Summary
In sexual species, gametes have to find and recognize one another. Signaling is thus central to sexual reproduction and involves a rapidly evolving interplay of shared and divergent interests [1-4]. Among Caenorhabditis nematodes, three species have evolved self-fertilization, changing the balance of intersexual relations [5]. Males in these androdioecious species are rare, and the evolutionary interests of hermaphrodites dominate. Signaling has shifted accordingly, with females losing behavioral responses to males [6, 7] and males losing competitive abilities [8, 9]. Males in these species also show variable same-sex and autocopulatory mating behaviors [6, 10]. These behaviors could have evolved by relaxed selection on male function, accumulation of sexually antagonistic alleles that benefit hermaphrodites and harm males [5, 11], or neither of these, because androdioecy also reduces the ability of populations to respond to selection [12-14]. We have identified the genetic cause of a male-male mating behavior exhibited by geographically dispersed C. elegans isolates, wherein males mate with and deposit copulatory plugs on one another's excretory pores. We find a single locus of major effect that is explained by segregation of a loss-of-function mutation in an uncharacterized gene, plep-1, expressed in the excretory cell in both sexes. Males homozygous for the plep-1 mutation have excretory pores that are attractive or receptive to copulatory behavior of other males. As excretory pore plugs are injurious and hermaphrodite activity is compromised in plep-1 mutants, the allele may be unconditionally deleterious, persisting in the population because the species' androdioecious mating system limits the reach of selection.
Results and Discussion
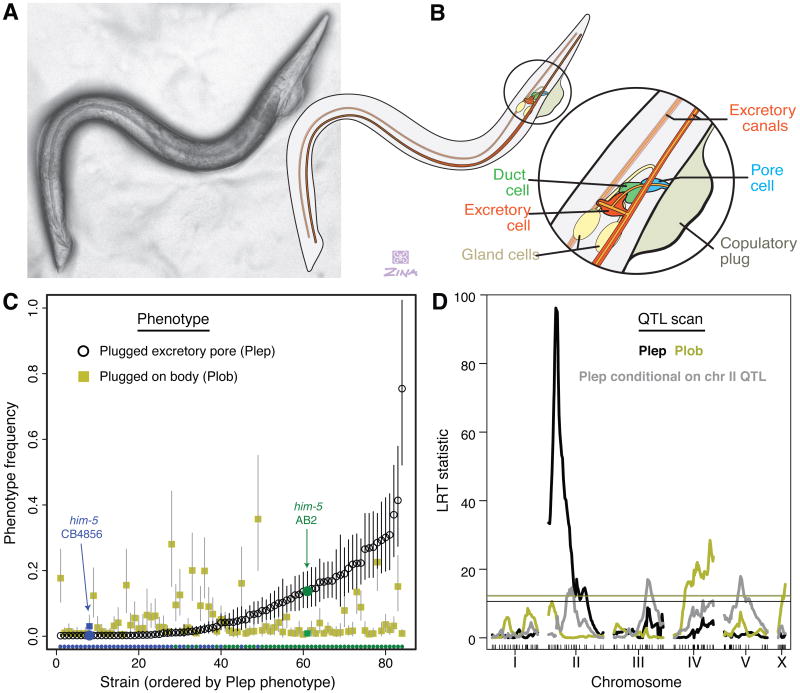
Gems and Riddle found that males of the Australian wild isolate AB2, maintained in male-only populations, deposit copulatory plugs on one another's excretory pores (Figures 1A and 1B) [10]. Most C. elegans strains, including the Hawaiian isolate CB4856, do not exhibit this behavior, although it also present and variable in the androdioecious C. briggsae [6]. The excretory system, which functions in osmoregulation, is essential for development and viability [15], and we found that pore plugs impinge on male function. Relative to their genetically identical brothers, plugged C. elegans adult males lived less long (Cox proportional hazard regression coefficient and standard error: -0.654 ± 0.189, P = 0.0005; see Experimental Procedures, Supplemental Data File, and Figure S1), and had lower mating success well before differences in mortality were apparent (Fisher's Exact Test, P = 0.0008).
Figure 1. Excretory system anatomy and the genetic architecture of excretory pore plugging.
(A) A male worm with a copulatory plug on its excretory pore (QG71, right lateral view). (B) Schematic of the excretory system. Paired bilaterally symmetric canals are drained through the excretory, duct and pore cells, immediately posterior and ventral to the pharyngeal bulb. The function of the gland cells is unknown. (C) Ectopic copulatory plugging exhibits transgressive segregation. Of the 82 recombinant inbred lines, ordered by Plep phenotype, many have higher Plep and Plob frequencies than either parental strain (blue and green). The plotted values and prediction intervals are derived from a mixed-effect generalized linear model (see Methods). Colors at the bottom represent strain genotype at the major-effect Plep QTL. (D) QTL mapping of ectopic copulatory plugging in the recombinant inbred lines. Single QTL scans for excretory pore (black) and body (gold) plugging show one and two significant linkages. After conditioning on the major chromosome II QTL, three additional Plep QTL are significant (gray). Horizontal lines show permutation-based thresholds for genome-wide significance at P = 0.05.
