Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most prevalent multidrug-resistant pathogens worldwide, exhibiting increasing resistance to the latest antibiotic therapies. Here we show that the triple β-lactam combination meropenem/piperacillin/tazobactam (ME/PI/TZ) acts synergistically and is bactericidal against MRSA N315 and 72 clinical MRSA isolates in vitro, and clears MRSA N315 infection in a mouse model. ME/PI/TZ suppresses evolution of resistance in MRSA via reciprocal collateral sensitivity of its constituents. We demonstrate that these activities also extend to other carbapenem/penicillin/β-lactamase inhibitor combinations. ME/PI/TZ circumvents the tight regulation of the mec and bla operons in MRSA, the basis for inducible resistance to β-lactam antibiotics. Furthermore, ME/PI/TZ subverts the function of penicillin-binding protein 2a (PBP2a) action via allostery, which we propose as the mechanism for both synergy and collateral sensitivity. Showing similar in vivo activity to linezolid, ME/PI/TZ demonstrates that combinations of older β-lactam antibiotics could be effective against MRSA infections in humans.
Introduction
Multidrug-resistant (MDR) pathogens represent a growing threat to human health, with many infectious diseases effectively regressing toward the pre-antibiotic era 1,2, exemplified by the dramatic rise of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infections. In the 1940’s, S. aureus infections were primarily treated with first-generation β-lactams (penicillins), which target the penicillin-binding proteins (PBPs), the critical transpeptidases for cell-wall synthesis 3. Four PBPs (PBP1–PBP4) perform these functions in S. aureus 3. Emergence of β-lactamase-producing strains led to development of β-lactamase-resistant second-generation penicillins, including methicillin. Soon after the introduction of methicillin in 1959, the first MRSA strains were reported 4. These strains acquired a highly regulated collection of genes from a non-S. aureus source that produced inducible resistance to β-lactam antibiotics 3. One of these genes, mecA, encodes penicillin-binding protein 2a (PBP2a). PBP2a performs the critical transpeptidase reaction that cross-links the cell wall, even under challenge by β-lactam antibiotics, when other PBPs are inhibited 5,6. The mechanistic basis for this outcome is complex, involving a closed conformation for the active site, whose function is regulated by allostery 7,8. The emergence of MRSA has virtually eliminated the use of β-lactams as therapeutic options against S. aureus. The recently developed β-lactam agent ceftaroline, which exhibits activity in treatment of MRSA infections, does so by binding to the allosteric site of PBP2a, triggering opening of the active site for inactivation by the drug 8,9; however, resistance to ceftaroline 10 and other antibiotics used to treat MRSA, including linezolid, vancomycin, and daptomycin, has been reported 11–13.
Use of multidrug combination therapy targeting orthogonal cellular processes has been successful in treating Mycobacterium tuberculosis, Helicobacter pylori, and other infections 14,15. However, resistance is increasing even against these therapies 16,17. We have identified a new potential therapy against MRSA consisting of a combination of clinically approved drugs from three distinct generations and subclasses of β-lactam antibiotics, all targeting cell-wall synthesis: meropenem, piperacillin, and tazobactam (ME/PI/TZ). This therapy uses elements from three strategies: 1) use of semi-synthetic antibiotic derivatives that target multiple nodes in the same cellular system 10,18, 2) use of combinations of these antibiotics that increase drug potency by utilizing drug synergy 19,20, and 3) use of collateral sensitivity between constituents of the combination to suppress resistance evolution 21,22. Each of these methods have been successfully employed against the major MDR Gram-negative and Gram-positive human pathogens 23,24. However, these strategies used individually have often been thwarted by the evolution of new resistance in MDR pathogens, leading to diminishing options for treating their infections 4,12,18.
We hypothesize that ME/PI/TZ operates through inhibition of PBP1 by meropenem, the targeting of PBP2 by piperacillin, protection of piperacillin from the PC1 class A β-lactamases by tazobactam 5,25–28, and allosteric opening of the active site of PBP2a by meropenem for inhibition by another molecule of antibiotic in the combination 9. This culminates in a synergistic response by simultaneous perturbation of multiple components of the cell-wall synthesis machinery in MRSA. We find that exposure of MRSA N315 to the components of ME/PI/TZ reveals reciprocal collateral sensitivities within this highly synergistic triple combination that suppress the evolution of resistance, in contrast to some synergistic combination therapies that instead accelerate resistance evolution 20,29. This effect is consistent with recent work showing that collateral sensitivity slows evolution of resistance in a non-pathogenic laboratory strain of Escherichia coli 21,30. Our results support renewed clinical use of older β-lactam antibiotics against MRSA when used in judiciously conceived synergistic combinations of collaterally sensitive components, opening a new treatment paradigm with existing drugs that are already approved for human use.
Results
Three β-lactams synergistically kill MRSA in vitro
Based on its high level of resistance against 23 diverse antibiotics (Supplementary Results, Supplementary Table 1), we selected S. aureus MRSA N315 31 from a group of fully genome-sequenced MDR strains of MRSA for this study. MRSA N315 contains the staphylococcal chromosome cassette mec (SCCmec) type II encoding the mec methicillin-resistance operon 32, as well as penicillinase plasmid pN315 containing the bla β-lactamase operon 33. From a focused combinatorial screen of these 23 antibiotic compounds, including representatives from every major drug class (Supplementary Table 1), we identified the combination of ME/PI/TZ to display highly synergistic, bactericidal activity against MRSA N315 in vitro, using the metric of the fractional inhibitory concentration index (FICI), FICI = 0.11 34,35 (Supplementary Table 2a). For any number of drugs in combination, a FICI less than 1 indicates synergy, a FICI equal to 1 indicates additivity, and a FICI greater than 1 indicates indifference or antagonism 34,35. Notably, these three drugs all belong to different sub-classes of the β-lactam drugs, which target the critical transpeptidase enzymes of cell-wall synthesis, though MRSA strains are typically highly resistant to most β-lactams 6. The general resistance to individual β-lactams results from the inability of these drugs to inhibit the transpeptidase active site of PBP2a, which compensates for β-lactam inhibition of the other transpeptidases in S. aureus 6.
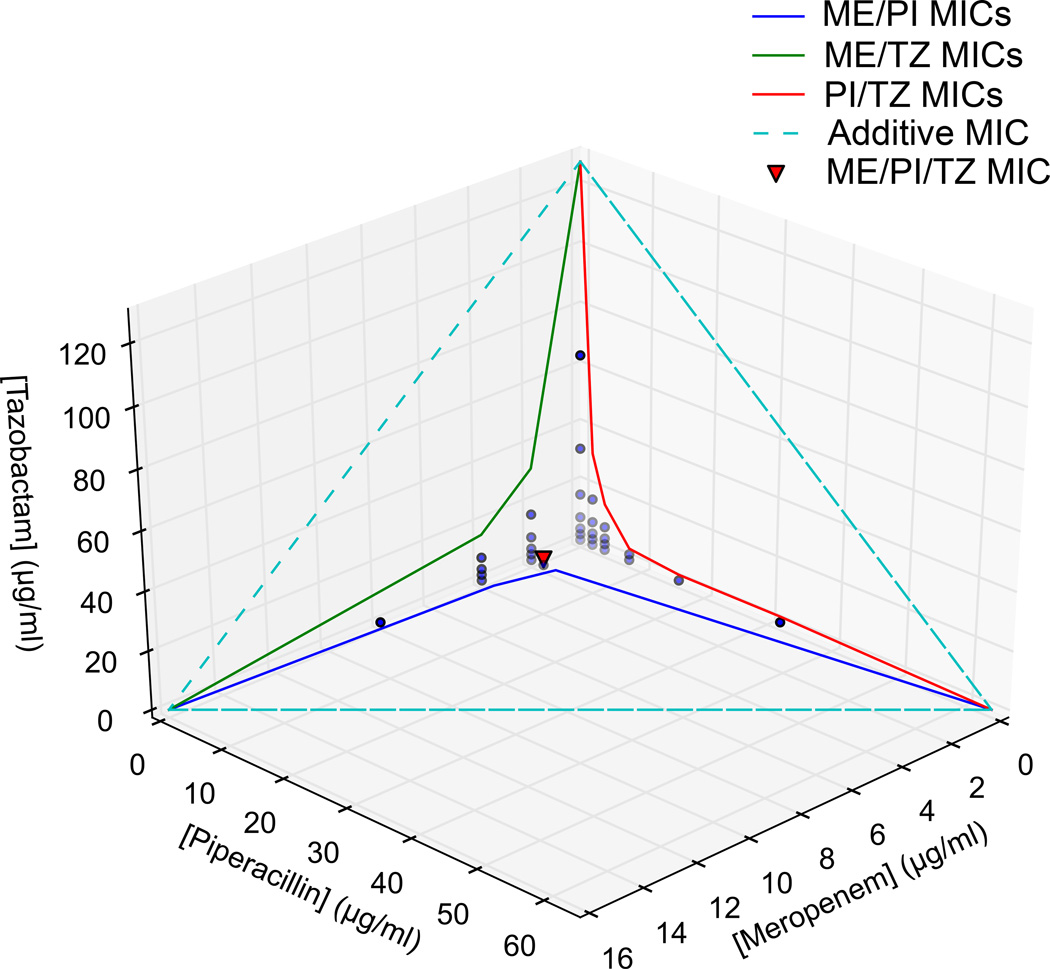
ME/PI/TZ exhibited increased synergy against MRSA N315 relative to its three constituent double combinations meropenem/piperacillin (ME/PI), meropenem/tazobactam (ME/TZ) and piperacillin/tazobactam (PI/TZ) at clinically relevant concentrations (Fig. 1, Supplementary Tables 2b, c). All three β-lactam compounds were tested for final MIC and FICI using a 3-D checkerboard with twofold dilution series of each compound from 128–2 µg/ml, and no-drug media. These allowed up to a 64-fold difference in component ratios to be explored for maximal synergy, as well as allowing for isolation of results for each single compound, all constituent double combinations, and the triple combination. Using the 3-D checkerboard, we determined the optimal ratio for ME/PI/TZ to be 1:1:1 for minimal drug input and maximal synergy against MRSA N315. The minimal-inhibitory concentrations (MICs) of the meropenem and piperacillin components in the combination against MRSA N315 (2 µg/ml each) were below the clinical susceptibility breakpoints for each of these drugs alone against methicillin-susceptible S. aureus (4–8 µg/ml), while tazobactam has no susceptibility breakpoint alone, and is given clinically at a 1:8 ratio with piperacillin 36. The constituent double combinations ME/PI and PI/TZ were also synergistic against N315 with FICI = 0.44 and 0.22, respectively, while ME/TZ is less synergistic at 0.67. Based on the Loewe additivity model of synergy, drugs cannot be synergistic with themselves 30. Though the β-lactams all target the cell-wall synthesis pathway, our use of the FICI method (Loewe additivity) confirms the non-additive nature of these interactions. In contrast to the high synergy of ME/PI/TZ seen in MRSA N315, the combination exhibits no additive activity (FICI = 1.12) in the methicillin-susceptible S. aureus (MSSA) reference strain ATCC 29213 36,37 (Supplementary Tables 2b, c), and we hypothesize the necessity of PBP2a for synergy to occur.
