Abstract
Noncoding RNAs regulate gene expression at both the transcriptional and post-transcriptional levels, and play critical roles in development, imprinting and the maintenance of genome integrity in eukaryotic organisms [1–3]. Therefore, it is important to understand how the production of such RNAs are controlled. In addition to the three canonical DNA dependent RNA polymerases (Pol) Pol I, II and III, two non-redundant plant-specific RNA polymerases, Pol IV and Pol V, have been identified and shown to generate noncoding RNAs that are required for transcriptional gene silencing via the RNA-directed DNA methylation (RdDM) pathway. Thus, somewhat paradoxically, transcription is required for gene silencing. This paradox extends beyond plants, as silencing pathways in yeast, fungi, flies, worms, and mammals also require transcriptional machinery [4,5]. As plants have evolved specialized RNA polymerases to carry out gene silencing in a manner that is separate from the essential roles of Pol II, their characterization offers unique insight into how RNA polymerases facilitate gene silencing. In this review, we focus on the mechanisms of Pol IV and Pol V function, including their compositions, their transcripts, and their modes of recruitment to chromatin.
Introduction
In Arabidopsis, DNA methylation is established and maintained via several pathways that together shape the DNA methylation landscape and repress the expression of transposable elements and some neighboring genes [6–9]. Here, we focus on an RNA polymerase-centric view of the RNA-directed DNA methylation (RdDM) pathway (Figure 1), and refer those interested in a more comprehensive picture to several recent reviews [6,8]. At its core, the RdDM pathway requires two types of noncoding RNAs, Pol IV-dependent 24-nucleotide (nt) small interfering RNAs (siRNA) [10–15] and Pol V-dependent intergenic noncoding (IGN) RNAs [16], that mediate cytosine methylation in all sequence contexts (CG, CHG and CHH, where H=A, C or T) by the de novo methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) [17] (Figure 1).
Figure 1. An RNA polymerase-centric view of the RNA-directed DNA methylation (RdDM) pathway highlighting several self-reinforcing loops.
Biogenesis of the methylation-targeting siRNAs is initiated by the activity of RNA polymerase IV (Pol IV). Pol IV transcripts are processed by the activities of RDR2, DCL3, and HEN1 into 24-nt siRNAs, which are loaded into argonaute (AGO) effector proteins. Independent of Pol IV activity, another RNA polymerase, Pol V, generates long intergenic noncoding (IGN) transcripts. Both Pol V itself, as well as the transcripts it produces, facilitated the recruitment of siRNA-loaded AGO proteins (and additional components not shown) to chromatin, ultimately leading to recruitment of the de novo DNA methyltransferase DRM2. Investigation into the mechanisms for Pol IV and Pol V recruitment to RdDM targets have identified several necessary proteins (and protein complexes) and uncovered interconnected and self-reinforcing loops between DNA and histone methylation as detailed below. The association of Pol IV at many genomic loci depends on SHH1 and its H3K9me binding activity, and H3K9 methylation requires a family of SET domain histone methyltransferases (SUVH4, SUVH5, and SUVH6) that also function as methyl-DNA binding proteins. Together these factors generate a self-reinforcing loop of DNA and histone methylation (peach dashed oval) wherein DNA methylation mediates the association of SUVH4/5/6 with chromatin, leading to the deposition H3K9 methylation that is bound by SHH1, facilitating Pol IV recruitment and siRNA production and ultimately leading to the establish DNA methylation to complete the loop. The association of Pol V at chromatin depends on members of the DDR complex and two associated methyl-DNA binding proteins, SUVH9 and SUVH2, generating a self-reinforcing loop in which DNA methylation deposited via the RdDM pathway is required for Pol V localization and the establishment of additional DNA methylation (grey dashed oval). Currently defined features of Pol IV and Pol V dependent transcripts are indicated in cartoon format above the dashed arrows in purple and in red, respectively. (
 ) Histone Methylation, (
) Histone Methylation, (
 ) DNA methylation, (
) DNA methylation, (
 ) RNA methylation.
) RNA methylation.
In RdDM, Pol IV is proposed to generate long single-strand RNAs (ssRNAs) that are rapidly converted into double-strand RNAs (dsRNAs) by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) [18]. This process also involves CLASSY 1 (CLSY1) [19], but the precise role of this putative chromatin remodeling factor remains unknown. These dsRNAs are then processed into 24-nt siRNAs by DICER-LIKE 3 (DCL3) [18], stabilized by methylation at their 3′end by HUA ENHANCER 1 (HEN1) [20] and loaded into ARGONAUTE (AGO) effector proteins [21], specifically, AGO4 [22,23], AGO6 [23–26] or AGO9 [23,27,28]. Unexpectedly, at least for AGO4, this loading occurs in the cytoplasm and the siRNA-bound AGO4 proteins are then re-imported into the nucleus [29] to target cytosine methylation at cognate DNA sequences within the genome. This downstream methylation-targeting portion of the pathway requires a second RNA polymerase, Pol V [12,30], which generates IGN transcripts [16] and is required for the recruitment of RdDM components including AGO4 and DRM2 to chromatin [31–35]. This Pol V-dependent recruitment is likely via a combination of both protein-protein interactions, mediated by the GW rich “AGO hook” [36] interaction domain within the Carboxy-Terminal Domain (CTD) of Pol V [31,32], as well as nucleic acid interactions involving the AGO-bound 24-nt siRNAs [34,35,37–39]. However, the relative contributions of these protein-protein and nucleic acid interactions remain unclear, as does the nature of the nucleic acid interactions (siRNA:IGN transcript vs siRNA:DNA) [8,16].
