) and PR-positive cells in the embryonic mammary glands (Ismail et al. 2002), hormone dependent morphological effects are primarily observed at and after the onset of puberty. ERα−/− mammary gland ducts fail to form TEBs and ramify in the fat pad (Mallepell et al. 2006). PR−/− mammary epithelium shows a less drastic phenotype but displays a reduction in side branching and impaired alveologenesis. Elegant experiments performed using wild type mammary epithelial cells mixed with ERα−/− or PR−/− (Brisken et al. 1998; Mallepell et al. 2006) mammary epithelial cells formed chimeric outgrowths that rescued the phenotypes initially observed in the absence of the wild type cells. Importantly, these studies dissected out a paracrine mechanism in which hormones act on ERα/PR+ cells, which then secrete growth factors such as Amphiregulin (estrogen effector), RANKL and Wnt4 (progesterone effectors) locally, causing their adjacent cells to proliferate (Brisken et al. 2000; Ciarloni et al. 2007; Beleut et al. 2010; Rajaram et al. 2015). Hormones induce downstream factors such as Wnt4 (Asselin-Labat et al. 2010) and R-spondin1 (Cai et al. 2014) that act on MaSCs, thus indirectly regulating stem cell activity. Additionally, progesterone has been implicated in inducing vast changes to the epigenome of the luminal cell compartment that may underlie this paracrine effect (Pal et al. 2013).
Several core regulators of the different mammary lineages have been identified. Transcriptional factor ΔNp63 is a master regulator of the basal/mammary stem cell lineage that acts by potentiating Wnt signaling in this compartment (Chakrabarti et al. 2014). p63+ basal cells can additionally regulate secretion of Neuregulin1 and effect luminal progenitor differentiation and lactogenesis (Forster et al. 2014). p53 has also been ascribed the role of a negative regulator of the mammary stem cell pool and modulates stem cell division dynamics by controlling the proportion of asymmetric cell division to symmetric cell division in vitro using mammosphere assays (Cicalese et al. 2009). A separate study observed an expansion in the basal and luminal mammary stem cell population in TP53 null mice (Chiche et al. 2013). Whether this in vivo stem cell expansion is a direct consequence of increased symmetric cell divisions upon TP53 loss remains to be determined. Gata3 on the other hand is considered a master regulator of the mature mammary luminal lineage. Loss of Gata3 leads to the accumulation of CD61+ luminal progenitors (Asselin-Labat et al. 2007). A similar accumulation of progenitors is also observed upon in tissues displaying a Brca1 loss of function mutation (Lim et al. 2009). Alveolar differentiation is regulated in part by the ETS family transcription factor Elf5 in a prolactin hormone-dependent manner (Oakes et al. 2008). Elf5 also seems to be a key regulator of basal to luminal cell fate decisions (Chakrabarti et al. 2012). At what point between gestation and puberty the K8+/K14+ embryonic bud cells diverge into basal and luminal specific lineages with their distinct transcriptional machinery and acquire their characteristic Notch and Wnt signaling polarity remains an outstanding question.
Lineage tracing studies
In an attempt to study mammary stem cell pools and their resultant progeny in their in vivo unperturbed stem cell niches, mammary gland biology has relied heavily on genetic lineage tracing studies. These elegant studies, more recently combined with high-resolution 3D microscopy techniques, have helped uncover the spatial and temporal dynamics that exist within the mammary epithelial hierarchy. For a detailed current review of lineage tracing methods in the mammary gland, see Sale and Pavelic (Sale & Pavelic 2015). Many lineage tracing studies have utilized Tamoxifen-inducible models, however results should be interpreted with caution in light of the now well understood effects of the ER antagonist on mammary gland homeostasis (Rios et al. 2014; Shehata et al. 2014).
