Abstract
Auxin is a key regulator of plant growth and development. Classical molecular and genetic techniques employed over the past 20 years identified the major players in auxin-mediated gene expression and suggest a canonical auxin response pathway. In recent years, structural and biophysical studies clarified the molecular details of auxin perception, the recognition of DNA by auxin transcription factors, and the interaction of auxin transcription factors with repressor proteins. These studies refine the auxin signal transduction model and raise new questions that increase the complexity of auxin signaling.
Graphical Abstract
Introduction
The phytohormone auxin (indole-3-acetic acid, IAA) is a master regulator of plant growth and development through control of cell division and expansion [1]. Because auxin potently impacts growth and development, auxin regulation must be precise. A variety of auxin regulatory strategies, ranging from biosynthesis and metabolism control [2–4] to transport and localization within the plant [5, 6], influence plant growth and development. Ultimately, auxin sensing triggers the myriad of gene expression changes required for plant growth and development.
Nuclear auxin response pathway components were identified using molecular and genetic approaches to uncover three major families of proteins that intimately link hormone perception and gene expression. Discovery of these protein families - e.g. the auxin binding F-box proteins (TIR1; TRANSPORT INHIBITOR RESPONSE 1 and AFB1–5; AUXIN SIGNALING F-BOX PROTEINS 1–5), the AUXIN RESPONSE FACTOR (ARF) transcription factors, and AUXIN/INDOLE 3-ACETIC ACID INDUCIBLE (Aux/IAA) repressor proteins - employed a combination of screens and phylogenetics [7–11].
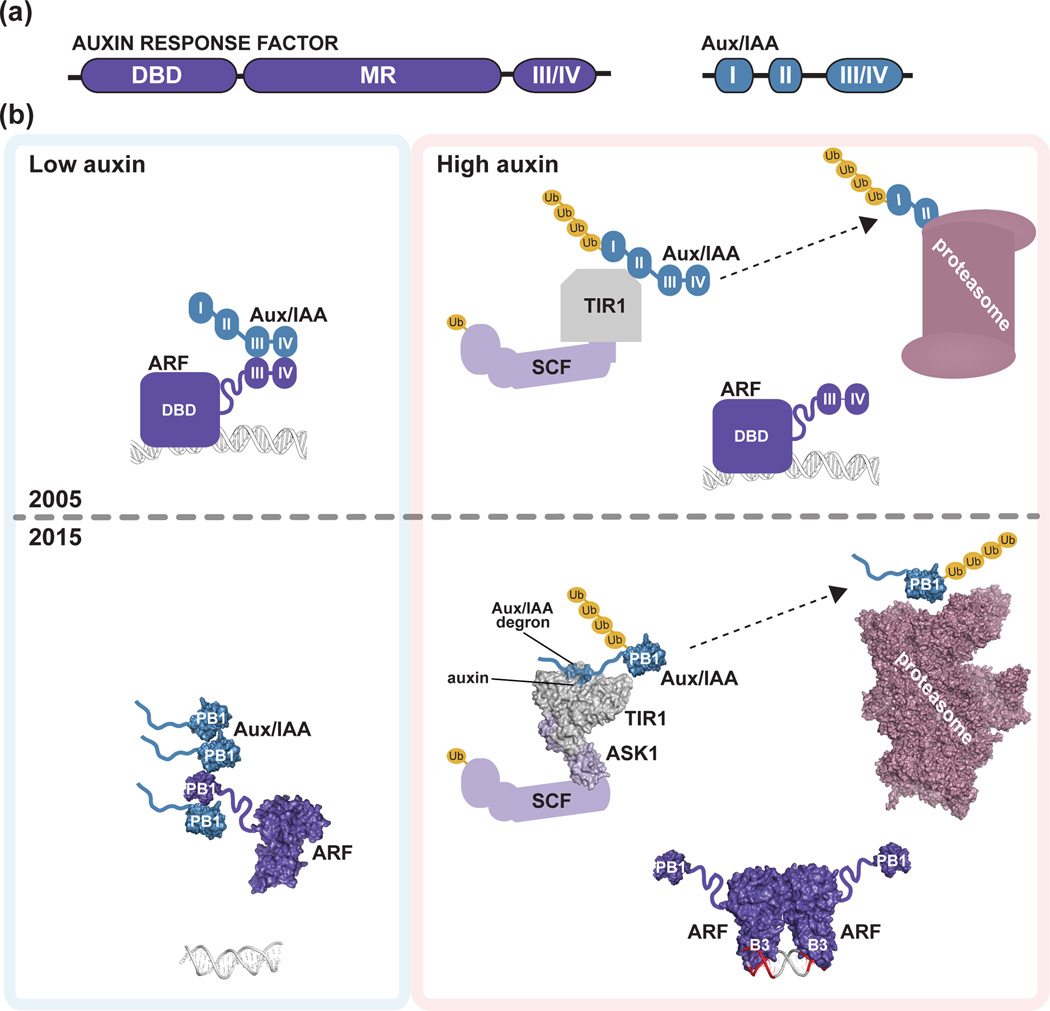
Integration of these proteins into a pathway from auxin perception to gene expression occurred through a series of creative studies. Protoplast and seedling-based assays, established the canonical 'domain' organization of the ARF transcription factors as an N-terminal DNA-binding domain (DBD), a middle region (MR) conferring either activation or repression properties, and a C-terminal region containing two sequence motifs (III/IV) [12] (Fig. 1a). Subsequent studies determined that the III/IV motif of ARF and Aux/IAA proteins mediates ARF•ARF, Aux/IAA•Aux/IAA, and ARF•Aux/IAA interactions [8, 13]; that the Aux/IAA motif I facilitates interaction with TOPLESS (TPL) co-repressors [14]; and that the Aux/IAA motif II contains a degron that controls protein stability [15]. These investigations also established Aux/IAA proteins as ARF repressors [16] and defined two ARF subfamilies - positive and negative transcriptional regulators [12]. Further, molecular biology-focused studies suggested TIR1 as an auxin receptor [17, 18].