Given the central roles of Pol IV and Pol V in the RdDM pathway, recent efforts have focused on gaining a better understanding of their functions. Here we present current insights into the mechanisms governing Pol IV and Pol V activity, focusing on (1) the composition and evolution of these plant specific polymerases, (2) their enzymatic activity and the nature of their transcripts, and (3) the mechanisms through which these polymerases associate with chromatin.
Composition and evolution of Pol IV and Pol V
RNA polymerases are large holoenzymes that contain both catalytic and regulatory/non-catalytic subunits [40,41]. The roles of these subunits are best characterized for Pol II, with the first and second largest subunits forming the catalytic center, and many of the other subunits governing important aspects of polymerase function that remain poorly defined or completely unknown in RNA Pol IV and Pol V, including template selection, initiation, elongation, transcriptional fidelity, termination, and RNA processing [40,41] (Figure 2A). Thus, defining the composition and diversity of RNA Pol IV and Pol V subunits is a critical step in understanding how they function.
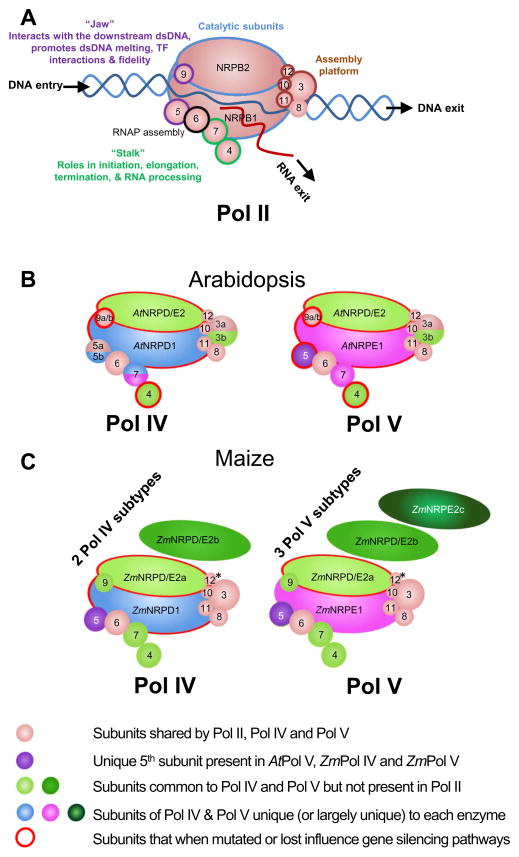
Figure 2. Subunit composition summary for Pol IV and Pol V in Arabidopsis and maize.
(A) Cartoon representation of RNA polymerase II, showing the 12 core subunits (1–12) along with the DNA entry, DNA exit and RNA exit channels. Polymerase features and associated functions in yeast Pol II are labeled much as described in [50]. (B and C) Cartoon representations of RNA Pol IV and Pol V in Arabidopsis (At) and maize (Zm), respectively, modeled after diagrams in [51]. Subunit compositions are based on the totality of mass spectrometry data from [42–46] and likely represent multiple polymerase subtypes with different combinations of subunits. The individual polymerase subunits are color coded as indicated in the legend. *Subunit 12: Although subunit 12 is conserved in Pol I, II, and III as well as in AtPol IV and AtPol V, no peptides were identified from the maize affinity purifications thus it remains unclear if this subunit is truly absent or was not detectable for technical reasons [46].
Consistent with the different functions of Pol II, Pol IV and Pol V, and with the different types of RNA transcripts they produce (detailed in the next section), affinity purifications of Pol IV and Pol V from Arabidopsis thaliana [42–44], cauliflower [45], and maize [46] have revealed significant differences in their composition (Figure 2). These polymerases, designated Nuclear RNA Polymerase (NRP) B, D, or E in reference to Pols II, IV, and V, respectively, are comprised of 12 core subunits and approximately half of these subunits are shared by all three polymerases [42,46] (Figure 2). Current data suggests that the unique subunits of Pol IV and Pol V arose from their Pol II counterparts via many independent duplication events, starting prior to the evolution of land plants, followed by “Escape from Adaptive Conflict” sub-functionalization [47–50]. Indeed, contrary to previous reports [47,48], all land plants have specialized Pol IV and Pol V complexes [49,50] with unique catalytic cores and 7th subunits [48–50]. On the other hand, additional 4th and 5th subunits appear to be unique to flowering [48–50] and seed plants [49], respectively. Within flowering plants, more recent events have generated even greater diversity between Pols II, IV and V [48–50], giving rise to the unique 7th subunits in Arabidopsis Pol IV and Pol V as well as the additional 2nd and 9th subunits shared in the maize Pol IV and Pol V subtypes [42,46] (Figure 2). While further characterization of these divergent polymerase subunits is required to begin assigning specific functions, they are likely playing key roles in shaping the functions of Pol IV and Pol V. Ongoing efforts to understand the diversity, evolution, and function of these RNA polymerases are described below.
Arabidopsis Pol IV and V catalytic and non-catalytic subunits
In Arabidopsis, the catalytic centers of Pol IV and Pol V are unique compared to each other and to Pol II [42]. Pol IV and Pol V share a common second largest subunit (NRPD/E2), but they have distinct largest subunits, NRPD1 and NRPE1, respectively [42]. Consistent with the identification of Pol IV [15] and Pol V [16] dependent transcripts, these polymerases (as well as maize Pol IV and Pol V) have maintained key residues associated with catalysis [47,51], and these residues are required for RdDM [52,53]. However, their sequences have diverged at many highly conserved locations invariant in Pols-I, -II, and -III [47,51], suggesting that Pol IV and Pol V are not experiencing the same evolutionary constraints as their essential counterparts [54].