To dissect the genetic causes of excretory pore plugging, we intercrossed AB2 and CB4856 (each carrying a male-generating him-5 mutation) and created recombinant inbred lines (RILs) from F2 hermaphrodites by selfing for 10 generations. We genotyped 82 RILs at 157 markers and scored their copulatory plugs in 4-day, 40-male assays (Supplemental Data File). RILs varied in the frequency of plugs deposited on excretory pores (Plep: plugged excretory pore), and also in frequency of plugs placed elsewhere on males' bodies (Plob: plugged on body), which occurs rarely in the parental strains (Movies S1 and S2). The phenotypes exhibit transgressive segregation, with many RILs showing higher plug frequencies than the parental strains (Figure 1C), and they implicate different genomic regions (Figure 1D).
A major-effect QTL on chromosome II largely discriminates between strains with some excretory-pore plugging and those with none (Figures 1C and 1D), while the residual variation in plug frequency among strains is genetically complex, with significant QTLs on chromosomes II, III and V. Plug deposition elsewhere on the body maps to chromosomes IV and X. However, after conditioning on the major-effect Plep QTL, residual variance for that phenotype is correlated with the variance in Plob (correlation of strain effects is 0.36, P = 0.00089), suggesting shared etiology. All QTL effects are as predicted by parental differences: AB2 alleles at the four Plep QTLs increase excretory pore plugging, CB4856 alleles at the two Plob QTLs increase male body plugging. However, transgressive segregation implies that additional undetected loci act in the reciprocal directions.
The Plep phenotype could be due to increased copulatory activity, or increased attractiveness or receptivity of Plep males [10]. To distinguish between these possibilities we introgressed a defective plg-1 allele into QG71, the most highly Plep RIL, rendering these animals incapable of making copulatory plugs [9]. We then assayed populations comprising 10 plg-1- RIL males and 30 GFP-marked CB4856 males. After 4 days, an average of 5.4 plg-1- males, and none of the CB4856 males, had received excretory pore plugs, indicating that males of the non-Plep strain CB4856 readily copulate with the excretory pores of other males, and pore attractiveness or receptivity underlies the Plep phenotype.
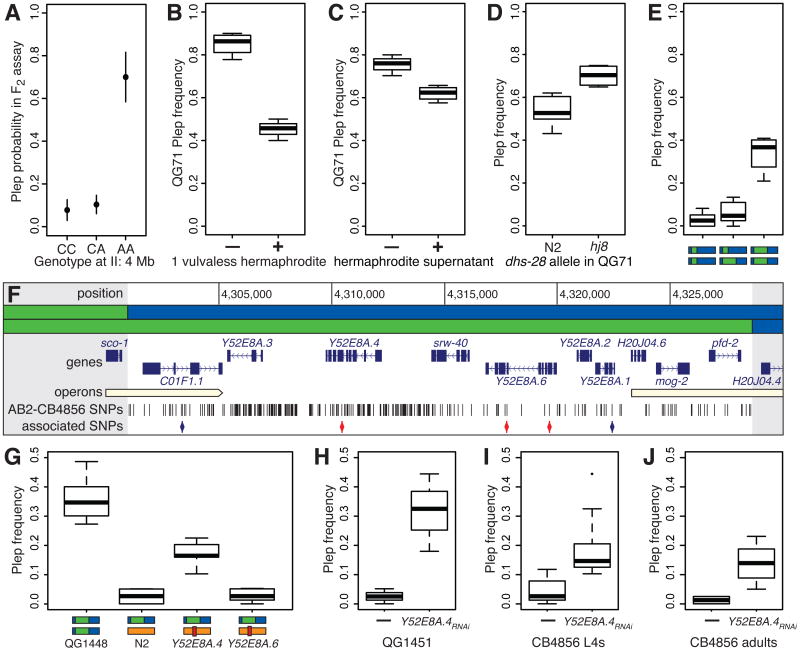
The major-effect chromosome II QTL is specific to the Plep phenotype and thus is predicted to confer pore-attractiveness to animals that carry the AB2 allele. We assayed genetically heterogeneous F2 male populations from a cross of AB2 and CB4856, and found that genetic markers closely linked to the QTL cosegregated, in a recessive pattern, with receipt of excretory pore plugs (P < 10-15 for linkage, P < 10-4 for dominance; Figure 2A; Supplemental Data File).
Figure 2. Identification of plep-1.
(A) The Chr II QTL confers recessive receptivity to excretory pore plugs in heterogeneous F2 population assays. Proportions of Plep males and their binomial confidence intervals are plotted by genotype at a QTL-linked marker (C for the CB4856 allele, A for AB2). (B) The frequency of Plep QG71 males is significantly reduced by the presence of a single vulvaless hermaphrodite and (C) by aqueous supernatant from hermaphrodites. (D) Excretory pore plugging is not mediated by ascaroside pheromones. QG71 males with an introgression of the dhs-28(hj8) loss of function allele are plugged at higher rates than those with a matched N2 introgression. (E) A 27kb interval defined by NILs behaves as a recessive locus. Strains QG1451, QG1448, and their F1 are depicted as bars representing chromosome II, with CB4856 DNA in blue and AB2 DNA in green, as in (F), which details the NIL-defined interval. The interval encompasses 10 protein-coding genes and 217 AB2-CB4856 nucleotide variants, of which 5 perfectly associate with Plep phenotype across 11 wild strains (blue: synonymous or intronic, red: non-synonymous). (G) In complementation tests for Mos1-insertion mutants of Y52E8A.4 and Y52E8A.6 (N2 background: orange), Y52E8A.4 fails to complement the Plep NIL QG1448. (H-J) RNAi against Y52E8A.4 induces headplugging when administered to L4 males of non-Plep NIL QG1451 and CB4856 him-5, and to CB4856 him-5 adult males.