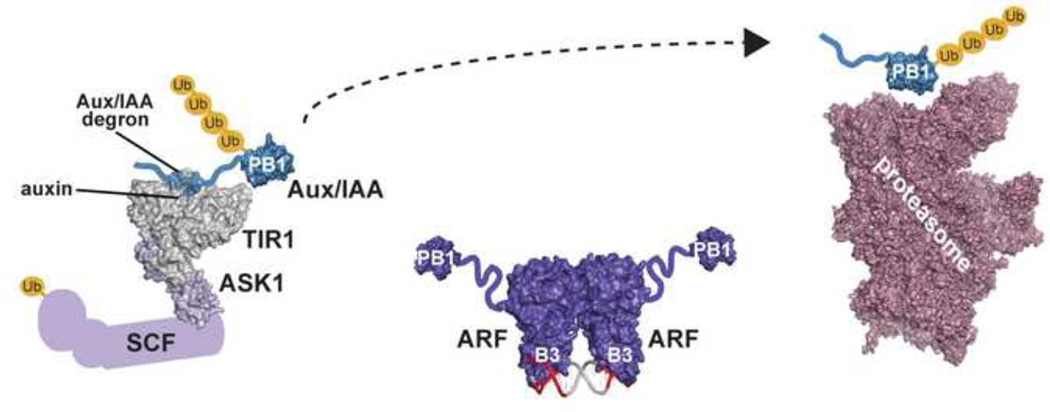
Figure 1. Initial and refined auxin response models.
(a) Schematic of homology motifs in Auxin Response Factor (ARF) and Aux/IAA proteins. The ARF DNA binding domain (DBD), middle region (MR), and III/IV interaction motif are indicated. The Aux/IAA sequence motifs I (TOPLESS interaction), II (degron), and III/IV (interaction motif) are shown. (b) The top panel depicts the auxin response model from the year 2005. Under low local auxin concentrations, Aux/IAA proteins interact with ARF proteins, thereby repressing their action. In the presence of auxin , TIR1/AFB perceives auxin, allowing for SCFTIR1/AFB interaction with Aux/IAA proteins. Subsequently, Aux/IAA repressors are degraded via the 26S proteasome to allow for ARF-mediated gene expression changes. The bottom panel integrates structural biology contributions to our understanding of auxin signaling, including the ‘molecular glue’ interaction of auxin with TIR1 that allows for interaction of SCFTIR1/AFB with Aux/IAA repressors (PDB: 2P1Q; [22]); the ARF5 DBD structure (PDB: 4LDU; [34]) providing the ‘molecular calipers’ model of AuxRE recognition (PDB: 4LDX; [34]), and the possibility of PB1 domain multimerization in ARF (PDB: 4NJ6; [39]) and Aux/IAA (PDB: 2MUK; [42]) proteins to regulate auxin signaling. The cryo-EM proteasome structure is also shown for reference ([49]; PDB: 4CR2).
Various features of these three protein families led to a general model for plant auxin responses (Fig. 1b). Under low auxin, Aux/IAA proteins repress ARF-mediated auxin-responsive gene transcription. Upon increased auxin, IAA binds an auxin-perceiving F-box protein, permitting Aux/IAA interaction. TIR1/AFB•auxin•Aux/IAA complex formation leads to Aux/IAA ubiquitylation and degradation and frees ARF proteins to regulate auxin-responsive gene expression [19].
A series of recent structural biology studies revealed salient features that guide the molecular interplay between IAA and auxin signaling components. This review summarizes current structural biology contributions to the establishment, dissection, and refinement of the auxin response pathway.
