Abstract
Lineage tracing is a method that delineates all progeny produced by a single cell or a group of cells. The possibility of performing lineage tracing initiated the field of Developmental Biology, and continues to revolutionize Stem Cell Biology. Here, I introduce the principles behind a successful lineage-tracing experiment. In addition, I summarize and compare different methods for conducting lineage tracing and provide examples of how these strategies can be implemented to answer fundamental questions in development and regeneration. The advantages and limitations of each method are also discussed.
Introduction
In a lineage-tracing experiment, the cells of interest are marked at one timepoint, and the progeny derived from these marked cells are revealed at a later timepoint. In the 1870s, Charles Otis Whitman observed the early division of leech embryos and followed the fate of individual cells from the one cell stage to the germ-layer stage [1, 2]. This seminal work suggested that a definite developmental fate could be assigned to each cell in the early cleavage eggs and its clonal progeny. As such, cell fate determination is not a stochastic process as previously speculated. Since the early 20th century, developmental biologists have developed numerous ways for tracking descendants produced by specific cells, with the desire to unravel how a complex organism develops from a single cell. The same principle has now been widely adapted by stem cell biologists, as the central theme of adult stem cell biology is to understand how a diverse array of cell types is formed and maintained. In fact, lineage tracing remains the most rigorous method to define adult stem cells for a given tissue.
Although the actual strategies evolve with time, a successful lineage-tracing experiment always needs to fulfill the following three requirements: (1) A careful assessment of the cells that are marked at the initial timepoint, so that the starting populations are clearly defined. (2) The markers used to mark the cells remain exclusively in the original cells and their progeny and will not diffuse to the neighboring cells. (3) These markers are sufficiently stable and are not toxic to the cells during the entire tracing period. Violation of any of these requirements can result in labeling of unrelated cells or alteration in cell behavior, thus leading to misinterpretation of the tracing results.
Below, some of the most commonly used lineage-tracing strategies are summarized, beginning with historical perspectives, followed by recent notable examples. Understanding the pros and cons and the underlying principles of each tracing method can greatly facilitate experimental design and data interpretation.
Nonselective Markers
Many membrane, cytoplasmic, and nuclear dyes have been developed for a wide variety of applications. Although these dyes are often nonselective in terms of which cells get labeled, when combined with carefully designed strategies, it is sometimes possible to label only a specific subset of cells. This nonselective nature can therefore become advantageous as it can be used when genetic labeling methods (see below) are not feasible. Since each marker has distinct properties, it is particularly important to keep in mind the three abovementioned requirements and evaluate if the cells of interest are indeed specifically labeled.
Vital Dye
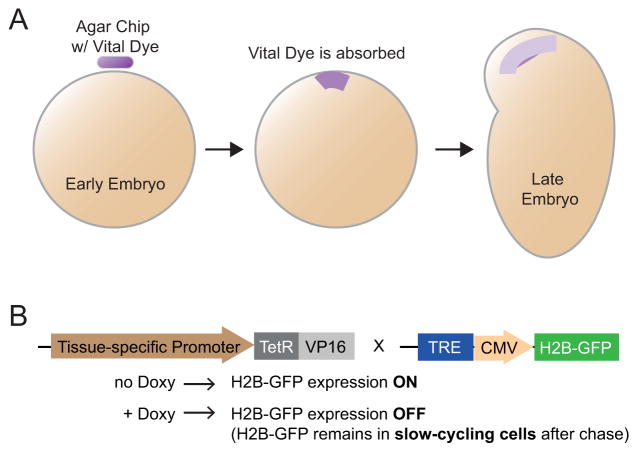
Using a colored substrate to label cells seems like an intuitive idea—a dyed cell can be distinguished easily from the rest of the tissues. However, how a dye can be specifically applied to a small number of cells and whether the dye is harmful to the cells are among the biggest challenges. In 1929, embryologist Walter Vogt pioneered the use of “vital dye” (a dye that stains but does not kill cells) to study cell fate in Xenopus embryos. He implanted a tiny agar chip containing Nile Blue on top of the cells of interests. The dye is absorbed by the cells underneath the chip, and the fate of the labeled cells can be followed over time (Fig. 1A). By altering the position of the chip, Vogt was able to label different areas of the cleavage embryo. The information gathered from this approach allowed Vogt to construct a “fate map” of the 32-cell blastula Xenopus embryos [3].
Figure 1.
