Abstract
The protein kinase D (PKD) family members, PKD1, PKD2 and PKD3 constitute a family of serine/threonine kinases that are essential regulators of cell migration, proliferation and protein transport. Multiple types of cancers are characterized by aberrant expression of PKD isoforms. In breast cancer PKD isoforms exhibit distinct expression patterns and regulate various oncogenic processes. In highly invasive breast cancer, the leading cause of cancer-associated deaths in females, the loss of PKD1 is thought to promote invasion and metastasis, while PKD2 and upregulated PKD3 have been shown to be positive regulators of proliferation, chemoresistance and metastasis. In this review, we examine the differential expression pattern, mechanisms of regulation and contributions made by each PKD isoform to the development and maintenance of invasive breast cancer. In addition, we discuss the potential therapeutic approaches for targeting PKD in this disease.
Keywords: Protein kinase D, PKD, Invasive breast cancer, Invasion, Multi-drug resistance
Introduction
Protein kinase D (PKD) encompasses a family of serine/threonine kinases that belong to the calcium/calmodulin-dependent kinase (CaMK) superfamily. The first family member to be identified was PKD1, initially designated as PKCµ, an atypical member of the protein kinase C (PKC) family [1, 2]. Two other PKD family members have since been identified, PKD2 [3] and PKD3 [4]. PKDs have multiple cellular targets and have therefore been implicated in diverse biological functions such as cell migration [5–7], proliferation [8, 9], protein transport [10], epithelial to mesenchymal transition (EMT) [11, 12], angiogenesis [13], and transcriptional regulation [14]. Given the vast array of cellular functions relevant to tumor development and progression regulated by PKD isoforms, it is not surprising that deregulated expression of PKD family members have been reported in multiple types of cancers. PKDs seem to have organ-specific tumor suppressive or tumor promoting functions. In pancreatic cancer and basal cell carcinoma, increased expression of PKD1 has been described [15, 16]. In other types of malignancies, opposing roles for PKD family members have been reported. For example, in androgen-independent prostate cancer, PKD1 is down-regulated while PKD3 has been shown to be a positive regulator of cancer cell growth and survival [17, 18]. Increasing recent evidence suggests that PKD isoforms are critical regulators of breast cancer and studies aimed at elucidating the functional roles and expression patterns of PKDs in this malignancy are an active area of ongoing research.
PKD isoforms in breast cancer
In females, breast cancer is the most frequently diagnosed malignancy and accounts for most cases of cancer-related deaths globally [19]. Although breast cancer is a very complex and heterogeneous disease, gene expression studies conducted over a decade ago have identified specific molecular traits associated with the disease [20]. Molecular subtypes of breast cancer include the luminal A and B cancers characterized by the high expression of the estrogen and/or progesterone hormone receptors; the HER2 type characterized by high expression of HER2, a receptor tyrosine kinase which belongs to the epidermal growth factor receptor (EGFR) family; and basal-like/triple-negative breast cancer (TNBC). TNBCs express neither HER2 nor the hormone receptors and as such they are unsuitable for molecular targeted therapy [21]. Despite improvements in therapeutic strategies and diagnostic techniques, mortality resulting from breast cancer, particularly TNBC is still high [19]. It is therefore imperative to understand the contribution made by molecules such as PKD family members to breast cancer progression. PKD1 is strongly expressed and active in ductal epithelial cells of the normal breast, but its expression is lost during tumor development. Analysis of human breast cancer samples have shown that in some of the most highly invasive tumors PKD1 expression is completely lost [5, 22]. On the other hand, PKD2 and PKD3 are only marginally expressed in the normal breast while both have been reported to be upregulated in advanced and aggressive breast cancer [23, 24].
Isoform specificity of the PKD family
The differential expression pattern of PKD family members in breast cancer indicates that each one is fulfilling a unique role during disease progression. Disparate functions of isoenzymes in the same protein family are not just restricted to the PKD family. A similar phenomenon is seen in the Akt protein kinase family where despite high structural homology among members, non-redundant functions have been reported [25]. Indeed, though PKD1, PKD2 and PKD3 have overlapping functions, isoform-specific functions are described in the literature. Despite high structural homology among the PKD isoforms, some structural variability exists and to a certain extent can help explain the different functions of each PKD that will be discussed below. For example, PKD1 and PKD2 contain a c-terminal PDZ-binding motif, while PKD3 does not (Fig. 1) [26]. This is functionally relevant since it was demonstrated that this particular characteristic allows PKD1 and 2, but not PKD3 to regulate the localization of Kidins220 at the surface of neural cells and its trafficking between the trans-golgi network and the plasma membrane [27]. Moreover, differences in localization within the cell likely contribute to the isoform-specific functions of PKD family members. In resting cells PKD1 is localized primarily within the cytosol [28], but upon stimulation can be found in other cellular structures such as the Golgi [29], nucleus [30] or mitochondria [31]. Like PKD1, in unstimulated cells PKD2 is predominantly cytoplasmic [32]. On the other hand, in the absence of stimulation, PKD3 is localized in both the nucleus and cytoplasm [33]. In summary, the level of expression, spatial distribution and activation status of each PKD may determine its impact on a specific biological function. For example, in gastric cancer cells, PKD2, compared to its counterparts PKD1 and PKD3 is particularly well positioned to phosphorylate HDAC7 and induce the expression of the transcription factor Nur77 [34]. There is not a lot of evidence identifying isoform-specific substrates for each PKD, but some studies have shown that it is possible. For example, G-protein-coupled receptor kinase-interacting protein 1 (GIT1) has been identified as a substrate specific for PKD3. PKD3-mediated phosphorylation of GIT1 on serine 46 induces cellular protrusive activity and paxillin trafficking [35]. Below we discuss in further detail the isoform specificity of the PKDs in the context of breast cancer.
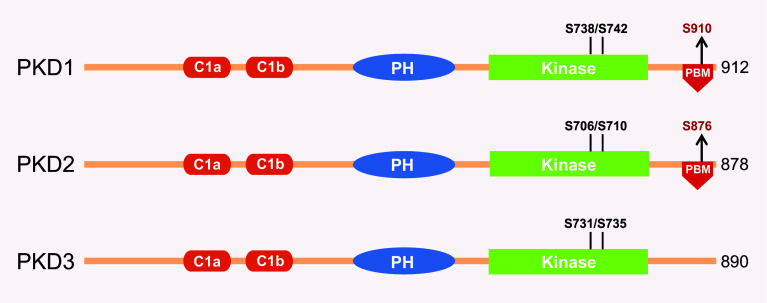
Fig. 1.
Schematic representation of PKD isoforms. PKD1, PKD2, and PKD3 exhibit high homology with the presence of cysteine-rich domains C1a and C1b, pleckstrin homology (PH) and kinase domains in all three isoforms. Only PKD1 and PKD2, but not PKD3 have a C-terminal PDZ-binding motif (PBM). Serine residues in the activation loop kinase domain critical for PKD activation by novel PKC (nPKC) isoforms are indicated. Autophosphorylation sites within the PDZ-binding motif are indicated in red
Regulation of PKD isoform expression
PRKD1 is epigenetically silenced during breast tumor progression
PKD isoforms often show different expression and activation status in tissues. In the normal breast, PKD1 is the major isoform present while PKD2 and PKD3 are moderately expressed [5, 22, 23, 36, 37]. PKD1 expression has been assessed in breast cancer cell lines as well as in a large number of breast cancer patient samples [5, 22, 23]. The same pattern of expression is always observed; PKD1 expression is gradually lost during breast cancer progression as the tumor becomes increasingly aggressive [22, 38]. The mechanisms underlying this regulation have been investigated and while missense mutations in the exon sequences of PRKD1 were described, these genetic variations do not account for the loss of PKD1 expression during breast tumorigenesis [39, 40]. Genetic alterations are not always the causative agents which govern changes in protein expression, and in this case epigenetic changes, specifically, promoter-specific DNA methylation is responsible for PKD1 silencing. It was shown that the loss of PKD1 expression in invasive breast cancer cell lines was directly correlated with hypermethylation of its promoter (Fig. 2). Comparing the PRKD1 promoter region in a subset of invasive and non- or minimally invasive breast cancer cell lines, Borges et al. found a very high percentage of methylated CpG sites in the invasive cells and as expected, no methylation in the non-invasive cells which also express a high level of PKD1 [22]. The same mechanism of regulation was observed in patient samples where increased PRKD1 gene promoter methylation was also detected only in the most aggressive types of breast cancer including invasive ductal carcinoma (IDC) ER+/HER2−, IDC ER−/HER+ and TNBC [22]. Moreover, this study showed an increase in PRKD1 promoter methylation in IDC patients with positive lymph nodes compared to patients with negative lymph nodes indicating that silencing of PKD1 by hypermethylation of its promoter may be associated with metastasis. Interestingly, it was also shown that there is a correlation between the expression of PKD1 and ERα in breast cancer cell lines [5, 22, 41]. This correlation most likely indicates that similar mechanisms lead to silencing of both signaling molecules. This is supported by a recent study, in which PKD1 protein expression was assessed in a large array of breast cancer patients diagnosed with TNBC and in the normal breast [23]. In this study, it was shown that loss of ER goes along with loss of PKD1, but this most likely is due to epigenetic silencing of both genes by DNA methyltransferases, and therefore may not be functionally related. To date, the mechanism of regulation underlying the possible association between the kinase and the hormone receptor is unknown and warrants further investigation.