Figure 1. 3D-Checkerboard synergy determination showing isoboles of minimal inhibitory concentrations (MIC) and in vitro growth in single-, double-, or triple-drug conditions for ME/PI/TZ.
Colored lines/isoboles within each panel indicate MICs of two drugs in combination. Dashed lines indicate theoretical concentrations of additive interactions. Points indicate top sub-inhibitory concentrations of meropenem (ME), piperacillin (PI) and tazobactam (TZ) for each tested condition. The red triangle indicates the MIC of all three drugs in combination (Each at 2 µg/ml).
We propose that the mechanism of synergy observed for ME/PI/TZ results from allosteric triggering of PBP2a by its constituents, akin to that reported for ceftaroline 8,9. Indeed, we determined that meropenem binds to the allosteric site of PBP2a with a dissociation constant (Kd) of 270 ± 80 µM (equivalent to 104 ± 31 µg/ml). The mean peak plasma concentration in healthy humans after a bolus intravenous injection of meropenem at the recommended 1 g dose is 112 µg/ml 38. The concentrations of meropenem achieved clinically are above the Kd; thus at these concentrations meropenem binding to the allosteric site of PBP2a would trigger opening of the active site of PBP2a, enabling access to its transpeptidase active site for acylation/inactivation either by another molecule of meropenem or by other β-lactams in the combination 6,8,39.
We recapitulated the highly synergistic activity of ME/PI/TZ seen in MRSA N315 against all in a panel of 72 clinical MRSA isolates with multiple SCCmec types represented (Supplementary Tables 3a, b). The MIC of the combination against the clinical isolates ranged from 0.4–33.3 µg/ml for each component, with a mean of 9.7 µg/ml, and an MIC50 and MIC90 of 3.7 µg/ml and 33.3 µg/ml, respectively (Supplementary Table 4a).
Class-specificity of β-lactam synergy against MRSA
We determined that the observed synergy is not limited to the antibiotics assayed, but can be generalized to their respective β-lactam classes, by testing MRSA N315 and representative clinical MRSA isolates against other carbapenem/penicillin/β-lactamase inhibitor combinations. We found that treatment of MRSA N315 with imipenem/piperacillin/clavulanate (IM/PI/CV) shows equal or greater synergism to ME/PI/TZ. Meropenem/amoxicillin/tazobactam (ME/AX/TZ) maintains high synergy in MRSA N315 only (FICI = 0.04), with a clinical MRSA isolate showing less synergy (FICI = 0.55) (Supplementary Table 2b). MICs for components of these substituted triples are all below the mean peak human plasma concentrations of these compounds in vivo 40,41. Similar to ME/PI/TZ, IM/PI/CV shows less-than-additive activity against MSSA ATCC 29213 (FICI = 1.14) (Supplementary Tables 2b, c). These results further support the necessity of the presence of the mecA gene product PBP2a with its attendant allosterism for synergy, due to lack of synergy of carbapenem/penicillin/β-lactamase inhibitor combinations in methicillin-susceptible S. aureus.
We also tested the effect of replacing the carbapenem component of the combination with either a monobactam or a cephalosporin, two other later-generation β-lactam derivatives. In contrast to ME/PI/TZ, the triple combinations aztreonam/piperacillin/tazobactam (AZ/PI/TZ) and cefepime/piperacillin/tazobactam (CP/PI/TZ) (FICI for both = 0.33) have lower levels of synergy than PI/TZ alone (FICI = 0.22) (Supplementary Table 2b), possibly because aztreonam (a monobactam) has selective binding to Gram-negative PBPs 42, while cefepime (a cephalosporin) preferentially targets PBP2 over PBP1 6.
We confirmed the targets of the constituents of ME/PI/TZ by reducing the expression of PBP1, PBP2, PBP2a or PBP3 using a xylose-inducible antisense-RNA strategy in the MRSA COL strain background 43. Using this system, complementary RNA targeting a PBP gene reduces its expression, and promotes hypersusceptibility to drugs that target its PBP 44. When expression levels of PBP2a were attenuated, the strain behaved as a methicillin-susceptible S. aureus and was sensitized to all tested β-lactams (Supplementary Table 5). When meropenem, piperacillin, and tazobactam were tested against the pbpA antisense strain, only meropenem showed larger zones of inhibition under xylose induction, confirming PBP1 as a target of meropenem (Supplementary Table 5). For the pbp2 antisense strain both meropenem and piperacillin showed increased effectiveness under xylose induction, demonstrating that they each have some activity against PBP2 (Supplementary Table 5). We did not observe any effect with the pbp3 antisense strain, consistent with our hypothesis that ME/PI/TZ activity is focused on disrupting PBP1, PBP2, and PBP2a (Supplementary Table 5). The antisense strains in all cases but that of pbp3 showed sensitization to the triple combination, underscoring the observed synergy.
ME/PI/TZ suppresses resistance evolution in MRSA N315
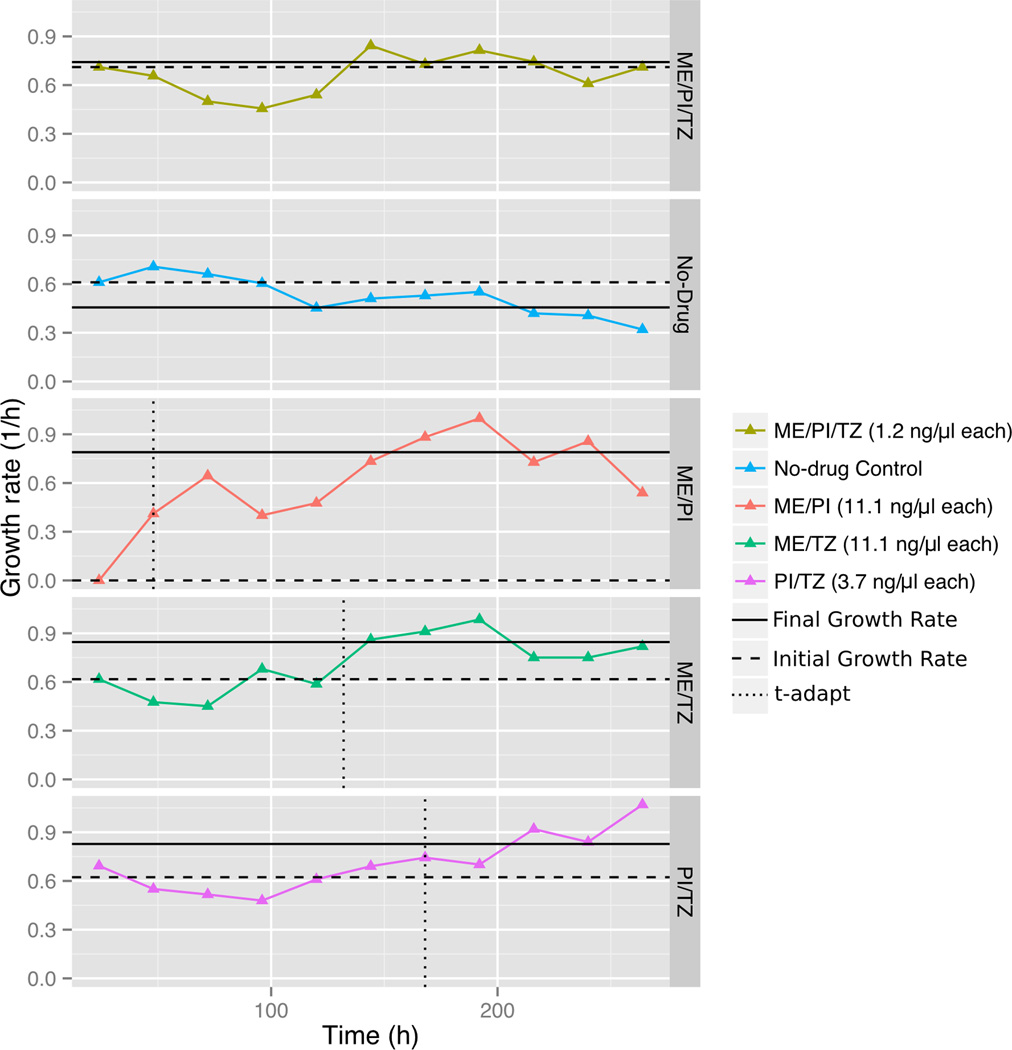
It is obvious that development and spread of resistance can dramatically dampen the effectiveness and longevity of an antimicrobial therapy. We demonstrated that ME/PI/TZ suppresses the evolution of resistance in MRSA using serial passaging in sub-inhibitory antibiotic concentrations of the triple combination and each of its constituents. To more accurately model a clinical treatment in vitro and in vivo, we applied these drugs at fixed dosages over extended periods as occurs in clinical treatment, not at increasing doses over time. During the 11-day experiment, we observed no evolution of resistance in MRSA N315 to ME/PI/TZ. In contrast, we observed resistance evolution against all double combinations and single constituents within 1–8 days, consistent with prior work 20,45 (Fig. 2). Viable cells were observed in all conditions above the initially determined MIC for the doubles and singles, but not for those conditions at or above the initial MIC for ME/PI/TZ. Increases in growth rates over time were noted in all doubles and singles, while the growth rate of N315 in sub-MIC ME/PI/TZ over time was unchanged throughout the experiment, equivalent to the no-drug control (Fig. 2) 20. Also, N315 exposed to the double combination ME/PI showed a threefold increase in MIC after day one, indicating that viable cells were present after day one, but did not grow until further passage and adaptation. Determination of the minimal-bactericidal concentration (MBC) confirmed that the triple combination ME/PI/TZ is bactericidal against MRSA N315 (Supplementary Table 4b). Together, these results demonstrate the suppression of emergence of new resistance against ME/PI/TZ in MRSA N315.