In addition to forming half of the catalytic center, the largest subunit of eukaryotic Pol II polymerases (NRPB1) possesses a highly conserved CTD with a repeating heptad motif that regulates Pol II function [55]. The largest subunits of Pol IV and Pol V completely lack these repeats [54] and, despite sharing a common ancestor [47], they have limited homology with each other outside a single conserved domain of unknown function, termed Defective Chloroplasts and Leaves (DeCLs) [49,54], that was likely acquired via an ancient gene fusion [50]. Perhaps the key functional difference between the CTDs of Pol IV and Pol V is due to the presence of a Pol V-specific GW-rich AGO-hook motif that interacts with AGO4 [31,32] and aids in the targeting of DNA methylation (Figure 1). Interestingly, although the presence of an AGO-hook in the NRPE1 CTD is conserved across land plants [49], the sequences of the GW-repeats and surrounding regions are rapidly evolving and very little sequence conservation is observed among a broad survey of plant species [32,46,49]. While retention of an AGO-hook motif is likely driven by a requirement for AGO interactions, factors driving the rapid evolution of the CTD as a whole remain unclear. One intriguing hypothesis suggests it might be driven by host-pathogen interactions, as several viral suppressor proteins with AGO-hook motifs have been identified [36,56,57]. However, there is currently no direct evidence that this phenomenon extends to AGOs that influence the RdDM pathway.
Non-catalytic and regulatory subunits
Of the 10 non-catalytic polymerase subunits, four subunits of Pol IV and Pol V are distinct from Pol II and half of these differ between Pol IV and Pol V (Figure 2B), making these subunits prime candidates for imparting Pol IV and Pol V specific functions. Notably, the subunits in question (the 3rd, 4th, 5th, and 7th) along with the unique catalytic subunits (1st and 2nd) of Pol IV and Pol V are predicted to cluster mainly on the “leading face” of the polymerase holoenzyme, surrounding functionally important areas including the catalytic center as well as the DNA entry and RNA exit channels [46]. Specifically, the 4th and 7th subunits form a sub-complex termed the “Stalk” [40] that is positioned near the RNA exit channel and is unique in Pol II, Pol IV, and Pol V (Figure 2). The Stalk is implicated in multiple steps of the Pol II transcription cycle [40] (Figure 2), and mutations in the 7th subunit of yeast Pol II specifically affect its role in small RNA biogenesis, demonstrating a precedence for the involvement of this sub-complex in gene silencing [58]. In Pol II, the 5th and 9th subunits interact with the downstream DNA duplex and basal transcription factors, respectively, and form the “Jaw”, which is implicated in several features poorly understood with respect for Pol IV and Pol V function—template selection, elongation and polymerase fidelity [40,59,60].
Notably, viable mutants in almost all of the aforementioned Pol IV and Pol V subunits have been identified. Initial characterization of these mutants reveal roles for NRPD/E4 [61], NRPE5 [42,45,53,62], and NRPB/D/E9b [62,63] but not NRPB/D/E9a [63] in gene silencing. Though this is clearly just the beginning, these studies have already provided insights into the functions of these subunits. First, these studies demonstrated that a short N-terminal extension in NRPE5 is required for its stability [42,53], suggesting a potential role for this region in facilitating incorporation of NRPE5 into Pol V. Second, they found that loss of NRPB/D/E/9b does not affect the accumulation of Pol V-dependent transcripts and speculate that rather than regulating Pol V transcription, this subunit might facilitate interactions with RNA processing components [62,63]. With increased accessibility to next generation sequencing technologies and the in vitro recapitulation of Pol IV and Pol V transcription (highlighted below), more details regarding the potential locus specificity and roles of these subunits in polymerase activity are likely to follow.
Maize Pol IV and V catalytic and non-catalytic subunits
Purification of the maize Pol IV and Pol V polymerases revealed subunit compositions that suggest maize and Arabidopsis employ different strategies to generate functional diversity within their plant-specific RNA polymerases. While in Arabidopsis Pol IV and Pol V differ in both their catalytic and non-catalytic subunits [42], maize Pol IV and Pol V share a common set of non-catalytic components and instead possess an expanded repertoire of catalytic subunits [46]. Specifically, they utilize unique largest subunits (ZmNRPD1 and ZmNPRE1) that associate with one of three second largest subunits (ZmNRPD/E2a, D/E2b and E2c), giving rise to 2 Pol IV subtypes and 3 Pol V subtypes [46] (Figure 2C). Like in Arabidopsis, these Pol IV and Pol V complexes utilize forms of the 4th, 5th, and 7th subunits that are unique from Pol II [46]. However, they differ from Arabidopsis in their utilization of a specialized 9th subunit [46], demonstrating they have a more divergent Jaw region that likely imparts additional functionality to these polymerases.
As in Arabidopsis, viable Pol IV/V mutants are available in maize, and despite the fact that only mutations in two subunits, NRPD1 and NRPD/E2a, have been identified (Figure 2), their phenotypes have already provided compelling evidence for functional diversity within polymerase subtypes. Specifically, maize nrpd1 [64] and nrpd/e2a mutants [65–67], which have defects in a silencing phenomenon termed paramutation [68,69] and display a myriad of developmental defects not observed in Arabidopsis [70], fail to phenocopy each other [65–67]. These findings strongly suggest that NRPD/E2a and 2b, as well as the polymerase subtypes they associate with [46], act in a non-redundant fashion. In addition to potential differences in activities of these subtypes, functional diversity is likely also mediated by interactions with accessory factors, for example MOP1 [71] (homolog of RDR2 in Arabidopsis) appears to specifically associate with the NRPD/E2a containing Pol IV subtype [46].