Structural identification of auxin signal perception by SCFTIR1/AFB
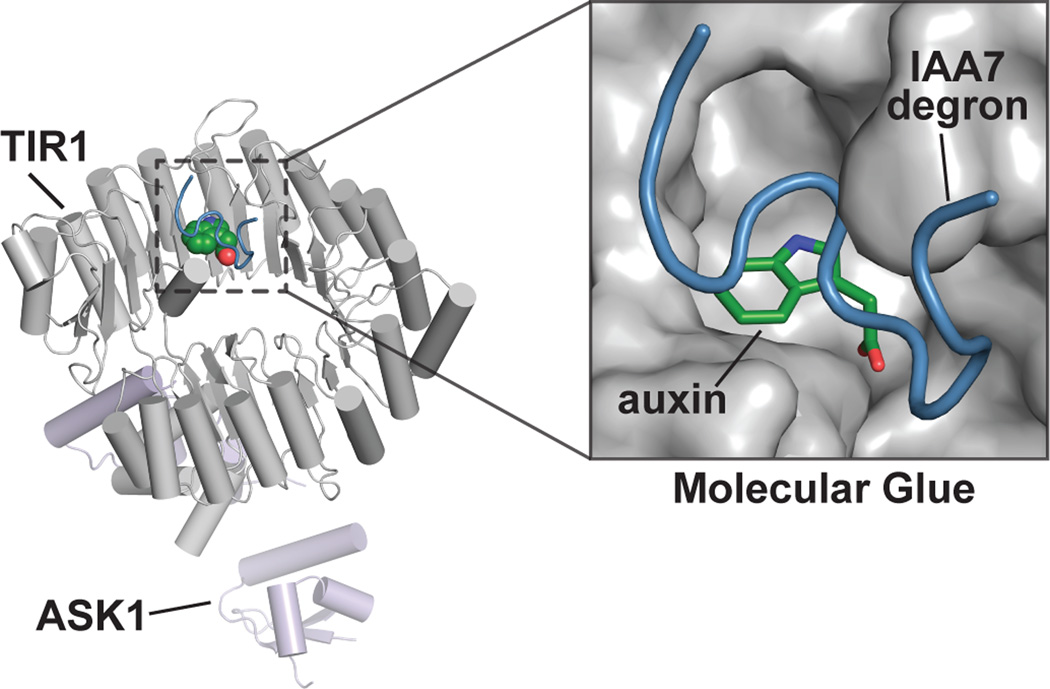
The TIR1/AFB F-box protein family provides a mechanism for auxin perception and mediates Aux/IAA protein ubiquitylation for proteasomal degradation [11, 20, 21]. After initial studies identified TIR1 as an auxin receptor [17, 18], the landmark structural study by Tan et al. [22] introduced a new mechanism for auxin perception and Aux/IAA degradation (Fig. 2). The TIR1•auxin•Aux/IAA complex x-ray crystal structure revealed how auxin binds to TIR1 to mediate interaction with the IAA7 degron motif. Auxin binding to TIR1 forms the 'molecular glue' allowing formation of the TIR1•auxin•Aux/IAA complex, which mediates Aux/IAA ubiquitylation for degradation (Fig. 1b). Later, crystallographic analysis of the E3 proteins CORONATINE INSENSITIVE1 [23] and Cereblon [24] in complex with ligands and substrates extended this 'molecular glue' model to jasmonate repressor degradation in plants and to Ikaros and Aiolos degradation in humans, respectively.
Figure 2. Molecular glue mechanism of auxin perception and Aux/IAA recognition by SCFTIR1/AFB.
The x-ray crystal structure of the SCFTIR1/AFB•auxin•IAA7 peptide complex (PDB: 2P1Q; [22]) revels the presence of an auxin binding pocket that mediates Aux/IAA protein interaction. The overall structure of the complex formed by AtTIR1 (gray), AtASK1 (light purple), IAA (green), and a peptide corresponding to the AtIAA7 degron (blue) is shown. The inset depicts the TIR1 auxin-binding pocket, in which auxin acts as a molecular glue to allow interaction with Aux/IAA protein degron motifs.
Importantly, the TIR1•auxin•Aux/IAA complex [22] crystal structure provided insight into auxin perception in plants. The structural details of auxin binding to TIR1 aided the development of chemical probes to study auxin biology. In particular, biochemical studies following TIR1 crystallographic analysis allowed for assessment of the agonist and antagonist activity of different auxins and auxin derivatives [25], leading to the rational design of a potent auxin antagonist (auxinole; α-[2,4-dimethylphenylethyl-2-oxo]-IAA) for use as a chemical probe of auxin action [26].
TIR1 structural studies also paved the way for systematic examination of ligand and substrate specificity of the TIR1/AFB family. Each TIR1/AFB family member targets specific Aux/IAA proteins for degradation [20] and, interestingly, Aux/IAA protein regions in addition to the degron contribute to this binding [15]. Moreover, different natural and synthetic auxins provide differential binding affinities for cognate F-box•Aux/IAA pairs [15, 27]. TIR1 structural information also guided subsequent studies of TIR1/AFB stability. TIR1 mutations in N-terminal α-helix residues abrogate CULLIN1 interaction and result in enhanced TIR1 stability [28], suggesting an autocatalytic mechanism of TIR1/AFB degradation. Understanding how this autocatalytic mechanism affects auxin signaling may provide important insight into TIR1/AFB roles in plant development. Further, integrating TIR1/AFB biochemical data with updated ARF and Aux/IAA expression profiling [29] and new studies involving additional auxins [15, 27] may provide important clues to further untangle the increasingly complex auxin signaling web.