Figure 2. Change in growth rates over time of MRSA N315 when challenged with antibacterial combinations.
Growth rates of MRSA N315 over an 11-day period were computed for each antibacterial combination tested at one threefold dilution below MIC. The differences in growth rate (Δr) between day one (Initial Growth Rate) and the averaged rate of the last six days of the assay (Final Growth Rate) were calculated. MRSA N315 in conditions whose change in growth rate Δr >0.2 were considered to be adapted. The adaptation time parameter t-adapt was calculated as the time at which change in growth rate was half-maximal. Adaptation rate, α = (Δr /2)/t-adapt (1/h2), was computed for strains meeting this criterion. Results are from two replicate experiments. Adaptation rate for ME/PI: α = 8.23×10−3 h−2; ME/TZ: α = 8.68×10−4 h−2; PI/TZ: α = 4.32×10−3 h−2. Only ME/PI/TZ at one threefold dilution below MIC (1.2 µg/ml each) and No-drug Control displayed lack of increase in growth rate and were non-adapted.
Collateral sensitivities underlie resistance suppression
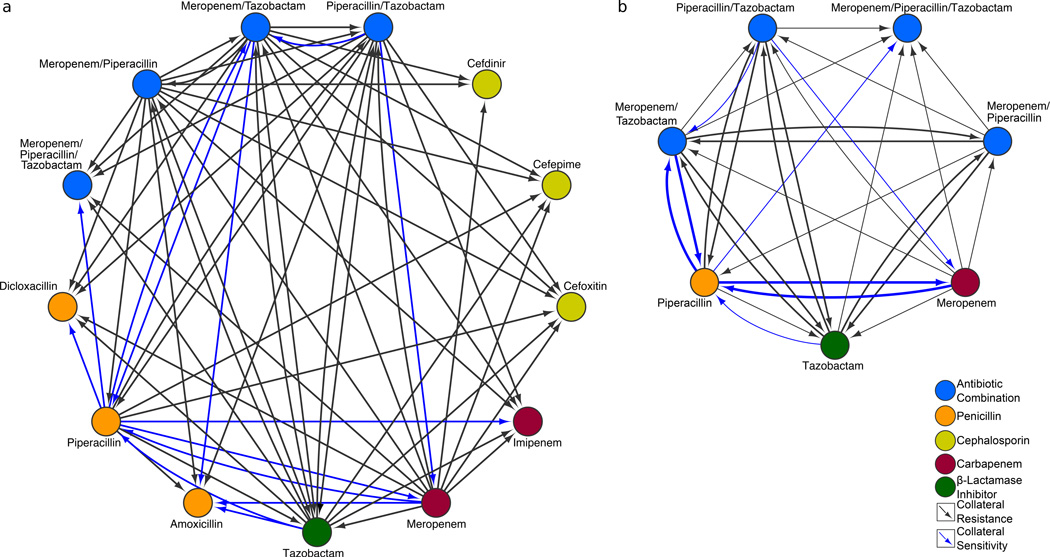
To determine whether collateral sensitivity was a factor in the suppression of adaptation of ME/PI/TZ, we analyzed the effects of prior exposure of MRSA N315 to a range of β-lactams on susceptibility to the other components (Fig. 3 and Supplementary Fig. 1). We observed that there was strong reciprocal collateral sensitivity between meropenem and piperacillin, and between piperacillin and ME/TZ, while PI/TZ sensitized MRSA N315 to meropenem, but not reciprocally. Collateral sensitivity to piperacillin was also conferred by prior exposure to tazobactam, but not vice-versa. Interestingly, we found no collateral sensitivity to tazobactam after exposure to any other single or double compounds. Collateral sensitivity and resistance profiles of amoxicillin and piperacillin are nearly identical, with adaptation to meropenem also sensitizing MRSA N315 to amoxicillin (Fig. 3 and Supplementary Fig. 1). Piperacillin also showed collateral sensitization to imipenem, an even more potent carbapenem against MRSA N315. However, none of the cephalosporins tested for collateral sensitivity by the carbapenem/penicillin/β-lactamase inhibitor combinations or constituents resulted in sensitivity. For this subclass of β-lactams, we noted only increased resistance or indifference. These results confirm that the observed suppression of resistance by collateral sensitivity is specific to the constituent drug classes of ME/PI/TZ.
Figure 3. Collateral sensitivities underlie suppression of adaptation to antibacterial combinations in MRSA N315.
a, MRSA N315 interaction network of collateral sensitivities and resistance between ME/PI/TZ, its single and double constituents, and other β-lactam compounds of various sub-classes (cephalosporins, penicillins, carbapenems, and β-lactamase inhibitors). Node colors indicate sub-classes of β-lactams, β-lactamase inhibitors, or combinations. Blue arrows indicate collateral sensitivities. Black lines indicate collateral resistance. For example, adaptation to piperacillin sensitizes MRSA N315 to meropenem and imipenem. Cephalosporins were not collaterally sensitive to any of the compounds we tested. Where pairs were not tested or no collateral effects were seen, no connecting arrows are shown. b, MRSA N315 interaction network of collateral sensitivities and resistance between ME/PI/TZ and its single and double constituents only. Bold blue arrows indicate reciprocal collateral sensitivities between two nodes, e.g., piperacillin and meropenem/tazobactam.
Genomic alterations of adapted MRSA N315
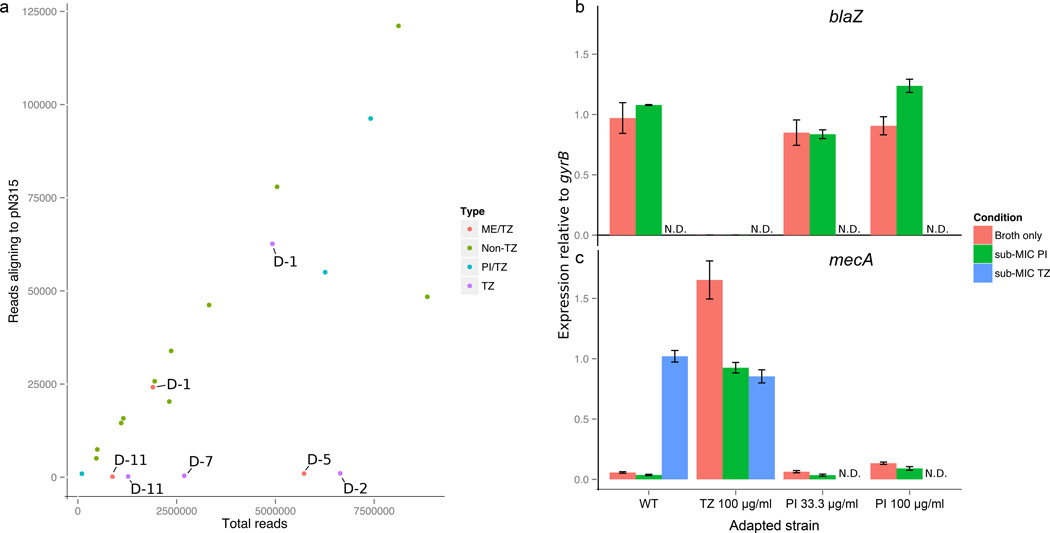
We used whole-genome sequencing to investigate the genomic basis of the sensitivity and resistance phenotypes of wild-type and adapted MRSA N315 strains. We found no mutations in PBP or β-lactamase genes within any of the adapted MRSA N315 isolates. However, absence of read coverage identified that the penicillinase plasmid pN315 was lost in isolates adapted to tazobactam-only (100 µg/ml) and ME/TZ (11.1 µg/ml each) (Fig. 4a). This plasmid loss occurred much more rapidly than with previously reported techniques for curing plasmids from MRSA, such as high heat and SDS treatment 46. In PI/TZ adapted isolates, we observed that approximately 400 kb of the MRSA N315 chromosome (GenBank ID: BA000018.3) was duplicated after analysis of read coverage depth, from approximate genomic positions 2,100,000 to 2,550,000 bp. Interestingly, this interval contains several putative and confirmed genes involved in cell-wall synthesis, including ddlA D-Ala-D-Ala ligase (Supplementary Fig. 2).
Figure 4. Genomic evidence for mechanisms of synergy and collateral sensitivity.
a, Adaptation of MRSA N315 to meropenem/tazobactam or tazobactam alone destabilizes plasmid pN315. Read coverage aligning to pN315 in MRSA N315 adapted to drug combinations containing tazobactam (TZ) or not containing tazobactam (non-TZ), versus total reads per sample. Days of adaptation under the given conditions are indicated, e.g., D-2 indicates isolate was sequenced after two days of adaptation. b, c, qRT-PCR confirms dysregulation of the bla and mec operons as causative mechanisms of some collateral sensitivities in MRSA N315. Expression of blaZ or mecA shown relative to gyrB in wild-type MRSA N315 or adapted strains (N315 adapted to TZ 100 µg/ml, and PI 33.3 or 100 µg/ml), subsequently grown in broth-only or broth + sub-MIC PI or TZ. N.D. = Not determined. ”−” indicates no expression. Loss of blaZ expression in MRSA N315 adapted to TZ confirms loss of blaZ and the bla operon, and is consistent with dysregulation of mecA expression. Data are from three replicate experiments. Error bars indicate standard error of measurement.