Pol IV- and Pol V-dependent transcripts
The existence of Pol IV- and Pol V-dependent transcripts, which started as just a hypothesis, has now been borne out with the identification of these RNAs in vivo [15,16] and the recapitulation of minimal transcriptional activity using purified Pol IV and Pol V in vitro [46,72]. Below we summarize how these transcripts were identified, what is known about the physical nature and function of these RNAs, and what template features are required for polymerase activity in vitro.
Pol V-dependent transcripts
Given the roles of long noncoding RNAs in targeting chromatin modifications in other systems [73], it was hypothesized that Pol V might act analogously in RdDM. To identify such transcripts, Wierzbicki et al. [16] searched a well characterized region of the genome that displays DNA methylation at intergenic loci largely devoid of known Pol I, II, or III transcripts. This search identified a set of six IGN transcripts that are present at low levels in wild-type plants but are undetectable or reduced in pol v mutants [16]. These IGN transcripts range in size, but can be up to 200-nt in length, placing them in the long noncoding RNA category [16]. They can initiate from multiple adjacent sites, lack poly-A tails and their 5′ ends appear to be a mixture of capped/5′ triphosphates and 5′ monophosphates, suggesting the presence of primary Pol V-dependent transcripts and processed Pol V-dependent transcripts, respectively [16] (Figure 1). Attesting to the importance of these IGN transcripts in facilitating RdDM (and further supporting a role for Pol V in their generation) catalytically dead versions of Pol V failed to produce IGN transcripts and failed to silence adjacent genes [16].
Based on the identification and initial characterization of Pol V-dependent transcripts, a new model was proposed [16] in which Pol V and its transcripts serve as a platform for the recruitment of additional RdDM factors, ultimately leading to the establishment of DNA methylation (Figure 1). As outlined in the introduction, this model is continuing to evolve as additional RdDM factors and IGN transcripts are identified [34,35,37–39,74,75]. However, our knowledge of Pol V-dependent transcripts remains limited to a handful of examples and thus likely represents just the tip of the iceberg. We will have to await a global identification and analysis of Pol V-dependent transcripts to more fully understand the nature and function of these noncoding RNAs.
Pol IV- and RDR2-dependent transcripts
Pol IV is required for the production of >90% of the Arabidopsis 24-nt siRNAs [13,14] and prevailing models suggest this polymerase generates long ssRNAs that act as substrates for siRNA production (Figure 1). Yet, until recently, no such transcripts had been identified. To detect these elusive transcripts, Li et al. [15] devised an elaborate genetic setup coupled with next generation sequencing that allowed Pol IV- and RDR2-dependent transcripts to be inferred by looking at the differences between the transcriptome profiles of dcl234 triple mutants, which accumulates Pol IV-dependent transcripts, and dcl234 nrpd1 (or rdr2) quadruple mutants, which fail to accumulate these transcripts. All told, Pol IV/RDR2-dependent transcripts corresponding to ~50% of known siRNA generating loci were recovered [15] and, like Pol II transcribed genic regions, these loci are flanked by A/T-rich sequences and depleted in nucleosomes, suggesting these features of Pol II function may be conserved in Pol IV [15]. The absence of detectable Pol IV/RDR2-dependent transcripts at many of the remaining loci can likely be attributed, at least in part, to low levels of siRNA production and/or DNA methylation that arise naturally (i.e. in a wild-type background) or artificially (i.e. in the dcl234 triple mutant) and thus are more difficult to detect [15].
Characterization of the nature of these Pol IV/RDR2-dependent RNAs, and the genetic requirements for their production, support some but not all aspects of the current model for siRNA biogenesis, suggesting the model should be revisited. Consistent with the model, Pol IV/RDR2-dependent transcripts are generated from both strands and, like Pol V-dependent transcripts [16], they lack a polyA-tail [15]. Unexpectedly, they also lack a 5′ cap, suggesting that they may represent processed forms of Pol IV/RDR2-dependent transcripts [15]. Alternatively, Pol IV might prime off an unusual RNA substrate that lacks a 5′ triphosphate [15]. Indeed Pol IV can extend off an RNA primer with a 5′ monophosphate in vitro [72], but the existence of such RNAs in vivo remains unknown. In either case, these findings suggest the involvement of specialized RNAs or unknown RNA processing events as an early step associated with Pol IV function. This notion is supported by a much earlier observation that Pol IV localization by immunofluorescence is disrupted upon RNase A treatment (which degrades ssRNA) [76]. Further highlighting our incomplete understanding of how siRNAs are generated, the Pol IV-dependent transcripts identified were equally dependent on RDR2 for their production [15]. This contradicts the current model in which RDR2 acts downstream of Pol IV function and is at odds with in vitro data showing Pol IV activity is unaffected in rdr2 mutants [72]. What role, if any, RDR2 plays in the aforementioned RNA processing events or in other aspects of Pol IV function, and what causes Pol IV to display different dependencies on RDR2 in vitro and in vivo, await further investigation. Finally, it warrants mentioning that the techniques used to identify Pol IV-dependent transcripts do not preserve information regarding the size of these RNAs [15]. Thus, while in silico assembly of these transcripts provides a useful means of assessing the genomic distribution of Pol IV-dependent transcripts, their length remains unclear. Taken together, the identification of Pol IV-dependent transcripts has perhaps raised more questions than answers, suggesting we are on the cusp of a new phase of discovery regarding the transcriptional activity of Pol IV and the biogenesis of 24-nt siRNAs.