Mechanistic clues of auxin responsive gene transcription through binding of AuxREs
ARF proteins are central players in the nuclear auxin response pathway. Initially discovered in a yeast one-hybrid screen for proteins that bind the canonical auxin response element (AuxRE) [7, 30], these proteins modulate auxin-responsive gene transcription. The Arabidopsis genome encodes 22 ARF proteins with a three-domain architecture (Fig. 1a), consisting of an N-terminal B3 type DBD, middle region, and interaction domain. B3 domains are plant-specific DBD [31]. The ARF MR can be enriched in either glutamine or serine, which respectively correspond to activation and repression properties [12, 32]. The C-terminal III/IV motif, described later as a PB1 domain, provides a versatile protein-protein interaction region [12, 33]. Recent structural studies reveal the mechanism of ARF recognition of the AuxRE [34].
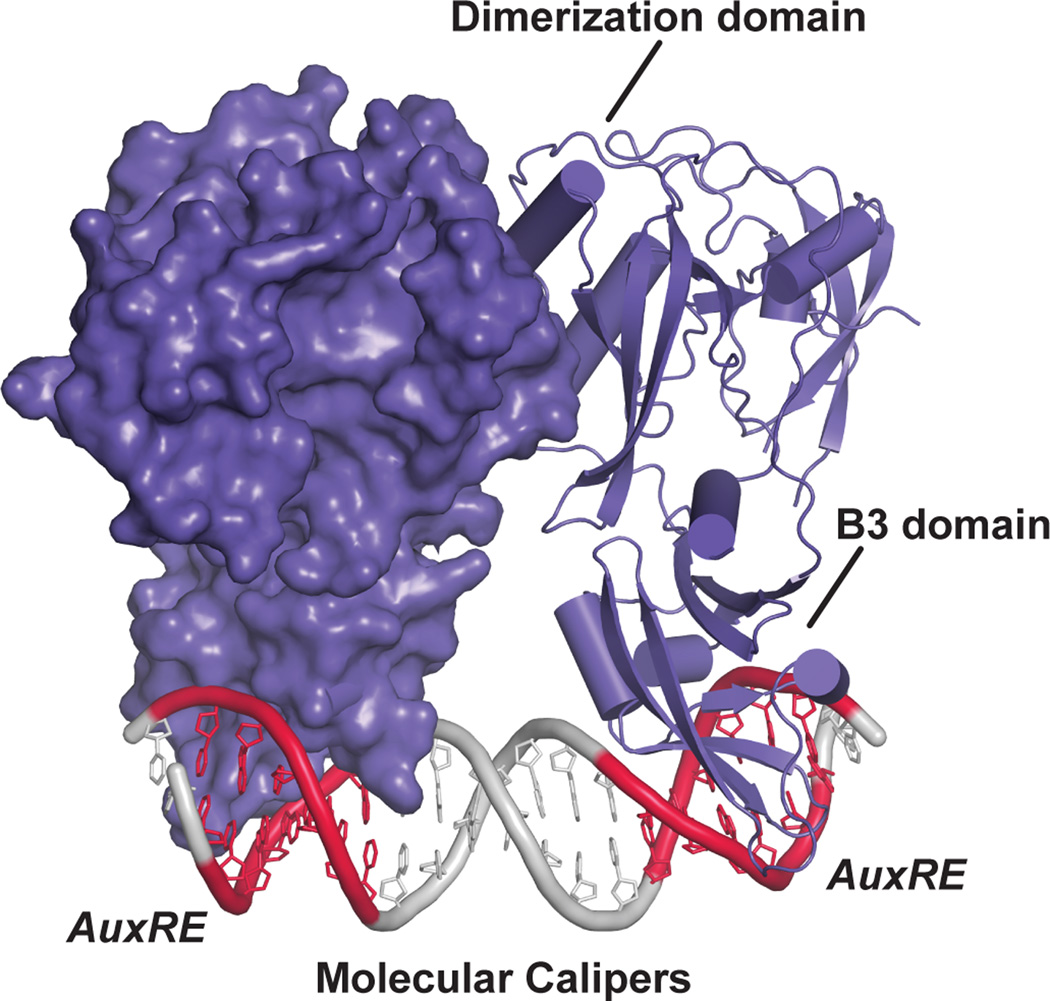
The ARF1 and ARF5 DBD X-ray crystal structures reveal a large dimerization domain, which adopts a novel fold, flanking the B3 DBD [34] (Fig. 3). Mutational analysis and small-angle X-ray scattering (SAXS) experiments verify that ARF DBD dimerization occurs in solution and is important in planta for transcriptional activity [34], as previously suggested [12]. Indeed, earlier domain deletion experiments are consistent with the dimeric ARF DBD structures [35–37].