Several lines of evidence suggest that basal mammary stem cells (some K5+ cells, Procr+ cells) possess an inherent capacity for bipotency, i.e. they can act as precursors of both basal and luminal cells during different stages of development (Rios et al. 2014; Wang et al. 2014). This ability is particularly striking in the embryonic bud and during pregnancy (Axin2+ basal cells) whereas mammary tissue homeostasis in the adult primarily relies on unipotent stem cell division (K5+, K14+, αSMA+ basal cells) (Van Keymeulen et al. 2011; van Amerongen et al. 2012; Prater et al. 2014). Certainly, in the case of both Lgr5+ (De Visser et al. 2012) and Axin2+ MaSCs (van Amerongen et al. 2012), a stage-dependent influence on the lineage contribution by these particular MaSC populations exists. Notch2 receptor-driven Cre based lineage tracing helped identify a novel putative cell population distributed periodically throughout the luminal mammary epithelial compartment that are hypothesized to potentially mark the epithelial branch points, but it is unclear if these are actually live cells that are detected since they do not conform to known mammalian cell or nuclear sizes (Šale et al. 2013). Interestingly, luminal progenitor cells marked by an Elf5-driven Cre are unipotent in their ability to generate the luminal population alone (Rios et al. 2014). Lineage tracing using WAP-driven Cre recombinase identified PI-MECs that are sustained through multiple rounds of pregnancy and repopulate alveoli in subsequent pregnancies with ERα/PR− cells (Boulanger et al. 2005). Indeed, a recent study using a Notch1 receptor-driven Cre that marks ERα/PR− luminal progenitors in the adult mammary gland only gives rise to ERα/PR− differentiated luminal and alveolar cells (Rodilla et al. 2015). The source and turnover of ERα/PR+ cells remains hitherto unknown. Thus, lineage tracing has helped answer some very critical questions regarding the mammary epithelial cell hierarchy but has also generated intriguing new questions to be addressed (Fig 1).
Cell(s) of Origin in Breast Cancer
While genetic lineage tracing studies have helped refine our understanding of mammary epithelial hierarchies, the broader question that still exists is which of these cell types can serve as the cell or cells of origin in the different subtypes of breast cancer. For an excellent recent review, see Visvader and Stingl (Visvader & Stingl 2014). Several instances of basal bipotent stem cells giving rise to luminal cells have now been described; however, the reverse process, i.e. K8+ luminal cells giving rise to basal cells has never been observed in vivo in the normal mammary gland. Alterations in the tumor suppressor Brca1, was observed to result in abnormal luminal to basal differentiation (Proia et al. 2011). Elegant studies have also shown that exogenous introduction of Slug alone could confer differentiated luminal cells with a basal cell fate. In addition, introduction of Slug together with Sox9 converted differentiated luminal cells into basal mammary stem cells with long-term reconstitution ability (Guo et al. 2012). Additionally, a recent study employing the use of retroviruses to target Polyoma middle T and Her2 oncogenes specifically to the luminal epithelial compartment demonstrated unequivocal luminal to basal transdifferentiation upon oncogenic insult (Hein et al. 2015). Along the same lines, acquiring properties of EMT reprograms cells to a more stem-like state (Mani et al. 2008). Could such a transcriptional network be epigenetically favored during tumorigenesis? Alternatively, could basal cells increase their usage of bipotent cell division at the cost of unipotent cell divisions that are prevalent during mammary tissue homeostasis to amplify a progenitor pool that contributes to tumorigenesis? Is oncogene-induced transdifferentiation of the luminal cells alone sufficient to explain the plasticity and heterogeneity observed in breast cancers?
The basal-like subtype of breast cancer is often triple-negative i.e. lacking ERα, PR and amplified Her2 and expresses basal cell markers (e.g. K5, K14, P-cadherin and Id4) (Vieira et al. 2012; Prat et al. 2013). This correlates with the profile in basal mammary stem cells, which were therefore intuitively thought to be a potential cell of origin for the basal-like tumor subtype. A model of Brca1 mutant basal tumors wherein Brca1 and p53 were altered in the luminal or basal cells alone using lineage specific Cre recombinases was employed to address this question. These studies showed that Brca1 and p53 conditional alterations in the luminal compartment reproduced the histogenesis of basal-like tumors more accurately than the same mutations in the basal compartment (Molyneux et al. 2010). These data combined with the observation that there is an expansion in the luminal progenitor pool upon Brca1 loss suggests, but does not prove in the absence of direct lineage tracing studies that luminal progenitors are likely to be the cell of origin for Brca1 mediated basal-like tumors (Lim et al. 2009). Another study focused on the cell of origin of the hormone receptor-negative pregnancy-associated breast cancer. The model utilized an RCAS retrovirus to target Her2 specifically in cells expressing the receptor for the retrovirus in WAP+ alveolar cells (Haricharan et al. 2013). The resultant tumors histologically resembled pregnancy associated breast cancers and confirmed that alveolar luminal cells (potentially PI-MECs) could act as the cellular origin for this tumor. This latter model also has significant advantages over genetically engineered mouse models (GEMMs) as targeted mutations to a handful of cells parallels tumorigenesis in humans better as compared to GEMMs where large swathes of cells are affected that may bias results.
Given the multitude of lineage tracing models that have shed light on normal mammary gland development, the elucidation of the cells of origin for breast cancer subtypes and the effect of cell specific alterations in critical regulatory pathways is in short order. However, one caveat is that these studies will all be performed in GEMMs and the conclusions from these studies will still be inferential with respect to the cell of origin for the different human breast cancer subtypes.