The attractiveness of Plep males is modulated by the presence of hermaphrodites. QG71 exhibits reduced Plep when a single vulvaless hermaphrodite is present among males (P < 10-11; Figure 2B), and aqueous extract from a hermaphrodite culture is sufficient to decrease male-male copulation (P = 0.015; Figure 2C; Supplemental Data File).
Water-soluble ascaroside pheromones mediate communication in nematode populations and are candidate factors governing male attractiveness [16, 17]. To test whether ascarosides underlie the Plep phenotype we introgressed dhs-28(hj8), a mutation that abrogates synthesis of the short-chain ascaroside pheromones [18], from the N2 lab background into QG71. In parallel, we introgressed the wild-type N2 genome into the same region to make dhs-28+ control strains. Pheromone-defective males exhibited slightly elevated Plep rates (P = 9.4×10-4; Figure 2D; Supplemental Data File), indicating pheromonal communication is not required for pore attractiveness, and wild-type signaling discourages plug receipt.
We attempted to Mendelize the chromosome II QTL by introgressing it from AB2 into the CB4856 background. Using visible marker mutations we narrowed the Plep-conferring interval (Figure S2) to 27kb, minimally defined by near-isogenic lines (NILs) QG1448 and QG1451 (Supplemental Data File). The interval behaves as a recessive Mendelian locus (Figure 2E; Supplemental Data File) and contains 10 protein-coding genes (Figure 2F).
We sequenced the genomes of AB2 and CB4856, identifying 217 segregating variants in the NIL-defined QTL interval, and next employed association mapping to prioritize variants. Genotypes were called at the 217 positions from Illumina sequencing of 9 additional wild isolates capable of making copulatory plugs (Supplemental Data File). One strain, PB306, exhibits a Plep phenotype similar to AB2 and is nearly identical across the interval, while the others exhibit diverse haplotypes and little or no Plep (Supplemental Data File). Of the 217 variants, 5 are perfectly Plep-associated across the 11 strains (Figure 2F), including 3 nonsynonymous polymorphisms in two genes, Y52E8A.4 and Y52E8A.6.
A MosI transposon-insertion allele of Y52E8A.6 complemented the NIL QG1448 (P = 0.79), but a Y52E8A.4 insertion allele failed to complement (P < 10-9; Figure 2G; Supplemental Data File). Y52E8A.4 carries a single variant in AB2 associated with Plep, a V>D replacement at position 278, which is predicted to fall within a conserved transmembrane helix and be highly damaging to protein function.
Knock-down of Y52E8A.4 by feeding RNAi to L4 males caused QG1451, the non-Plep NIL, to exhibit excretory pore plugging (P < 10-9; Figure 2H; Supplemental Data File). RNAi in the CB4856 background also induced excretory pore plugging (P = 10-7; Figure 2I), though at a lower level than in QG1451 (P = 0.006). Moreover, RNAi administered to adult CB4856 males was also effective (P = 2×10-5; Figure 2J), implying Plep is independent of any role for Y52E8A.4 in development and that wild-type function involves rapid protein turnover. We assigned the name plep-1 to Y52E8A.4.
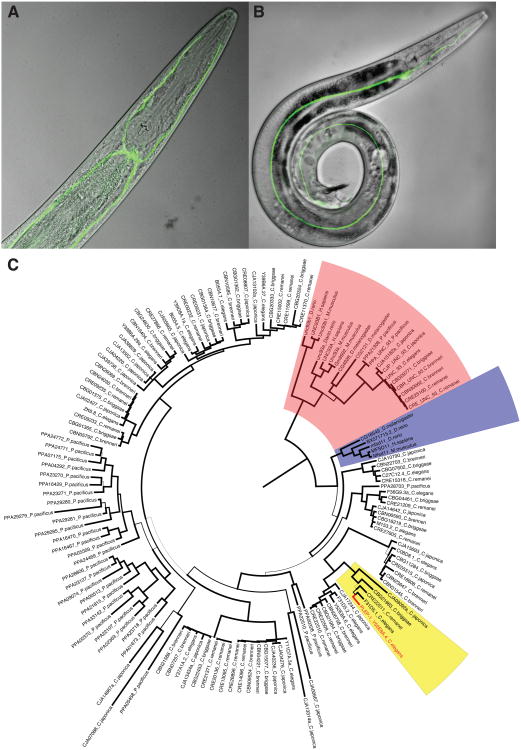
A plep-1 transcriptional reporter was expressed consistently only in the excretory cell in both males and hermaphrodites (Figures 3A and 3B). The excretory cell comprises bilaterally symmetric canals and a cell body ventral to the terminal pharyngeal bulb, connected to the excretory pore by a duct cell (Figure 1B). Pore, duct, and excretory gland cells all fail to express pplep-1∷GFP. We hypothesize that alteration of excretory cell function through loss of plep-1 activity may render the chemical milieu of the excretory pore more attractive to other males, or the pore more accessible to intromission.
