Abstract
To overcome endoplasmic reticulum (ER) stress, ER-localized stress sensors actuate distinct downstream organelle-nucleus signaling pathways to invoke a cytoprotective response, known as the unfolded protein response (UPR). Compared to yeast and metazoans, plant UPR studies are more recent but nevertheless fascinating. Here we discuss recent discoveries in plant UPR, highlight conserved and unique features of the plant UPR as well as critical yet-open questions whose answers will likely make significant contributions to the understanding plant ER stress management.
Introduction
In growth and stress situations, cell homeostasis depends on the functional integrity of the major biosynthetic organelle, the ER, which synthesizes one third of the cellular proteome and essential lipids, and assembles complex-protein associated carbohydrates. When the cell's biosynthetic demands overcome the ER biosynthetic capacity, a potentially lethal condition, known as ER stress, is initiated. In multicellular eukaryotes if the UPR fails, ER stress-induced apoptosis occurs. Along eukaryotic lineages, specialized sensors have evolved to manage the UPR and control life-or-death outcomes in ER-stress conditions [1,2]. For ER-stress protection, yeast relies exclusively on the ER-associated ribonuclease and kinase IRE1. Plants and metazoans have evolved a complex signaling network based on membrane-tethered transcription factors (MTTFs) (Box 1), some of which are plant-specific (Figure 1). In metazoans, the ER-associated PERK also operates in UPR signaling (Box 1). Although PERK-like activity has been detected is plants [3], plant PERK-like proteins have not been identified yet (Box 1).
Box 1. Multicellular UPR signaling strategy in multicellular eukaryotes.
Membrane-tethered bZIP TFs
In response to ER stress in metazoans, the ER membrane-tethered activating transcription factor 6 (ATF6) moves to the Golgi where the site 1 and site 2 proteases (S1P, S2P) release the TF domain from the membrane anchor. This process is followed by nuclear translocation of the TF domain to modulate gene transcription. The Arabidopsis genome encodes functional homologs of ATF6: AtbZIP28 [44,45] and AtbZIP17 [40]. Functional equivalent TFs have been reported in other plant species [7,46]. The mechanisms of activation of AtbZIP28 are similar to those of ATF6 with COPII-dependent ER-to-Golgi translocation [47] and subsequent activation through proteolysis at the Golgi by S1P [48] and S2P [38], which recognize cleavage sites in the lumenal face and in the transmembrane domain of AtbZIP28, respectively [47,48]. Activation of AtbZIP17 relies on S2P [38], but reliance on S1P is yet unknown.
PERK
In metazoans, the protein kinase RNA-like ER kinase (PERK) attenuates general translation but favors selective translation of the ATF4 TF through selective phosphorylation of the eukaryotic elongation factor eIF2α [2,49]. In plants a functional homologue of PERK has not been identified yet, despite the evidence that a PERK-like phosphorylation of transcription initiation factors (eIF) occurs in ER stress. AteIF2α hyper-phosphorylation occurs in a loss-of-function mutant background of the Arabidopsis SPIKE1 (SPK1), a DOCK family GEF involved in cell morphogenesis [3,50]. Intriguingly, spk1 has constitutively elevated expression levels of UPR target genes, and shows hypersensitivity to tunicamycin [3]. The evidence that simultaneous loss of AtIRE1 and AtbZIP28 is lethal at embryonic level [18] supports that if PERK-like proteins were involved in the UPR in plant cells, they would likely function in specific stress situations. Yet it cannot be excluded that the kinase activity of AtIRE1, which has been experimentally validated [18], may be responsible for the observed AteIF2α hyper-phosphorylation.