Breast cancer stem cells
Breast tumors show a great deal of intertumoral heterogeneity and are classified into several subsets with varied patient outcomes and implications for treatment. One model that has been used to explain this heterogeneity suggests that an epithelial hierarchy akin to that observed in the normal mammary gland exists in tumors. This model places a cell described as a cancer stem cell (CSC) or tumor initiating cell, with self-renewal and differentiation abilities, on top of a hierarchy of cells that forms the tumor bulk. These CSCs have the ability to seed new tumors upon transplantation at limiting dilutions and can also be grown as mammospheres. CSCs are additionally proposed to be slow dividing or quiescent (Pece et al. 2010) which allows them a means to escape chemotherapy and seed tumor recurrence. This idea is cemented by the correlation that basal-like and claudin-low breast cancer subtypes that are often refractory to conventional therapy typically display an undifferentiated, stem cell-enriched signature (Prat & Perou 2010). The simplistic assumption is that targeting this CSC pool along with standard-of-care treatments may thus eliminate the tumor and the source of tumor recurrence.
The first clues suggesting the existence of CSCs came from studies by Al Hajj et al. demonstrating that human breast cancer cells grown as xenografts contain a subset of CD44+CD24− cells with increased tumorigenic capacity when compared to the remaining population (Al-Hajj et al. 2003). Additionally, aldehyde dehydrogenase (ALDH) has been identified as a potential marker for human CSCs (Ginestier et al. 2007). Pertinently, a recent study shows that the CD44+CD24− cells are distinct from the ALDH+ CSCs with the former population representing a quiescent, mesenchymal-like population and the latter representing a cycling, epithelial-like CSC population (Liu et al. 2014). Studies using mouse-derived tumors identified different surface markers e.g. CD29hiCD24+ for CSCs in TP53 null tumor models (Zhang et al. 2008). These CSCs displayed resistance to radiotherapy and resulted in a model for combinatorial therapy using radiotherapy in conjunction with an Akt inhibitor that decreased survival and inhibited the Wnt/β-catenin pathway. These results suggest that targeting resistant CSCs in combination with standard therapies may have value (Zhang et al. 2010). In other models such as in the MMTV-Wnt1 model, the thymocyte antigen Thy1+CD24+ population enriches for CSCs (Cho et al. 2008). However, this seems to contrast with a recent study demonstrating that MMTV-Wnt1 basal-like tumor growth is strictly dependent on a luminal source of Wnt1 (Cleary et al. 2014). One possibility that can explain this apparent discrepancy is that CSCs can divide asymmetrically to give rise to the luminal Wnt1 supporting population, analogous to the transplantation of a single normal MaSC that engenders its own niche. Furthermore, critical regulators of the basal/mammary stem cell populations during normal development, such as ΔNp63, regulate CSCs through Hedgehog (Maria et al. 2015) and Wnt dependent pathways (Chakrabarti et al. 2014). The lack of consistent markers of CSCs in different models has catalyzed an alternative approach utilizing pathway specific reporters to identify a subset of cells with CSC-like properties. This method has successfully been utilized in some tumors using Wnt (Zhang et al. 2010) and Stat3 specific reporters (Wei et al. 2014). Particularly using the Wnt pathway reporter, CSCs were identified that are resistant to radiotherapy through efficient DNA damage repair machinery (Zhang et al. 2010). Other studies have focused on integrin signaling. Inhibition of Focal Adhesion Kinase (FAK), that mediates integrin signaling, results in a decline in CSCs (Luo et al. 2013). Several other pathways such as Notch and TGFβ are actively being investigated. A multitude of targeted therapies are being developed in an attempt to specifically eliminate the CSC population including Wnt (Blagodatski et al. 2014) and integrin antagonists (Goodman & Picard 2012). These therapeutic strategies rely on the presence of an immutable CSC target whereas the emerging evidence suggests that this may be an oversimplification.
The CSC state has been intertwined with the acquisition of EMT properties. This links CSCs with metastasis, thus creating additional challenges for the course of therapy. Human mammary epithelial cells undergoing EMT acquire CSC properties and similar to the normal mammary gland (Mani et al. 2008), exogenous expression of EMT inducers can induce plasticity in the non-CSC pool allowing them to acquire CSC-like properties (Hollier et al. 2013). However, EMT is not absolutely associated with the gain of stem cell properties. A deeper understanding of the dynamics and regulation of these processes will help inform treatment strategies in the future. The CSC hypothesis is now more complex than the simple linear model with a CSC situated on top of the hierarchy. One of the biggest challenges to this model is the issue of plasticity, potentially related to dynamic EMT processes. Several studies have now demonstrated that the non-CSCs can acquire CSC-like properties either as a result of genetic mutations resulting in dedifferentiation (Chaffer et al. 2011) or in a microenvironment dependent manner (Iliopoulos et al. 2011; Malanchi et al. 2012). The evolving model of tumorigenesis now departs from this single hierarchical model and incorporates a multi-clonal heterogenous perspective with continual crosstalk between the different populations. Thus, the revised CSC model is not mutually exclusive of earlier models of clonal evolution, and hypothesizes that there may be more than a single CSC population.