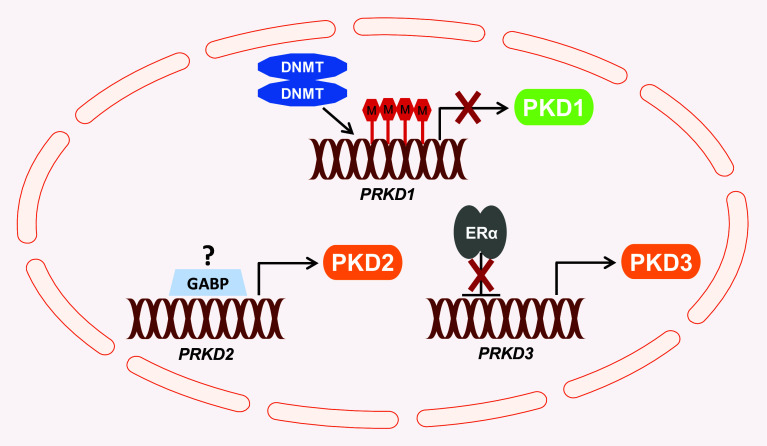
Fig. 2.
Regulation of expression of PKD isoforms in invasive breast cancer. In invasive breast cancer, the PRKD1 gene is silenced by hypermethylation of its promoter region which leads to inhibition of transcriptional activity and loss of PKD1 expression. The potential regulators of PKD2 expression in the breast have not yet been identified. However, the transcription factor GABP, a positive regulator of PRKD2 gene expression in CML, could also possibly modulate its expression during breast tumor progression. ERα represses PKD3 expression by direct binding to the PRKD3 promoter. In ER-negative breast cancer, the loss of ER expression allows the upregulation of PKD3
Is GABP a potential regulator of PKD2 expression in the breast?
In numerous reports, the expression pattern of PKD2 in breast cancer cell lines is very uniform and cannot be linked to tumor subtypes [5, 22, 36, 37]. In samples from patients with TNBC, PKD2 was weakly expressed but still showed a slight but significant decrease in expression when compared to less aggressive cancers or normal tissue [23]. However, in vitro studies have shown its presence in invasive breast cancer cell lines including MDA-MB-231, MDA-MB-468, BT20 and HCC1806 [23, 36]. Furthermore, PKD2 in some of these cell lines has been shown to drive proliferation, migration and invasion and mediate multi-drug resistance [36, 42, 43]. The information available about the genetic regulation of PKD2 is limited, but a recent study on chronic myelogenous leukemia (CML) has identified PRKD2 as a candidate target gene for the GA-binding protein (GABP), a tetrameric transcription factor involved in cell cycle regulation [44]. The same study showed that PRKD2 was among the GABP-bound genes upregulated in mice and human CML and further concluded with chromatin immunoprecipitation and gene expression data that PRKD2 expression was regulated by direct binding of GABP to its promoter (Fig. 2). To date, this is the only study showing how PKD2 can be regulated at the gene expression level; however, it is still not clear if a similar mechanism of regulation occurs in normal breast tissue or during breast carcinogenesis.
Estrogen receptor negatively regulates PKD3 expression
PKD3 is only weakly detected in the normal breast [23], but has been shown to be highly upregulated in invasive breast cancer [23, 24, 36]. Furthermore, PKD3 was associated with metastatic progression to distant organs in vivo, but also poor prognosis for patients with advanced breast cancer [23]. Elevated levels of PKD3 protein were detected in TNBC cells and analysis of microarray gene expression data sets showed increased expression of PKD3 mRNA in basal-like breast cancer [24]. This is in concordance with another study by Hao et al. where the expression of PKD2 and PKD3 was investigated in a subset of breast cancer cell lines. Here, the authors found that unlike the expression of PKD2 which remained relatively homogeneous, an increase in PKD3 expression was detected in TNBC cell lines as well as in EGFR-overexpressing breast cancer cells [36]. In a more recent study, the same pattern of PKD3 expression was observed on a greater scale in TNBC as well as in basal breast cancer using large cohorts of patient biopsies or breast cancer cell lines [23, 45, 46]. Here, a reverse correlation between the expression of PKD3 and ERα was described, and elevated PKD3 levels were associated with the loss of hormone receptor expression (Fig. 2). Interestingly, analysis of the PRKD3 promoter sequence led to the identification of a new mechanism of regulation, showing that ERα inhibits PRKD3 expression through direct binding to its promoter at two distinct estrogen response element (ERE) sites [23]. ER is generally described as an activator of transcription but in some cases it can also act as a repressor of gene expression [47, 48], so it is not surprising that its presence in the less aggressive and invasive forms of breast cancer prevents or blocks the expression of oncogenic PKD3.
Roles of PKD isoforms in invasive breast cancer
PKD1 blocks migration and metastatic progression
In normal epithelial cells of the breast PKD1 is the main isoform present [5, 22]. In breast cancer, PKD1 affects both cell proliferation and invasive behavior (Fig. 3). In ER-positive MCF-7 breast cancer cells PKD1 has been shown to sensitize cells to β-estradiol-induced proliferation and to enhance cell cycle transition from G0/G1 to S phase through MEK/ERK-dependent signaling [41, 49]. However, loss of PKD1 expression has been shown to lead to an acquisition of metastatic characteristics. It is now well-established that PKD1 acts as a negative regulator of the motile and invasive phenotype by targeting multiple signaling pathways [5, 11, 22, 50, 51]. Actin-driven directed cell migration is regulated by a cycle of phosphorylation and dephosphorylation of cofilin [52]. The balance between LIM kinase (LIMK) and slingshot phosphatase 1L (SSH1L) activities regulating cofilin phosphorylation at serine residue S3 is essential to ensure proper cofilin-mediated actin reorganization. Both, the accumulation of either unphosphorylated or hyperphosphorylated cellular cofilin can lead to decreased migration. Downstream of RhoA, PKD1 affects the cofilin cycle by mediating inactivation of SSH1L [7, 38, 53, 54] and activation of p21-activated kinase 4 (PAK4) which is upstream of LIMK [55, 56]. PKD1 phosphorylates SSH1L in its actin-binding motif. This phosphorylation generates a binding site for 14-3-3 proteins which facilitates the translocation of slingshot from the lamellipodium to the cell cytoplasm [38]. PKD1-mediated phosphorylation of PAK4 facilitates the activation of LIMK, which directly phosphorylates cofilin [55]. In sum, such signaling results in an accumulation of S3-phosphorylated inactive cofilin. Consequently, actin severing activities are attenuated, free barbed ends are not generated and a concomitant decrease in cell migration is observed. Inhibition of directed cell migration has also been shown to be dependent on PKD1-mediated phosphorylation of cortactin [6] and Ras and Rab interactor 1 (RIN1) [51]. PKD1-mediated phosphorylation of cortactin attenuates WAVE2-driven actin polymerization [6] while PKD1 inhibition of cell migration via RIN1 was shown to be dependent on the tyrosine kinase Abl [51].
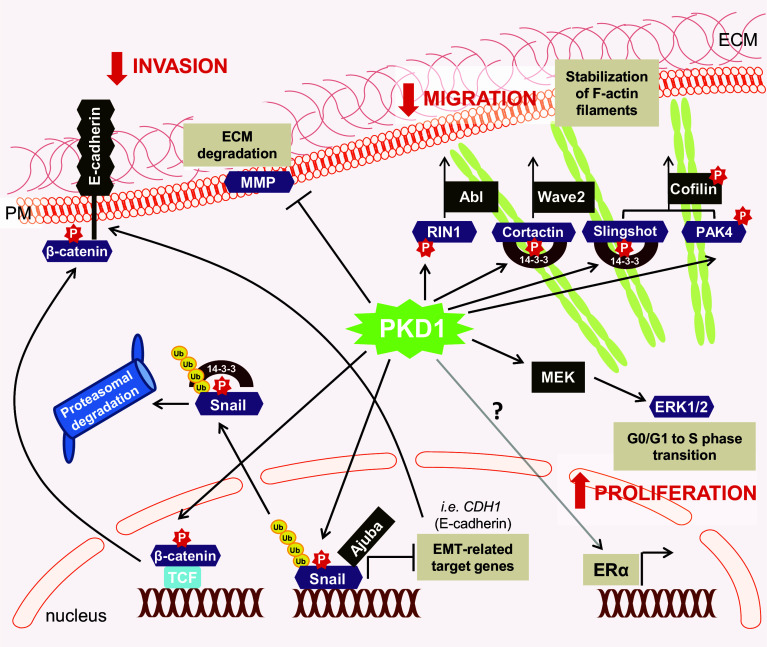
Fig. 3.