Lineage tracing with nonselective dyes. (A): Strategies employed by Walter Vogt to mark small areas of embryos with vital dyes. (B): Schematic representation of the bitransgenic strategy to mark slow-cycling cells (Doxy: Doxycyclin; TetR: Tet Repressor; TRE: Tetracycline Response Element)
Carbocyanine Dyes and Dextrans
Vital dyes are water-soluble and might diffuse to neighboring cells, which violates the second principle of lineage tracing. As a result, a dye-positive cell can either be the progeny of the cells labeled originally or an unrelated cell that absorbs the diffused dye. The invention of carbocyanine dyes (lipid-soluble but water-insoluble dyes) such as DiI or DiO and high-molecular-weight dextrans (large molecules that cannot pass through cell junctions) conjugated with fluorochromes circumvented this issue. These molecules are often directly injected into specific cell(s) of interests, which requires substantial technical skills. In addition, they are diluted through each round of cell division. Therefore, these dyes are less effective for labeling highly proliferating cells. Nevertheless, these dyes were successfully applied to studying the development of trunk neural crests and the endothelial cell lineage [4, 5] and were used to construct an impressive fate map of zebrafish neural plate [6].
Nucleotide Pulse-Chase
Initially designed as a method to identify slow-cycling cells within a tissue, nucleotide pulse-chase experiments can also be used to determine lineages. The idea is simple: when providing a Thymidine analog (3H-Thymidine, BrdU, or EdU) to an organism at a specific time (pulse), all the proliferating cells will incorporate the labeled Thymidine into their newly synthesized DNA. However, labeled cells that continue to proliferate after the pulse period will dilute out the incorporated Thymidine analog through each round of division. On the other hand, cells that do not proliferate much after the initial pulse will retain the labeled Thymidine analog even after a long period of time (chase). The fate of these label-retaining cells (LRCs) can therefore be followed using the incorporated nucleotide as a marker. Inspired by this idea, Tumbar et al. further developed a bi-transgenic strategy that uses GFP-labeled histone H2B (H2B-GFP). In this approach, H2B-GFP is initially induced in all of the cells intended to be monitored (pulse). During the chase period, the synthesis of new H2B-GFP is turned off. Cells that are slow-cycling retain their H2B-GFP, while fast-cycling cells dilute their H2B-GFP and become GFP negative [7] (Fig. 1B).
By contrast, if only the cells of interest are proliferative and all the other cells are quiescent at a specific timepoint, it is also possible to introduce a short pulse of BrdU and follow the fate of these proliferating cells before BrdU becomes too diluted to detect. For this strategy to work effectively, a thorough understanding of proliferation timing and division pattern within a tissue is essential. One successful example is seen in determining the fate of outer root sheath (ORS) cells in the hair follicle [8]. By first performing a detailed proliferation analysis of the hair follicle, specific parts of ORS cells can be labeled with either H2BGFP or BrdU by altering the timing of pulse and chase. Since different parts of ORS cells share the same genetic markers, it would otherwise be impossible to determine their fate using genetic strategies.
One major pitfall of using radioactive 3H-Thymidine or BrdU/EdU to label cells is its potential cytotoxicity. While the toxicity of short-term BrdU labeling is minimal, 10-day continuous BrdU labeling was shown to force normally dormant hematopoietic stem cells (HSCs) into cycle, possibly by damaging downstream fast-cycling hematopoietic lineages that become heavily loaded with BrdU [9]. Nonradioactive isotope, on the other hand, has no known toxicity and may be particularly suitable for long-term labeling. For example, Senyo et al. administrated 15N Thymidine to mice for over 8 weeks to label the rare-dividing cardiomyocytes [10]. With multi-isotope imaging mass spectrometry to visualize 15N Thymidine positive cells, they showed that newly generated cardiomyocytes are generated from existing cardiomyocytes, not from a putative progenitor population.
Carbon Dating
Because of its invasive nature, performing lineage-tracing experiments in humans may seem like wishful thinking. The nontoxic nature of stable isotopes opens exciting potential to perform lineage analysis in healthy individuals [11]. However, this approach is restricted to tissues that can be harvested. In this regard, an innovative carbon dating strategy might provide some possibilities to birthdate and trace cell fate in other human tissues.
The level of atmospheric isotope Carbon14 (14C) was elevated during mid-1950s to early 1960s because of intensive nuclear weapon testing and declined dramatically after that due to diffusion and equilibration with the biosphere. This peak and decline of 14C levels resulted in different amounts of 14C being incorporated into the DNA depending on the time when DNA was synthesized. Since 14C is extremely stable, Frisén and colleagues developed a method that uses this different amount of 14C to retrospectively birthdate cells in the human body [12].
Lineage relationships deduced from carbon dating data are based on the fact that a cell population with an earlier birthdate can be the origin of cells with a later birthdate but not vice versa. Its application in lineage tracing is limited and correlative at best. That said, this strategy already provided some important clues in understanding neurogenesis in humans. Frisén’s group found that continued neurogenesis occurs in the adult human hippocampus, just as it does in mice. However, a sharp difference between these two species is seen in the subventricular zone (SVZ) [13]. In mice, continued neurogenesis in the SVZ generates neurons that migrate to the olfactory bulb. While in humans, neurogenesis can be detected in the SVZ, but instead of migrating into the olfactory bulb, these newborn neurons are likely to move to the striatum [12–14].