Figure 3. Recognition of auxin response elements (AuxRE) by the ARF DNA binding domain using a molecular calipers mechanism.
The structure of the AtARF1 dimerized DNA binding domain bound to the ER7 everted repeat containing two AuxREs (PDB: 4LDX; [34]) is shown. One monomer is shown as a surface rendering and the other as a ribbon diagram. The dimerization domain allows for spacing of the B3 DNA binding domain into the major groove of the DNA to interact with the AuxRE (red), leading to a suggested molecular calipers mechanism of DNA recognition by ARF proteins [34].
Boer et al. [34] also delivered an in vitro mechanistic dissection of DNA recognition by ARF proteins. ARF DBDs preferentially bind a TGTCGG AuxRE [34] rather than the canonical TGTCTC [30]. Intriguingly, surface plasmon resonance experiments suggest that the ARF1 (repressing ARF) and ARF5 (activating ARF) DBD display similar AuxRE binding affinity; however, the preferred spacing between AuxRE elements differ for ARF1 and ARF5 [34]. The authors hypothesize that this spacing confers specificity, and, combined with the dimerization requirement, reveals a new ‘molecular calipers’ mechanism of ARF DBD interaction, DNA binding, and transcriptional regulation [34] (Figs. 1b & 3).
Future work is required to determine whether ARF binding site preferences exist in planta to provide a mechanism for auxin response modulation. Because in planta studies are confounded by multiple ARF proteins, in vivo studies using a recently-developed yeast-based system that recapitulates the auxin signaling pathway may be useful in probing the relationship between promoter architecture and ARF transcriptional control [38]. This technology could address new questions in ARF•DNA interaction and regulation. For instance, what are the implications of AuxRE degeneracy? Also, does the spacing of tandem AuxREs drive specific ARF interaction pairs? Gaining further insight will provide mechanistic insight into ARF DBD interactions modulate auxin signaling.
ARF and Aux/IAA PB1 domain structures introduce a multimerization option
ARF and Aux/IAA proteins interact through two C-terminal regions of sequence homology (i.e., sequence motifs III and IV) [8]. X-ray crystal structures of the C-terminal regions of Arabidopsis ARF7 [39] and ARF5 [40] demonstrate that region III/IV adopts a type I/II Phox/Bem1p (PB1) domain structure (Fig. 4a), as suggested by bioinformatic analysis [41]. Likewise, NMR structures of the PB1 domains of Arabidopsis IAA17 [42] and Pisum sativum IAA4 (PDB: 2M1M) confirm a similar architecture in these repressor proteins. Thus, the III/IV region of ARF and Aux/IAA proteins fold into canonical type I/II PB1 domains.
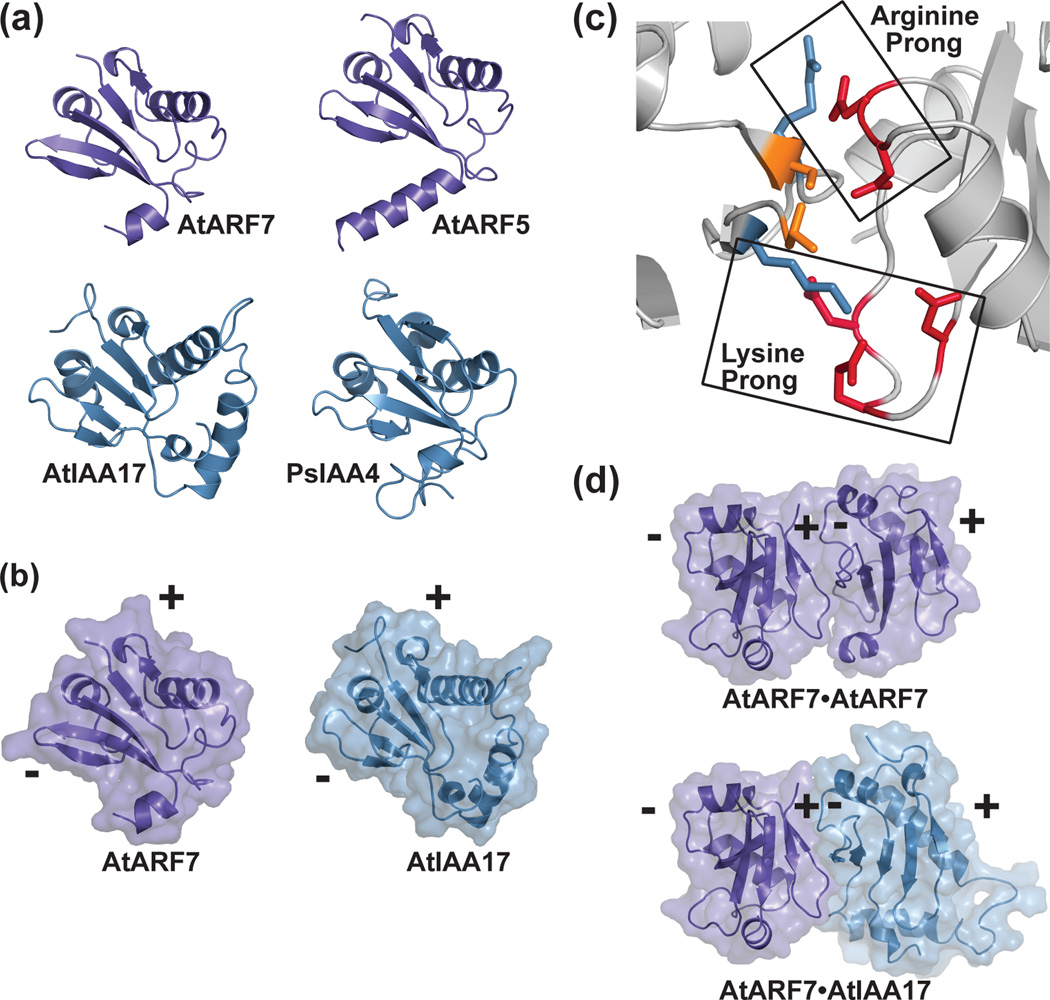
Figure 4. Crystal structures of the ARF and Aux/IAA PB1 domains and the versatility of protein-protein interaction.