PKD1 signaling in breast cancer and normal breast. In non-invasive breast cancers PKD1 promotes cell proliferation by induction of the MEK/ERK signaling cascade and by sensitizing cells to β-estradiol possibly through positive regulation of ERα expression. Phosphorylation of Cortactin, Slingshot, PAK4 and RIN1 by PKD1 leads to the stabilization of F-actin filaments and decreased cell migration. Phosphorylation of Snail by PKD1 diminishes its ability to inhibit its target genes (e.g., E-cadherin) by losing its interaction with its co-repressor Ajuba; PKD1-mediated phosphorylation also induces nuclear export and targets Snail for proteasomal degradation. Phosphorylation of β-catenin by PKD1 facilitates its nuclear export and re-localization to the plasma membrane. PKD1 negatively regulates MMP expression which results in decreased invasion abilities. Loss of PKD1 occurs in invasive breast cancer and this promotes cell migration, invasion and EMT
In addition, PKD1 also can inhibit EMT, a biological process that allows a polarized epithelial cell to undergo multiple biochemical changes that enable it to assume a mesenchymal phenotype which includes enhanced migratory capacity and invasiveness. EMT is central to the metastasis process of multiple types of cancers including breast cancer [57–59]. An EMT is characterized by increased expression of matrix metalloproteinases (MMPs); and upregulation of various MMPs has been shown to occur in breast cancer [60]. Eiseler et al. showed that expression of an active PKD1 was able to reduce the mRNA level of several MMPs including MMP-2, MMP-7, and MMP-9 in MDA-MB-231 cells which subsequently diminished the ability of these cells to invade [5].
Recently, other studies have shed light on additional mechanisms that explain how PKD1 maintains the epithelial phenotype of normal breast cells in vitro [11]. E-cadherin is an important marker for epithelial cells which plays a critical role in epithelial cell–cell contacts and has thus been implicated in the maintenance of mammary gland tissue architecture and homeostasis. The loss of its expression is also associated with EMT and the acquisition of metastatic characteristics. The transcription factor Snail (SNAI1) acts as a repressor of E-cadherin expression and its upregulation in several types of cancers promotes the EMT process [61–65]. Snail forms a complex with its co-repressor Ajuba, which is indispensable for the repression of E-cadherin transcription [66]. PKD1-mediated phosphorylation of Snail at S11 results in the positive regulation of E-cadherin expression [11, 12, 50]. Phosphorylation of Snail at S11 which occurs in the nucleus decreases its interaction with Ajuba [11] and facilitates its interaction with the E3 ubiquitin ligase FBXO11 in the nucleus [50]. FBXO11 acts as a classical E3 ligase by targeting Snail for proteasomal degradation [50]. Nuclear export of S11-phosphorylated Snail can be mediated by 14-3-3 proteins [12]. Interestingly, the active form of PKD1 as well as Snail phosphorylated at S11 were detected in epithelial cells of the normal breast, and both were decreased in IDC [11]. Together, these data not only confirm that PKD1 is a negative regulator of cell motility and invasion but that it also contributes to the maintenance of the epithelial phenotype of breast cells by preventing E-cadherin loss.
In addition to its impact on E-cadherin expression, PKD1 has been shown to phosphorylate and regulate the subcellular localization of β-catenin. In prostate and colon cancer, PKD1 is associated with the trafficking of β-catenin out of the nucleus and its decreased transcriptional activity which in turn leads to reduced expression of oncogenes such as c-Myc and cyclin D1 [67–69]. There is no evidence so far that the same mechanism of regulation of β-catenin occurs during breast tumor progression, but it is highly possible.
PKD2 mediates drug resistance and affects cancer cell proliferation and motility
Unlike PKD1 (or PKD3), expression levels of PKD2 are not correlating with certain breast cancer subtypes [22, 43]. Using MDA-MB-231 as a model system, PKD2 has been shown to be a critical mediator of multi-drug resistance in breast cancer cells (Fig. 4). The level of phosphorylated PKD2 increased in a time-dependent manner in response to stimulation with paclitaxel. Furthermore, depletion of PKD2 decreased the paclitaxel-induced expression of P-glycoprotein (P-gp), a protein that functions as an efflux pump for multiple types of drugs [43]. Increased levels of auto-phosphorylated PKD2 have also been detected in doxorubicin-resistant MCF-7 when compared to the parental cell lines [42]. In this case, however, PKD2 did not alter multi-drug-resistance as no changes in multi-drug resistance- or apoptosis-related gene expression were detected [42]. Interestingly, doxorubicin-resistant MCF-7 cells were previously characterized as more mesenchymal than the parental epithelial MCF-7 cells [70, 71].
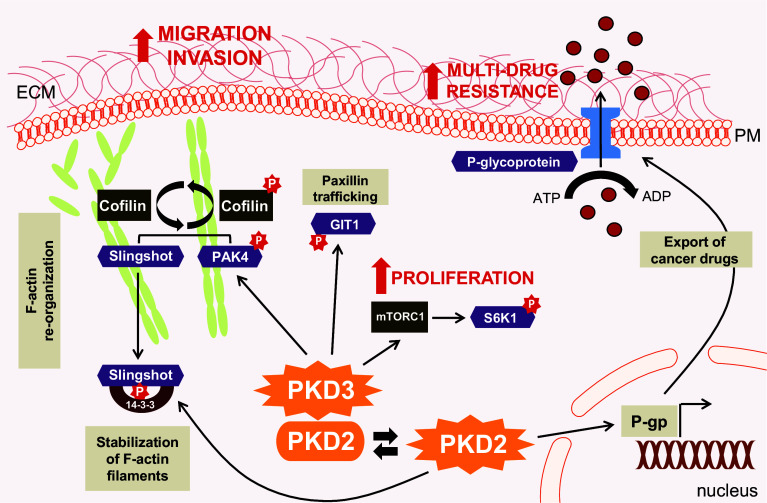
Fig. 4.
Pathways by which PKD2 and PKD3 promote the oncogenic all major phenotype. PKD3 drives aspects of tumorigenesis, including tumor cell proliferation, migration and invasion. PKD3 mediates activation of S6K1 through mTORC1 leading to cell proliferation. Phosphorylation of GIT1 regulates Paxillin trafficking and contributes to cell spreading. Furthermore, endogenous high PKD3 activity contributes to PAK4 activation, while SSH1L activity is unaffected. This allows a functional cofilin cycle that facilitates F-actin re-organization and directed cell migration. PKD2, when active, has been shown to induce the expression of p-glycoprotein, a transmembrane protein which mediates the export of multiple types of cancer drugs from cells
Additionally, PKD2 was shown to positively regulate migration of the drug-resistant cells, and under the same conditions to negatively regulate migration of the parental MCF-7 cells. Such pro- and anti-migratory properties attributed under different conditions long-time made it difficult to establish PKD2’s impact on cell migration and invasion [36, 42, 55, 72, 73]. A recent study now has shown that highly migratory breast cancer cells express PKD2 in complex with PKD3. In these complexes PKD3 is basally active and promotes pro-migratory cofilin function by activating PAK4/LIMK. This can be altered when PKD2 is additionally activated, shifting the cofilin pool into the inactive S3-phosphorylated state by inactivating its phosphatase slingshot [74]. Alternatively, the decrease in basal PKD3 activity can shift the cofilin pool into the dephosphorylated active state (basal activity of SSH1L and decreased PAK4/LIMK activity), also having a negative impact on cell motility. This model now would implicate that even if PKD3 is inactive, an activation of PKD2 (i.e., by exposure to above chemotherapeutics) may increase directed cell migration [74].
In TNBC cell lines such as HCC1806, inhibition of PKD2 led to a reduction in cell proliferation [36]. Even though PKD2 has been shown to promote proliferation in other cancer cells like colorectal cancer [75] and glioma [76], the exact mechanisms by which PKD2 promotes cancer cell growth are still elusive.