Sequencing-Based Technology
Random somatic mutations that occurred in individual cells are transmitted into progeny. These naturally occurring sequence variants can also be used as inherited genomic signatures that allow lineage relationships to be reconstructed. One important consideration is that somatic mutations can be used to construct cell lineages only if the mutations do not confer a growth advantage or disadvantage since such changes can bias lineage contributions. Based upon this idea, Ehud Shapiro’s group conducted DNA sequencing of the highly mutated DNA satellite regions and used this information to build cell lineage trees [15, 16]. They demonstrated that mouse lymphoma likely begins from a single aberrant cell [17]. In addition, they demonstrated that in acute myeloid leukemia, leukemia cells at relapse were closely related to their stem cell subpopulation, implying that relapse might have originated from the rarely dividing stem cells [18].
In principle, this strategy allows lineage trees to be constructed from any organisms and tissues, thereby representing one of the rare opportunities to analyze lineages in human organs. At the same time, it should be noted that, by itself, it cannot provide information regarding the exact cellular origin. Rather, it serves as a mean to understand the lineage relationships among all the sampled cells only.
Genetic Labeling
Rapid development of genetic tools since the 1990s completely transformed our approaches toward lineage tracing. By introducing genetic markers like fluorescent proteins or enzymes (such as beta-galactosidase and alkaline phosphatase) into cells, a wide variety of lineages have now been delineated.
Transplantation
Using genetic markers to trace cell fate is rooted in the generation of chimera embryos (embryos containing tissues from more than one genetic source). By transplanting trunk neural crest from an embryo of a pigmented strain into an unpigmented strain of chickens, Mary Rawles demonstrated elegantly that pigment-producing melanocytes that give the feathers their distinctive colors actually originate from the trunk neural crests[19].
In the 1970s, to study the developmental fate of cells in the avian embryos, Nicole Le Douarin pioneered the use of chick-quail chimeras. Chick and quail are similar in their development. Transplanted quail cells can be integrated into the chick embryos and participate in the development of the host tissues. With Feulgen DNA stain, condensed nucleolar-associated heterochromatin found only in quail cells can be used to distinguish quail cells from chick cells. A fine map of the central nervous system and skeletal system were constructed using this approach [20]. Le Douarin et al. also demonstrated the extraordinary migration ability of the neural crest cells [21].
One of the most notable applications of transplantation studies is the isolation of hematopoietic stem cells (HSCs). Weissman and colleagues used antibodies to enrich a population of bone marrow cells. By transplanting these cells into lethally irradiated mice, they demonstrated that these cells are able to reconstitute all blood cell types in the recipients. As such, the cells they isolated should comprise HSCs. This pioneer study demonstrated the fundamental characteristics of adult stem cells: self-renewal and multi-potency. Their work also ignited a large number of investigations aimed at identifying, characterizing, and purifying adult stem cells from a wide variety of tissues and organs [22].
It is also worth noting that many new studies demonstrated that cell behavior under transplantation and under steady state can be different. For example, when thymus epithelial cells are transplanted into the skin, they form epidermis and hair follicles instead of thymus [23]. In the mammary epithelium, transplantation studies suggest the presence of multipotent stem cells that can give rise to both luminal and myoepithelial lineages, while genetic lineage-tracing studies suggest that these two populations are maintained by distinct populations of unipotent stem cells [24]. In the corneal epithelium, grafting results indicate that grafted limbus cells do not migrate toward the corneal center [25], while in recent lineage-tracing advances, limbal cells were observed to migrate and participate in corneal homeostasis under steady state [26, 27]. A recent study also suggests that endogenous HSCs and transplanted HSCs may behave differently [28]. That said, although transplantation is no longer viewed as a “gold standard” way to define stem cells, it certainly has its indispensable historical values and remains clinically important.
Cre-Lox-Based Strategy
Site-specific genetic recombination, particularly through Cre-lox-based strategy, has become the most widely used method. Expression of Cre recombinase is under the control of a cell-type or tissue-specific promoter. This line is then crossed to a reporter line carrying enzymes or fluorescent proteins inserted in a ubiquitously expressed locus such as Rosa26. Reporter expression is prevented by a stop cassette flanked by two loxP sites. Upon Cre activation, the stop cassette is excised, allowing reporter genes to express in a cell-type specific manner (Fig. 2). Since the removal of the stop cassette is permanent, the reporter genes are expressed in all the progeny produced by the initial cells where the Cre is once activated [29–32]. The simplicity and unambiguous nature of this method and the wide variety of Cre lines available nowadays have made this approach even more accessible.