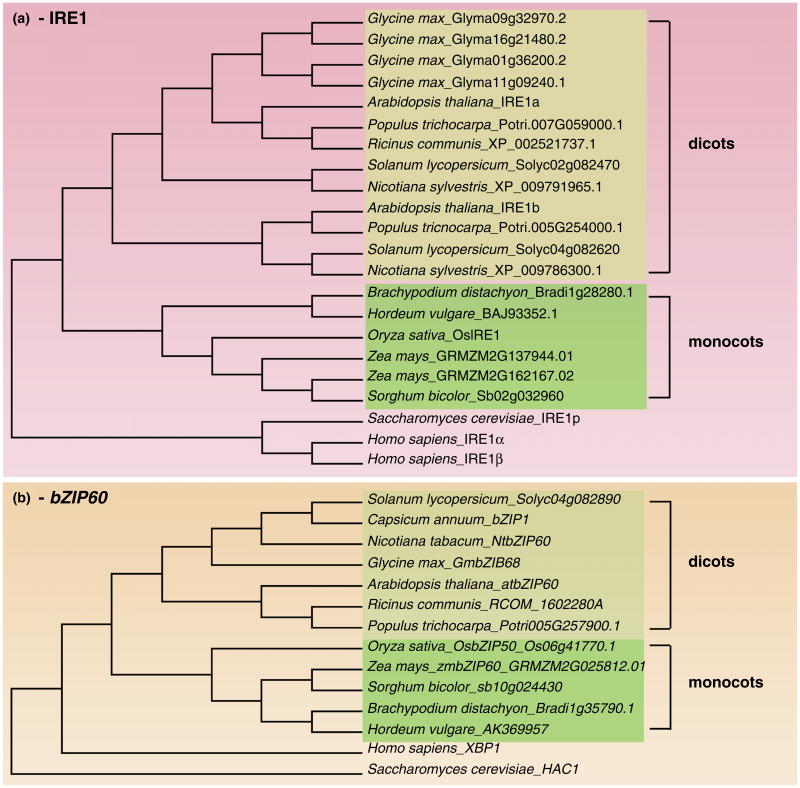
Figure 1.
Phylogenetic analysis of IRE1-like proteins and AtbZIP60-like transcripts from different plant species, yeast and human. (a) Phylogenic tree analysis of IRE1-like proteins from Plantae (eudicots and monocots), Metazoa (Homo sapiens) and Fungi (Saccharomyces cerevisiae) was generated with MEGA6 program with 1000 replicates in the bootstrap test [51] using the neighbor-joining method [52]. The IRE1-like amino acid sequences were collected from NCBI database. Branch length is not to scale. (b) Phylogenic tree analysis of AtbZIP60-like transcripts from Plantae (dicots and monocots), Metazoan (Homo sapiens) and Fungi (Saccharomyces cerevisiae) was generated with MEGA6 program with 1000 replicates in the bootstrap test [51] using the neighbor-joining method [52]. The AtbZIP60-like transcripts were collected from NCBI database. Branch length is not to scale.
How the sensors input stress to modulate an effective output in life-or-death decisions is largely unknown, especially in plants. Here we discuss plant UPR strategies that recent exciting results have brought to light in support of convergence of signaling strategies among multicellular eukaryotes as well as plant-specific features of the UPR.
Diversification of IRE1 signaling in the UPR: activation of a transcription factor
Plants encode one or more IRE1 isoforms (Figure 1a). IRE1-mediated splicing of a transcription factor (TF) mRNA is a conserved strategy for UPR gene regulation (Figure 2). Among eukaryotes the IRE1-TF targets are members of the large bZIP-TF family: Hac1 in yeast and Xbp1 in metazoans. In flowering plants, AtbZIP60, Osb-ZIP74/50 and ZmbZIP60 are the targets identified in Arabidopsis thaliana (dicot), Oryza sativa and Zea mays (monocots), respectively [4–9] (Figure 1b). The consensus mRNA structure that is recognized by IRE1 (i.e. twin-stem loops with conserved bases in each loop) is largely conserved across eukaryotes. The excised intron sequence is slightly different among monocots but conserved in dicots [8]. However, there are noticeable differences in the splicing output among eukaryotes. In plants, the IRE1-mediated mRNA splicing causes an intron-excising frameshift that occurs closely upstream of the region encoding a putative transmembrane domain and leads to elimination of the membrane anchor in the translated active TF [4,7,8]. In metazoans, the IRE1-spliced Xbp1 loses the ER membrane anchor and gains the TF domain located in the new C-terminal tail [10,11]. In yeast, the splicing of Hac1 mRNA is followed by transcriptional joint of the DNA-binding domain to the newly encoded TF transcription activation domain [12]. The lack of a close conservation of the sequence of the mature TFs and the diversification of the splicing outputs among eukaryotes support that, despite IRE1 being an ancient sensor, IRE1-TF targets may have evolved independently. Under this light, the notion that Hac1 and Xbp1 are the sole direct TF targets of IRE1 may not apply in plants where IRE1 may have other direct TF targets that may fine-tune IRE1 signaling in the UPR management in specific stress situations. This possibility is predicated by the evidence that even in the simultaneous absence of the Arabidopsis ER-stress activated TFs (AtbZIP28 and AtbZIP60), one of the major UPR gene markers, AtBiP3 is mildly induced in conditions of ER stress [13••].