PKD3 is associated with all tumorigenic functions
In breast cancer PKD3 is involved in all aspects of oncogenic signaling (Fig. 4). Multiple studies have confirmed overexpression of PKD3 protein in invasive breast cancer cell lines [23, 24, 36]. Moreover, in patient samples of basal-like and TN breast cancers, increased levels of PKD3 mRNA and protein have been reported [23, 24]. PKD3 appears to have typical oncogenic functions in invasive breast cancer. In the TNBC cell line HCC1806, depletion of PKD3 attenuated cell proliferation by up to 65 % [36]. The same effect on cell proliferation was also observed in another TNBC cell line (MDA-MB-231) as well as in the ER−/HER+ cell line, HCC1954 [23]. One of the possible mechanisms responsible for the effects of PKD3 on cell proliferation seems to involve S6 Kinase 1 (S6K1). S6K1 was identified as a major downstream target of PKD3 which mediates its pro-oncogenic functions. S6K1 is a substrate of mammalian target of rapamycin complex 1 (mTORC1) [24]. The mTOR signaling cascade is a principal regulator of cell homoeostasis, metabolism and survival. mTORC1-mediated activation of S6K1 promotes protein synthesis, a process central to cell growth [77]. Using MDA-MB-231 cells as a model for TNBC, treatment of these cells with the PKC/PKD activator phorbol-12,13-dibutyrate (PDBu) induced phosphorylation of S6K1. Stimulation of cells with the mTORC1 inhibitor rapamycin attenuated the PDBu-induced phosphorylation of S6K1 which suggests that mTORC1 is necessary for this PKD3-mediated effect. Furthermore, depletion of PKD3 in two cell lines, MDA-MB-231 and MDA-MB-468, diminished serum-induced phosphorylation of S6K1. As expected, depletion of PKD3 in these cell lines also blocked proliferation [24].
As indicated above, PKD3 regulates the cofilin phosphorylation status, but mainly through activation of PAK4/LIMK. Under normal growth conditions basally active PKD3 does not inhibit slingshot, with the net effect to guarantee a functional cofilin cycle and directed cell migration [74]. Another mechanism of how PKD3 regulates cell motility and cell spreading is through phosphorylation of GIT1. The incorporation of GIT1 into motile cytoplasmic paxillin-positive adhesion complexes is dependent on PKD3-mediated phosphorylation of serine 46. A GIT1.S46A mutant failed to localize to these motile cytoplasmic paxillin-positive adhesion complexes, but was instead observed at focal adhesions. MCF-7 cells expressing GIT1.S46D, a phospho-mimetic mutant, were more migratory than cells expressing wild-type GIT1, and cells expressing GIT1.S46A were less migratory than cells expressing the wild-type protein [35].
Above effects are in accordance with the data reported by Borges et al. showing that a knockdown or inhibition of PKD3 decreased ER-breast cancer cell migration, but also increased cell spreading and altered F-actin organization. This study further showed the dramatic impact of PKD3 on cell invasion not only in vitro but also in vivo in an orthotopic model of metastatic breast cancer [23]. In this in vivo model, whether PKD3 was inhibited using a specific PKD inhibitor or down-regulated with a reverse genetics approach, both led as expected to a reduction in tumor growth but also more importantly to a dramatic decrease in lymph node infiltration and number and size of lung metastases [23].
The pro-oncogenic functions of PKD3 have been observed in other types of cancers and are not just limited to invasive breast cancer. PKD3 has also been shown to be crucial for prostate cancer cell growth and survival. In prostate cancer, increased nuclear localization of PKD3 correlates with higher tumor grade [17]. In highly aggressive androgen-independent prostate cancer cell lines (PC3 and DU145), increased expression of PKD3 augmented the basal levels of both phospho-Akt and phospho-ERK1/2, suggesting anti-apoptotic and proliferation signaling [17]. The significance of PKD3-mediated activation of Akt and ERK1/2 in invasive breast cancer remains unexplored.
Protein kinase D enzymes as therapeutic targets for advanced breast cancer
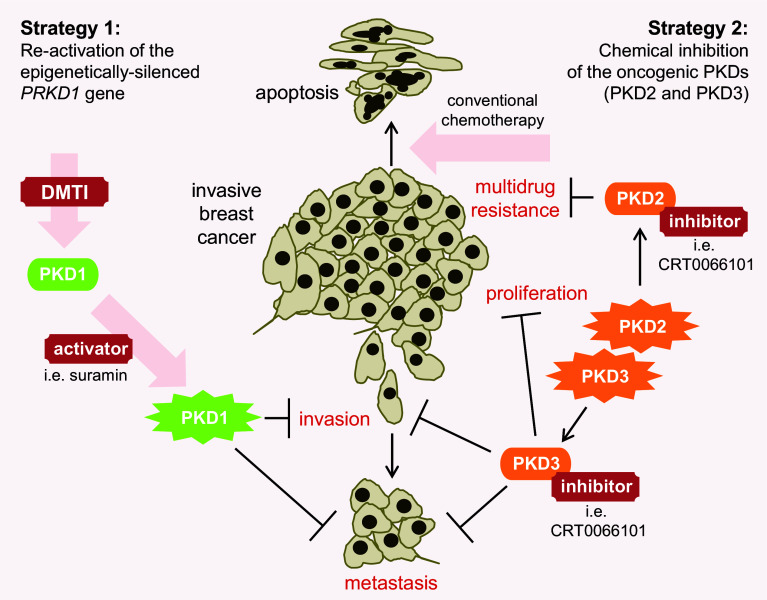
The PKD family members represent significant diversity in respect to their functions in development and progression of breast cancer. This is not unlike other family of kinases in which different isoforms can have different, sometimes opposite functions despite sharing a high structural homology [25]. PKD isoforms mainly differ in their contribution to cell migration, invasion and chemoresistance [5, 11, 12, 22–24, 35, 36, 43, 78]. This difference in function between isoforms is not restricted to just breast cancer as they were also described in colon, gastric and prostate cancers [12, 17, 69, 73, 79–81]. The molecular mechanisms underlying PKD-mediated effects across multiple types of cancers are still unclear and more in-depth investigation is required. Generally, opposite functions among members of the same kinase family make it difficult to utilize therapeutic agents such as pan inhibitors. However, the particular expression pattern of each PKD in different types of breast cancer renders their therapeutic targeting possible. Clearly, loss of PKD1 expression in context of PKD2/PKD3 expression drives oncogenic processes associated with metastatic progression [50, 78, 82]. This setting occurs in ER-negative IDC, which are the most aggressive breast cancers and associated with the worst outcome [83–86]. Preclinical studies using orthotopic animal models have shown that these invasive tumors which are characterized by the absence of PKD1 and high levels of PKD2/PKD3 can be targeted by two different strategies (Fig. 5): (1) the re-activation of the epigenetically silenced PRKD1 gene to block metastatic progression of highly invasive breast cancers; or (2) the chemical inhibition of the oncogenic PKD2/PKD3 to inhibit tumor growth, metastasis and chemoresistance [22, 23, 78]. Bona fide targets for both approaches are triple-negative, or more generally, ER-negative breast cancers.
Fig. 5.
Strategies to target PKD isoforms in invasive breast cancer. Invasive breast cancers express PKD2 and PKD3, but not PKD1. Two strategies have been shown to be successful in preclinical animal models. First, PKD1 re-expression can be initiated by treatment with DNA methyltransferase inhibitors (DMTIs). Upregulation of PKD1 bocks invasion and metastasis, and additional activation with activators such as suramin further enhances these effects. A second strategy is the use of pan PKD inhibitors to target oncogenic functions of PKD2 and PKD3. Inhibition of PKD3 decreases tumor cell proliferation, invasion and metastasis. Inhibition of PKD2 in cancer cell lines has been shown to decrease multi-drug resistance, and therefore it is predicted that PKD inhibitors will act synergistically with currently used conventional chemotherapy, by sensitizing cancer cells to these agents
Re-activation of PRKD1 as an approach to block metastatic progression
PKD1 is present in non-invasive breast cancer, but inhibits the ability of these cancer cells to acquire a more aggressive and invasive phenotype [5, 11, 12, 22]. Apparently, among all PKD isoforms, it is the presence or absence of PKD1 which seems to be the decisive factor that determines cell invasiveness. There are two options for different types of breast cancer. Non-invasive breast cancers in which PKD1 is expressed and contributes to cell proliferation can be targeted with PKD inhibitors (which are described in the next paragraph). It is expected that although cells treated with pan PKD inhibitors lose PKD1’s inhibitory function on EMT at the same time PKD2 and PKD3 oncogenic signaling are also blocked. An expected outcome of PKD inhibition with pan inhibitors in breast cancers that are PKD1, PKD2 and/or PKD3 positive should be a decrease in primary tumor growth as well as metastasis. However, so far this scenario has not been confirmed by in vivo studies using orthotopic animal models.