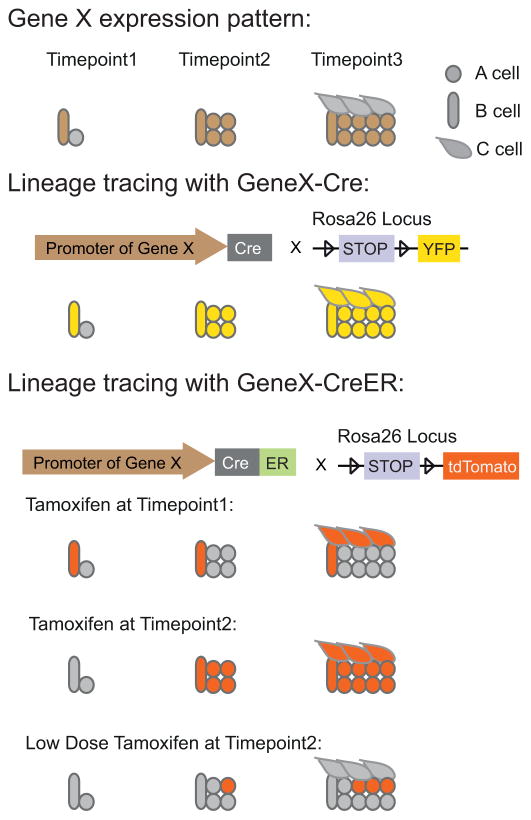
Figure 2.
A hypothetical example illustrating the different results obtained by using Cre and CreER driven by the same promoter. Gene X is expressed only in B cells at time point 1, and starts to be expressed in both A and B cells from time point 2. Now consider the lineage tracing results with GeneX-Cre in comparison with GeneX-CreER. If GeneX-Cre is used, the precise origin of C cells cannot be identifiable since both A and B cells are marked by time point 2. If GeneX-CreER is used, and tamoxifen is given at time point 1, B cells are specifically marked; the results indicate that B cells can generate C cells. If GeneX-CreER is used, and tamoxifen is given at time point 2, both A and B cells are marked at time point 2, while A, B, and C cells are all marked at time point 3. If GeneX-CreER is used, but with only a low dosage of tamoxifen given at time point 2, then, since there are many more A cells than B cells, only A cells are marked at the start of tracing; at time point 3, newly generated A cells are marked, but none of the C cells are marked. All together, the results from GeneX-CreER experiments suggest that B cells are progenitors of C cells, while A cells are unipotent and only generate more A cells.
Initial experiments were often performed using Cre lines that are constitutively expressed. However, gene expression can be dynamically regulated. Many of the Cre lines can express in cells of interests at one point and in some other cells at a later timepoint, complicating the end results. This hurdle was overcome with the development of inducible Cre such as CreER, in which Cre recombinase is fused with a mutated form of estrogen receptor that binds Tamoxifen but not its endogenous ligands. Cre recombinase is kept in the cytoplasmic compartment until the application of Tamoxifen, which then allows Cre to enter the nucleus transiently.
Using an inducible Cre has two additional advantages: First, constitutive Cre activity produces a constant supply of reporter-expressing cells, while transient Cre activity allows production of reporter positive cells only at a defined timepoint when Tamoxifen is applied. This difference becomes essential when trying to determine whether the promoter marks bona fide stem cells. Both stem cells and their transit-amplifying progeny (cells that proliferate rapidly to generate differentiated cells but can do so for a finite number of times) can give rise to downstream lineages. Nevertheless, only stem cells can self-renew over an extended period of time. Transient Cre induction allows the distinction between transit-amplifying cells and stem cells, as reporter-expressing cells can be found in the tissue even long after Cre induction if stem cells are marked but will diminish overtime if transit-amplifying cells are marked.
Second, one can achieve mosaic or preferential labeling with CreER but not Cre by titrating the amount of Tamoxifen. It can be challenging to find a Cre line that exclusively labels one population. However, if Cre has significantly stronger activity in the cells of interest than in other cells or if the intended area comprises many more cells than other areas that are also marked by the same Cre, lowering the amount of Tamoxifen can bias labeling toward the intended cells. This method has been applied to label bulge over hair germ [7] or to label outer root sheath (ORS) cells over bulge cells to delineate the fate of hair follicle cells [8].
A hypothetical example outlined in Fig. 2 illustrates some major differences between using a Cre and a CreER line in lineage tracing: An organ is composed of A and B cells at timpoint1 and develops into an organ composed of A, B, and C cells at timpoint3. GeneX is only expressed in A cell at timepoint1 but starts to express in both A and B cells from timepoint2 onward. If lineage tracing is performed using GeneX-Cre, the cell origin of C cells cannot be determined unequivocally.
On the other hand, if GeneX-CreER is used, one can selectively label only B cells (if Tamoxifen is applied at timepoint1) or only A cells (if low dosage Tamoxifen is applied at timepoint2 since there are much more A cells than B cells), thereby determining the exact cell origin of C cells.