(a) The X-ray crystal structures of the PB1 domains of AtARF7 (PDB: 4NJ6; [39]) and AtARF5 (PDB: 4CHK; [40]) and the NMR solution structures of AtIAA17 (PDB: 2MUK; [42]) and PsIAA4 (PDB: 2MIM) reveal a common fold. (b) The PB1 domain scaffold presents conserved residues on opposing positive (+) and negative (-) interaction surfaces. A mixed surface/ribbon view of the PB1 domains of AtARF7 [39] and AtIAA17 [42] are shown. (c) PB1-mediated interaction of ARF and/or Aux/IAA proteins use a two-pronged binding motif. Biophysical studies of the ARF7 PB1 domain (PDB: 4NJ6; [39]) revealed how conserved lysine and arginine residues on the positive-face interact with two structurally distinct groups of acidic residues. (d) PB1 domains of ARF and Aux/IAA provide a versatile scaffold for protein interaction. Mixed surface/ribbon diagram showing the crystallographically-observed head-to-tail orientation of two AtARF7 PB1 domains (PDB: 4NJ6; [39]). Modeling of a hypothetical AtARF7•AtIAA17 complex, based on the AtARF7•AtARF7 structure, predicts the overall scaffold allowing for mixed-protein interaction.
Type I/II PB1 domain interactions are facilitated by opposing electrostatic faces provided by two conserved sequence motifs: a positively-charged invariant lysine and a cluster of negatively-charged residues, termed the OPCA motif [43]. These differential electrostatic faces allow for directional protein interaction (Fig. 4b) that may result in protein multimerization.
Biophysical characterization of PB1 domain interactions revealed a preference for the mixed ARF5•IAA17 (Kd = 0.073 µM) interaction [42] compared to the ARF5•ARF5 (Kd = 0.86 µM) self-interaction [42], the ARF7•ARF7 (Kd = 0.18 µM) self-interaction [44], or the IAA17•IAA17 self-interaction (Kd = 6.6 µM) [42]. Mutagenesis and calorimetric analysis to map the ARF7 interaction interface further defines the role of two decentralized hot spots for these interactions [44]. These two electrostatic prongs (Fig. 4c) serve to stabilize ARF7 PB1 interaction and are highly conserved across the ARF and Aux/IAA [44]. Structural studies of the PB1 domains of ARF7 [39] and ARF5 [40] reveal how these domains self-interact (Fig. 4d) and suggest the mixed ARF•Aux/IAA interaction uses the same interface (Fig. 4d). The versatility of the PB1 domain in ARF and Aux/IAA interactions derives from two features in the interface: 1) a core two-pronged interaction that provides the energetic driving force for binding and 2) co-variation of residues between the ARF and Aux/IAA proteins, which likely provide binding specificity.
Biochemical and structural studies indicate that ARF7 [39], ARF5 [40], and IAA17 [42] multimerize in vitro and that mutations in either face of the PB1 domain abrogates multimerization [39, 40]. PB1-mediated multimerization may play a role in auxin responses (Fig. 1b). For example, overexpression of a stabilized IAA16 variant bearing mutations in either the positive or negative PB1 domain face ameliorates IAA16 repressive activity, suggesting that ARF•Aux/IAA dimerization is insufficient for ARF repression [39]. Further, mutation of each ARF7 electrostatic face individually increased DR5 auxin reporter activity in the presence of auxin in protoplast assays, whereas mutating both ARF7 electrostatic faces lead to DR5 reporter activity in the absence of auxin [40], suggesting that these electrostatic faces are required for ARF repression by Aux/IAA proteins. Single electrostatic face IAA17 and IAA19 mutants only partially repressed auxin-responsive reporter activity in protoplasts [40], consistent with the possibility that Aux/IAA•ARF dimerization is insufficient to fully repress ARF activity. Together, these reports suggest that Aux/IAA oligomerization, rather than simple dimerization, may be important for ARF activity repression. Additionally, recent mathematical modeling supports the possibility that ARF multimerization may modulate auxin sensitivity [45]. A critical next step is to further investigate the existence, effect, and role of ARF and/or Aux/IAA multimerization in vivo.