Pol IV and V in vitro activity
Using affinity purified Pol IV and Pol V complexes from Arabidopsis and maize, Haag et al. [46,72] have demonstrated Pol IV and Pol V transcriptional activity in vitro. These experiments revealed several unusual template features, including the requirement of an RNA primer, that undoubtedly contributed to the delay in the identification of in vitro polymerase activity [72]. Like Pol II, Pol IV will transcribe a tripartite substrate that includes an RNA primer and mimics a transcription bubble, but its activity is more robust using a bipartite substrate, suggesting that Pol IV does not efficiently displace the non-template DNA strand [72]. In addition to these DNA-templated Pol IV-dependent products, shorter RDR2-dependent products were also observed [72], which is consistent with data showing that RDR2 co-purifies with Pol IV [43,44,46,72]. Further investigation into the connections between Pol IV and RDR2 activities revealed that Pol IV transcription is independent of RDR2, while RDR2 activity requires the presence of Pol IV, but not its polymerase activity [72]. As discussed above, this one-way dependence in vitro, is at odds with what appears to be a complete interdependency of these activities in vivo [15]. Intriguingly, Pol IV is also able to transcribe an all-RNA substrate, demonstrating that it is not a strictly DNA-dependent RNA polymerase [72]. In comparison with Pol IV, the template requirements for Pol V are even more restrictive; Pol V is only active using a bipartite substrate, suggesting this polymerase is even less efficient at displacing the non-template DNA strand, and revealing a more strict dependence on a DNA template [72]. Notably, the observed activities for both Pol IV and Pol V are weak relative to Pol II, which could be an intrinsic feature of these polymerases as they show poor conservation within two regions (the trigger loop and the bridge helix) that are required for efficient nucleotide incorporation [77], or could indicate sub-optimal template features or missing accessory components.
Despite great progress in identifying Pol IV and Pol V-dependent transcripts in vivo and establishing in vitro transcription assays, it remains unclear what templates these polymerases utilize in vivo and it is also unclear how their transcription is regulated to prevent the production of aberrant noncoding RNAs. Regulation of transcription occurs at all stages of the transcription cycle (initiation, elongation, termination) and controls important aspects of polymerase activity including, template selection, fidelity and processivity [40,54,78]. It is unclear how Pol IV and Pol V initiation occurs, but if they do indeed require an RNA primer in vivo, they may completely bypass this step by hijacking RNAs produced by other polymerases, like Pol II, and proceed directly to elongation [54]. The elongation phase involves displacement of the non-template DNA strand and extension of the RNA transcript. Potential roles for the components of the Pol V-associated DDR complex (Figure 1) in template strand unwinding are discussed in Pikaard et al. [78]. Regarding termination, we are completely in the dark. However, armed with new biochemical assays and available genetic mutants, the field is now primed to address these open questions in both Arabidopsis and maize using a combination of in vitro and in vivo studies. As additional parameters are revealed, it will be of great interest to understand the extents to which specialized polymerase subunits influence these features of Pol IV and Pol V activity.
Association of Pol IV and Pol V with chromatin
Given the lack of defined promoter or other conserved sequence features at known targets of the RdDM pathway, the question of how Pol IV and Pol V are specifically recruited to these genomic loci has been a topic of intense interest over the last decade. Recently, several studies employing genetic, genomic and biochemical techniques have begun shedding light on this question and, although they have not revealed any common sequence determinants, they have revealed a dependence on specific chromatin modifications, establishing a series of self-reinforcing loops between DNA and histone modifications (Figure 1).
Pol V association with chromatin requires the DDR complex and two methyl-DNA binding proteins
The first insights into the recruitment of Pol V to chromatin were nearly coincident with the discovery of Pol V-dependent transcripts [16]. Two known RdDM components, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1) [79] and DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) [80,81], were shown to be required for both the accumulation of Pol V-dependent transcripts and Pol V occupancy at several IGN producing loci [16,35]. Shortly after these discoveries, a third component, RNA-DIRECTED DNA METHYLATION 1 (RDM1), was found to stably associate with both DRD1 and DMS3 [82] and to be required for the production of IGN transcripts [82,83]. This protein complex, termed DDR (after its three main components) also associates Pol V [82], suggesting that the DDR complex facilitates Pol V association with chromatin via a direct interaction [82] (Figure 1).
Until recently it was unclear whether the DDR complex functioned in a locus specific or a more global manner, nor was it clear how this complex itself was recruited to chromatin. The path towards answering these questions began with the identification of Pol V binding sites on a genome-wide scale, which, consistent with the role of Pol V in RdDM, are highly correlated with DNA methylation, siRNAs, and IGN transcripts [74,75]. In drd1, dms3, or rdm1 single mutants, Pol V enrichment at binding sites identified in wild-type plants were lost, demonstrating that the DDR complex acts as a global regulator of Pol V chromatin association [75]. In addition to the DDR complex, two methyl-DNA binding proteins, SU(VAR)3-9 HOMOLOG 2 (SUVH2) and SUVH9 [84], that function in RdDM in a partly redundant manner [84,85] and interact with components of the DDR complex [86,87], were also shown to be required for Pol V-dependent transcripts and Pol V chromatin association [86,87]. This finding suggested a link between DNA methylation and Pol V occupancy and indeed, Pol V enrichment at chromatin is nearly abolished in a met1 mutant [86], which causes global decreases in DNA methylation [88]. Thus, a self-reinforcing loop model has emerged wherein Pol V recruitment depends on pre-existing DNA methylation. In this model, DNA methylation is bound by SUVH2 and SUVH9, which in turn interacts with the DDR complex, thus promoting the association of Pol V with chromatin and leading to the generation of noncoding RNAs that facilitate additional DNA methylation (Figure 1).