In contrast to non-invasive breast cancers, highly invasive breast cancers do not express PKD1 due to hypermethylation of the PRKD1 gene promoter [22]. A similar silencing mechanism of downregulation of PKD1 expression has also been observed in gastric cancer [22, 81]. Epigenetic alterations such as abnormal DNA methylation of certain gene promoter regions is a well-known silencing mechanism associated with tumor formation and progression [87, 88]. Furthermore, hypermethylation of gene promoters is highly associated with the transcriptional repression of tumor-suppressor genes [22, 89–91]. Interestingly, unlike genetic mutations, DNA methylation is a reversible process and silenced tumor-suppressor genes have been successfully re-activated using DNA methyltransferase inhibitors (DMTI). DMTIs include 5-azacytidine, 5-aza-2′-deoxycytidine (decitabine) and RX-3117, all of which have been approved by the Food and Drug Administration (FDA) [22, 78, 92, 93]. It was shown that in gastric and invasive breast cancer cell lines PKD1 re-expression can be induced using low doses of decitabine or RG108, another DMTI [22, 78, 81]. More importantly, treatment with DMTI in breast cancer cells led to re-expression of PKD1 at physiological levels, resulting in a PKD1-dependent reversion of their invasive phenotype in vitro. In vivo in a xenograft model for metastatic progression such reactivation of PKD1 almost completely blocked metastasis [22]. A caveat with such a treatment strategy is that DMTIs target multiple genes and allow their re-activation. For example, the most common and known decitabine-activated genes in breast cancer are TP53, CDKN1A and the gene encoding for the estrogen receptor [94, 95]. This lack of specificity could raise concern for any clinical application especially since re-activation of dormant oncogenes could potentially occur in normal cells. However, it was shown that while decitabine dramatically decreased proliferation of different types of human cancer cell lines, the growth of the control normal cells was unaffected [96]. A more recent study where gene re-expression was measured in response to decitabine in normal cells compared to tumor cells indicated that normal cells were less sensitive to such drug-induced gene-re-activation [97]. Based on multiple animal models and cell line studies, DMTIs such as decitabine have also been shown to induce the re-expression of genes involved in the regulation of apoptosis, cell cycle regulation or the major histocompatibility complex (MHC) and to therefore promote the anti-tumor immune response and to block tumor progression [98–101]. Finally, clinical trials for patients with leukemia or myelodysplastic syndrome treated with decitabine were promising and patients showed little side effects [97]. All these data together indicate that a treatment strategy implicating DMTIs would not only lead to re-expression of PKD1 and subsequent inhibition of local invasion and metastasis, but also upregulate the expression of other genes targeting tumor growth, both of which are beneficial for patients diagnosed with invasive breast cancers and limited treatment options.
It also has been suggested that combination treatments with other chemotherapeutic agents could increase efficiency of DMTIs. In invasive breast cancer cells (MDA-MB-231, BT20, HCC1954) a combination of DMTIs with PKD1 activators has been shown to significantly reduce their invasive potential in 2D and 3D cell culture, as compared to when drugs were used alone [78]. In this study, PKD1 activation was achieved using suramin (8,8′-{carbonylbis[imino-3,1-phenylenecarbonylimino(4-methyl-3,1-phenylene)carbonyl-imino]}di(1,3,5-naphthalenetrisulfonic acid). Suramin, a compound that previously had been shown to inhibit tumor growth and angiogenesis of metastatic breast cancer, is also a direct activator of PKD1 in vitro [102] and was used successfully in breast cancer cells in combination with DMTIs [103, 104]. Other PKD1 activators that potentially could be used include curcumin [105], a natural polyphenolic compound found in turmeric that has anti-inflammatory, anti-tumor and anti-oxidant functions [106, 107]. The additional use of PKD activators raises the question of how these compounds impact the two other isoforms in invasive breast cancer cells. However, published data regarding the predominant role of PKD1 as an inhibitor of cell motility, suggest that whether or not PKD2 or PKD3 are present, once PKD1 is re-expressed and active it will block the invasive phenotype. It is imperative that PKD activators are only used in combination with DMTIs, since activation of PKD2 and PKD3 in absence of PKD1 expression could lead to a complete opposite phenotype and promote tumor progression.
Chemical inhibition of oncogenic PKD2/PKD3 signaling to inhibit tumor growth, metastasis and chemoresistance
Since PKD2 and PKD3 in invasive breast cancers have been associated with increased tumor growth, metastatic progression, multi-drug resistance as well as decreased metastasis-free survival [23, 24, 36, 42, 43, 108], an alternative to a PKD1 re-expression strategy, is to target these kinases with small molecule inhibitors. For example, CID755673 and its analogs [109, 110], 3,5-diarylazoles [111], 2,6-naphthyridine and bipyridyl inhibitors and analogs [112], as well as CRT0066101 [113] and CRT5 [114] have been shown to inhibit PKD activity in vitro and in different cell lines. An issue with most of these compounds is that although they are effective in vitro in blocking proliferation, migration and invasion [115], they are metabolized too quickly when administered to mouse xenograft models. Among all these compounds only CRT0066101 has been used successfully in vivo in xenograft models of pancreatic [113] and colorectal [75] cancer as well as in a model of metastatic progression of breast cancer [23]. In mice with orthotopic ER-negative breast cancers, CRT0066101 significantly reduced primary tumor growth, local invasion, and metastasis to distant organs in vivo without showing any side effects. More importantly, the same results were obtained with specific knockdown of PKD3 suggesting that this kinase is the main target of CRT0066101 in ER-negative breast cancer cells [23]. It is however possible, since CRT0066101 is administered orally, that some of the additional anti-tumorigenic effects observed in the CRT0066101-treated model are due to systemic inhibition of PKD-mediated angiogenesis [79, 116, 117]. Of course, a potential problem with using pan inhibitors is the management of detrimental off-target effects and to combat this problem the specificity of each compound must be fully investigated. CRT0066101 appears to be a promising candidate since no detrimental effects have been observed in all the models tested. For the treatment of advanced breast cancers that do not express PKD1, pan PKD inhibitors could be even more effective if used in combination with current chemotherapeutic agents for which PKD2 or PKD3 have been associated to mediate multi-drug resistance [42, 43, 108].
Alternative approaches to chemical inhibition, could include nucleic acid-based therapies such as micro-RNA replacement therapy or systemic delivery of siRNA that could allow to specifically target the oncogenic PKDs [118–120]. Such strategies have been used successfully in animals suggesting that the therapeutic applications in humans could be very promising [121, 122]. Using the miRGen 2.0 database (http://diana.cslab.ece.ntua.gr/mirgen/) [123], 18 potential miRNAs were found to be complementary with the PRKD1 mRNA sequence but none with PRKD2 and PRKD3 mRNA. Thus, the development of miRNA mimics specific for both kinases, as well as a stable systemic or targeted delivery system, need further investigation. However, targeting breast cancer cells with siRNA specific for PKD isoforms seams feasible. For example, shRNA-mediated depletion of PKD3 had a strong impact on growth and the metastatic potential of estrogen receptor negative tumors, similar as results obtained with CRT0066101 [23].
Conclusions
Evidence from multiple studies investigating the role of PKD isoforms in the development and metastatic progression of breast cancer suggests disparate functions. In summary, PKD1 contributes to proliferation but blocks EMT and inhibits invasion, while the two other isoforms promote proliferation, migration and multi-drug-resistance of breast cancer cells. These incongruent functions of PKD family members in breast cancer can in part be attributed to substrate specificity and structural differences. Because PKD1 expression is lost in the most aggressive type of breast cancer, whereas the oncogenic version PKD3 is upregulated, PKDs can be valid therapeutic targets. The efficacy of the aforementioned treatment options (reactivation of PKD1 or use of pan PKD inhibitors) would require reliable diagnostic tools to detect the expression of each PKD isoform and to predict response. Patients with down-regulated PKD1 are the ideal candidates for both treatment strategies, and data obtained from large cohorts of breast cancer cell lines, breast tissue biopsies as well as from clinical datasets suggests that patients with TNBC or ER-breast cancer would benefit the most from PKD-based therapies. Collectively, the results obtained with preclinical orthotopic animal models using both PKD-based treatment regimens have been promising. Nonetheless, additional work is needed to determine the efficacy of pan PKD inhibitors in conjunction with currently used chemotherapeutics and to elucidate in even further detail the mechanisms which govern the expression and effects of PKD isoforms in breast cancer.