Variations of Cre Lox–Based Strategy
Split-Cre
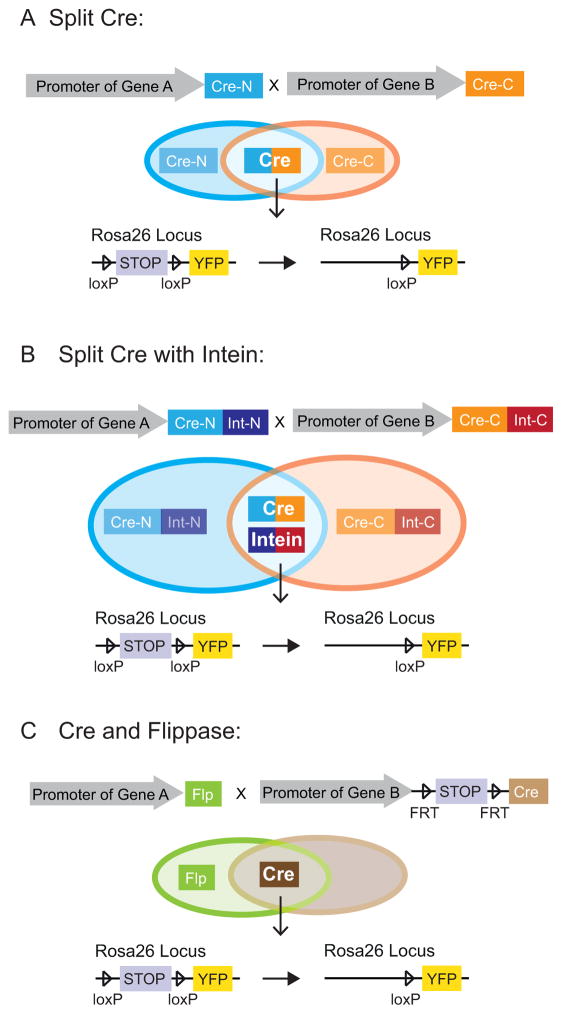
Additional strategies have been designed to overcome the challenges of finding a Cre line that expresses exclusively in the desired cell type. If the cells of interest are the only area where both gene promoters are active, a split-Cre strategy can be used. In this approach, the coding sequence of Cre recombinase is separated into two complementation-competent fragments driven by two independent promoters. Each Cre fragment remains inactive by itself but can assemble into a functional enzyme when present in the same cells. Therefore, although each promoter might express in other unrelated areas, active Cre will only be produced in the cells of interest. The first generation of this split-Cre approach relied on the spontaneous association of these two fragments [33] (Fig. 3A). A modified version added in an intein peptide [34], which excises itself and promotes rejoining of the remaining Cre protein in a process called protein splicing [35], thereby enhancing association efficiency (Fig. 3B). This strategy has been used to mark and trace adult Neural Stem Cells in the dentate gyrus [36].
Figure 3.
Schematic diagrams of lineage tracing strategies employing split-Cre or two recombinases. (A): In a split-Cre approach, each promoter controls the expression of either the N-terminus of Cre (Cre-N) or the C-terminus of Cre (Cre-C). A functional Cre protein is assembled only in cells where both promoters are active. (B): An intein is a fragment of a protein which catalyzes its own excision and promotes the rejoining of remaining portions. When the split-Cre protein (split into Int-N and Int-C fragments) is combined with intein, the rejoining efficiency of Cre-N and Cre-C is increased. (C): Two separate recombinases, Cre and flippase, can also be combined in lineage tracing to achieve cell-type specificities. In the cells where both promoters are active, flippase excises a “stop” codon flanked by two FRT sites, allowing the expression of Cre. Activated Cre further mediates the excision of a “stop” codon flanked by two loxP sites located downstream of the Rosa26 promoter, thereby turning on the expression of reporter genes such as YFP.
Two Recombinases
Similarly, using two different recombinases, Cre and Flip, can also achieve the same effect as the split-Cre strategy. In this case, one promoter is used to drive Flip recombinase (Flippase), while the other one controls the expression of Cre with a frt-stop-frt cassette in front of it. In the cells in which both promoters are active, the stop cassette is excised by Flippase, allowing the expression of Cre (Fig. 3C). Jensen et al. have successfully used this approach to trace subpopulations of serotonin-producing neurons [37].
It should be noted that in both the split-Cre and the two recombinases approaches, three transgenes have to be brought together for a single lineage tracing (two transgenes are needed to activate the Cre, plus the Rosa26 reporter). This laborious nature should be taken into consideration before designing an experiment.