The loss of pN315 in MRSA N315 correlates with increased sensitivity to piperacillin and amoxicillin, both penicillins that should be sensitive to the blaZ (PC1) class A β-lactamase encoded on the plasmid. However, the loss of pN315 also results in increased resistance to tazobactam-only and ME/PI/TZ (Fig. 3, Supplementary Fig. 1, Supplementary Table 6a). One possible link between the presence of pN315 and ME/PI/TZ activity is the known regulatory crosstalk between MecI and BlaI repressors and their shared mec operon target 47–49. To test the effect of the loss of pN315 on expression of genes known to be important for ME/PI/TZ activity, we performed qRT-PCR analysis of the adapted and wild-type MRSA N315 strains (Fig. 4b). We determined that expression of the blaZ β-lactamase in the pN315 plasmid within wild-type MRSA N315 is constitutive, but in clones adapted to tazobactam we saw no expression of blaZ, consistent with loss of pN315 in these clones. We also found that expression of mecA is constitutive in the blaZ-null MRSA N315 isolate that was adapted to tazobactam at 100 µg/ml, consistent with dysregulation of the mec operon via loss of pN315 and the bla operon. Finally, we found tazobactam to be a strong inducer of mecA in wild-type MRSA N315, at levels similar to the constitutive expression of mecA seen in the blaZ-null condition.
ME/PI/TZ has synergy against MRSA with evolved resistance
We then examined the role that resistance to components of ME/PI/TZ has on its effectiveness against MRSA (Supplementary Table 6a). Previous exposure of MRSA N315 to piperacillin at either 33.3 or 100 µg/ml showed subsequent sensitization of the strain to ME/PI/TZ, from 3.7 to 1.2 µg/ml for each component. However, prior exposure of MRSA N315 to ME/TZ (11.1 µg/ml each) or meropenem-only (33.3 µg/ml) showed a nine-fold increase in levels of resistance to ME/PI/TZ (increasing from 3.7 to 33.3 µg/ml for each component). Exposure to tazobactam-only gave intermediate gains in resistance to ME/PI/TZ up to day 7 (11.1 µg/ml each), and higher resistance at day 11 (33.3 µg/ml each). Prior exposure to ME/PI or PI/TZ generated only a threefold increase in MIC (from 3.7 to 11.1 µg/ml) over the 11 days.
Despite the elevated MICs to ME/PI/TZ in the isolates adapted to the component drugs, the triple-drug combination still maintained synergy in all adapted isolates (Supplementary Table 6b). This is consistent with synergistic drug activity within the range of ME/PI/TZ MICs observed for the 72 clinical MRSA isolates (Supplementary Table 4), relative to their single-drug MICs. These results show that even when genomic changes enabling sub-component resistance can be selected, the overall synergistic activity of the triple-drug combination is maintained. In contrast to recent work with a non-pathogenic E. coli strain 30, we observed no change in the overall drug interaction profile of ME/PI/TZ regarding synergy with increased resistance to any component drug.
ME/PI/TZ is effective as linezolid against MRSA in vivo
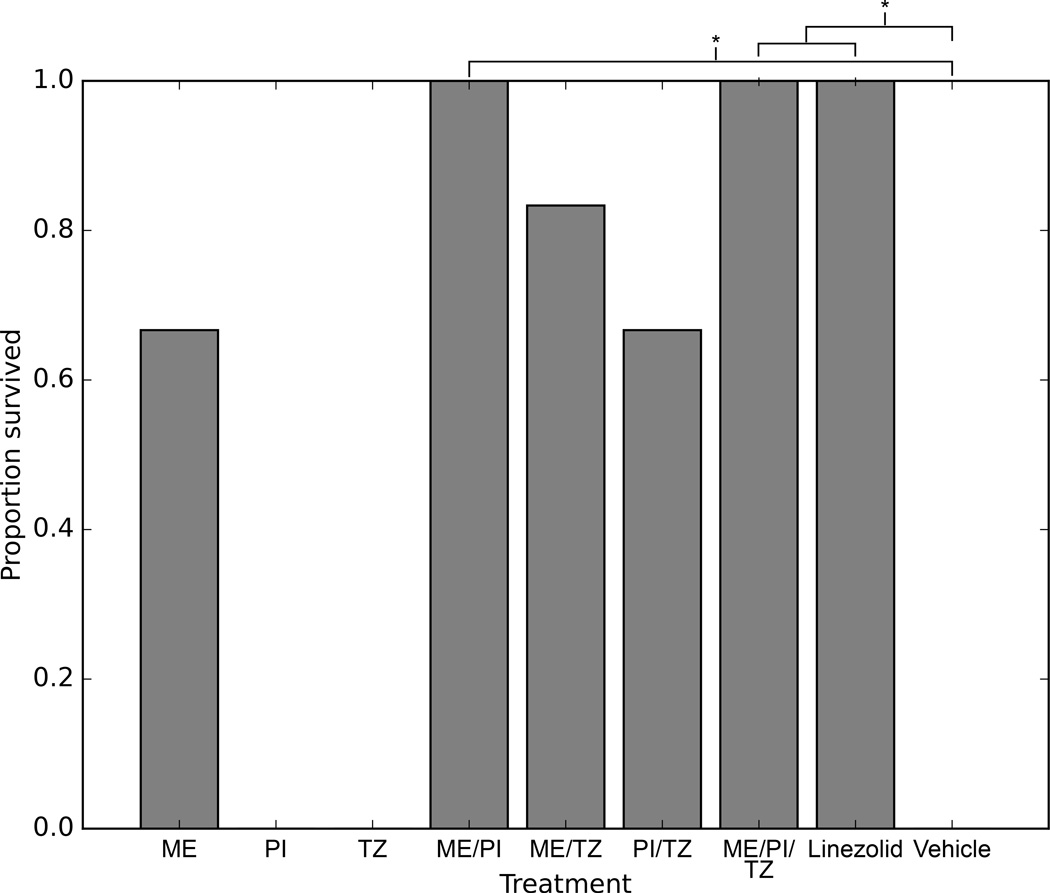
Next we tested if ME/PI/TZ or its constituents can be effective in treating MRSA infections in vivo using a neutropenic mouse model of peritonitis. Blood taken at 11 h post-infection from mice that were treated with either ME/PI/TZ, ME/PI (67 mg/kg each) or linezolid (30 mg/kg) yielded zero plated colonies and no growth in liquid cultures, indicating clearance of infection (Fig. 5, Supplementary Fig. 4, Supplementary Dataset 1). All mice (n = 6/group) from each of these treatments survived for six days post-infection (total duration of the mouse study). The efficacy of ME/PI/TZ and ME/PI was similar to linezolid monotherapy based on clearance of MRSA infection and survival of all treated mice compared to vehicle (p = 0.02, Fisher’s exact test).
Figure 5. Efficacy of ME/PI/TZ treatment in a neutropenic mouse peritonitis model of MRSA N315.
Neutropenic (cyclophosphamide-treated), 6–8 week old, outbred, ICR female mice were infected intraperitoneally with a 0.5 ml inoculum of 1 × 107 CFU/ml MRSA N315 in 5% mucin. Replicate mice (n = 6 per treatment) received one of 8 antibiotic treatments or vehicle. Proportional survival of mice from each drug treatment after 6 days is shown. Treatment with ME/PI/TZ, ME/PI, and linezolid are significantly different than vehicle (*p = 0.02, Fisher’s exact test, Bonferroni correction). ME, meropenem; PI, piperacillin; TZ, tazobactam.
In contrast to the complete rescue of the infected mice by ME/PI/TZ, ME/PI, or linezolid, several mice treated with ME/TZ, PI/TZ, or meropenem-alone, and all mice treated singly with piperacillin or tazobactam succumbed to the infection, most within 48 h (Fig. 5, Supplementary Dataset 1). Treatment with these other drug regimens was not significantly different than treatment with vehicle (p > 0.05, Fisher’s exact test) (Supplementary Table 7a), where all mice also succumbed to the infection within 48 h.
We tested MRSA N315 cultures from blood drawn from mice treated with meropenem, piperacillin, or vehicle for their in vitro MICs against ME/PI/TZ and its constituent single drugs to determine whether adaptation occurred during passage in vivo. All four tested isolates of MRSA N315 had identical MICs for the triple ME/PI/TZ and all constituent drugs, and thus identical synergy (Supplementary Table 7b). These data suggest no adaptation occurred within these strains to overcome the triple ME/PI/TZ tested within the 11-h passage in vivo.
Discussion
We have shown that triple antibacterial combinations containing carbapenems, penicillins, and β-lactamase inhibitors target multiple nodes in the same cellular system (cell-wall synthesis) and are highly synergistic and bactericidal against diverse MRSA strains in vitro, at clinically achievable concentrations. This contrasts with recent work showing collateral sensitivity and synergy to arise from combinations of drug classes working against orthogonal cellular targets in non-pathogenic lab strains only 22,30. Because carbapenems and other drugs at high concentration could have toxic effects, reduced per-drug dosages via synergy mitigate potential toxicities 50. Our 3-D checkerboard testing confirmed the optimal input concentrations for ME/PI/TZ to be given in a 1:1:1 ratio (2 µg/ml each) against MRSA N315, which is below the susceptibility breakpoints for these compounds against methicillin-susceptible S. aureus, and is an 8–64-fold reduction in input concentrations for these formerly inactive drugs against this highly resistant MRSA strain. Our mechanistic analyses support our hypothesis that targeting of PBP1 by meropenem, targeting of PBP2 by piperacillin, protection of piperacillin by tazobactam from β-lactamase cleavage, and allosteric opening of the active site of PBP2a by meropenem for inhibition by another molecule of antibiotic in the combination, result in synergy by simultaneously perturbing multiple components of the MRSA cell-wall synthesis system (Supplementary Fig. 3).
We have also shown that this combination has activity in a highly lethal neutropenic MRSA in vivo model, demonstrating that this triple combination of clinically approved β-lactams can clear infection similar to substantially more expensive monotherapies like linezolid. The plasma levels of meropenem observed in mice correlate well with plasma drug levels in healthy humans 51, and meropenem would attain the Kd at these clinically achievable concentrations to trigger allostery for opening of the active site of PBP2a, providing accessibility for inhibition by meropenem and other β-lactams in the combination 7,8.