Acknowledgments
This work was supported by the NIH grants GM086435, CA184527 and a Pilot Project grant from the Mayo Clinic Breast Cancer SPORE (CA116201-03DR4) to PS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The funders had no role in decision to publish, or preparation of the manuscript.
Abbreviations
- CaMK
Calcium/calmodulin-dependent kinase
- CML
Chronic myelogenous leukemia
- S6K1
S6 Kinase 1
- DMTI
DNA methyltransferase inhibitors
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial to mesenchymal transition
- ER
Estrogen receptor
- ERE
Estrogen response element
- GABP
GA-binding protein
- GIT1
G-protein-coupled receptor kinase-interacting protein 1
- IDC
Invasive ductal carcinoma
- LIMK
LIM domain kinase
- MMP
Matrix metalloproteinase
- mTORC1
Mammalian target of rapamycin complex 1
- PAK4
p21-activated kinase 4
- PDZ
Postsynaptic density 95/Discs large/zona occludens 1
- PKC
Protein kinase C
- PKD
Protein kinase D
- RIN1
Ras and Rab interactor 1
- SSH1L
Slingshot 1L
- TNBC
Triple-negative breast cancer
References
- 1.Johannes FJ, Prestle J, Eis S, Oberhagemann P, Pfizenmaier K. PKCu is a novel, atypical member of the protein kinase C family. J Biol Chem. 1994;269:6140–6148. [PubMed] [Google Scholar]
- 2.Valverde AM, Sinnett-Smith J, Van Lint J, Rozengurt E. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc Natl Acad Sci USA. 1994;91:8572–8576. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturany S, Van Lint J, Muller F, Wilda M, Hameister H, Hocker M, Brey A, Gern U, Vandenheede J, Gress T, Adler G, Seufferlein T. Molecular cloning and characterization of the human protein kinase D2. A novel member of the protein kinase D family of serine threonine kinases. J Biol Chem. 2001;276:3310–3318. doi: 10.1074/jbc.M008719200. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi A, Seki N, Hattori A, Kozuma S, Saito T. PKCnu, a new member of the protein kinase C family, composes a fourth subfamily with PKCmu. Biochim Biophys Acta. 1999;1450:99–106. doi: 10.1016/S0167-4889(99)00040-3. [DOI] [PubMed] [Google Scholar]
- 5.Eiseler T, Doppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11:R13. doi: 10.1186/bcr2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K. Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J Biol Chem. 2010;285:18672–18683. doi: 10.1074/jbc.M109.093880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–5638. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 8.Wong C, Jin ZG. Protein kinase C-dependent protein kinase D activation modulates ERK signal pathway and endothelial cell proliferation by vascular endothelial growth factor. J Biol Chem. 2005;280:33262–33269. doi: 10.1074/jbc.M503198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhukova E, Sinnett-Smith J, Rozengurt E. Protein kinase D potentiates DNA synthesis and cell proliferation induced by bombesin, vasopressin, or phorbol esters in Swiss 3T3 cells. J Biol Chem. 2001;276:40298–40305. doi: 10.1074/jbc.M106512200. [DOI] [PubMed] [Google Scholar]
- 10.Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ, Malhotra V. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastea LI, Doppler H, Balogun B, Storz P. Protein kinase D1 maintains the epithelial phenotype by inducing a DNA-bound, inactive SNAI1 transcriptional repressor complex. PLoS One. 2012;7:e30459. doi: 10.1371/journal.pone.0030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res. 2010;70:7810–7819. doi: 10.1158/0008-5472.CAN-09-4481. [DOI] [PubMed] [Google Scholar]
- 13.Qin L, Zeng H, Zhao D. Requirement of protein kinase D tyrosine phosphorylation for VEGF-A165-induced angiogenesis through its interaction and regulation of phospholipase Cgamma phosphorylation. J Biol Chem. 2006;281:32550–32558. doi: 10.1074/jbc.M604853200. [DOI] [PubMed] [Google Scholar]
- 14.Ha CH, Wang W, Jhun BS, Wong C, Hausser A, Pfizenmaier K, McKinsey TA, Olson EN, Jin ZG. Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J Biol Chem. 2008;283:14590–14599. doi: 10.1074/jbc.M800264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br J Dermatol. 2006;154:586–593. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
- 16.Trauzold A, Schmiedel S, Sipos B, Wermann H, Westphal S, Roder C, Klapper W, Arlt A, Lehnert L, Ungefroren H, Johannes FJ, Kalthoff H. PKCmu prevents CD95-mediated apoptosis and enhances proliferation in pancreatic tumour cells. Oncogene. 2003;22:8939–8947. doi: 10.1038/sj.onc.1207001. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68:3844–3853. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 18.Jaggi M, Rao PS, Smith DJ, Hemstreet GP, Balaji KC. Protein kinase C mu is down-regulated in androgen-independent prostate cancer. Biochem Biophys Res Commun. 2003;307:254–260. doi: 10.1016/S0006-291X(03)01161-6. [DOI] [PubMed] [Google Scholar]
- 19.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 20.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 21.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010;23(Suppl 2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 22.Borges S, Doppler H, Perez EA, Andorfer CA, Sun Z, Anastasiadis PZ, Thompson E, Geiger XJ, Storz P. Pharmacologic reversion of epigenetic silencing of the PRKD1 promoter blocks breast tumor cell invasion and metastasis. Breast Cancer Res. 2013;15:R66. doi: 10.1186/bcr3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges S, Perez EA, Thompson EA, Radisky DC, Geiger XJ, Storz P. Effective targeting of estrogen receptor-negative breast cancers with the protein kinase D inhibitor CRT0066101. Mol Cancer Ther. 2015;14:1306–1316. doi: 10.1158/1535-7163.MCT-14-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huck B, Duss S, Hausser A, Olayioye MA. Elevated protein kinase D3 (PKD3) expression supports proliferation of triple-negative breast cancer cells and contributes to mTORC1-S6K1 pathway activation. J Biol Chem. 2014;289:3138–3147. doi: 10.1074/jbc.M113.502633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toker A, Marmiroli S. Signaling specificity in the Akt pathway in biology and disease. Adv Biol Regul. 2014;55:28–38. doi: 10.1016/j.jbior.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, Rubin CS. Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 2011;12:785–796. doi: 10.1038/embor.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Jean-Mairet RM, Higuero AM, Iglesias T. Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem. 2006;281:18888–18900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- 28.Matthews SA, Iglesias T, Rozengurt E, Cantrell D. Spatial and temporal regulation of protein kinase D (PKD) EMBO J. 2000;19:2935–2945. doi: 10.1093/emboj/19.12.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nhek S, Ngo M, Yang X, Ng MM, Field SJ, Asara JM, Ridgway ND, Toker A. Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol Biol Cell. 2010;21:2327–2337. doi: 10.1091/mbc.E10-02-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldron RT, Rozengurt E. Protein kinase C phosphorylates protein kinase D activation loop Ser744 and Ser748 and releases autoinhibition by the pleckstrin homology domain. J Biol Chem. 2003;278:154–163. doi: 10.1074/jbc.M208075200. [DOI] [PubMed] [Google Scholar]
- 31.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–8530. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rey O, Yuan J, Rozengurt E. Intracellular redistribution of protein kinase D2 in response to G-protein-coupled receptor agonists. Biochem Biophys Res Commun. 2003;302:817–824. doi: 10.1016/S0006-291X(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 33.Rey O, Yuan J, Young SH, Rozengurt E. Protein kinase C nu/protein kinase D3 nuclear localization, catalytic activation, and intracellular redistribution in response to G protein-coupled receptor agonists. J Biol Chem. 2003;278:23773–23785. doi: 10.1074/jbc.M300226200. [DOI] [PubMed] [Google Scholar]
- 34.von Blume J, Knippschild U, Dequiedt F, Giamas G, Beck A, Auer A, Van Lint J, Adler G, Seufferlein T. Phosphorylation at Ser244 by CK1 determines nuclear localization and substrate targeting of PKD2. EMBO J. 2007;26:4619–4633. doi: 10.1038/sj.emboj.7601891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huck B, Kemkemer R, Franz-Wachtel M, Macek B, Hausser A, Olayioye MA. GIT1 phosphorylation on serine 46 by PKD3 regulates paxillin trafficking and cellular protrusive activity. J Biol Chem. 2012;287:34604–34613. doi: 10.1074/jbc.M112.374652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao Q, McKenzie R, Gan H, Tang H. Protein kinases D2 and D3 are novel growth regulators in HCC1806 triple-negative breast cancer cells. Anticancer Res. 2013;33:393–399. [PubMed] [Google Scholar]