Heterogeneity of breast cancers: lessons from the mammary epithelial hierarchy
The emergence of high-throughput technologies has helped classify breast cancers into at least five molecular subtypes based on the analysis of gene expression patterns among individual tumors, paving a new path in the subclassification of breast cancers and the immense heterogeneity present within them (Perou et al. 2000; Sorlie et al. 2001). These intrinsic subtypes, commonly referred to as Luminal A, Luminal B, HER2-enriched, Basal-like, Claudin-low, and Normal Breast-like, are also recapitulated in several GEMMs, highlighting their utility in providing an additional toolbox for unraveling the complexities surrounding tumor heterogeneity (Herschkowitz et al. 2007; Pfefferle et al. 2013). The convergence of developmental and cancer biology fields occurred upon the identification of similarities between the transcriptional profiles of the intrinsic subtypes and individual epithelial subpopulations of the normal breast (Prat & Perou 2010). Intriguingly, the luminal progenitor fraction of the normal breast epithelium exhibited striking similarities in gene expression to that of the basal-like subtype of breast cancers (Lim et al. 2009), indicating that the luminal progenitor might be the cell-of-origin for BRCA-1 mutant carriers (Molyneux et al. 2010). The aMaSC signature, on the other hand, correlated with the Claudin-low and Normal Breast-like subtypes, while the mature luminal signature was closely associated with Luminal A and B subtypes. The overlap in aMaSC and Claudin-low signatures corroborates other studies that demonstrate mesenchymal features of this particular subtype (i.e. lack of claudins and tight junctions) and the fact that an EMT confers the transition of a cell to an undifferentiated stem-like state (Mani et al. 2008; Taube et al. 2010). While most studies rely on the alignment of the adult human epithelial hierarchy to breast cancer subtype, Spike et al. identified a signature from the isolation of the murine embryonic MaSCs, that showed significant enrichment of the fMaSC signature in basal-like and Her2+ tumors (Spike et al. 2012). A separate study also demonstrated that embryonic mammary signature subsets were enriched in basal-like breast cancers and those of BRCA1-null murine tumors (Zvelebil et al. 2013). Collectively, these data may implicate the potential cell-of-origin for a particular subtype based upon the association with a lineage-restricted gene signature. Alternatively, they may simply reflect the differentiation status of a given tumor, which correlates with a defined epithelial subpopulation within the hierarchy. Future lineage-tracing studies will likely resolve these outstanding questions. Nonetheless, the identification of subtype specificity mirroring the normal breast epithelial cell hierarchy is an intriguing correlation.
Aside from the molecular signatures of epithelial cell types instructing the biology and heterogeneity present within breast cancers, these signatures have potential clinical applications with respect to breast cancer etiology, prognostic capabilities, and therapeutic insights. For instance, Pfefferle et al. identified consensus mouse and human signatures across multiple independent datasets and compiled ‘enriched’ and ‘refined’ signatures of the epithelial subpopulations to determine whether certain characteristics derived from the normal epithelial hierarchy could predict a pathologically complete response (pCR) of breast cancer patients to neoadjuvant chemotherapy (Pfefferle et al. 2015). Indeed, both ‘refined’ LumProg-HsEnriched signatures and one ‘refined’ fMaSC-MmEnriched signature were capable of predicting pCR across all breast cancer patients analyzed. Additionally, the prognostic application of a different aMaSC signature derived from FACS-sorted mouse epithelial cells on the basis of CD24+lowSca1−CD49fhighc-Kit−, was used to predict the likelihood of triple-negative breast cancers (TNBCs) to metastasize (Soady et al. 2015). Unlike Pfefferle et al., this aMaSC signature was enriched in the Normal-like instead of Claudin-low subtype, suggestive of differences present within the aMaSC signatures derived. Although not all lineage-restricted epithelial gene expression patterns possess prognostic capabilities, signatures defining the aMaSC-assocated Claudin-low group were enriched in residual tumors following either endocrine therapy or chemotherapy in the neoadjuvant setting, implying that resistant disease may involve an EMT-like aMaSC/Claudin-low population exhibiting less differentiated characteristics (Creighton et al. 2009). Collectively, such studies emphasize the value of understanding the normal developmental hierarchy to provide a foundation for unraveling subtype specificity and heterogeneity in breast cancers.