Figure 3. plep-1 expression and phylogeny.
(A) Maximum intensity projection along the dorsoventral axis showing pplep-1∷GFP expression in the excretory cell body, just posterior to the terminal pharyngeal bulb, and symmetric canals in a young adult hermaphrodite. (B) Lateral view of a young adult male. In addition to consistent expression in the excretory cell, fluorescence in two dorsal cells in the male tail, shown here, was observed inconsistently. (C) Phylogeny for UNC-93 domain (IPR010291) homologs. UNC-93, MFSD11 and PLEP-1 clades are highlighted (red, blue and yellow). Unboxed genes are MFSD11 paralogs in Caenorhabditis and Pristionchus nematodes.
plep-1 is one of 16 paralogs in C. elegans, part of a recurrent gene-family expansion extending across rhabditid phylogeny (Figure 3C). These genes have no known function but share a Major Facilitator Superfamily-like domain with a single-copy ortholog, MFSD11, in flies and mammals. The domain also occurs in a more distantly related worm gene, unc-93, and its orthologs in other species. UNC-93 is a presumed regulatory component of a TWIK (Tandem of P-domains in a Weakly Inward rectifying K+ channel) complex [19], and PLEP-1 and its paralogs are inferred to have similar roles. TWIK channels have also undergone gene-family expansions in rhabditids, with 47 genes in the C. elegans genome.
Few genes show altered expression in plep-1 mutant whole young adult hermaphrodites (Supplemental Data File). Of 46 genes differentially expressed between N2 and a plep-1 Mos1 insertion line (False Discovery Rate < 0.1), the two most significant are implicated in neurotransmitter biosynthesis or activity (see Supplemental Experimental Procedures), and a third, unc-64 (syntaxin), is expressed in neurons and secretory cells including the excretory gland cells [20]. Gene set enrichment analysis identified 6 ontology terms overrepresented in genes with lower expression in the plep-1 mutant (P < 0.005, FDR < 0.1), including aromatic amino acid metabolism, neuropeptide Y receptor activity, and a large group of mostly unstudied genes annotated by homology with transmembrane transport. While causally ambiguous, these associations are consistent with the view that loss of plep-1 function might subtly alter the excreted small-molecule profile.
A survey of 55 wild isolates from around the world revealed that the derived 278D allele occurs in strains from Australia, Western North America, and Chile (Supplemental Data File). Sets of isolates from localities in Santa Barbara, Salt Lake City, and Adelaide were polymorphic, including an instance of polymorphism among worms inhabiting a single piece of fruit. None of 15 strains from Europe, 4 from Africa, or 4 from the Azores carried the D allele. We assayed a subset of plg-1+ isolates in our standard 40-worm assays. Among these strains, 278D is necessary but not sufficient for a strong Plep phenotype (Supplemental Data File), indicating that genetic background affects plugging, consistent with our findings in the RILs (Fig. 1C). Further, at least one isolate carrying 278D also carries a loss-of-function mutation in plg-1, and so is incapable of depositing copulatory plugs.
A role for male-male competition in evolution of excretory pore plugging, whether through direct injury to the plug recipient or through distraction of rival males in mating clumps, is unlikely due to the rarity of unrelated males in natural populations [9]; consistent with this, C. briggsae males deposit plugs on their own excretory pores [6]. Alternatively, resource dependent male-male copulation could serve to attract receptive hermaphrodites, alleviating sperm-limited fecundity and reinforcing habitat quality cues. Given male scarcity and the low frequency of outcrossing seen in wild C. elegans populations [21, 22], selection on male function is expected to be a weak force in androdioecious mating systems, however. With hermaphrodite fitness generally maximised by early-life selfing, and multiple examples of hermaphrodites eschewing male attention [6, 23, 24], a role in sexual signaling conferring a net benefit to hermaphrodites also appears unlikely.
More credibly, excretory pore plugging is compatible with mutation accumulation, due to relaxed selection on male function, or sexual antagonism, favoring alleles beneficial to hermaphrodites despite harm to males. The sexual antagonism model predicts that loss of plep-1 function benefits hermaphrodites, while the mutation accumulation model predicts that it is neutral. We measured hermaphrodite activity, a proxy for viability and a component of fitness, as a function of plep-1 genotype under taxing hypoosmotic conditions of starvation in water. Mutants for genes expressed in the excretory system, including TRP channel gtl-1 and aquaporin aqp-8, show compromised osmotic homeostasis under similar conditions [25, 26]. We found significantly reduced activity for multiple plep-1 loss-of-function alleles (Figure S3; Supplemental Data File; Generalized linear mixed model, genotype effect P < 1×10-12 after 24 hours in H2O).