The high affinity for ARF•ARF and ARF•Aux/IAA interactions suggests the need for regulating these interactions. In mammals, phosphorylation regulates PB1 interactions in p62/SQSTM1 signaling [46]. In plants, phosphorylation of the ARF MR regulates ARF2 repression of transcription [47], ARF7 activation of transcription [48] and ARF19 activation of transcription [48]. Although in vivo phosphorylation of ARF or Aux/IAA PB1 domains has not been demonstrated, in vitro data suggest the threonine, which is a predicted phosphorylation site, near the invariant lysine of the ARF7 PB1 domain stabilizes ARF7•ARF7 interaction [44]. Understanding the control of PB1 interaction may lead to regulatory systems in auxin signaling.
Structural and functional studies of ARF and Aux/IAA PB1 domain interfaces [39, 40, 42, 44] raise further questions regarding interaction specificity. Does the specificity of ARF•ARF, ARF•Aux/IAA, or Aux/IAA•Aux/IAA PB1 interactions play a functional role in auxin signaling? Can variation among key PB1 interaction residues guide inference of interaction partners? Do these proteins multimerize in vivo, and if so, does this multimerization play a role in auxin response regulation? Answers to these questions may provide further insight into the function of distinct ARF and Aux/IAA proteins.
Conclusions
Structural biology provided new insights that define and refine the auxin response pathway. The first views of the TIR1•auxin•Aux/IAA complex established the ‘molecular glue’ mechanism for auxin perception; ARF DBD structural analyses revealed a molecular basis for AuxRE recognition; and ARF and Aux/IAA PB1 domain structures led to insight into how protein interactions attenuate auxin responses in plants and raises the possibility of ARF and/or Aux/IAA multimerization. Together, these studies have refined the auxin response pathway from what was known in 2005 to an updated view in 2015 (Fig. 1b). These studies provide molecular snapshots of the auxin response mechanism and reinforce the core pieces of the canonical pathway; however, they also suggest new directions for dissecting the complexity and diversity of the auxin web.
Biophysical research is critical to understanding and dissecting complex systems in plants, such as auxin signaling. Importantly, structural and biophysical analysis of TIR1/AFB1-5, ARF proteins, and Aux/IAA proteins provide a foundation for translation to in vivo studies. With structural data in hand, we can move forward with additional genetic and biochemical tests to validate and expand upon these results.
Highlights.
Structural biology studies refined the auxin response pathway
The TIR1 E3 ubiquitin ligase structure reveals the auxin perception mechanism
ARF DNA binding domain structures establish the ‘molecular calipers’ mechanism
ARF and Aux/IAA PB1 interaction domain structures increase pathway complexity
Acknowledgements
Support from the National Science Foundation Graduate Research Fellowship Program (2011101911 to D.A.K.), the United States Department of Agriculture – National Institute of Food and Agriculture Fellowship Program (MOW-2014-01877 to D.A.K.), the National Institutes of Health (R00 GM089987-03 to L.C.S.), and the National Science Foundation (MCB-1157771 to J.M.J. and IOS-1453750 to L.C.S.) is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Enders TA, Strader LC. Auxin activity: Past, present, and future. Am J Bot. 2015;102:180–196. doi: 10.3732/ajb.1400285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. Journal of Experimental Botany. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westfall CS, Muehler AM, Jez JM. Enzyme action in the regulation of plant hormone responses. J Biol Chem. 2013;288:19304–19311. doi: 10.1074/jbc.R113.475160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peer WA. From perception to attenuation: auxin signalling and responses. Curr Opin Plant Biol. 2013;16:561–568. doi: 10.1016/j.pbi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Adamowski M, Friml J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- 10.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 15.Calderón-Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 18.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 19.Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-Based Auxin Perception: Mechanism and Role in Plant Growth and Development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demaegdt H, Smitz L, De Backer JP, Le MT, Bauwens M, Szemenyei E, Toth G, Michotte Y, Vanderheyden P, Vauquelin G. Translocation of the insulin-regulated aminopeptidase to the cell surface: detection by radioligand binding. Br J Pharmacol. 2008;154:872–881. doi: 10.1038/bjp.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. **The TIR1 auxin receptor structure provided the first mechanistic insight into auxin perception. This study further provided evidence that auxin directly allows the interaction of Aux/IAA repressor proteins with SCFTIR1, thereby establishing the 'molecular glue' mechanism of auxin perception.