In addition to the aforementioned major players that regulate Pol V recruitment, several other RdDM components have also been shown to play more modest roles in association with Pol V function, including SU(VAR)3-9-RELATED PROTEIN 2 (SUVR2) [89,90], DOMAINS REARRANGED METHYLTRANSFERASE 3 (DRM3) [91–93], CHROMATIN REMODELING 27 (CHR27) and CHR28 (a.k.a. SNF2-RING-HELICASE LIKE 1 (FRG1) and FRG2) [89,94]. While the precise functions of these factors remain to be elucidated, they likely function to fine-tune rather than globally regulate the activity and distribution of Pol V.
Pol IV association with chromatin requires SHH1, an H3K9me reader
In addition to the core Pol IV subunits, affinity purification of this polymerase have identified several other accessory factors [43,44,46,72], and characterization of one such component, SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) [43] (a.k.a. DNA-BINDING TRANSCRIPTION FACTOR 1 (DTF1) [95]) has provided mechanistic insight into the association of Pol IV with chromatin [96]. Initial characterization of shh1 mutants demonstrated defects in gene silencing, DNA methylation, and siRNA accumulation [43,95], but not in the production of Pol V-dependent transcripts, suggesting a specific role for SHH1 in connection with Pol IV [43]. Subsequent genome-wide characterization of shh1 mutants revealed this factor acts in a highly locus specific manner, phenocopying pol iv mutants at approximately half of the siRNA producing loci genome-wide, but having no significant affect at the remaining loci [44,96]. Mechanistically, the observed defects in shh1 stem from loss of Pol IV occupancy specifically at SHH1-regulated genomic loci [96]. This role of SHH1 requires its SAWADEE domain [96], which recognizes H3K9 methylation [44,96] and adopts a unique tandem-tudor domain fold that interrogates the methylation status of both the H3K4 and H3K9 positions [96]. In Arabidopsis, H3K9 methylation maps to regions of the genome that harbor DNA methylation, including those targeted by the RdDM pathway, and is redundantly controlled by a family of histone methyltransferases, SUVH4, SUVH5 and SUVH6 [97–102]. Thus, SHH1 connects the DNA methylation machinery with H3K9 methylation, establishing yet another self-reinforcing loop between repressive chromatin modifications. Alas, several key questions remain unanswered, including how Pol IV recruitment to the remaining RdDM targets is controlled and how SHH1 exerts its affects only at a subset of RdDM targets, as H3K9me is a general feature of RdDM targets.
In regards to Pol IV and Pol V targeting, two additional topics warrant a brief discussion. First, purification of Pol IV and Pol V from maize revealed these polymerases associate with components involved in the recruitment of Arabidopsis Pol IV and Pol V to chromatin, namely, ZmSHH2a as well as ZmDMS3 and ZmCHR127 (related to AtDRD1), respectively [46]. Thus, while functional studies have not yet been completed, it appears that similar mechanisms are likely in place to facilitate the recruitment of maize Pol IV and Pol V to chromatin. Second, while updated models of Pol IV and Pol V targeting have provided key sights into the pathway (Figure 1), the built in reinforcing loops raises the question of how DNA methylation is initially established at a naïve locus. It is still early days, but genetic evidence suggests a role for Pol II in DNA methylation pathways [90,103–107] and several groups have begun characterizing non-canonical RdDM pathways that are excellent candidates for establishing DNA methylation at naïve loci [108–112].
Conclusion and outlook
Plants encode specialized RNA polymerases that act non-redundantly in the de novo DNA methylation pathway, differ in their subunit compositions and in the types of noncoding RNAs they generate, and employ unique machinery for their recruitment to chromatin. Recent advances in our understanding of the evolution and composition of these polymerases has confirmed the existence of specialized Pol IV and Pol V machinery across land plants, and has revealed functional diversity both between and within Pol IV and Pol V subtypes. The identification of Pol IV and Pol V transcripts in vitro and in vivo has provided the first clues into the activities of these polymerases and suggests that they are not playing by the same rules governing Pol II activity. Finally, a quite detailed view of the machinery required to facilitate recruitment of these polymerases to chromatin has emerged, revealing a dependence on previously established chromatin modifications as part of an intricate network of self-reinforcing loops.
Paper Highlights.