Although the possibility that the plep-1 mutation may benefit hermaphrodites under other conditions cannot be excluded, and the ecological relevance of extended exposure to water is unknown, the deleterious effect of plep-1(V278D) on hermaphrodite activity weighs against mutation accumulation impinging solely on male function. These data instead suggest the allele may be unconditionally deleterious for both sexes. The evolution of androdioecy, in addition to changing selection on male function, has reduced C. elegans' effective population size and recombination rate, such that deleterious mutations are less likely to be eliminated, and more likely to hitchhike in linkage with beneficial alleles [5, 13, 14, 27, 28].
Though ascarosides underlie the attractive nature of hermaphrodite secretions [29], increasing evidence points to alternative mechanisms of mate attraction [30-32] and retention [6]. While the worm's excretions are not well understood, our data suggest a role for plep-1 in either the excreted chemical profile or in excretory system homeostasis, such that loss-of-function causes the male excretory pore to resemble the vulval microenvironment.
Experimental Procedures
RIL construction and genotyping
We generated recombinant inbred lines from a cross of QX1199 (him-5 (e1490) > CB4856) males and QG5 (him-5 (e1490) > AB2) hermaphrodites. We singled 140 F2 L4 hermaphrodites and inbred these through 10 generations of selfing, after which the 98 surviving lines were frozen. We genotyped 82 RILs at 157 SNPs by GoldenGate assay at the University of Utah (Supplemental Data File).
RIL phenotyping
Each strain was assayed an average of 2.1 times, randomized across 14 blocks (days). Assays were performed on NGM-agarose plates, which differ from the recipe in [33] by the addition of 1.25% agarose. Forty L4 males were transferred to NGM-agarose plates seeded one day earlier with 50μl OP50 E. coli bacteria. The males were picked from plates that had been seeded with 2 plugged hermaphrodites 3 days earlier, to control population density. Four days after the 40 L4s were plated, each worm was scored for the presence of copulatory plugs on its excretory pore and elsewhere on its body (Supplemental Data File). All assays were performed at 20°C.
Quantitative genetic analysis
We fitted the observed counts of worms with and without plugs with a mixed-effect generalized linear model with binomial error, using the function glmer in R [34, 35]. Assay date and strain were included as random effects, and we tested for an improvement of fit by incorporating marker genotypes as fixed effects. As a test statistic, we use the chi-square value from difference in log likelihood between the full model and a reduced model with intercept as the only fixed effect. We performed a single-QTL scan of the genome by testing multiple imputations of genotypes at 2cM spacing as well as at the observed markers [36], taking the average Chi-square value at each marker across imputations, inversely weighted by the residual deviance. Genome-wide significance thresholds were estimated by permuting the genotype vectors, preserving the original strain and date structure [37, 38]. Genome-wide thresholds at P = 0.05 were estimated from 200 permutations for each analysis.
For plotting the phenotypes in Figure 1C, we extracted estimates of each strain's plugging frequency from the random strain effect of a mixed-effect generalized linear model that incorporated assay date, as described above. The strain effects, conditional modes from the mixed-effect linear model, were extracted using the ranef function, and the prediction intervals are 1.96 times the standard errors derived from the posterior variances (ranef argument condVar=T). These values were then converted into frequencies.
Genetic correlation between Plep and Plob was tested by analyzing the Pearson correlation between the estimated strain effects.
Trans-plugging assays
We set up assays with 10 L4 QG2288 (plg-1(N2) > QG71) males and 30 GFP-marked QG693 (mIs12 II; him-5 V > CB4856) males. After four days, we scored each worm for the Plep phenotype, blind to genotype, and then scored each worm for pharyngeal GFP to determine its genotype.
F2 assays
F2 males derived from a cross of AB2 males and CB4856 hermaphrodites and sibmating of F1s were subjected to our standard 40-male 4-day assay. Individual males were then scored for excretory pore plugs and lysed for genotyping. Each worm was genotyped by PCR at an indel polymorphism at 4.057 Mb on chromosome II, close to the QTL peak at the SNP marker at 4.317 Mb (Supplemental Data File). Linkage and dominance were assessed with a logistic regression including additive and dominance terms and significance was tested with a likelihood ratio test. Figure 2A plots the simple phenotypic proportions and their 95% confidence intervals calculated by normal approximation.
Effect of hermaphrodites
We generated vulvaless hermaphrodites by RNAi by feeding, targeting lin-3 and mex-3 in AB2 larvae. Knockdown of mex-3 results in dead embryos, which prevents embryos from hatching inside their vulvaless mothers (bagging). Standard 4-day assays were set up with 40 QG71 L4 males and 1 young adult Vul hermaphrodite (Supplemental Data File).
Hermaphrodite supernatant was generated by incubating ∼150 QG71 hermaphrodites in 10μl of water for an hour, then freezing and thawing the tubes. 5μl of the supernatant was spotted on 6cm plates prior to the addition of males. 5μl of water was added to control plates (Supplemental Data File).
We tested significance of each treatment in a logistic regression in R: anova(glm(data ∼ treatment, family = “binomial”), test = “LRT”), with p-values estimated from the Chi-square distribution.
dhs-28 introgressions
We tested QG2285 (dhs-28 (hj8) X > QG71) and two introgression-control strains, QG2286 and QG2287 (dhs-28 (N2) X > QG71) in standard 4-day 40 worm assays (Supplemental Data File). We tested significance of each genotype in a logistic regression in R. We detected no difference in phenotype between the two control strains (P = 0.59), and we therefore pooled their data to test for an effect of dhs-28 genotype and to plot the results in Figure 2D.