The mammary epithelial hierarchy can inform on many aspects of subtype specificity (i.e intertumoral heterogeneity); however, the transcriptomic profiles of breast cancers fail to take into account the intratumoral heterogeneity present within a given tumor. With the advent of massive parallel sequencing, the heterogeneous landscape present within tumors can be interrogated with greater resolution (Banerji et al. 2012; Ellis et al. 2012; Shah et al. 2012; Stephens et al. 2012; TCGA 2012). For instance, from a subtype specific stance, basal-like tumors possess a transcriptional signature similar to the luminal progenitor population of the normal hierarchy (Lim et al. 2009); however, basal-like tumors harbor a greater mutational repertoire and intratumoral heterogeneity relative to non-basal-like tumors within TNBCs (Shah et al. 2012), inferring an inherently more complex tumor architecture among the basal-like subtype. Aside from the mutational repertoire and identification of driver and passenger mutations, an outstanding question lies in how mosaic networks of cells, likely representing some degree of hierarchical organization, cooperate within a given tumor. Thus, investigation of the heterogeneous cell populations present, along with the pathways regulating these, is another approach to identify and target specific cell clones. Recently, Cleary et al., discovered interclonal cooperation between Hras-mutant basal cells and Wnt1-secreting luminal cells of the MMTV-Wnt1 transgenic model, suggesting that even cells with inferior fitness can provide a supportive role in an ever-changing tumor landscape (Cleary et al. 2014). Along the same lines, in a TP53 null transplantable murine model, which accurately reflects the heterogeneity and subtype-specificity of human breast cancers (Herschkowitz et al. 2012), mesenchymal-like niche cells and tumor-initiating cells were found to cooperate within a diverse cellular landscape, where the niche factors Wnt2 and CXCL12 were secreted by the mesenchymal cells and required by the tumor-initiating cells to sustain tumor growth (Zhang et al. 2015). Collectively, these data highlight the importance of not only identifying the cellular constituents within a given tumor, but also the signaling pathways required by specific cell populations to propagate the inherent heterogeneity observed. Unraveling this complexity may provide new insights into the mechanisms driving metastatic behavior of particular subpopulations and the ability of some of these subpopulations to resist therapy.
Discussion
Progress over the last decades in gathering a nuanced understanding of mammary gland biology has greatly honed our research approach to tackle the challenges of breast cancer. Markers of different epithelial subpopulations and stem cells are continually being refined. Notwithstanding, use of these markers in a lineage tracing setting would allow us a window into early tumorigenesis that may prove key to our understanding of the cellular dynamics in tumors. Additionally, the identification of pathway-specific regulators of these subpopulations could help provide an opportunity to gain a better grasp of potential mediators inter- and intratumoral heterogeneity among breast cancers. Importantly, as the evidence emerges for multiple clonal populations in tumors that mutually communicate and depend on each other, similar to the heterotypic interactions in the normal mammary gland, an exciting prospect is that targeting this critical communication would starve the interacting clones and thus impede tumor progression. Moving forward, this crosstalk may partially be deconstructed based on our understanding of the paracrine mechanisms that orchestrate normal homeostasis. Intrinsic to this problem is also determining the fitness of several combinatorial clones and if specific clonal populations are selected for during metastasis or post-treatment. A number of outstanding questions remain to be addressed.
What intrinsic and extrinsic cues specify a quiescent versus a cycling MaSC fate? How do these stem cell pools communicate? Are there similar stem cell pools in tumors?
How are the hormone receptor-positive cells derived and patterned during development?
How does spatial distribution of the different MaSC populations and other cells of the hierarchy contribute to normal development? Are cellular hierarchies organized in a spatial manner within breast cancers?
What is the extent of plasticity among the different epithelial subsets in the mammary gland i.e. can cells transiently acquire MaSC properties, or functionally differentiated characteristics based on developmental stage and the hormonal milieu?
Do the different mutations and/or cells of origin observed in breast cancer subtypes specify fitness in a handful of clones in heterogeneous tumors? Do these clones evolve stochastically or coordinately as an ecosystem?
How does the heterogeneous landscape of a metastasis relate to that observed in the primary tumor? Are these differences primarily due to genetic selection or epigenetic regulation by microenvironmental pressures at the secondary site?
Acknowledgments
The authors would like to thank Dr. Michael Toneff for critical reading of this manuscript. These studies were supported by grant CA16303 from the National Cancer Institute. A.S. is supported by the Baylor College of Medicine Comprehensive Training Program grant RP140102 from CPRIT. We apologize to those investigators whose work we were unable to cite due to space limitations.