- 37.Onodera Y, Nam JM, Hashimoto A, Norman JC, Shirato H, Hashimoto S, Sabe H. Rab5c promotes AMAP1-PRKD2 complex formation to enhance beta1 integrin recycling in EGF-induced cancer invasion. J Cell Biol. 2012;197:983–996. doi: 10.1083/jcb.201201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eiseler T, Doppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–556. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 40.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van ’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR (2012) The landscape of cancer genes and mutational processes in breast cancer. Nature 486:400–404 [DOI] [PMC free article] [PubMed]
- 41.Karam M, Bieche I, Legay C, Vacher S, Auclair C, Ricort JM. Protein kinase D1 regulates ERalpha-positive breast cancer cell growth response to 17beta-estradiol and contributes to poor prognosis in patients. J Cell Mol Med. 2014;18:2536–2552. doi: 10.1111/jcmm.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alpsoy A, Gunduz U (2015) Protein kinase D2 silencing reduced motility of doxorubicin-resistant MCF7 cells. Tumour Biol. doi:10.1007/s13277-015-3081-3 [DOI] [PubMed]
- 43.Chen J, Lu L, Feng Y, Wang H, Dai L, Li Y, Zhang P. PKD2 mediates multi-drug resistance in breast cancer cells through modulation of P-glycoprotein expression. Cancer Lett. 2011;300:48–56. doi: 10.1016/j.canlet.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Yang ZF, Zhang H, Ma L, Peng C, Chen Y, Wang J, Green MR, Li S, Rosmarin AG. GABP transcription factor is required for development of chronic myelogenous leukemia via its control of PRKD2. Proc Natl Acad Sci USA. 2013;110:2312–2317. doi: 10.1073/pnas.1212904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marques M, Laflamme L, Gaudreau L. Estrogen receptor alpha can selectively repress dioxin receptor-mediated gene expression by targeting DNA methylation. Nucleic Acids Res. 2013;41:8094–8106. doi: 10.1093/nar/gkt595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karam M, Legay C, Auclair C, Ricort JM. Protein kinase D1 stimulates proliferation and enhances tumorigenesis of MCF-7 human breast cancer cells through a MEK/ERK-dependent signaling pathway. Exp Cell Res. 2012;318:558–569. doi: 10.1016/j.yexcr.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Zheng H, Shen M, Zha YL, Li W, Wei Y, Blanco MA, Ren G, Zhou T, Storz P, Wang HY, Kang Y. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell. 2014;26:358–373. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziegler S, Eiseler T, Scholz RP, Beck A, Link G, Hausser A. A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol Biol Cell. 2011;22:570–580. doi: 10.1091/mbc.E10-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuno K. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal. 2013;25:457–469. doi: 10.1016/j.cellsig.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 53.Olayioye MA, Barisic S, Hausser A. Multi-level control of actin dynamics by protein kinase D. Cell Signal. 2013;25:1739–1747. doi: 10.1016/j.cellsig.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Storz P. Protein kinase D1: a novel regulator of actin-driven directed cell migration. Cell Cycle. 2009;8:1975–1976. doi: 10.4161/cc.8.13.9016. [DOI] [PubMed] [Google Scholar]
- 55.Spratley SJ, Bastea LI, Doppler H, Mizuno K, Storz P. Protein kinase D regulates cofilin activity through p21-activated kinase 4. J Biol Chem. 2011;286:34254–34261. doi: 10.1074/jbc.M111.259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bastea LI, Doppler H, Pearce SE, Durand N, Spratley SJ, Storz P. Protein kinase D-mediated phosphorylation at Ser99 regulates localization of p21-activated kinase 4. Biochem J. 2013;455:251–260. doi: 10.1042/BJ20130281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 59.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 60.Kohrmann A, Kammerer U, Kapp M, Dietl J, Anacker J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: new findings and review of the literature. BMC Cancer. 2009;9:188. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 62.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 63.Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C, Garcia de Herreros A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol. 2003;23:5078–5089. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 65.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 66.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Du C, Jaggi M, Zhang C, Balaji KC. Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res. 2009;69:1117–1124. doi: 10.1158/0008-5472.CAN-07-6270. [DOI] [PubMed] [Google Scholar]
- 68.Jaggi M, Chauhan SC, Du C, Balaji KC. Bryostatin 1 modulates beta-catenin subcellular localization and transcription activity through protein kinase D1 activation. Mol Cancer Ther. 2008;7:2703–2712. doi: 10.1158/1535-7163.MCT-08-0119. [DOI] [PubMed] [Google Scholar]
- 69.Sundram V, Ganju A, Hughes JE, Khan S, Chauhan SC, Jaggi M. Protein kinase D1 attenuates tumorigenesis in colon cancer by modulating beta-catenin/T cell factor activity. Oncotarget. 2014;5:6867–6884. doi: 10.18632/oncotarget.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iseri OD, Kars MD, Arpaci F, Atalay C, Pak I, Gunduz U. Drug resistant MCF-7 cells exhibit epithelial-mesenchymal transition gene expression pattern. Biomed Pharmacother. 2011;65:40–45. doi: 10.1016/j.biopha.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 71.Tezcan O, Gunduz U. Vimentin silencing effect on invasive and migration characteristics of doxorubicin resistant MCF-7 cells. Biomed Pharmacother. 2014;68:357–364. doi: 10.1016/j.biopha.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 72.Tan M, Hao F, Xu X, Chisolm GM, Cui MZ. Lysophosphatidylcholine activates a novel PKD2-mediated signaling pathway that controls monocyte migration. Arterioscler Thromb Vasc Biol. 2009;29:1376–1382. doi: 10.1161/ATVBAHA.109.191585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou Z, Zeng F, Xu W, Wang C, Ke Z, Wang QJ, Deng F. PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci. 2012;125:4800–4811. doi: 10.1242/jcs.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doppler H, Bastea LI, Borges S, Spratley SJ, Pearce SE, Storz P. Protein kinase d isoforms differentially modulate cofilin-driven directed cell migration. PLoS One. 2014;9:e98090. doi: 10.1371/journal.pone.0098090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei N, Chu E, Wipf P, Schmitz JC. Protein kinase d as a potential chemotherapeutic target for colorectal cancer. Mol Cancer Ther. 2014;13:1130–1141. doi: 10.1158/1535-7163.MCT-13-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X, Xue P, Yang M, Shi H, Lu D, Wang Z, Shi Q, Hu J, Xie S, Zhan W, Yu R. Protein kinase D2 promotes the proliferation of glioma cells by regulating Golgi phosphoprotein 3. Cancer Lett. 2014;355:121–129. doi: 10.1016/j.canlet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 77.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borges S, Doppler HR, Storz P. A combination treatment with DNA methyltransferase inhibitors and suramin decreases invasiveness of breast cancer cells. Breast Cancer Res Treat. 2014;144:79–91. doi: 10.1007/s10549-014-2857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Azoitei N, Pusapati GV, Kleger A, Moller P, Kufer R, Genze F, Wagner M, van Lint J, Carmeliet P, Adler G, Seufferlein T. Protein kinase D2 is a crucial regulator of tumour cell-endothelial cell communication in gastrointestinal tumours. Gut. 2010;59:1316–1330. doi: 10.1136/gut.2009.206813. [DOI] [PubMed] [Google Scholar]
- 80.Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, Balaji KC. E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 2005;65:483–492. [PubMed] [Google Scholar]
- 81.Kim M, Jang HR, Kim JH, Noh SM, Song KS, Cho JS, Jeong HY, Norman JC, Caswell PT, Kang GH, Kim SY, Yoo HS, Kim YS. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 2008;29:629–637. doi: 10.1093/carcin/bgm291. [DOI] [PubMed] [Google Scholar]
- 82.Doppler H, Bastea LI, Eiseler T, Storz P. Neuregulin mediates F-actin-driven cell migration through inhibition of protein kinase D1 via Rac1 protein. J Biol Chem. 2013;288:455–465. doi: 10.1074/jbc.M112.397448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Auvinen P, Rilla K, Tumelius R, Tammi M, Sironen R, Soini Y, Kosma VM, Mannermaa A, Viikari J, Tammi R. Hyaluronan synthases (HAS1-3) in stromal and malignant cells correlate with breast cancer grade and predict patient survival. Breast Cancer Res Treat. 2014;143:277–286. doi: 10.1007/s10549-013-2804-7. [DOI] [PubMed] [Google Scholar]