Fine-mapping with NILs
NILs were constructed according to the scheme in Figure S2, using visible markers sqt-2 (sc108), mIs12, and dpy-2(e8). Phenotypes were scored in the standard 40-worm 4-day assay (Supplemental Data File). These NILs narrow the QTL to the interval between 4,301,062 and 4,328,486, defined by NILs QG1448 (Plep) and QG1446 (nonPep). We subsequently used QG1451, which has a breakpoint between CB4856 and AB2 DNA in the same interval as QG1446, as our nonPep NIL for comparisons with QG1448.
QG1448, QG1451, and their F1 male progeny were assayed in the standard assay to generate Figure 2E (Supplemental Data File).
Identifying associated variants
We generated Illumina sequencing data for 10 strains capable of producing copulatory plugs [9]. Reads were mapped with bwa mem (0.7.5a) [39] to the reference N2 genome (WS220) and processed with Picard (broadinstitute.github.io/picard) (1.103) and samtools (samtools.sourceforge.net) (0.1.18) utilities. Variants were called with the GATK HaplotypeCaller (2.6-4) [40] and filtered on mapping quality (MQ ≥ 40.0), depth (4 ≤ DP ≤ mean coverage × 3), read strand bias (FS ≤ 60.0), position within read (ReadPosRankSum ≥ -8.0), and variant quality: depth (QD ≥ 4.0). Regions of the 27kb interval are highly repetitive and we were unable to make reliable calls in those regions. Genotype data are reported in the Supplemental Data File (WS220 coordinates), where the causal plep-1 variant is 4310427 A>T (WBVar01538101). Variant functional impact was predicted using Sift, PolyPhen and TMHMM [41-43].
We assayed excretory pore plugging in each of these strains in triplicate 4-day assays, each including 20 L4 males of the wild isolate and 20 QX1199 males to serve as plug donors in case the wild isolate is a poor mater (Supplemental Data File).
Complementation Tests
We received a population segregating ttTi36640, a Mos1 insertion in Y52E8A.6, from Maite Carre-Pierrat, and a population segregating ttTi58321, a Mos1 insertion in Y52E8A.4, from Patricia Kuwabara [44]. From each population we homozygosed the insertion allele with validation by PCR.
We crossed QG1448 males with hermaphrodites of QG1448, N2, ttTi36640, and ttTi58321, and picked the resulting F1 L4 male progeny to set up standard 4-day 40-worm assays (Supplemental Data File).
RNAi experiments
We inoculated LB-Ampicillin with colonies of HT115(DE3) strains carrying pL4440 (empty vector negative control) and pY52E8A.4RNAi and seeded the resulting overnight cultures on NGM-agarose plates containing IPTG and Carbenicillin [45]. We used these plates for our standard 4-day 40-male assays with strain QX1199 and QG1451 (Supplemental Data File). The QX1199 L4 experiments were conducted on two dates, and we included date as an effect in the logistic regression (though it was not significant). The QX1199 experiments and QG1451 experiments were performed in single batches.
Transcriptional Reporter Analysis
The plep-1 transcriptional reporter comprises 1,169bp of upstream sequence from the start codon to a HindIII site (4,313,385-4,312,216 in WS240 coordinates) amplified from N2, digested with HindIII, and cloned into HindIII/SmaI sites of pSM-GFP [46]. The reporter was injected into N2 hermaphrodites with a Pmyo-3∷mCherry marker (pCFJ104 [47]) to obtain multicopy array F1 animals. Staged worms from two stably expressing lines (QG2465 and QG2466) were mounted on 2% agar pads in 10μM levamisole and imaged on a Leica SP5 confocal microscope over four independent experiments.
plep-1 gene family phylogenetics
UNC-93 domain (IPR010291) protein sequences were downloaded from Interpro (March 2014), aligned (Mafft 6.864 [48], linsi mode) and subject to Bayesian phylogenetic inference (MrBayes 3.2.2 [49]) with 10 independent 4-chain runs of 10M generations (GTR model with gamma-distributed rate variation, mixed amino acid model prior).
Male longevity and reproductive success
Developmentally synchronized QG71 young adult males were placed in groups of 40 on 6cm plates and left to mate for 3 days at 20°C. Four males, blocked according to the presence or absence of a plugged excretory pore, were placed on a 6cm plate seeded with 150μl of a saturated culture of OP50-GFP [50], along with eight virgin young adult fog-2 females (hermaphrodites that cannot produce self sperm) to minimize subsequent headplugging and leaving. A total of 93 plates (47 with plugs, 46 without) were initiated. A single treatment of 50μl 1M KCl was added to 23 plates within each block to test for an effect of osmotic stress. Mating proceeded for two days at 20°C, then males were removed to fresh plates and young females were replenished (continued every two days thereafter to measure longevity, see below). Over the initial two days, mating failed for 15 of 47 plates with Plep males and 2 of 46 plates for unplugged males. The effect of KCl on mating success was not significant (Fisher's Exact Test, P = 0.28).