Notably, the double combination ME/PI cleared the MRSA N315 infection in vivo similarly to ME/PI/TZ and linezolid within 11 h. In vitro we observed high synergy scores and reciprocal collateral sensitivity for this combination, similar to what we saw for ME/PI/TZ, but ME/PI did not suppress evolution of resistance to the same extent that ME/PI/TZ did. This property may not have been relevant to this aggressive infection model, but may be important for longer treatment times seen in human infections with MRSA. ME/PI/TZ is also likely to be effective at lower total concentrations than ME/PI because of its higher synergy. Longer exposure of the N315 strain to the tazobactam component of ME/PI/TZ in vivo may also promote ejection of pN315 plasmid with concomitant sensitization to the penicillin component, in line with the in vitro results for collateral sensitivity and suppression of adaptation. Indeed, to more adequately address this question, potential longer-term in vivo resistance evolution would need to be tested under sub-lethal concentrations of the drugs in important follow-up mouse experiments.
Our robust mechanistic in vitro results and preliminary in vivo results for ME/PI/TZ activity suggest this combination may be made immediately available for use in the clinic, since it includes currently FDA-approved drugs, which had met their obsolescence as therapies against MRSA decades ago. However, further mechanistic features of the combination that were shown in vitro (synergy, resistance suppression over longer periods of dosing, collateral sensitivity, etc.) will require substantially more in vivo testing to support the promising but preliminary activity observed in our highly aggressive neutropenic mouse model.
We note that high resistance to meropenem or tazobactam slightly reduces the effectiveness of ME/PI/TZ, while maintaining its synergy, and our resistance evolution analysis cannot account for resistance genes acquired horizontally that could break the relationship between meropenem, piperacillin, and tazobactam. Despite these caveats, we believe the ME/PI/TZ combination is an immediately viable anti-MRSA therapeutic, and endorse further mechanistic exploration into the putative superior efficacy of high-order antibiotic combinations that are both synergistic and encoded by collaterally sensitive constituents. Having similar activity to linezolid against MRSA in vivo, the potential efficacy of ME/PI/TZ reopens broad prospects for the clinical use of β-lactams against the staphylococci. It also suggests that this line of research into repurposing existing antibiotics in carefully designed synergistic combinations would address immediate clinical needs, as these agents are already approved for human use. Emergence of resistance to any antibiotic or any antibiotic combination is inevitable. Yet, as evidenced in our study, combinations composed of key drug-drug interaction features may be a tool in mitigating the emergence of antibiotic resistance by preserving the usefulness of existing agents available to us in our pharmacological armamentarium.
Online Methods
Microbiological Studies
MRSA N315 was a gift from Dr. Steven Gill, University of Rochester, Rochester, NY, USA. S. aureus ATCC 29213 was acquired from the American Type Culture Collection. 72 de-identified clinical MRSA isolates were selected at random from the clinical isolate strain bank at Barnes-Jewish Hospital, St. Louis, MO, USA. Minimal inhibitory concentration (MIC) assays for inhibition of growth were performed following the recommendations of the Clinical and Laboratory Standards Institute (CLSI) 37. Briefly, 23 antibacterial compounds (Supplementary Table 1) were selected based on coverage of all major drug classes, including three compounds not classified as antibiotics for human use, but with known antibacterial properties. Compounds were dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 50 mg/ml. Exceptions: Sulfometuron at 20 mg/ml in DMSO; Tobramycin, D-cycloserine, and colistin at 50 mg/ml in H2O and filtered at 2 µm. The 23 compounds were formulated into all 253 possible unique pairwise combinations at fixed ratios and at 100× concentrations in solvent. To increase the range of concentrations assayed for possible synergistic or antagonistic drug interactions (>2,000-fold), the drug stocks were arrayed at 20 µl volumes into threefold dilution series down eight rows in 96-well (Costar) master drug plates, using a BioMek FX robotic liquid handler (Beckman Coulter, Inc.). Drugs were then mixed 1:100 into 96-well plates containing 200 µl/well of cation-adjusted Mueller-Hinton broth (CAMHB). All drug susceptibility assay wells were inoculated with ~1 µl of mid-log phase MRSA N315 bacterial culture at 0.5 McFarland standard (~2 × 108 CFU/ml) using a sterile 96-pin replicator (Scinomix), the plates sealed with Breathe-Easy membranes (Sigma-Aldrich Co.) and placed into a sealable plastic bag containing a moist paper towel, and grown at 37 °C without shaking for 24 h. Endpoint growth at 37 °C after 24 h was determined by optical density at 600 nm ≥0.1 using a Synergy H1 reader (BioTek, Inc.).
Synergy of antibiotic combinations was determined using the fractional inhibitory concentration index (FICI) method 52,53. By this method, the MIC of the antibiotic compound in combination is divided by the MIC of the compound alone, yielding the fractional contribution of each drug component in the combination. Quotients for all compounds in a combination are summed and drug interactions scored using the formula:
Select pairwise combinations against MRSA were then combined with each of the 21 remaining single drugs to make triple combinations, formulated and tested in identical fashion to the double combinations. Synergy of combinations was confirmed via triplicate measurements of drug conditions at the MIC. Based on its high synergy against MRSA N315 in the sparse screening, ME/PI/TZ and its constituents were selected for further characterization. Final susceptibility testing of ME/PI/TZ and its components was performed in triplicate 96-well plates with a 3-D checkerboard assay. Master plates of each single drug were made at 300× concentrations in solvent, combined into 8 final master plates at 100×, with each of the master plates comprising a standard 2-D checkerboard of meropenem and piperacillin at increasing twofold concentrations across wells, and containing a fixed twofold dilution concentration of tazobactam in all wells. These master plates were then mixed 1:100 into deep-well 96-well plates (USA Scientific, Inc.) containing 600 µl/well of cation-adjusted Mueller-Hinton broth (CAMHB), and split into triplicate plates resulting in 8 × twofold dilutions for each drug component from 128 to 0 µg/ml in CAMHB, at 200 µl/well. In this manner, varying concentrations of all three drugs are tested against each other, generating MIC determinations of three-, two-, and single-drug MICs. Plates were inoculated with MRSA N315 culture as above using a sterile 96-pin replicator, plates sealed with Breathe-Easy membranes and placed into a sealable plastic bag containing a moist paper towel, and grown at 37 °C without shaking for 24 h. Endpoint growth at 37 °C after 24 h was determined by optical density at 600 nm ≥0.1 using a Synergy H1 reader.
Minimal bactericidal concentration (MBC) for ME/PI/TZ in MRSA N315 was determined via duplicate wells of ME/PI/TZ at indicated concentrations in CAMHB media, inoculated with MRSA N315 in mid-log phase as above using a sterile 96-pin replicator, plates sealed with Breathe-Easy membranes and placed into a sealable plastic bag containing a moist paper towel, and grown at 37 °C without shaking for 24 h. Endpoint growth at 37 °C after 24 h was determined by optical density at 600 nm ≥0.1 using a Synergy H1 reader. 100 µl of a 1:100 dilution of 50 µl drawn from duplicate ME/PI/TZ wells was plated on non-selective Mueller-Hinton agar (MHA) plates and incubated overnight for 24 h. No colony growth at or two dilutions above the MIC confirmed bactericidal activity, as defined by CLSI 54. Meropenem (CAS 96036-03-2) and clavulanate (CAS 61177-45-5) were obtained from AK Scientific, Inc. Piperacillin (CAS 59703-84-3), tazobactam (CAS 89786-04–9), imipenem (CAS 74431-23-5), amoxicillin (CAS 26787-78-0), cefdinir (CAS 91832-40-5), cefepime (CAS 123171-59-5), cefoxitin (CAS 33564-30-6), and dicloxacillin (CAS 13412-64-1) were obtained from Sigma-Aldrich Co.
Antibiotic Hypersusceptibility Assays Against MRSA PBPs
Assays were performed as previously described 26. Briefly, antisense-RNA constructs targeting PBPs in MRSA COL (PBP2a, PBP1, PBP2, and PBP3), were each cloned into vector pTET15 with a xylose-inducible promoter, and selected with 34 µg/ml of chloramphenicol. LB-Miller agar plates ± 50 mM xylose were seeded at 48 °C with 107 CFU MRSA COL containing vector + antisense construct, were cooled and then were exposed to several concentrations of single antibiotic drugs (meropenem, piperacillin, tazobactam), fixed double combinations of same drugs, or ME/PI/TZ. Zones of inhibition (ZOI) were measured in triplicate. Differential ZOI between + xylose and − xylose were scored as follows: +++, more than twofold increase in zone diameter with xylose induction; ++, twofold increase in zone diameter; +, less than twofold increase; −, no change in zone diameter.