- 84.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 85.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 86.Wang WJ, Lei YY, Mei JH, Wang CL. Recent progress in HER2 associated breast cancer. Asian Pac J Cancer Prev. 2015;16:2591–2600. doi: 10.7314/APJCP.2015.16.7.2591. [DOI] [PubMed] [Google Scholar]
- 87.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 88.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 89.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 90.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg962. [DOI] [PubMed] [Google Scholar]
- 91.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 92.Cooper SJ, von Roemeling CA, Kang KH, Marlow LA, Grebe SK, Menefee ME, Tun HW, Colon-Otero G, Perez EA, Copland JA. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol Cancer Ther. 2012;11:2105–2115. doi: 10.1158/1535-7163.MCT-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thummuri D, Kumar S, Surapaneni SK, Tikoo K (2015) Epigenetic regulation of protein tyrosine phosphatase PTPN12 in triple-negative breast cancer. Life Sci 130:73–80 [DOI] [PubMed]
- 94.Skliris GP, Munot K, Bell SM, Carder PJ, Lane S, Horgan K, Lansdown MR, Parkes AT, Hanby AM, Markham AF, Speirs V. Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003;201:213–220. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- 95.Zhu WG, Hileman T, Ke Y, Wang P, Lu S, Duan W, Dai Z, Tong T, Villalona-Calero MA, Plass C, Otterson GA. 5-aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J Biol Chem. 2004;279:15161–15166. doi: 10.1074/jbc.M311703200. [DOI] [PubMed] [Google Scholar]
- 96.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 97.Yang AS, Estecio MR, Garcia-Manero G, Kantarjian HM, Issa JP (2003) Comment on “Chromosomal instability and tumors promoted by DNA hypomethylation” and “Induction of tumors in mice by genomic hypomethylation”. Science 302:1153 author reply 1153 [DOI] [PubMed]
- 98.Adair SJ, Hogan KT. Treatment of ovarian cancer cell lines with 5-aza-2′-deoxycytidine upregulates the expression of cancer-testis antigens and class I major histocompatibility complex-encoded molecules. Cancer Immunol Immunother. 2009;58:589–601. doi: 10.1007/s00262-008-0582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nie J, Liu L, Li X, Han W. Decitabine, a new star in epigenetic therapy: the clinical application and biological mechanism in solid tumors. Cancer Lett. 2014;354:12–20. doi: 10.1016/j.canlet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 100.Vijayaraghavalu S, Dermawan JK, Cheriyath V, Labhasetwar V. Highly synergistic effect of sequential treatment with epigenetic and anticancer drugs to overcome drug resistance in breast cancer cells is mediated via activation of p21 gene expression leading to G2/M cycle arrest. Mol Pharm. 2013;10:337–352. doi: 10.1021/mp3004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA, Brown R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–4576. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 102.Gschwendt M, Kittstein W, Johannes FJ. Differential effects of suramin on protein kinase C isoenzymes. A novel tool for discriminating protein kinase C activities. FEBS Lett. 1998;421:165–168. doi: 10.1016/S0014-5793(97)01530-5. [DOI] [PubMed] [Google Scholar]
- 103.Gradishar WJ, Soff G, Liu J, Cisneros A, French S, Rademaker A, Benson AB, 3rd, Bouck N. A pilot trial of suramin in metastatic breast cancer to assess antiangiogenic activity in individual patients. Oncology. 2000;58:324–333. doi: 10.1159/000012120. [DOI] [PubMed] [Google Scholar]
- 104.Lustberg MB, Pant S, Ruppert AS, Shen T, Wei Y, Chen L, Brenner L, Shiels D, Jensen RR, Berger M, Mrozek E, Ramaswamy B, Grever M, Au JL, Wientjes MG, Shapiro CL. Phase I/II trial of non-cytotoxic suramin in combination with weekly paclitaxel in metastatic breast cancer treated with prior taxanes. Cancer Chemother Pharmacol. 2012;70:49–56. doi: 10.1007/s00280-012-1887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sundram V, Chauhan SC, Ebeling M, Jaggi M. Curcumin attenuates beta-catenin signaling in prostate cancer cells through activation of protein kinase D1. PLoS One. 2012;7:e35368. doi: 10.1371/journal.pone.0035368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goel A, Jhurani S, Aggarwal BB. Multi-targeted therapy by curcumin: how spicy is it? Mol Nutr Food Res. 2008;52:1010–1030. doi: 10.1002/mnfr.200700354. [DOI] [PubMed] [Google Scholar]
- 107.Yallapu MM, Othman SF, Curtis ET, Bauer NA, Chauhan N, Kumar D, Jaggi M, Chauhan SC. Curcumin-loaded magnetic nanoparticles for breast cancer therapeutics and imaging applications. Int J Nanomedicine. 2012;7:1761–1779. doi: 10.2147/IJN.S29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen J, Shen Q, Labow M, Gaither LA. Protein kinase D3 sensitizes RAF inhibitor RAF265 in melanoma cells by preventing reactivation of MAPK signaling. Cancer Res. 2011;71:4280–4291. doi: 10.1158/0008-5472.CAN-10-3761. [DOI] [PubMed] [Google Scholar]
- 109.George KM, Frantz MC, Bravo-Altamirano K, Lavalle CR, Tandon M, Leimgruber S, Sharlow ER, Lazo JS, Wang QJ, Wipf P. Design, synthesis, and biological evaluation of PKD inhibitors. Pharmaceutics. 2011;3:186–228. doi: 10.3390/pharmaceutics3020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem. 2008;283:33516–33526. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gamber GG, Meredith E, Zhu Q, Yan W, Rao C, Capparelli M, Burgis R, Enyedy I, Zhang JH, Soldermann N, Beattie K, Rozhitskaya O, Koch KA, Pagratis N, Hosagrahara V, Vega RB, McKinsey TA, Monovich L. 3,5-diarylazoles as novel and selective inhibitors of protein kinase D. Bioorg Med Chem Lett. 2011;21:1447–1451. doi: 10.1016/j.bmcl.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 112.Meredith EL, Ardayfio O, Beattie K, Dobler MR, Enyedy I, Gaul C, Hosagrahara V, Jewell C, Koch K, Lee W, Lehmann H, McKinsey TA, Miranda K, Pagratis N, Pancost M, Patnaik A, Phan D, Plato C, Qian M, Rajaraman V, Rao C, Rozhitskaya O, Ruppen T, Shi J, Siska SJ, Springer C, van Eis M, Vega RB, von Matt A, Yang L, Yoon T, Zhang JH, Zhu N, Monovich LG. Identification of orally available naphthyridine protein kinase D inhibitors. J Med Chem. 2010;53:5400–5421. doi: 10.1021/jm100075z. [DOI] [PubMed] [Google Scholar]
- 113.Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, Kelland L, Jamieson S, Sutherland R, Raynham T, Charles M, Bagherzadeh A, Foxton C, Boakes A, Farooq M, Maru D, Diagaradjane P, Matsuo Y, Sinnett-Smith J, Gelovani J, Krishnan S, Aggarwal BB, Rozengurt E, Ireson CR, Guha S. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2010;9:1136–1146. doi: 10.1158/1535-7163.MCT-09-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Evans IM, Bagherzadeh A, Charles M, Raynham T, Ireson C, Boakes A, Kelland L, Zachary IC. Characterization of the biological effects of a novel protein kinase D inhibitor in endothelial cells. Biochem J. 2010;429:565–572. doi: 10.1042/BJ20100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lavalle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, Wipf P, Wang QJ. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5. doi: 10.1186/1472-6769-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ochi N, Tanasanvimon S, Matsuo Y, Tong Z, Sung B, Aggarwal BB, Sinnett-Smith J, Rozengurt E, Guha S. Protein kinase D1 promotes anchorage-independent growth, invasion, and angiogenesis by human pancreatic cancer cells. J Cell Physiol. 2011;226:1074–1081. doi: 10.1002/jcp.22421. [DOI] [PubMed] [Google Scholar]
- 117.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda) 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nourbakhsh M, Jaafari MR, Lage H, Abnous K, Mosaffa F, Badiee A, Behravan J. Nanolipoparticles-mediated MDR1 siRNA delivery reduces doxorubicin resistance in breast cancer cells and silences MDR1 expression in xenograft model of human breast cancer. Iran J Basic Med Sci. 2015;18:385–392. [PMC free article] [PubMed] [Google Scholar]
- 120.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 121.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 122.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, Zeng YX, Shao JY. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:R2. doi: 10.1186/bcr2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alexiou P, Vergoulis T, Gleditzsch M, Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T, Hatzigeorgiou AG. miRGen 2.0: a database of microRNA genomic information and regulation. Nucleic Acids Res. 2010;38:D137–D141. doi: 10.1093/nar/gkp888. [DOI] [PMC free article] [PubMed] [Google Scholar]