Multicolor Labeling
Initially designed to tackle the complexity of neuronal circuitry [38], the “brainbow” or “confetti” mice have proven to be a tool for analyzing clonal dynamics within a cell population. This method utilizes incompatible flox sites or multiple fluorescent proteins arranged in a specific manner (Fig. 4A, 4B). Upon Cre activation, each cell expresses a random combination of fluorescent proteins that gives the cell a distinct color. Using this strategy, Snippert et al. demonstrated that although multiple Lgr5+ stem cells exist in the small intestinal crypt base, the crypts drift toward clonality with time, suggesting that progeny from one stem cell might outcompete the rest [39]. Tabansky et al. showed that in cleavage-stage mouse embryos, strong developmental bias already exists in the seemingly equivalent blastomeres [40]. Similar approaches have also been applied to study lineage relationships in zebrafish [41]. Because of the design of the constructs, more excision events are naturally required for some fluorescent proteins to express over others. Therefore, Cre activity should be tittered to obtain optimal multicolor labeling in a given population.
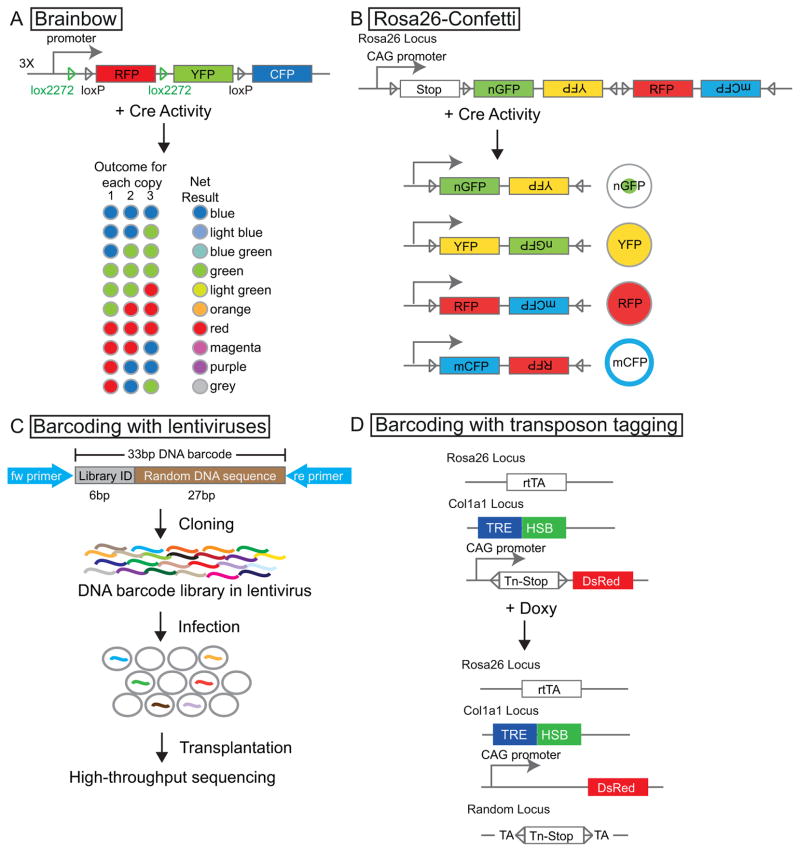
Figure 4.
Lineage tracing strategies designed to study heterogeneity within a given population. (A): Brainbow. Consider a Brainbow line carrying three copies of Brainbow cassettes. Ten different hues can be produced with a random combination of colors resulting from recombination events at each of the loci. (B): Rosa26-Confetti. Four different fluorescent proteins are inserted into the Rosa26 locus. Upon Cre activation, a cell will express one of the four fluorescent colors. (C): Lentiviruses carrying a complex DNA barcode library can be used to transduce cells and trace their fate; this is particularly useful in the hematopoietic system. (D): Sleeping Beauty transposon-mediated random insertion generates a unique genetic tag for each cell, making it possible to conduct lineage tracing in the hematopoietic system without the need for transplantation.
Barcoding
Unlike solid tissues, the hematopoietic system is unique in its physical organization. HSCs are mostly localized to the bone marrow, while their differentiated progeny circulate through the entire body. A genetic barcoding strategy has therefore been developed to investigate HSC heterogeneity. HSCs are transduced with lentiviruses containing a DNA barcode library transplanted into recipients, and the differentiated progeny produced by these HSCs are analyzed with high-throughput sequencing [42] (Fig. 4C). Although this is a powerful approach, transplantation is still required.
To understand the clonal contribution of endogenous HSCs, Sun et al. developed a barcoding strategy to mark each HSC with a distinct genetic tag. This new method involves triple-transgenic mice carrying (1) a hyperactive sleeping beauty transposase under the control of Doxycyclin (Doxy)-responsive element, (2) a DNA transposon (Tn), and (3) a Doxy-dependent transactivator rtTA-M2. When the mice are fed a Doxy-containing diet, rtTA-M2 is activated. This in turn drives the expression of transposase, which mobilizes the transposon Tn. Since the insertion of Tn is random in the genome, each cell will carry a single and distinct insertion site. Upon Doxy withdrawal, these insertion sites serve as permanent genetic tags in stem cells and will transmit to all of their progeny (Fig. 4D). With this uniquely designed method, Sun et al. showed that under steady state conditions, long-lived progenitors are responsible for renewing the hematopoietic system, while HSCs have very limited contributions. This finding suggests that HSCs behave very differently under homeostasis and upon transplantation [28].