Plants encode extra RNA polymerases (Pol IV and V) that facilitate DNA methylation
Pol IV and V subunits evolved from Pol II subunits, and are continuing to diversify
Pol IV and V differ in their composition and in the noncoding RNAs they produce
In vitro, Pol IV and V utilize non-canonical templates and require an RNA primer
Pol IV and V employ unique machinery to facilitate their recruitment to chromatin
Acknowledgments
J.A.L. is supported by the US National Institutes of Health grant GM112966 and the Jacobs Foundation and M.Z is supported by a Salk Pioneer Fellowship. We thank R. Mosher and the editorial staff for comments and suggestions. The authors apologize to colleagues whose work was not mentioned due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15:517–522. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 3.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cecere G, Grishok A. A nuclear perspective on RNAi pathways in metazoans. Biochim Biophys Acta. 2014;1839:223–233. doi: 10.1016/j.bbagrm.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature reviews Genetics. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 7.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matzke MA, Kanno T, Matzke AJ. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu Rev Plant Biol. 2015;66:243–267. doi: 10.1146/annurev-arplant-043014-114633. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Zhu JK. Active DNA demethylation in plants and animals. Cold Spring Harb Symp Quant Biol. 2012;77:161–173. doi: 10.1101/sqb.2012.77.014936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. This publication, along with references #11 and #12, identified a role for RNA polymerase IV in RdDM through the mapping of genetic mutants defective in DNA methylation and gene silencing. [DOI] [PubMed] [Google Scholar]
- 11*.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. This publication identified a role for RNA polymerase IV in RdDM through the mapping of genetic mutants defective in DNA methylation and gene silencing. [DOI] [PubMed] [Google Scholar]
- 12*.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. This publication identified a role for RNA polymerase IV in RdDM through the mapping of genetic mutants defective in DNA methylation and gene silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015;25:235–245. doi: 10.1101/gr.182238.114. This publication was the first to identify Pol IV-dependent transcripts in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. This publication was the first to identify Pol V-dependent transcripts in vivo and to demonstrate that Pol V transcription activity is required for DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 23.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eun C, Lorkovic ZJ, Naumann U, Long Q, Havecker ER, Simon SA, Meyers BC, Matzke AJ, Matzke M. AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS One. 2011;6:e25730. doi: 10.1371/journal.pone.0025730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan CG, Zhang H, Tang K, Zhu X, Qian W, Hou YJ, Wang B, Lang Z, Zhao Y, Wang X, et al. Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J. 2015;34:581–592. doi: 10.15252/embj.201489453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duran-Figueroa N, Vielle-Calzada JP. ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signal Behav. 2010;5:1476–1479. doi: 10.4161/psb.5.11.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell. 2012;46:859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 30*.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. This publication identified a role for RNA polymerase V in RdDM through the mapping of genetic mutants defective in DNA methylation and gene silencing. [DOI] [PubMed] [Google Scholar]
- 31.Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 32.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157:1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohmdorfer G, Rowley MJ, Kucinski J, Zhu Y, Amies I, Wierzbicki AT. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014;79:181–191. doi: 10.1111/tpj.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azevedo J, Cooke R, Lagrange T. Taking RISCs with Ago hookers. Curr Opin Plant Biol. 2011;14:594–600. doi: 10.1016/j.pbi.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Q, Rowley MJ, Bohmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2012 doi: 10.1111/tpj.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 41.Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 42*.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. This publication employed affinity purification to reveal the subunit compositions of the Pol II, Pol IV and Pol V holoenzymes in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 46*.Haag JR, Brower-Toland B, Krieger EK, Sidorenko L, Nicora CD, Norbeck AD, Irsigler A, LaRue H, Brzeski J, McGinnis K, et al. Functional diversification of maize RNA polymerase IV and V subtypes via alternative catalytic subunits. Cell Rep. 2014;9:378–390. doi: 10.1016/j.celrep.2014.08.067. This publication includes the affinity purification of maize Pol II, IV, and IV and, along with reference #72 in Arabidopsis, demonstrates Pol IV and Pol V transcriptional activity in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J, Hall BD. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 2007;64:101–112. doi: 10.1007/s00239-006-0093-z. [DOI] [PubMed] [Google Scholar]
- 48.Tucker SL, Reece J, Ream TS, Pikaard CS. Evolutionary history of plant multisubunit RNA polymerases IV and V. subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol. 2010;75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- 49*.Huang Y, Kendall T, Forsythe ES, Dorantes-Acosta A, Li S, Caballero-Perez J, Chen X, Arteaga-Vazquez M, Beilstein MA, Mosher RA. Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol Biol Evol. 2015;32:1788–1799. doi: 10.1093/molbev/msv060. This paper, along with reference #50, provides the most current views regarding the ancient origins and ongoing evolution of RNA Pol IV and Pol V across all land plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Wang Y, Ma H. Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits. New Phytol. 2015;207:1198–1212. doi: 10.1111/nph.13432. This paper, along with reference #49, provides the most current views regarding the ancient origins and ongoing evolution of RNA Pol IV and Pol V across all land plants. [DOI] [PubMed] [Google Scholar]
- 51.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 52.Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 2009;106:941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ream TS, Haag JR, Pikaard CS. Plant Multisubunit RNA Polymerases IV and V. Nucleic Acid Polymerases. 2014;30:289–308. [Google Scholar]
- 55.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 56.Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giner A, Lakatos L, Garcia-Chapa M, Lopez-Moya JJ, Burgyan J. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010;6:e1000996. doi: 10.1371/journal.ppat.1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knippa K, Peterson DO. Fidelity of RNA polymerase II transcription: Role of Rbp9 in error detection and proofreading. Biochemistry. 2013;52:7807–7817. doi: 10.1021/bi4009566. [DOI] [PubMed] [Google Scholar]
- 60.Hemming SA, Jansma DB, Macgregor PF, Goryachev A, Friesen JD, Edwards AM. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J Biol Chem. 2000;275:35506–35511. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- 61.He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eun C, Lorkovic ZJ, Sasaki T, Naumann U, Matzke AJ, Matzke M. Use of forward genetic screens to identify genes required for RNA-directed DNA methylation in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol. 2012;77:195–204. doi: 10.1101/sqb.2012.77.015099. [DOI] [PubMed] [Google Scholar]