Figure 2.
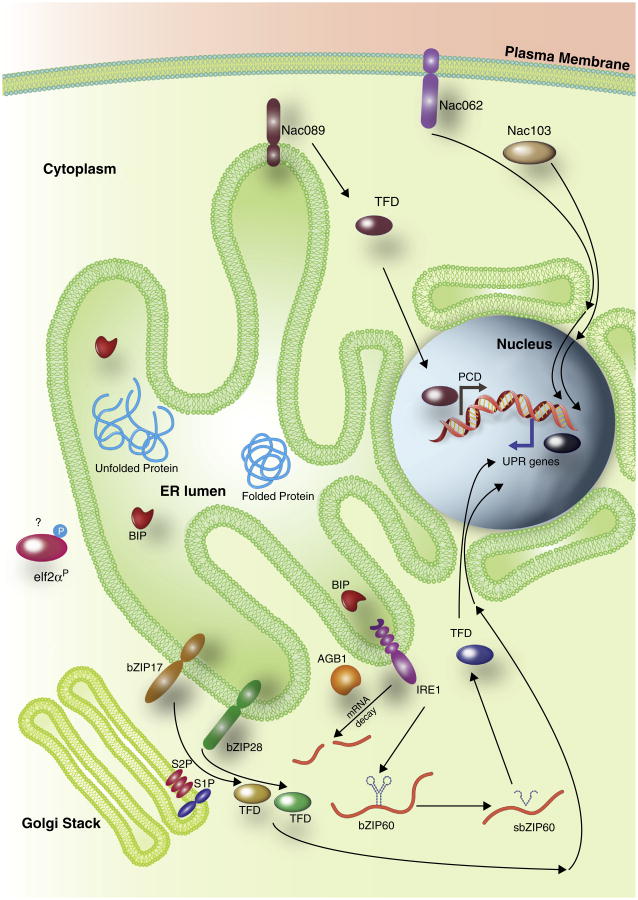
Players of the plant UPR known to date. The diagram illustrates that in addition to IRE1, bZIP28 and bZIP60, also bZIP17 plays a role in the control of UPR target genes in Arabidopsis, as recently proposed [27]. A role for the beta subunit of the heterotrimeric G-protein complex, AGB1, in modulating the UPR has also been reported [17••,53]. Furthermore, recent work has shown that NAC TFs participate to the plant UPR. NAC are plant-specific transcription factors, among which at least 13 members are predicted to be membrane-anchored in Arabidopsis. In condition of stress, the plasma membrane NAC062/ANAC062/NTL6 relocates to the nucleus, and the overexpression of a protein deletion devoid of the membrane anchor leads to activation of UPR genes [54••]. Another membrane-anchored NAC TF, NAC089, participates to ER stress responses by relocating from the ER to the nucleus and regulating PCD-related gene expression [55•]. NAC089 expression is regulated by bZIP28 and bZIP60; bZIP60 also regulates directly the expression of NAC062. Finally the cytosolic NAC103, whose expression is directly regulated by bZIP60, is involved in controlling UPR gene expression [26•]. The evidence that plant-specific NAC TFs have a role in stress responses support that in addition to common routes plants have evolved unique strategies for ER stress defense that, as in the case of NAC062, may also depend on the activity of TFs that are localized to the membrane organelles other than the ER. Interestingly also, it has been recently suggested that unspliced AtbZIP60 may have a role in modulating the expression of BiP3 in conditions of ER stress [27]. Phosphorylation of the elf2α may occur in a hypothetical PERK-like pathway for ER stress responses. See also the main text for additional details. TFD: transcription factor domain
Diversification of IRE1 signaling in the UPR: RIDD
In addition to TF-mRNA splicing, metazoan IRE1 degrades mRNAs through a site-specific cleavage process, known as regulated IRE1-dependent decay (RIDD) [14], which is uncoupled from the site-specific splicing activity of IRE1 [15]. RIDD activity is required for ER homeostasis in non ER-stress conditions and increases progressively with ER-stress intensity. RIDD also occurs in Arabidopsis [16••], but its physiological relevance without ER-stress induction is yet to be defined [16••]. In normal conditions of growth, loss of AtIRE1 function causes defects in root elongation [17••], independently of Atb-ZIP60 [18••]. It is possible therefore that in the absence of induced ER stress RIDD may be important to maintain growth homeostasis in specific plant organs by controlling the levels of mRNA of proteins involved in cell growth.