Both multicolor labeling and barcoding strategies can be used to study heterogeneity among a given cell population. However, there are some important differences: barcoding strategies allow unlimited number of tags to be generated in theory, while multicolor labeling is ultimately limited by the amount of hues offered by the color cassettes a transgenic mouse carries. On the other hand, multicolor labeling provides visual information, while barcoding strategies rely on DNA sequencing to determine the tag each cell contains. It will not be possible to visualize these barcodes in situ. As such, one needs to be able to purify putative downstream cells with other markers such as surface antigens for barcoding strategy to work.
Direct Observation
Theoretically, the fate of a specific cell can also be followed by direct observation. In reality, this method is restricted by the observer’s ability to identify the cells of interests at the starting timepoint and all the progeny produced by these cells at successive timepoints. On the other hand, since direct observation does not perturb cells or organisms, it has the unique advantage of looking at lineage progression in the same animal. From a historical perspective, Whitman’s work on leech embryos is an excellent demonstration of the power of direct observation. Another impressive example is represented by the construction of a complete lineage tree of Caenorhabditis elegans (C. elegans), which was done by observing live embryos without any markers under a Nomarski microscope [43].
Live Imaging
With the advance in imaging techniques and tools, direct observation of cell fate can also be achieved through live imaging. This works the best when combined with additional marking strategies to unambiguously identify the cells of interests. Because of their transparency, C. elegans and zebrafish embryos are the two most popular model systems for this approach [5, 44].
Two-photon microcopy provides great depth and resolution into live animals and has greatly extended live-imaging capacity to tissues and organisms that are traditionally less accessible. Greco and colleagues have successfully adapted two-photon microscopy and genetic markings to trace the fate of bulge and hair germ cells in the same hair follicle throughout hair cycles [45, 46]. Ritsma et al. showed that in the small intestine, Lgr5+ cells in the border of stem cell and transit-amplifying cells can be pushed into the transit-amplifying zone after proliferation of Lgr5+ cells at the crypt base [47]. This type of information is particularly challenging to obtain with traditional lineage tracing approaches that take snapshots of timepoints from different animals.
Depending on the tissues being imaged, live imaging can sometimes be an invasive procedure and often cannot be performed over a long period of time. However, the idea of watching cell behavior in situ is attractive and can provide unambiguous real-time information on how a cell contributes to its downstream lineages.
Conclusions and Future Outlook
From these examples, one can appreciate that although there is no universal method for conducting lineage tracing, there is likely to be an effective strategy suitable for the questions of choice. While our knowledge of tissue biology continues to grow, lineage tracing will continue to play an important role in our quest for understanding development and regeneration. With many exciting developments in probing single-cell dynamics, it is interesting to see how lineage tracing strategies integrate with single-cell analysis to address heterogeneity within a given cell population. Rapid development of sequencing technology and analysis might also shed insights into lineage tracing in human tissues. It will also be important to tackle how different lineages integrate and interact with one another and what dictates the choice of cell fate.
Acknowledgments
I thank Y. Fong, O. Chung, L. Demas, D. Tan, and B. Zhang for discussions and critical reading of the manuscript. I apologize to those colleagues whose work could not be referenced because of space constraints. Y. H. is a recipient of the NIH K99-R00 pathway to independence award (R00-AR063127), the Milton Fund Award, and the Smith Family New Investigator Award.