Adaptation and Cross-resistance Assays
Meropenem, piperacillin, and tazobactam stocks were dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 10 mg/ml. Combinations of doubles ME/PI, ME/TZ and PI/TZ, and the triple ME/PI/TZ were formulated into final stock concentrations of 3.33 mg/ml each (100×) in DMSO. The drug stocks were arrayed at 20 µl volumes into threefold dilution series down eight rows in 96-well (Costar) master drug plates, using a BioMek FX robotic liquid handler (Beckman Coulter, Inc.). Drugs were then mixed 1:100 into deep-well 96-well plates (USA Scientific, Inc.) containing 1,980 µl/well of cation-adjusted Mueller-Hinton broth (CAMHB). All combinations were arrayed in duplicate, and all single drugs singly, plus allowance for (+) and (−) bacterial growth controls in quadruplicate. Top concentrations of all drug combinations were 33.3 µg/ml for each component, while top concentrations for single drugs was 100 µg/ml. From the 96-well 2 ml deep-well plate, 13 × 96-well (Costar) plates (150 µl/well) were aliquoted, sealed with sterile foil seals (VWR International LLC), and stored at –80 °C. For each day of the adaptation assay, one 96-well (Costar) plate (150 µl/well) was thawed at ~25 °C, shaken for 1 min on a tabletop plate shaker (Thermo Fisher Scientific, Inc.), and briefly centrifuged before removing the sterile foil. On day zero, all drug susceptibility assay wells from Plate 1 were inoculated with ~1 µl of mid-log phase MRSA N315 bacterial culture at 0.5 McFarland standard (~2 × 108 CFU/ml) using a sterile 96-pin replicator (Scinomix), the plate sealed with Breathe-Easy membrane (Sigma-Aldrich Co.) and grown at 37 °C with constant fast-linear shaking (562 cpm) for 24 h using a PowerWave HT reader (BioTek, Inc.). Growth was determined by optical density at 600 nm every 24 min. At 24 h (end of passage-day one), the plate was removed from the reader, unsealed with sterile technique, and ~1 µl of the well contents transferred to an identical thawed 96-well plate, using a sterile 96-pin replicator. Each well of the 24 hour growth plate was filled with 150 µl of sterile CAMHB+30% glycerol and the plate stored at –80 °C for later analysis. This process was repeated for a total of 11 days, generating 11 × 96-well plates of MRSA N315 under adaptation to all combinations and single drugs comprising ME/PI/TZ. To test for cell viability, at the end of the assay on day 11, all wells from the final plate were pinned with a sterile 96-pin replicator and transferred to a 96-well (Costar) plate containing CAMHB only.
From each plate over the 11-day adaptation assay, the absorbance at OD600 from each 24-min time point for duplicate wells from each drug combination at each threefold dilution was averaged, and the natural logarithm (ln) was calculated for each averaged absorbance. The logarithmic growth phase of the cultures was linearized by this method and plotted in Excel. Growth rate r was determined by determining difference in linearized absorbance between start and end of logarithmic growth phase (ΔA), determining time points of those absorbances (Δt), and taking the quotient [ΔA/Δt = r]. Growth rate over time was plotted in Excel. The differences in growth rate (Δr) between day one (Initial Growth Rate) and the averaged rate of the last six days of the assay (Final Growth Rate) were calculated. Following Hegreness et al 20, (Δr/2) was used to determine the adaptation time parameter t-adapt, the time at which change in growth rate was half-maximal. Those wells containing cells in drug conditions whose differences in growth rates were >0.2 h–1 between day one and the average of the last six days of growth were considered significantly adapting to conditions and an adaptation rate, α = (Δr /2)/t-adapt (1/h2), was computed for strains meeting this criterion.
For the antibiotic cross-resistance/susceptibility analysis, adapted isolates were chosen from the appropriate wells for each combination or single compound of the day-11 plate. Frozen isolates were streaked out on MHA plates and grown at 37 °C for 24 h to obtain single colonies, then distinct single colonies were picked, and re-grown at 37 °C for 24 h in antibiotic-containing broth conditions identical to those in which they grew originally on the day-11 plate. The adapted MRSA N315 cultures in mid-log phase were diluted to 0.5 McFarland standard, then re-inoculated in sterile antibiotic-containing 96-well Costar plates as with the original the adaptation plates, containing CAMHB with triplicate threefold dilutions of ME/PI/TZ, its constituent double combinations and single drugs, as well as the following beta-lactam drugs: amoxicillin (AX), cefdinir (CF), cefepime (CP), cefoxitin (CX), dicloxacillin (DC), and imipenem (IM), using a sterile 96-pin replicator, plates sealed with Breathe-Easy membranes and placed into a sealable plastic bag containing a moist paper towel, and grown at 37 °C without shaking for 24 h. Endpoint growth at 37 °C after 24 h was determined by optical density at 600 nm ≥0.1 using a Synergy H1 reader. As MRSA N315 grown with ME/PI/TZ showed no increase in growth rate, no adapted isolates were present for that combination. MICs of the adapted MRSA N315 with these drug combinations or single compounds were compared to the MIC of the drug(s) for wild-type MRSA N315, and the fold-differences in MIC were plotted in a heatmap using Plotly.
Expression profiling with qRT-PCR
Wild-type and adapted MRSA N315 isolates were grown in triplicate in 100 ml flasks to mid-log phase in CAMHB ± piperacillin at 11.1 µg/ml or tazobactam at 33.3 µg/ml. To harvest cells at mid-log phase, each culture flask was split into 2 × 50 ml screw-cap tubes, spun down at 4 °C for 10 min at 3500 rpm, supernatant removed, and pellets combined carefully with a 2 ml serological pipette. 1 ml RNAprotect Bacteria Reagent (Qiagen) was added to pellets to stabilize the RNA, vortexed briefly, and incubated for 5 min at RT. After incubation, tubes were spun again at 4 °C for 10 min at 3500 rpm, supernatant removed, and the pellets were stored at –80 °C.
Total RNA was extracted by resuspending cell pellets in Buffer B (200 mM NaCl, 20 mM EDTA), addition of 20% SDS, acid-washed sterile glass beads (Sigma, Inc.), phenol:chloroform:IAA, and bead-beating on ‘high’ for 5 min. Extraction mix was spun at 8000 rpm at 4 °C for 3 min (to separate the phases), and top aqueous phase was transferred into a new tube. RNA was precipitated with isopropanol and 3M NaOAc, mixed thoroughly by inversion, then spun at 4 °C at max rpm for 10 min, and supernatant was aspirated. The pellet was washed with ice-cold 70% EtOH, spun at max rpm at 4 °C for 5 min, then supernatant was aspirated and pellet was air-dried. The pellet was resuspended in nuclease-free water and incubated in a 50 °C heat block, vortexing periodically. To remove DNA contamination, TURBO-DNase buffer (Ambion, Inc.) and RNase-free TURBO-DNase was added to each sample, incubated at 37 °C for 30 min, and purified using MEGAClear columns and kit per manufacturer protocol. Samples were repurified using Baseline-ZERO DNase buffer (Epicentre, Inc.) and Baseline-ZERO DNase, following manufacturer protocol. Final RNA samples were eluted with 30 µl TE buffer, pH 7.0.
First-strand cDNA was synthesized from total RNA with SuperScript First-Strand Synthesis System for RT-PCR (Life Technologies). qRT-PCR of pbp2, mecA and blaZ in MRSA N315 was performed against gyrB using SYBR Select Master Mix for CFX (Life Technologies) on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.). Primer sequences used (0.3 µM each): pbp2_F: CGTGCCGAAATCAATGAAAGACGC, pbp2_R: GGCACCTTCAGAACCAAATCCACC; mecA_F: TGGAACGATGCCTATCTCATATGC, mecA_R: CAGGAATGCAGAAAGACCAAAGC; blaZ_F: TTTATCAGCAACCTTATAGTCTTTTGGAAC, blaZ_R: CCTGCTGCTTTCGGCAAGAC, gyrB_F: CGATGTGGATGGAGCGCATATTAG, gyrB_R: ACAACGGTGGCTGTGCAATATAC. CFX protocol: 2 min @ 50 °C, 2 min @ 95 °C, (15 s @ 95 °C, 1 min @ 60 °C) × 40 cycles. Gene expression was determined using the ΔΔCt method of normalized quantitation 55, where Ct indicates the cycle number at which exponential growth phase increases above threshold fluorescence signal.
Sequencing library preparation
Genomic DNA (gDNA) was extracted from wild-type and adapted MRSA N315 using lysostaphin digestion and phenol:chloroform:IAA extraction. 1 ml aliquots from overnight 5 ml shaking cultures of S. aureus strains were spun down at 13,000 rpm for 3 min, media poured off, and repeated with an additional 1 ml of culture. Cell pellets were resuspended in Buffer A (NaCl 200 mM, Tris 200 mM, EDTA 20 mM) at 4 °C and vortexed briefly. 2.5 µl of 10 mg/ml (200×) lysostaphin (Sigma-Aldrich, Inc.) was added to tubes, flick-mixed and spun down, then incubated in a 37 °C dry bath for 1 h. 0.1 mm zirconium beads (BioSpec Products, cat# 1107910), 20% SDS, and phenol:chloroform:IAA (25:24:1, pH 7.9) were added to the tubes, and samples chilled on ice. Cells were lysed by bead-beating on the "homogenize" setting for 4 min (beat 2 min, ice 2 min, beat 2 min), spun at 6800 rcf (4 °C) for 3 min, and aqueous phase (~500 µl) transferred to pre-spun phase-lock gel tubes (5Prime, cat#2302820). An equal amount (500 µl) of phenol:chloroform:IAA (25:24:1, pH 7.9) was added to tubes and mixed by inversion. Tubes were spun at max speed (20,800 rcf) (RT) for 5 min, aqueous phase transferred (~500 µl) to a new Eppendorf tube, and precipitated with –20 °C isopropanol and 1/10 volume of 3M NaOAc at pH 5.5 (Ambion, AM9740), and mixed thoroughly by inversion. Mixture was stored at –20 °C overnight, then spun at max speed at 4 °C for 20 min. Pellet was washed with 100% EtOH (RT) and spun down at 4 °C for 3 min, EtOH was carefully drawn off, and pellet air-dried for 15 min. Pellet was resuspended in 30 µl of TE (Ambion, AM 9861), incubated at 50 °C for 5 min, then DNA was purified with QIAGEN QIAQuick PCR purification columns with the following modifications: RNase A treatment at beginning of column clean-up with 4 µl Qiagen RNase (100 mg/ml) combined with every 300 µl buffer PB used, incubating in buffer PB/RNase for 15 min at RT. PE wash buffer holding in column at RT for 2 min, eluting gDNA with 35 µl of EB buffer pre-heated to 55 °C, letting sit for 1 min before final spin.