To assay longevity, males were gently prodded on the head daily and scored as dead if unresponsive. Worms dead at the plate edge due to desiccation or otherwise unaccounted for at census each day were censored. A Cox proportional hazard model was fit with the R survival package [51]. The main effect of initial KCl treatment on lifespan was significant and negative (P = 0.0001). The final longevity model also incorporated a suggestive interaction between Plep status and KCl treatment (marginally preferable to a model with additive effects only by likelihood ratio test, P = 0.068). All plugged males had perished by 21 days, leaving 12.7% of unplugged males alive (log 95% confidence interval 8.1-20.1%). Raw data are in the Supplemental Data File, and model fit is plotted in Figure S1.
Wild Isolate Genotypes
To characterize the geographic distribution of the plep-1 V278D allele, we genotyped 55 wild isolates of C. elegans by Illumina short-read genome sequencing or PCR-RFLP. C. elegans population structure is such that a random sample of strains is not a particularly meaningful concept. Our sample is enriched for strains that resemble AB2 on chromosome II, according to RAD-seq data from Andersen et al. [13], though it also includes many additional strains collected subsequent to that study. Genotypes, and phenotype data for a subset of plg-1+ strains, are reported in the Supplemental Data File.
Hermaphrodite Activity Assays
The activity of hermaphrodites in liquid culture was assessed for three mutant strains: QG2458, a Mos1 insertion (ttTi53821) in the N2 background; QG2457, an R13* nonsense point mutation (gk140536) in the N2 background; and QG2462, the AB2 plep-1 locus introgressed into the N2 him-5 background. In parallel, we assayed N2 and QG2456, an N2 derivative that controls for background effects introduced by the construction of the mutant strains (see Supplemental Experimental Procedures).
Developmentally synchronized young adult hermaphrodites were placed 10 to each well of a 96-well plate containing 100μl of H2O. A census was taken immediately and activity was scored blind over 24 hours as the number of active worms (swimming or making deep body bends) per well. Inspection at 48 hours confirmed that lack of activity at 24 hours is correlated with death.
Data were collected from 10 assays (n = 5,849 worms after limiting to wells with 7-13 worms, with mean = 10, standard deviation = 1.59; see the Supplemental Data File). A binomial generalized mixed effect linear model was fit (R package lme4 [35]) to worm activity counts per well at 24 hours with strain (or genotype as a binary variable) as fixed effect and a random assay effect. All plep-1 mutants were significantly less active than N2 (P < 0.01), while the two controls strains were indistinguishable (P = 0.4).
Gene expression analysis
RNA was extracted from young adult hermaphrodites (N2 and a Mos1 insertion line, QG2458) from cultures synchronized by L1 arrest (20 hours in M9 buffer). Labeled cRNA was in vitro transcribed from 1μg total RNA (Agilent Low Input Quick Amp Labeling Kit), hybridized to Agilent V2 4×44k microarrays against a pooled reference sample and scanned on an Agilent G256CA Scanner (Supplemental Data File). Four biological replicates and three technical replicates (after stripping microarrays by treatment with RNAseH) were hybridized. Raw data were imported into the R package limma [52, 53], background corrected (method=“normexp”, offset=50), and normalized within (method= “loess”, span = 0.05) and across (method= “Gquantile”) arrays. Multiple probes are present on the array for many, but not all, transcripts. We averaged over probes targeting the same set of transcript isoforms before statistical analysis, reducing the data to 22,837 probe sets for 20,143 genes. Technical replicate correlation was estimated with the function duplicateCorrelation, and differential expression for the N2-QG2458 contrast was estimated by linear model. Moderated statistics were computed with an intensity-dependent fit to prior variances, and p-values were adjusted by controlling the false discovery rate (Benjamini and Hochberg method). Raw data are accessible under NCBI GEO accession GSE68351, processed data are in the Supplemental Data File. Functional enrichments were analyzed with the Serial Pattern of Expression Levels Locator (SPELL 2.0.3r71) [54], generating rank correlation statistics against published data and testing for enrichment of gene ontology terms among the top 100 correlated genes. Gene set enrichment analysis (GSEA 2.0.14) [55] used WS234 gene ontology annotations, filtered to terms with 10-500 genes. For genes targeted by multiple probe sets, the set with the highest F-statistic was taken. FDR was estimated by permuting gene sets 10,000 times. Microarray data are available from NCBI GEO under accession GSE68351.
Supplementary Material
Highlights.
Wild isolates of C. elegans vary in a male-male mating behavior.
In some isolates, males deposit copulatory plugs on each other's excretory pores.
A mutation in a conserved gene renders pores attractive or receptive to matings.
Excretory pore plugging may be a side-effect of mating system evolution.
Acknowledgments
This work was supported by the NIH (R01 GM089972 to MVR and F32 HD065442 to ASC), the Zegar Family Foundation, and New York University. We thank N. Ringstad and A. Paaby for comments and Zina Deretsky for the illustration in Figure 1B.