References
- 1.Whitman CO. The embryology of Clepsine. Q J Microscop Sci (NS) 1887;18:215–315. [Google Scholar]
- 2.Whitman CO. A contribution to the history of the germ-layers in Clepsine. Journal of Morphology. 1887;1:105–182. [Google Scholar]
- 3.Vogt W. Gestaltungsanalyse am Amphibienkeim mit Örtlicher Vitalfärbung. II. Teil Gastrulation und Mesodermbildung bei Urodelen und Anuren. Wilhelm Roux Arch Entwicklungsmech Org. 1929;120:384–706. doi: 10.1007/BF02109667. [DOI] [PubMed] [Google Scholar]
- 4.Serbedzija GN, Bronner-Fraser M, Fraser SE. A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development. 1989;106:809–816. doi: 10.1242/dev.106.4.809. [DOI] [PubMed] [Google Scholar]
- 5.Woo K, Fraser SE. Order and coherence in the fate map of the zebrafish nervous system. Development. 1995;121:2595–2609. doi: 10.1242/dev.121.8.2595. [DOI] [PubMed] [Google Scholar]
- 6.Kugelberg E, Norstrom T, Petersen TK, et al. Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrobial agents and chemotherapy. 2005;49:3435–3441. doi: 10.1128/AAC.49.8.3435-3441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 10.Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinhauser ML, Bailey AP, Senyo SE, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spalding KL, Bhardwaj RD, Buchholz BA, et al. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 13.Spalding KL, Bergmann O, Alkass K, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst A, Alkass K, Bernard S, et al. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156:1072–1083. doi: 10.1016/j.cell.2014.01.044. [DOI] [PubMed] [Google Scholar]
- 15.Wasserstrom A, Frumkin D, Adar R, et al. Estimating cell depth from somatic mutations. PLoS computational biology. 2008;4:e1000058. doi: 10.1371/journal.pcbi.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frumkin D, Wasserstrom A, Kaplan S, et al. Genomic variability within an organism exposes its cell lineage tree. PLoS computational biology. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frumkin D, Wasserstrom A, Itzkovitz S, et al. Cell lineage analysis of a mouse tumor. Cancer Res. 2008;68:5924–5931. doi: 10.1158/0008-5472.CAN-07-6216. [DOI] [PubMed] [Google Scholar]
- 18.Shlush LI, Chapal-Ilani N, Adar R, et al. Cell lineage analysis of acute leukemia relapse uncovers the role of replication-rate heterogeneity and microsatellite instability. Blood. 2012;120:603–612. doi: 10.1182/blood-2011-10-388629. [DOI] [PubMed] [Google Scholar]
- 19.Rawles ME. Origin of melanophores and their role in development of color patterns in vertebrates. Physiological reviews. 1948;28:383–408. doi: 10.1152/physrev.1948.28.4.383. [DOI] [PubMed] [Google Scholar]
- 20.Le Douarin N. A biological cell labeling technique and its use in expermental embryology. Developmental biology. 1973;30:217–222. doi: 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- 21.Le Douarin NM, Teillet MA. The migration of neural crest cells to the wall of the digestive tract in avian embryo. Journal of embryology and experimental morphology. 1973;30:31–48. [PubMed] [Google Scholar]
- 22.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 23.Bonfanti P, Claudinot S, Amici AW, et al. Microenvironmental reprogramming of thymic epithelial cells to skin multipotent stem cells. Nature. 2010;466:978–982. doi: 10.1038/nature09269. [DOI] [PubMed] [Google Scholar]
- 24.Van Keymeulen A, Rocha AS, Ousset M, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 25.Majo F, Rochat A, Nicolas M, et al. Oligopotent stem cells are distributed throughout the mammalian ocular surface. Nature. 2008;456:250–254. doi: 10.1038/nature07406. [DOI] [PubMed] [Google Scholar]
- 26.Amitai-Lange A, Altshuler A, Bubley J, et al. Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem Cells. 2015;33:230–239. doi: 10.1002/stem.1840. [DOI] [PubMed] [Google Scholar]
- 27.Di Girolamo N, Bobba S, Raviraj V, et al. Tracing the fate of limbal epithelial progenitor cells in the murine cornea. Stem Cells. 2015;33:157–169. doi: 10.1002/stem.1769. [DOI] [PubMed] [Google Scholar]
- 28.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 30.Ito M, Liu Y, Yang Z, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature medicine. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 31.Tata PR, Mou H, Pardo-Saganta A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen KB, Collins CA, Nascimento E, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casanova E, Lemberger T, Fehsenfeld S, et al. Alpha complementation in the Cre recombinase enzyme. Genesis. 2003;37:25–29. doi: 10.1002/gene.10227. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Chen T, Sakurai K, et al. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Scientific reports. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anraku Y, Mizutani R, Satow Y. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB life. 2005;57:563–574. doi: 10.1080/15216540500215499. [DOI] [PubMed] [Google Scholar]
- 36.Beckervordersandforth R, Deshpande A, Schaffner I, et al. In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem cell reports. 2014;2:153–162. doi: 10.1016/j.stemcr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen P, Farago AF, Awatramani RB, et al. Redefining the serotonergic system by genetic lineage. Nature neuroscience. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 39.Snippert HJ, van der Flier LG, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Tabansky I, Lenarcic A, Draft RW, et al. Developmental bias in cleavage-stage mouse blastomeres. Current biology : CB. 2013;23:21–31. doi: 10.1016/j.cub.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan YA, Freundlich T, Weissman TA, et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu R, Neff NF, Quake SR, et al. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nature biotechnology. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulston JE, Schierenberg E, White JG, et al. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental biology. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 44.Thomas C, DeVries P, Hardin J, et al. Four-dimensional imaging: computer visualization of 3D movements in living specimens. Science. 1996;273:603–607. doi: 10.1126/science.273.5275.603. [DOI] [PubMed] [Google Scholar]
- 45.Rompolas P, Deschene ER, Zito G, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritsma L, Ellenbroek SI, Zomer A, et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507:362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]