Diversification of IRE1 signaling in the UPR: miRNA cleavage
In mammalian cells, IRE1 also cleaves sequences of specific miRNAs [19]. miRNAs operate to translationally repress gene expression [20]. Whether plant IRE1 has a similar control of miRNA levels is yet unknown. Selective miRNA cleavage could be a strategy to either overcome stress, modulate tissue growth, or both. Addressing this question may lead to new insights into the functions of plant IRE1, and to establishing whether IRE1 activity is conserved among eukaryotes. Indeed the findings that loss of IRE1 is lethal in conditions of prolonged ER stress support a function as ER-stress protector [17••]. The recent evidence that the mammalian IRE1 may not activate caspases to induce cell death in prolonged ER stress, contrarily to a previous notion [19], but rather delay the onset of apoptosis via decay of the pro-apoptotic Death Receptor 5 (DR5) [21], poses that plant and animal IRE1s may be functionally close in protecting the organism from death.
How may multiple IRE1 help plants in stress defense?
An intriguing question relates to the diversification of the ER-stress sensors in plants both qualitatively and quantitatively. In Arabidopsis there are two IRE1 isoforms, AtIRE1a and AtIRE1b [22]. Whether IRE1 isoforms may be functionally equivalent is a matter of debate. Both genes are expressed throughout the plant, but AtIRE1a is more abundant in embryos and seeds [22,23]. It has been proposed that unlikely AtIRE1a, AtIRE1b is relevant to AtbZIP60 splicing in conditions of DTT-induced stress [4], supporting the possibility of functional diversification of AtIRE1 proteins. This is further reinforced by the observations that while AtIRE1b has a preponderant role in AtbZIP60 splicing upon tunicamycin-induced ER-stress, AtIRE1a seems preferentially required for AtbZIP60 splicing upon pathogen and salicylic acid (SA) infection [5]. Although functional diversification of the SA response has been recently questioned [24], the evidence that AtIRE1b but not AtIRE1a is required for AtbZIP60 splicing [4] supports that AtIRE1 proteins may have undergone functional specialization. These observations raise the question how the multiple IRE1-functions may be executed in species with one IRE1 isoform. The observation that in Arabidopsis AtIRE1b but not AtbZIP60 is required for ER stress-induced autophagy [25••] hints to the possibility that, in species containing only one IRE1 gene, some of the IRE1-roles may be executed by other proteins. Nonetheless the evidence in Arabidopsis that in induced-stress conditions, unlike a double atire1a/atire1b mutant, single IRE1 loss-of-function mutants survive prolonged ER stress supports that at least the pro-survival role of AtIRE1 is shared by the two proteins. Whether the roles of the IRE1 isoforms are spatially and/or temporally regulated is yet to be established. It is possible that in certain tissues IRE1 may have a specific relevance and other ER stress sensors might operate predominately in other tissues.
How may multiple bZIPs help plants in stress defense?