500 ng of total DNA from each genome was sheared to ~300 bp fragments in nine rounds of shearing of ten min each on the BioRuptor XL. In each round the power setting was ‘H’ and samples were treated for 30 s and allowed to rest for 30 s. Each sample was concentrated using the Qiagen MinElute PCR purification kit per the manufacturer’s protocol. End Repair of the sheared DNA fragments was initiated with the addition of 2.5 µl of T4 DNA ligase buffer with 10 mM ATP (NEB, B0202S), 1 µl of 1 mM dNTPs (NEB), 0.5 µl T4 Polymerase (NEB, M0203S), 0.5 µl T4 PNK (NEB M0201S), and 0.5 µl Taq Polymerase (NEB, M0267S). This mixture was incubated at 25 °C for 30 min, then at 75 °C for 20 min. Barcoded adapters were then added to the solution along with 0.8 µl of T4 DNA ligase (NEB, M0202M), for the purpose of ligating the adapters to the DNA fragments. This solution was then incubated at 16 °C for 40 min, then 65 °C for 10 min. The adapter-ligated DNA was then purified using the Qiagen MinElute PCR purification kit per the manufacturer’s protocol.
The DNA fragments were then size selected on a 2% agarose gel in 1X TBE buffer stained with Biotium GelGreen dye (Biotium). DNA fragments were combined with 2.5 µl 6× Orange loading dye before loading on to the gel. Adapter-ligated DNA was extracted from gel slices corresponding to DNA of 250–300 bp using a QIAGEN MinElute Gel Extraction kit per the manufacturer’s protocol. The purified DNA was enriched by PCR using 12.5 µl 2× Phusion HF Master Mix and 1 µl of 10 µM Illumina PCR Primer Mix in a 25 µl reaction using 1 µl of purified DNA as template. DNA was amplified at 98 °C for 30 s followed by 18 cycles of 98 °C for 10 s, 65 °C for 30 s, 72 °C for 30 s with a final extension of 5 min at 72 °C. The DNA concentration was then measured using the Qubit fluorometer and 10 nmol of each sample (up to 106 samples per lane of sequencing) were pooled. Subsequently, samples were submitted for Illumina HiSeq-2500 Paired-End (PE) 101 bp sequencing at GTAC (Genome Technology Access Center, Washington University in St. Louis) at 9 pmol per lane.
DNA Sequence analysis
Alignment and variant calling
For the wild-type and adapted MRSA N315, all sequencing reads for each genome were de-multiplexed by barcode into separate genome bins. Reads were quality trimmed to remove adapter sequence and bases on either end with a quality score below 19. Any reads shorter than 31 bp after quality trimming were not used in further analysis. All reads were mapped to the Staphylococcus aureus subsp. aureus N315 chromosome (GenBank ID: BA000018.3) and pN315 plasmid (GenBank ID: AP003139). (Command: bowtie2 –x <reference_genome_index_name> −1 <forward_read_file> −2 <reverse_read_file> -q --phred33 --very-sensitive-local -I 200 -X 1000 -S <sam_output>). Variants from the reference were called using samtools 56 (Commands: samtools view -buS <sam_file> | samtools sort -m 4000000000 - <sample_prefix> ### samtools index <bam_file> ### samtools mpileup -uD -f <reference_genome> <bam_file> | bcftools view -bcv - > <bcf_file> ### bcftools view <bcf_file>). The variant call format (VCF) file was then filtered to remove SNPs with a quality score lower than 70 or coverage greater than twice the average coverage expected per base. Absence of read coverage or overabundant read coverage indicated plasmid loss or large duplication respectively. Any variant position found from the wild-type alignment was determined to be a result of alignment error or to be derived from lab specific drift in N315 and was removed from all other VCF files. Each variant position was then compared to known ORF locations in N315 to search for causal variants.
Code availability
All of the software used in this study is free and publicly available, except for Geneious, which requires a license from Biomatters Ltd (http://www.biomatters.com).
In vivo mouse model of MRSA infection
Animals
Outbred ICR female mice (6–8 weeks old, 17–25 g body weight; Harlan Laboratories, Inc.) were used. Mice were given Teklad 2019 Extruded Rodent Diet (Harlan Laboratories, Inc.) and water ad libitum. Mice were maintained in polycarbonate shoebox cages containing corncob (The Andersons, Inc.) and Alpha-dri (Shepherd Specialty Papers, Inc.) bedding under 12-h light/12-h dark cycle at 22 ± 1 °C. All procedures involving animals were approved by the University of Notre Dame Institutional Animal Care and Use Committee.
Neutropenic mouse peritonitis model of MRSA infection
Doses of cyclophosphamide (100 µl of 50 mg/ml in 0.9% saline corresponding to 200 mg/kg; Alfa Aesar, Inc.) were given intraperitoneally (IP) at 4 days and 1 day prior to infection. The S. aureus strain N315 was streaked onto Brain-Heart Infusion (BHI; Becton Dickson and Company) agar and grown overnight at 36 °C. The MRSA N315 bacterial inoculum was adjusted to approximately 1 × 108 CFU/ml (corresponding to OD540 = 0.5), then diluted to give 2 × 107 CFU/ml. A 10% porcine mucin (Sigma-Aldrich Co.) suspension was prepared and adjusted to pH 7. Immediately prior to infection, the bacterial inocula were diluted 1:1 with 10% mucin to a final concentration of 1 × 107 CFU/ml in 5% mucin. The mice were then infected IP with 0.5 ml of this inoculum. In vivo dosing of compounds in mice was compared with mean or range peak human plasma concentrations of studied β-lactams 38,40,41,57,58.
Antibiotic preparation
Meropenem was obtained from AK Scientific, Inc. Piperacillin and tazobactam were obtained from Sigma-Aldrich Co. Linezolid (CAS 165800-03-3) was obtained from AmplaChem. Antibiotics were dissolved at a concentration of 16.67 mg/ml in 30% DMSO/30% propylene glycol/40% water. Linezolid was used as positive control and was prepared at 7.5 mg/ml. Vehicle (30% DMSO/30% propylene glycol/40% water) was included as negative control. The dosing formulations were sterilized by passing through 0.2 µm filter prior to injection.
Bacterial isolation from blood
Blood samples were checked for bacterial growth by plating and liquid culture. Whole blood (100 µl, three samples per group) was spread onto Brain-Heart Infusion (BHI) agar plates and incubated at 36 °C overnight. Colonies were counted and three colonies were selected, grown overnight in liquid BHI culture at 36 °C, then mixed 1:1 with 30% LB-glycerol and stored at –80 °C. The remaining three blood samples of each group (50 µl) was added to 5 ml BHI broth and incubated overnight at 36 °C. When growth was noted, cultures were mixed 1:1 with 30% LB-glycerol and stored at –80 °C.
Statistical Analysis
Data for minimal inhibitory concentrations (MICs) of drugs against bacteria are derived from triplicate measurements in plate and broth culture. Adaptation data are taken from two replicate experiments for each drug combination condition. Data for qRT-PCR expression profiling are derived from three replicate experiments taken from three biological replicates each, with standard error of measurement calculated. Mice were treated with antibiotics or vehicle in groups of six (n = 6), as prior 59. Animal studies were not randomized or blinded, as the end point is either survival or death. Fisher’s exact test with Bonferroni correction was used for 8 independent tests (comparing each treatment to vehicle). Fisher’s exact test was used because planned experiments (with n = 6 mice) used small sample sizes, with independent tests.
Supplementary Material
Acknowledgments
We thank Rob Mitra for discussions regarding SNP calling and NGS data analysis; Meghan Wallace for MRSA SCCmec typing; Bin Wang for technical advice on genomic preparations and sequencing; Christian Munck, Morten Sommer, and Joseph Lehár regarding prior discussions of 23 antibiotics chosen for combinatorial screening; Jayne Marasa for screening optimization; Jason Fries for optimization of plate reader assays; Kevin Forsberg for discussions on mechanisms of reciprocal collateral sensitivity; Terence Crofts for antibiotic structures in figures; and members of the Dantas lab for helpful general discussion of the project and manuscript. We thank Dr. Terry Roemer of Merck Research Laboratories for the kind gift of the antisense strains. This work was supported in part by the NIH Director’s New Innovator Award (http://commonfund.nih.gov/newinnovator/), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK: http://www.niddk.nih.gov/), and the National Institute of General Medical Sciences (NIGMS: http://www.nigms.nih.gov/), of the National Institutes of Health (NIH) under award numbers DP2DK098089 and R01GM099538 to G.D. It is also supported in part by the National Institute of Allergy and Infectious Diseases (NIAID: http://www.niaid.nih.gov/) of the NIH under award numbers AI90818 to M.C. and S.M. and AI104987 to S.M. M.W.P. was supported by the NIGMS Cell and Molecular Biology Training Grant (GM007067). R.B. was supported by T32 GM075762 and by an individual Ruth L. Kirschstein National Research Service Award F31 AI115851 from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Accession codes
Sequence data reported in this paper are deposited at NCBI GenBank (BioProject ID PRJNA288150).
Author Contributions
P.R.G. designed the study, performed in vitro experiments, analyzed results, and wrote the paper. M.W.P. and C.-A.D.B. analyzed results and wrote the paper. R.B. performed in vivo experiments, analyzed results, and wrote the paper. A.B. and B.A.B. performed in vitro experiments and analyzed results. M.A.S., W.R.W., and V.A.S. performed in vivo experiments. S.M. and M.C. designed in vivo experiments, analyzed results, and wrote the paper. G.D. designed the study, analyzed results, and wrote the paper.
Competing Financial Interests Statement
The authors have submitted a Provisional U.S. Patent Application, Serial No. 62/190,588, based on the antibiotic combination results described in this study.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Sequence data reported in this paper are deposited at NCBI GenBank (BioProject ID PRJNA288150).