Footnotes
Note that it is not possible to directly compare absolute stem cell frequencies reported between different studies and laboratories, often performed in different mouse strains, to assess the relative purity of these enriched populations. Indeed, variations in cell surface marker expression e.g. CD61 have been reported in different mouse strains (Shehata et al. 2012). This will require more stringent single cell analyses and direct comparisons in the same mouse strain.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat M-L, Shackleton M, Stingl J, Vaillant F, Forrest NC, Eaves CJ, Visvader JE, Lindeman GJ. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M-L, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat M-L, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, Lindvall C, Williams BO, Alexander CM. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS ONE. 2009;4(8):e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleut M, Rajaram RD, Caikovski M, Ayyanan A, Germano D, Choi Y, Schneider P, Brisken C. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc Natl Acad Sci U S A. 2010;107:2989–2994. doi: 10.1073/pnas.0915148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagodatski A, Poteryaev D, Katanaev VL. Targeting the Wnt pathways for therapies. Mol and Cell Therap. 2014;2:28. doi: 10.1186/2052-8426-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Boulanger CA, Wagner K-U, Smith GH. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 2005;24:552–560. doi: 10.1038/sj.onc.1208185. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Brisken C, O’Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2:a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C, Park S, Vass T, Lydon JP, O’Malley BW, Weinberg RA. A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc Natl Acad Sci U S A. 1998;95:5076–5081. doi: 10.1073/pnas.95.9.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, Mcmahon JA, Mcmahon AP, Weinberg RA. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14:650–654. [PMC free article] [PubMed] [Google Scholar]
- Cai C, Yu QC, Jiang W, Liu W, Song W, Yu H, Zhang L, Yang Y, Zeng YA. R-spondin1 is a novel hormone mediator for mammary stem cell self-renewal. Genes & Dev. 2014;28:2205–2218. doi: 10.1101/gad.245142.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins Pa, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. 2011 Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012;30:1496–1508. doi: 10.1002/stem.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A, Tiede B, Romano R-A, DeCoste C, Mercatali L, et al. ΔNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nat Cell Biol. 2014;16:1004–1015. doi: 10.1038/ncb3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH-T, Kunasegaran K, Tarulli Ga, De Silva D, Voorhoeve PM, Pietersen AM. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res. 2014;16:R1. doi: 10.1186/bcr3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche A, Moumen M, Petit V, Jonkers J, Medina D, Deugnier M-A, Faraldo MM, Glukhova MA. Somatic loss of p53 leads to stem/progenitor cell amplification in both mammary epithelial compartments, basal and luminal. Stem Cells. 2013;31:1857–1867. doi: 10.1002/stem.1429. [DOI] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci U S A. 2007;104:5455–5460. doi: 10.1073/pnas.0611647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel CW. Regulation of cell division in aging mouse mammary epithelium. Adv Exp Med Biol. 1975;61:1–19. doi: 10.1007/978-1-4615-9032-3_1. [DOI] [PubMed] [Google Scholar]
- Daniel CW, Aidells BD, Medina D, Faulkin LJ., Jr Unlimited division potential of precancerous mouse mammary cells after spontaneous or carcinogen-induced transformation. Fed Proc. 1975;34:64–67. [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- De Visser KE, Ciampricotti M, Michalak EM, Tan DWM, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J. Developmental stage-specific contribution of LGR5+ cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos CO, Rebbeck C, Rozhkova E, Valentine A, Samuels A, Kadiri LR, Osten P, Harris EY, Uren PJ, Smith AD, et al. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc Natl Acad Sci U S A. 2013;110:1–8. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, Eaves CJ. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster N, Saladi S, vanBragt M, Sfondouris M, Jones F, Li Z, Ellisen L. Basal Cell Signaling by p63 Controls Luminal Progenitor Function and Lactation via NRG1. Dev Cell. 2014;28:147–160. doi: 10.1016/j.devcel.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin BJ, McMahon AP. Differential regulation of the Wnt gene family during pregnancy and lactation suggests a role in postnatal development of the mammary gland. Mol Cell Biol. 1992;12:2418–2423. doi: 10.1128/mcb.12.5.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, Pinkas J, Branstetter D, Dougall WC. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010;468:103–107. doi: 10.1038/nature09495. [DOI] [PubMed] [Google Scholar]