Loss of either AtbZIP60 or AtbZIP28 (Box 1) attenuates the expression of some common transcriptional targets such as BiP3 and calnexin, an effect that is additive when both genes are simultaneously deleted [26•]. This evidence supports overlapping functions of these two TFs for which a precise extent of overlap is yet to be completely understood both at a temporal resolution as well as target identity. Specifically, it is yet to be defined whether at the onset of ER stress, activation of AtbZIP28 and AtbZIP60 occurs simultaneously or sequentially. One may predict that mild stress might be resolved by the function of one of the two TFs and that the activation of the other TF may be actuated in situations of unresolved ER stress to potentiate an ER stress-mitigating response. It is also known however that activation of bZIP-TFs may lead to responses that are qualitatively distinct. For example, in conditions of salt stress, AtbZIP17 modulates only the expression of BiP3 among the known ER stress markers [27], which, on the other hand, is controlled by both AtbZIP28 and AtbZIP60 in conditions of ER stress induced by heat or by drugs such as tunicamycin and DTT. Furthermore, heat, tunicamycin and DTT but not salt stress can induce activation of AtbZIP60 and AtbZIP28, and a role of AtIRE1-AtbZIP60 but not AtbZIP28 has been demonstrated in virus-plant interactions [28]. These findings support that plant stress response relies on functional specialization of the UPR effectors for which the underlying mechanisms remain poorly understood. In this view the UPR sensors might be specialized in perceiving specific classes of unfolded proteins, which may be more sensitive than others to specific stresses.
Requirements for the UPR sensors
In yeast and metazoans, a model has emerged that the binding of unfolded proteins to IRE1 dimers allows further oligomerization and trigger the IRE1 activation, while the BiP binding to IRE1 may have a modulatory role in buffering the pool of IRE1 monomers [2,29]. In plants the mechanisms of activation of IRE1 are largely unknown. Nonetheless, binding to AtBiP is a mechanism that constrains AtbZIP28 to the ER and therefore modulates AtbZIP28 traffic to the Golgi for activation [30]. Such a mechanism, which is similar to the activation mechanism of ATF6 [31] could suggest also conservation of IRE1 activation. Similarly, AtbZIP17 may be retained in the ER via interactions with other proteins that specifically respond to salt stress. One alternative that cannot be excluded is that one branch of the UPR may be activated as consequence to the operation of another ER-stress response, such as production of radicals or unfolded proteins, which could lead to the activation of other ER stress defenses.
Interaction with unfolded proteins might not be the only upstream requirement for plant IRE1 activation. Functional and genetic evidence have shown that the ER-shaping protein AtRHD3 is a critical factor that acts upstream of AtIRE1 in the AtbZIP60 splicing in conditions of ER stress [32•], but the underlying causes are not known. In conditions of ER stress yeast and metazoan IRE1s are known to cluster in ER subdomains where stress signaling is believed to occur [2]. In the absence of induced ER stress, fluorescent protein fusions to plant IRE1s have been localized to the perinuclear ER [22]. It is possible that, if plant IRE1 is also mobilized into signaling clusters in ER-stress conditions, then alteration of the ER network structure or lipid composition, known to occur with RHD3 deletions [33,34], may affect mobilization of plant IRE1 into signaling hubs.
Besides factors acting upstream the UPR sensors in plants, we know very little about downstream regulating factors. Unlike metazoan IRE1, which is known to interact with multiple proteins which may or may not regulate its function [35], the identity of plant IRE1-interacting proteins is still unknown. The situation is markedly different for the bZIP-TFs. In vitro AtbZIP28 can homodimerize but also heterodimerize with AtbZIP60, Atb-ZIP49 or AtbZIP17 [36]. Validation of these findings in vivo coupled with functional analyses may provide insights into the relevance of the TF interactions in the UPR. AtbZIP28 has also been found to interact with NF-Y subunits to form a transcriptional complex that upregulates the expression of ER stress-induced genes [36]. Furthermore, AtbZIP28 and AtbZIP60 interact with AtAsh2 and AtWDR5a, which are core components of the COMPASS-like complex that regulates gene expression through histone H3K4 trimethylation [13••]. These findings support that the protein-protein interactions of the UPR bZIP-TFs may have important roles in the regulation of gene expression and that a diversification of such interactions may be needed to modulate responsive outputs that are different both at quantitative and qualitative levels. One critical yet unanswered question is how the activated sensors are restored to basal levels once the ER stress is resolved. In other words, how is the plant UPR tamed once ER homeostasis is obtained?