References for main text
- 1.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. The Lancet Infectious Diseases. 2011 doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 2.Davies J, Davies D. Origins and Evolution of Antibiotic Resistance. Microbiology and Molecular Biology Reviews. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuda CCS, Fisher JF, Mobashery S. Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cellular and molecular life sciences : CMLS. 2005;62:2617–2633. doi: 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature reviews. Microbiology. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malouin F, Bryan LE. MINIREVIEWS Modification of Penicillin-Binding Proteins of Beta-Lactam Resistance. Antimicrobial agents and chemotherapy. 1986;30:1–5. doi: 10.1128/aac.30.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuda C, Suvorov M, Vakulenko SB, Mobashery S. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. The Journal of biological chemistry. 2004;279:40802–40806. doi: 10.1074/jbc.M403589200. [DOI] [PubMed] [Google Scholar]
- 7.Fuda C, et al. Activation for Catalysis of Penicillin-Binding Protein 2a from Methicillin-Resistant Staphylococcus aureus by Bacterial Cell Wall. J Am Chem Soc. 2005;127:2056–2057. doi: 10.1021/ja0434376. [DOI] [PubMed] [Google Scholar]
- 8.Otero LH, Rojas-Altuve A, Llarrull LI, Carrasco-López C, Kumarasiri M. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16808–16813. doi: 10.1073/pnas.1300118110. http://doi:10.1073/pnas.1300118110/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1300118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villegas-estrada A, Lee M, Hesek D, Vakulenko SB, Mobashery S. Co-opting the Cell Wall in Fighting Methicillin-Resistant Staphylococcus aureus : Potent Inhibition of PBP 2a by Two Anti-MRSA β-Lactam Antibiotics. J Am Chem Soc. 2008;130:9212–9213. doi: 10.1021/ja8029448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long SW, et al. PBP2a mutations causing high-level Ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrobial agents and chemotherapy. 2014;58:6668–6674. doi: 10.1128/AAC.03622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid-resistant Staphylococcus. The Journal of antimicrobial chemotherapy. 2012 doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Hal SJ, Paterson DL, Gosbell IB. Emergence of daptomycin resistance following vancomycin-unresponsive Staphylococcus aureus bacteraemia in a daptomycin-naïve patient-a review of the literature. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2010 doi: 10.1007/s10096-010-1128-3. [DOI] [PubMed] [Google Scholar]
- 13.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. The New England journal of medicine. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 14.Bhusal Y, Shiohira CM, Yamane N. Determination of in vitro synergy when three antimicrobial agents are combined against Mycobacterium tuberculosis. International journal of antimicrobial agents. 2005;26:292–297. doi: 10.1016/j.ijantimicag.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Sjolund M, Wreiber K, Andersson DI, Blaser M, Engstrand L. Long-Term Persistence of Resistant Enterococcus Species after Antibiotics To Eradicate Helicobacter pylori. Ann Intern Med. 2013;139:483–488. doi: 10.7326/0003-4819-139-6-200309160-00011. [DOI] [PubMed] [Google Scholar]
- 16.Tupin A, et al. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. International journal of antimicrobial agents. 2010;35:519–523. doi: 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nature Genetics. 2011;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science (New York, N.Y.) 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmermann GR, Lehár J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug discovery today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Hegreness M, Shoresh N, Damian D, Hartl DL, Kishony R. Accelerated evolution of resistance in multidrug environments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lázár V, et al. Bacterial evolution of antibiotic hypersensitivity. Molecular Systems Biology. 2013;9 doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamovic L, Sommer MOA. Use of Collateral Sensitivity Networks to Design Drug Cycling Protocols That Avoid Resistance Development. Science Translational Medicine. 2013;5:204ra132–204ra132. doi: 10.1126/scitranslmed.3006609. [DOI] [PubMed] [Google Scholar]
- 23.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 24.Rice LB. Antimicrobial resistance in gram-positive bacteria. American journal of infection control. 2006;34:S11–S19. doi: 10.1016/j.ajic.2006.05.220. discussion S64-73. [DOI] [PubMed] [Google Scholar]
- 25.Waxman DJ, Strominger JL. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annual review of biochemistry. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 26.Lee SH, et al. Antagonism of chemical genetic interaction networks resensitize MRSA to β-lactam antibiotics. Chemistry & biology. 2011;18:1379–1389. doi: 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Koga T, et al. Affinity of Tomopenem (CS-023) for penicillin-binding proteins in Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy. 2009;53:1238–1241. doi: 10.1128/AAC.01433-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Bhachech N, Bush K. Biochemical comparison of imipenem, meropenem and biapenem: permeability, binding to penicillin-binding proteins, and stability to hydrolysis by beta-lactamases. The Journal of antimicrobial chemotherapy. 1995;35:75–84. doi: 10.1093/jac/35.1.75. [DOI] [PubMed] [Google Scholar]
- 29.Campbell EM, Chao L. A population model evaluating the consequences of the evolution of double-resistance and tradeoffs on the benefits of two-drug antibiotic treatments. PLoS ONE. 2014;9:e86971–e86971. doi: 10.1371/journal.pone.0086971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munck C, Gumpert HK, Wallin AIN, Wang HH, Sommer MOA. Prediction of resistance development against drug combinations by collateral responses to component drugs. Science Translational Medicine. 2014;6:262ra156–262ra156. doi: 10.1126/scitranslmed.3009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroda M, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 32.Goldstein F, et al. Identification and phenotypic characterization of a β-lactam-dependent, methicillin-resistant Staphylococcus aureus strain. Antimicrobial agents and chemotherapy. 2007;51:2514–2522. doi: 10.1128/AAC.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arêde P, Ministro J, Oliveira DC. Redefining the role of the β-lactamase locus in methicillin-resistant Staphylococcus aureus: β-lactamase regulators disrupt the MecI-mediated strong repression on mecA and optimize the phenotypic expression of resistance in strains with constitutive mecA. Antimicrobial agents and chemotherapy. 2013;57:3037–3045. doi: 10.1128/AAC.02621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berenbaum MC. What is Synergy. Pharmacological Reviews. 1989;1989 [PubMed] [Google Scholar]
- 35.Saiman L. Clinical utility of synergy testing for multidrug-resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis: 'the motion for'. Paediatric respiratory reviews. 2007;8:249–255. doi: 10.1016/j.prrv.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 36.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Nineteenth Informational Supplement. CLSI document M100-S19. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 37.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Eighth Edition. CLSI document M07-A8. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- 38.AstraZeneca Pharmaceuticals, L. P. Merrem IV (meropenem) for injection for intravenous use only prescribing information. Wilmington, DE: 2013. pp. 1–7. [Google Scholar]
- 39.Fishovitz J, et al. Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics. Journal of the American Chemical Society. 2014;136:9814–9817. doi: 10.1021/ja5030657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somani P, Freimer EH, Gross ML, Higgins JT. Pharmacokinetics of Imipenem-Cilastatin in Patients with Renal Insufficiency Undergoing Continuous Ambulatory Peritoneal Dialysis. Antimicrobial agents and chemotherapy. 1988;32:4–9. doi: 10.1128/aac.32.4.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinzig M, Brismar B, Nord CE. Pharmacokinetics and Tissue Penetration of Tazobactam and Piperacillin in Patients Undergoing Colorectal Surgery. Antimicrobial agents and chemotherapy. 1997;36:1997–2004. doi: 10.1128/aac.36.9.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stutman HR, Welch DF, Scribner RK, Marks MI. In vitro antimicrobial activity of aztreonam alone and in combination against bacterial isolates from pediatric patients. Antimicrobial agents and chemotherapy. 1984;25:212–215. doi: 10.1128/aac.25.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, et al. Antagonism of chemical genetic interaction networks resensitize MRSA to beta-lactam antibiotics. Chem. Biol. 2011;18:1379–1389. doi: 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Bouley R, et al. Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-cyanostyryl)quinazolin-4(3H)-one. Journal of the American Chemical Society. 2015;137:1738–1741. doi: 10.1021/jacs.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ankomah P, Johnson PJT, Levin BR. The pharmaco and population evolutionary dynamics of multi-drug therapy: experiments with S aureus E. coli and computer simulations. PLoS Pathogens. 2013;9:e1003300–e1003300. doi: 10.1371/journal.ppat.1003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonstein SA, Baldwin JN. Loss of the penicillinase plasmid after treatment of Staphylococcus aureus with sodium dodecyl sulfate. Journal of Bacteriology. 1972;109:262–265. doi: 10.1128/jb.109.1.262-265.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hackbarth CJ, Chambers HF. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrobial agents and chemotherapy. 1993;37:1144–1149. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowy FD. Antimicrobial resistance : the example of Staphylococcus aureus. Journal of Clinical Investigation. 2003;111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blazquez B, et al. Regulation of the Expression of the β Lactam Antibiotic-Resistance Determinants in Methicillin-Resistant Staphylococcus aureus (MRSA) Biochemistry. 2014;53:1548–1550. doi: 10.1021/bi500074w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craig WA. The pharmacology of meropenem, a new carbapenem antibiotic. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1997;24(Suppl 2):S266–S275. doi: 10.1093/clinids/24.supplement_2.s266. [DOI] [PubMed] [Google Scholar]
- 51.DeRyke CA, Banevicius MA, Fan HW, Nicolau DP. Bactericidal activities of meropenem and ertapenem against extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a neutropenic mouse thigh model. Antimicrobial agents and chemotherapy. 2007;51:1481–1486. doi: 10.1128/AAC.00752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y, Wang R, Pei F, Liang B-b. Antibacterial Activity of Allicin Alone and in Combination with Beta-Lactams against Staphylococcus spp. and Pseudomonas aeruginosa. Journal of Antibiotics. 2007;60:335–338. doi: 10.1038/ja.2007.45. [DOI] [PubMed] [Google Scholar]
- 53.Berenbaum MC. A method for testing for synergy with any number of agents. The Journal of infectious diseases. 1978;137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 54.NCCLS. Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline. NCCLS document M26-A. Wayne, PA: NCCLS; 1999. [Google Scholar]
- 55.Life_Technologies. SYBR® Select Master Mix for CFX User Guide. 2012. pp. 1–34. [Google Scholar]
- 56.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfizer. Zosyn (Piperacillin and Tazobactam for Injection, USP) for injection for intravenous use only prescribing information. Philadelphia, PA: 2012. pp. 1–26. [Google Scholar]
- 58.Merck. PRIMAXIN I.V. (IMIPENEM AND CILASTATIN FOR INJECTION) Whitehouse Station, NJ: 2014. pp. 1–17. [Google Scholar]
- 59.Ford CW, et al. In Vivo Activities of U-100592 and U-100766, Novel Oxazolidinone Antimicrobial Agents, against Experimental Bacterial Infections. Antimicrobial agents and chemotherapy. 1996;40:1508–1513. doi: 10.1128/aac.40.6.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.