- 23.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, Rychak E, Corral LG, Ren YJ, Wang M, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–809. doi: 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc Natl Acad Sci U S A. 2008;105:5632–5637. doi: 10.1073/pnas.0711146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol. 2012;7:590–598. doi: 10.1021/cb200404c. [DOI] [PubMed] [Google Scholar]
- 27. Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM. Defining binding efficiency and specificity of auxins for SCF(TIR1/AFB)- Aux/IAA co-receptor complex formation. ACS Chem Biol. 2014;9:673–682. doi: 10.1021/cb400618m. *Biochemical and biophysical approaches were employed in this study to quantify the affinity and specificity of TIR1/AFB family members for various natural and synthetic auxins in combination with Aux/IAA interaction specificity, uncovering specific TIR1/AFB•auxin•Aux/IAA interaction partners.
- 28.Yu H, Zhang Y, Moss BL, Bargmann BOR, Wang R, Prigge M, Nemhauser JL, Estelle M. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants. 2015;1 doi: 10.1038/nplants.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Piya S, Shrestha SK, Binder B, Stewart CN, Jr, Hewezi T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci. 2014;5:744. doi: 10.3389/fpls.2014.00744. *This study described the co-expression profile of all ARF and Aux/IAA proteins. Combined with previous large scale interaction data, this research may be important to untangling certain aspects of the auxin signaling web.
- 30.Ulmasov T, Liu Z-B, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13:647–655. doi: 10.1016/j.tplants.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guilfoyle TJ. The PB1 Domain in Auxin Response Factor and Aux/IAA Proteins: A Versatile Protein Interaction Module in the Auxin Response. Plant Cell. 2015;27:33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. **Structures of the ARF1 and ARF5 DNA binding domains revealed that these domains contain not only a B3-type DNA binding domain, but also a dimerization domain and an ancillary domain of unknown function. The authors further speculate a 'molecular calipers' mechanism of AuxRE recognition by ARF DNA binding domains to provide DNAbinding specificity by different ARF•ARF pairs.
- 35.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- 36.Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012;194:391–401. doi: 10.1111/j.1469-8137.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang S, Hagen G, Guilfoyle TJ. ARF-Aux/IAA interactions through domain III/IV are not strictly required for auxin-responsive gene expression. Plant Signal Behav. 2013;8:e24526. doi: 10.4161/psb.24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci U S A. 2014;111:9407–2412. doi: 10.1073/pnas.1324147111. *This report describes a simple and elegant synthetic system for studying and quantifying auxin signaling in yeast. Importantly, this system allows for studying the fine-tuning of auxin responses by varying auxin receptor, repressor, transcription factor, and reporter components.
- 39. Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A. 2014;111:5427–5432. doi: 10.1073/pnas.1400074111. **Along with [40], this structural study revealed the presence of a type I/II PB1 domain at the C-terminus of ARF proteins. It also provides evidence that multimerization between ARF and Aux/IAA proteins may be necessary for modulating auxin response repression.
- 40. Nanao MH, Vinos-Poyo T, Brunoud G, Thevenon E, Mazzoleni M, Mast D, Laine S, Wang S, Hagen G, Li H, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5:3617. doi: 10.1038/ncomms4617. **Along with [39], this structural study revealed the presence of a type I/II PB1 domain at the C-terminus of ARF proteins. It provides additional evidence that ARF and Aux/IAA multimerization may be a key manner of attenuating auxin response.
- 41.Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 42. Han M, Park Y, Kim I, Kim EH, Yu TK, Rhee S, Suh JY. Structural basis for the auxin-induced transcriptional regulation by Aux/IAA17. Proc Natl Acad Sci U S A. 2014;111:18613–18618. doi: 10.1073/pnas.1419525112. **In this study, NMR analysis revealed that the C-terminus of an Aux/IAA protein folds into a PB1 domain. Further, it provided the first calorimetric analysis of ARF•ARF, ARF•Aux/IAA, and Aux/IAA•Aux/IAA interactions, revealing that the ARF•Aux/IAA interaction holds higher affinity than ARF•ARF or Aux/IAA•Aux/IAA interactions.
- 43.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007:re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 44.Korasick DA, Chatterjee S, Tonelli M, Dashti H, Lee SG, Westfall CS, Fulton DB, Andreotti AH, Amarasinghe GK, Strader LC, et al. Defining a Two-Pronged Structural Model for PB1 Domain Interaction in Plant Auxin Responses. J Biol Chem. 2015 doi: 10.1074/jbc.M115.648253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farcot E, Lavedrine C, Vernoux T. A Modular Analysis of the Auxin Signalling Network. PLoS One. 2015;10:e0122231. doi: 10.1371/journal.pone.0122231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christian F, Krause E, Houslay MD, Baillie GS. PKA phosphorylation of p62/SQSTM1 regulates PB1 domain interaction partner binding. Biochim Biophys Acta. 2014;1843:2765–2774. doi: 10.1016/j.bbamcr.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. *This study provided important evidence demonstrating that phosphorylation, as a response to other developmental cues, is a means of regulating and modulating the activity of activating ARF proteins.
- 49.Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Forster F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]