- 63.Tan EH, Blevins T, Ream TS, Pikaard CS. Functional consequences of subunit diversity in RNA polymerases II and V. Cell Rep. 2012;1:208–214. doi: 10.1016/j.celrep.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erhard KF, Jr, Stonaker JL, Parkinson SE, Lim JP, Hale CJ, Hollick JB. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 65.Sidorenko L, Dorweiler JE, Cigan AM, Arteaga-Vazquez M, Vyas M, Kermicle J, Jurcin D, Brzeski J, Cai Y, Chandler VL. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009;5:e1000725. doi: 10.1371/journal.pgen.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stonaker JL, Lim JP, Erhard KF, Jr, Hollick JB. Diversity of Pol IV function is defined by mutations at the maize rmr7 locus. PLoS Genet. 2009;5:e1000706. doi: 10.1371/journal.pgen.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sloan AE, Sidorenko L, McGinnis KM. Diverse gene-silencing mechanisms with distinct requirements for RNA polymerase subunits in Zea mays. Genetics. 2014;198:1031–1042. doi: 10.1534/genetics.114.168518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arteaga-Vazquez MA, Chandler VL. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev. 2010;20:156–163. doi: 10.1016/j.gde.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Erhard KF, Jr, Hollick JB. Paramutation: a process for acquiring trans-generational regulatory states. Curr Opin Plant Biol. 2011;14:210–216. doi: 10.1016/j.pbi.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Pikaard CS, Tucker S. RNA-silencing enzymes Pol IV and Pol V in maize: more than one flavor? PLoS Genet. 2009;5:e1000736. doi: 10.1371/journal.pgen.1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, Sikkink K, Chandler VL. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 72**.Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Molecular cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. This publication, along with reference #46 in maize, is the first demonstration of Pol IV and Pol V transcriptional activity on a non-canoncial DNA templates that include an RNA primer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74**.Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes & Development. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. This publication, along with reference #75, revealed the genomewide distribution of Pol V, and demonstrated that the DDR complex is required for Pol V chromatin association genomewide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75**.Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. This publication, along with reference #74, revealed the genomewide distribution of Pol V, and demonstrated that the DDR complex is required for Pol V chromatin association genomewide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 77.Landick R. Functional divergence in the growing family of RNA polymerases. Structure. 2009;17:323–325. doi: 10.1016/j.str.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol. 2012;77:205–212. doi: 10.1101/sqb.2013.77.014803. [DOI] [PubMed] [Google Scholar]
- 79.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 80.Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJM. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nature Genetics. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 81.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuhlmann M, Mette MF. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol Biol. 2012;79:623–633. doi: 10.1007/s11103-012-9934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86**.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. This publication, along with reference #87, demonstrated roles for two methyl-DNA binding proteins, SUVH2 and SUVH9, in Pol V chromatin association genomewide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87**.Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. This publication, along with reference #86, demonstrated roles for two methyl-DNA binding proteins, SUVH2 and SUVH9, in Pol V chromatin association genomewide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han YF, Dou K, Ma ZY, Zhang SW, Huang HW, Li L, Cai T, Chen S, Zhu JK, He XJ. SUVR2 is involved in transcriptional gene silencing by associating with SNF2-related chromatin-remodeling proteins in Arabidopsis. Cell Res. 2014;24:1445–1465. doi: 10.1038/cr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE. The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001182. doi: 10.1371/journal.pgen.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhong X, Hale CJ, Nguyen M, Ausin I, Groth M, Hetzel J, Vashisht AA, Henderson IR, Wohlschlegel JA, Jacobsen SE. Domains rearranged methyltransferase3 controls DNA methylation and regulates RNA polymerase V transcript abundance in Arabidopsis. Proc Natl Acad Sci U S A. 2015;112:911–916. doi: 10.1073/pnas.1423603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Costa-Nunes P, Kim JY, Hong E, Pontes O. The cytological and molecular role of domains rearranged methyltransferase3 in RNA-dependent DNA methylation of Arabidopsis thaliana. BMC Res Notes. 2014;7:721. doi: 10.1186/1756-0500-7-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Groth M, Stroud H, Feng S, Greenberg MV, Vashisht AA, Wohlschlegel JA, Jacobsen SE, Ausin I. SNF2 chromatin remodeler-family proteins FRG1 and -2 are required for RNA-directed DNA methylation. Proc Natl Acad Sci U S A. 2014;111:17666–17671. doi: 10.1073/pnas.1420515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Bai G, Zhang C, Chen W, Zhou J, Zhang S, Chen Q, Deng X, He XJ, Zhu JK. An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res. 2011;21:1691–1700. doi: 10.1038/cr.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96**.Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. This publication reveals the SHH1 SAWADEE domain is a novel H3K9me reader that adopts a tandem tudor-like fold, and that this domain is required for the role of SHH1 in facilitating Pol IV occupancy at chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. Embo J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 101.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.You WH, Lorkovic ZJ, Matzke AJM, Matzke M. Interplay among RNA polymerases II, IV and V in RNA-directed DNA methylation at a low copy transgene locus in Arabidopsis thaliana. Plant Molecular Biology. 2013;82:85–96. doi: 10.1007/s11103-013-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sasaki T, Lee TF, Liao WW, Naumann U, Liao JL, Eun C, Huang YY, Fu JL, Chen PY, Meyers BC, et al. Distinct and concurrent pathways of Pol II- and Pol IV-dependent siRNA biogenesis at a repetitive trans-silencer locus in Arabidopsis thaliana. Plant J. 2014;79:127–138. doi: 10.1111/tpj.12545. [DOI] [PubMed] [Google Scholar]
- 106.He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23:2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, Lind M, Schmitt MJ, Simon SA, Gurazada SGR, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. Embo Reports. 2010;11:65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108*.Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2015;112:917–922. doi: 10.1073/pnas.1413053112. This publication, along with reference #110, provide evidence for alternative forms of RNA-directed DNA methylation with different genetic requirements that are proposed to act before the canonical establishment RdDM pathway in a phase being termed “Initiation”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McCue AD, Panda K, Nuthikattu S, Choudury SG, Thomas EN, Slotkin RK. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015;34:20–35. doi: 10.15252/embj.201489499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110*.Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013;162:116–131. doi: 10.1104/pp.113.216481. This publication, along with reference #108, provide evidence for alternative forms of RNA-directed DNA methylation with different genetic requirements that are proposed to act before the canonical establishment RdDM pathway in a phase being termed “Initiation”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mari-Ordonez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 112.Panda K, Slotkin RK. Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal Behav. 2013;8:e25206. doi: 10.4161/psb.25206. [DOI] [PMC free article] [PubMed] [Google Scholar]