- Goodman SL, Picard M. Integrins as therapeutic targets. Trends in Pharm Sci. 2012;33:405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Gu B, Watanabe K, Sun P, Fallahi M, Dai X. Chromatin effector Pygo2 mediates Wnt-notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell. 2013;13:48–61. doi: 10.1016/j.stem.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haricharan S, Hein SM, Dong J, Toneff MJ, Aina OH, Rao PH, Cardiff RD, Li Y. Contribution of an alveolar cell of origin to the high-grade malignant phenotype of pregnancy-associated breast cancer. Oncogene. 2013;33:5729–5739. doi: 10.1038/onc.2013.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein SM, Haricharan S, Johnston aN, Toneff MJ, Reddy JP, Dong J, Bu W, Li Y. Luminal epithelial cells within the mammary gland can produce basal cells upon oncogenic stress. Oncogene. 2015:1–7. doi: 10.1038/onc.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, Knezevic J, Greene SB, Darr D, Troester MA, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2012;109:2778–2783. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L, Silberstein GB. Key stages in mammary gland development: the mammary end bud as a motile organ. Breast Cancer Res. 2005;7:245–251. doi: 10.1186/bcr1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollier BG, Tinnirello AA, Werden SJ, Evans KW, Taube JH, Sarkar TR, Sphyris N, Shariati M, Kumar SV, Battula VL, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73:1981–1992. doi: 10.1158/0008-5472.CAN-12-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Macara IG. The Par3-like polarity protein Par3L is essential for mammary stem cell maintenance. Nat Cell Biol. 2014;16:529–537. doi: 10.1038/ncb2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch Ha, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108:1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: beta-catenin. Breast Cancer Res. 2010;12:213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- Ismail PM, Li J, DeMayo FJ, O’Malley BW, Lydon JP. A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol Endocrinol. 2002;16:2475–2489. doi: 10.1210/me.2002-0169. [DOI] [PubMed] [Google Scholar]
- Jackson HW, Waterhouse P, Sinha A, Kislinger T, Berman HK, Khokha R. Expansion of stem cells counteracts age-related mammary regression in compound Timp1/Timp3 null mice. Nat Cell Biol. 2015;17:217–227. doi: 10.1038/ncb3118. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, Stingl J, Waterhouse PD, Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Waterhouse PD, Kannan N, Narala S, Fang H, Di Grappa MA, Jackson HW, Penninger JM, Eaves C, Khokha R. RANK Signaling Amplifies WNT-Responsive Mammary Progenitors through R-SPONDIN1. Stem Cell Reports. 2015;5 doi: 10.1016/j.stemcr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Dijkgraaf GJ, Lawson DA, Littlepage LE, Shahi P, Pieper U, Werb Z. A role for matrix metalloproteinases in regulating mammary stem cell function via the Wnt signaling pathway. Cell Stem Cell. 2013;13:300–313. doi: 10.1016/j.stem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dyn. 2006;235:3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmen JG, Broekhof JL, Kuiper GG, Gustafsson JA, van der Saag PT, van der Burg B. Expression of estrogen receptor alpha and beta during mouse embryogenesis. Mech Dev. 1999;81:163–167. doi: 10.1016/s0925-4773(98)00223-8. [DOI] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Zhao X, Chen S, Liu S, Wicha MS, Guan JL. Distinct FAK activities determine progenitor and mammary stem cell characteristics. Cancer Res. 2013;73:5591–5602. doi: 10.1158/0008-5472.CAN-13-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr H-A, Delaloye J-F, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- Mallepell S, Krust A, Chambon P, Brisken C. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc Natl Acad Sci U S A. 2006;103:2196–2201. doi: 10.1073/pnas.0510974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria E, Giulia A, Giacobbe A, Peschiaroli A, Frezza V. p63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2015;112:3499–3504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes & Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, et al. Global Changes in the Mammary Epigenome Are Induced by Hormonal Cues and Coordinated by Ezh2. Cell Rep. 2013;3:411–426. doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Polyak K. Stem Cells in the Human Breast. Cold Spring Harb Perspect Biol. 2010;2:15. doi: 10.1101/cshperspect.a003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle AD, Spike BT, Wahl GM, Perou CM. Luminal progenitor and fetal mammary stem cell expression features predict breast tumor response to neoadjuvant chemotherapy. Breast Cancer Res Treat. 2015;149:425–437. doi: 10.1007/s10549-014-3262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2010;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Adamo B, Cheang MCU, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. The Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prater MD, Petit V, Alasdair Russell I, Giraddi RR, Shehata M, Menon S, Schulte R, Kalajzic I, Rath N, Olson MF, et al. Mammary stem cells have myoepithelial cell properties. Nat Cell Biol. 2014;16:942–50. doi: 10.1038/ncb3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram RD, Buric D, Caikovski M, Ayyanan A, Rougemont J, Shan J, Vainio SJ, Yalcin-Ozuysal O, Brisken C. Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. Embo J. 2015;34:641–652. doi: 10.15252/embj.201490434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JL, Sourisseau T, Soady K, Kendrick H, McCarthy A, Tang C, Brennan K, Linardopoulos S, White DE, Smalley MJ. Aurora a kinase regulates mammary epithelial cell fate by determining mitotic spindle orientation in a notch-dependent manner. Cell Rep. 2013;4:110–123. doi: 10.1016/j.celrep.2013.05.044. [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Roarty K, Shore AN, Creighton CJ, Rosen JM. Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J Cell Biol. 2015;208:351–366. doi: 10.1083/jcb.201408058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V, Dasti A, Huyghe M, Lafkas D. Luminal Progenitors Restrict Their Lineage Potential during Mammary Gland Development. PLoS Biol. 2015;13:e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Russo IH. Influence of Differentiation and Cell Kinetics on the Susceptibility of the Rat Mammary Gland to Carcinogenesis Influence of Differentiation and Cell Kinetics on the Susceptibility of the Rat Mammary Gland to Carcinogenesis. Cancer Res. 1980;40:2677–2687. [PubMed] [Google Scholar]
- Russo J, Ao X, Grill C, Russo IH. Pattern of distribution of cells positive for estrogen receptor a and progesterone receptor in relation to proliferating cells in the mammary gland. Breast Cancer Res Treat. 1999;53:217–227. doi: 10.1023/a:1006186719322. [DOI] [PubMed] [Google Scholar]
- Sale S, Pavelic K. Mammary lineage tracing: the coming of age. Cell and Mol Life Sci. 2015;72:1577–1583. doi: 10.1007/s00018-014-1817-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nature Cell Biol. 2013;15:1–11. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagroves TN, Lydon JP, Hovey RC, Vonderhaar BK, Rosen JM. C/EBPbeta (CCAAT/enhancer binding protein) controls cell fate determination during mammary gland development. Mol Endocrinol. 2000;14:359–368. doi: 10.1210/mend.14.3.0434. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, van Amerongen R, Zeeman AL, Giraddi RR, Stingl J. The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res. 2014;16:411. doi: 10.1186/s13058-014-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular Definition of Breast Tumor Heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Skibinski A, Breindel JL, Prat A, Galvan P, Smith E, Rolfs A, Gupta PB, Labaer J, Kuperwasser C. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GH, Medina D. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J Cell Sci. 1988;90 (1):173–183. doi: 10.1242/jcs.90.1.173. [DOI] [PubMed] [Google Scholar]
- Soady KJ, Kendrick H, Gao Q, Tutt A, Zvelebil M, Ordonez LD, Quist J, Tan DW, Isacke CM, Grigoriadis A, et al. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. 2015;17:31. doi: 10.1186/s13058-015-0539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10:183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67:93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Tanos T, Sflomos G, Echeverria PC, Ayyanan A, Gutierrez M, Delaloye JF, Raffoul W, Fiche M, Dougall W, Schneider P, Yalcin-Ozuysal O, Brisken C. Progesterone/RANKL is a major regulatory axis in the human breast. Sci Transl Med. 2013;5:182ra55. doi: 10.1126/scitranslmed.3005654. [DOI] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation. 2003;71:1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- Vieira AF, Ricardo S, Ablett MP, Dionísio MR, Mendes N, Albergaria A, Farnie G, Gerhard R, Cameselle-Teijeiro JF, Seruca R, et al. P-cadherin is coexpressed with CD44 and CD49f and mediates stem cell properties in basal-like breast cancer. Stem Cells. 2012;30:854–864. doi: 10.1002/stem.1075. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes & Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang X-O, Yang L, Zeng YA. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2014;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- Wei W, Tweardy DJ, Zhang M, Zhang X, Landua J, Petrovic I, Bu W, Roarty K, Hilsenbeck SG, Rosen JM, et al. STAT3 signaling is activated preferentially in tumor-initiating cells in claudin-low models of human breast cancer. Stem Cells. 2014;32:2571–2582. doi: 10.1002/stem.1752. [DOI] [PubMed] [Google Scholar]
- Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Williams JM, Daniel CW. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci U S A. 2010;107:3522–3527. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Tsimelzon A, Chang CH, Fan C, Wolff A, Perou CM, Hilsenbeck SG, Rosen JM. Intratumoral heterogeneity in a Trp53-null mouse model of human breast cancer. Cancer Discov. 2015;5:520–533. doi: 10.1158/2159-8290.CD-14-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvelebil M, Oliemuller E, Gao Q, Wansbury O, Mackay A, Kendrick H, Smalley MJ, Reis-Filho JS, Howard BA. Embryonic mammary signature subsets are activated in Brca1−/− and basal-like breast cancers. Breast Cancer Res. 2013;15:R25. doi: 10.1186/bcr3403. [DOI] [PMC free article] [PubMed] [Google Scholar]