Footnotes
Contributions: LMN, ASC, DM, MK, MY, and MVR designed experiments. LMN, ASC, DM, MK, MY, JPN, DDR, PA, MP, TI, and MVR performed experiments. LMN and MVR analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith JM, Harper D. Animal Signals. Oxford University Press; 2003. [Google Scholar]
- 2.Halliday TR. The study of mate choice. In: Bateson P, editor. Mate Choice. University of Cambridge; 1983. pp. 3–32. [Google Scholar]
- 3.Johansson BG, Jones TM. The role of chemical communication in mate choice. Biological Reviews. 2007;82:265–289. doi: 10.1111/j.1469-185X.2007.00009.x. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt TD. Pheromones and Animal Behaviour. Cambridge University Press; 2003. [Google Scholar]
- 5.Thomas CG, Woodruff GC, Haag ES. Causes and consequences of the evolution of reproductive mode in Caenorhabditis nematodes. Trends in Genetics. 2012;28:213–220. doi: 10.1016/j.tig.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia LR, LeBoeuf B, Koo P. Diversity in mating behavior of hermaphroditic and male-female Caenorhabditis nematodes. Genetics. 2007;175:1761–1771. doi: 10.1534/genetics.106.068304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleemann GA, Basolo AL. Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self-sperm depletion. Animal Behaviour. 2007;74:1339–1347. [Google Scholar]
- 8.Ting JJ, Woodruff GC, Leung G, Shin NR, Cutter AD, Haag ES. Intense sperm-mediated sexual conflict promotes reproductive isolation in Caenorhabditis nematodes. PLoS Biol. 2014;12:e1001915. doi: 10.1371/journal.pbio.1001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chasnov JR. The evolutionary role of males in C. elegans. worm. 2013;2:e21146. doi: 10.4161/worm.21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genetics. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Félix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altun ZF, Hall DH. Excretory system. Herndon LF, editor. WormAtlas. 2009 Available at: http://www.wormatlas.org/hermaphrodite/excretory/Excframeset.html.
- 16.Ludewig AH, Schroeder FC. Ascaroside signaling in C elegans. The C. Elegans Research Community, editor. WormBook. 2013:1–22. doi: 10.1895/wormbook.1.155.1. Available at: http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 17.Chute CD, Srinivasan J. Chemical mating cues in C. elegans. Seminars in Cell & Developmental Biology. 2014;33:18–24. doi: 10.1016/j.semcdb.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proceedings of the National Academy of Sciences. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz IP, Levin JZ, Cummins C, Anderson P, Horvitz HR. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J Neurosci. 2003;23:9133–9145. doi: 10.1523/JNEUROSCI.23-27-09133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saifee O, Wei L, Nonet ML. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol Biol Cell. 1998;9:1235. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrière A, Félix MA. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chasnov JR, So WK, Chan CM, Chow KL. The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proc Natl Acad Sci USA. 2007;104:6730–6735. doi: 10.1073/pnas.0608050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipton J. Mate Searching in Caenorhabditis elegans: A Genetic Model for Sex Drive in a Simple Invertebrate. J Neurosci. 2004;24:7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CG, Lamitina T, Agre P, Strange K. Functional analysis of the aquaporin gene family in Caenorhabditis elegans. AJP: Cell Physiology. 2006;292:C1867–C1873. doi: 10.1152/ajpcell.00514.2006. [DOI] [PubMed] [Google Scholar]
- 26.Teramoto T, Lambie EJ, Iwasaki K. Differential regulation of TRPM channels governs electrolyte homeostasis in the C. elegans intestine. Cell Metabolism. 2005;1:343–354. doi: 10.1016/j.cmet.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–376. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cutter AD. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol Biol Evol. 2003;20:665–673. doi: 10.1093/molbev/msg072. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PEA, Malik RU, Edison AS, Sternberg PW, Schroeder FC. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morsci NS, Haas LA, Barr MM. Sperm status regulates sexual attraction in Caenorhabditis elegans. Genetics. 2011;189:1341–1346. doi: 10.1534/genetics.111.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markert M, García LR. Virgin Caenorhabditis remanei females are attracted to a coital pheromone released by con-specific copulating males. worm. 2013;2:e24448. doi: 10.4161/worm.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stiernagle T. Maintenance of C. elegans. The C. Elegans Research Community, editor. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. Available at: http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 34.R Core Team. R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org/
- 35.Bates DM. lme4: Mixed-effects modeling with R. 2010 http://lme4r-forge r-projectorg/book.
- 36.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Churchill GA, Doerge RW. Naive application of permutation testing leads to inflated type I error rates. Genetics. 2008;178:609–610. doi: 10.1534/genetics.107.074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 44.Vallin E, Gallagher J, Granger L, Martin E, Belougne J, Maurizio J, Duverger Y, Scaglione S, Borrel C, Cortier E, et al. A genome-wide collection of Mos1 transposon insertion mutants for the C. elegans research community. PLoS ONE. 2012;7:e30482. doi: 10.1371/journal.pone.0030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahringer J. Reverse genetics. The C. Elegans Research Community, editor. WormBook. 2006 Available at: http://www.wormbook.org.
- 46.McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Kuma KI, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 50.Labrousse A, Chauvet S, Couillault C, Léopold Kurz C, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Current Biology. 2000;10:1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 51.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; 2010. [Google Scholar]
- 52.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 54.Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.