Conclusions
In the last few years, numerous significant studies have contributed to the understanding of the UPR; this trend is likely to keep steady. Numerous fundamental questions are still open (Box 2). Understanding the mechanisms of the plant UPR is important to define conserved and unique strategies for ER stress responses, and it offers exciting perspectives to improve plant productivity. While we are beginning to understand how the UPR intersects with hormone-regulated growth [37••,38], defense [5,24,39], and resistance to hostile environments [4,40], further insights into the temporal and spatial complexity of the UPR regulation will support biotechnological applications. For example, overexpression of rice IRE1 may lead to suppression of endogenous IRE1 expression, which may compromise endogenous defense [41]; on the other hand, overexpression of BiP proteins renders them more resistant to stress [42,43]. Exploration of plant ER-stress responses in the field using multiple species and ecotypes adapted to different environmental conditions could provide further insights into the design of novel strategies for plant ER stress management outside standard laboratory conditions.
Box 2. Major unsolved questions in the plant UPR.
What is the trigger for a life-to-apoptosis switch in ER stress?
What are the triggers of ER-stress induced autophagy?
Is plant age a contributing factor to the amplitude of the UPR?
Do the UPR sensors have specific functions in different cell types?
Do the various UPR sensors respond to the levels and/or duration of the accumulation of specific unfolded proteins?
How is plant IRE1 activated and inactivated?
What terminates the activity of activated bZIP-factors once ER stress is resolved and cell homeostasis is restored?
Does plant IRE1 cluster similar to the non-plant IRE1 proteins? And how does the ER shape specifically affect the UPR response?
Are there interacting proteins on the cytosolic region of plant IRE1?
What is the role of RIDD in normal conditions of growth in plants?
Does plant IRE1 cleave specific miRNAs as occurs in mammals?
What are the downstream factors that IRE1 utilizes to control organ development?
Does AtIRE1 have non-self-phosphorylation targets?
Does unspliced bZIP60 protein have a functional role in the absence of ER stress?
Are PERK-like proteins functional in plant UPR?
Acknowledgments
We apologize to those colleagues that we have not cited due to space constrains. This work was primarily supported by National Institutes of Health (GM101038) with contributing support from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021), National Aeronautics and Science Administration (NNX12AN71G), National Science Foundation (MCB 1243792) and DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 2013;77:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C, Kotchoni SO, Samuels AL, Szymanski DB. SPIKE1 signals originate from and assemble specialized domains of the endoplasmic reticulum. Curr Biol. 2010;20:2144–2149. doi: 10.1016/j.cub.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:7247–7252. doi: 10.1073/pnas.1102117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLOS ONE. 2012;7:e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 2011;1:29. doi: 10.1038/srep00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T, Takaiwa F. Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 2012;69:946–956. doi: 10.1111/j.1365-313X.2011.04844.x. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Humbert S, Howell SH. ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res Notes. 2012;5:144. doi: 10.1186/1756-0500-5-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu SJ, Yang ZT, Sun L, Sun L, Song ZT, Liu JX. Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol Plant. 2012;5:504–514. doi: 10.1093/mp/ssr115. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci U S A. 2015;112:2900–2905. doi: 10.1073/pnas.1419703112. This work shows that the ER stress causes the assembly of the preinitiation complex as well as the histone H3K4 trimethylation on specific promoters through the interaction between ER stress-related transcription factors (i.e. bZIP60 and bZIP28) and COMPASS-like components (i.e. Ash2 and WDR5a) regulating the gene expression in conditions of ER stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam AB, Koong AC, Niwa M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014;9:850–858. doi: 10.1016/j.celrep.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci U S A. 2013;110:5713–5718. doi: 10.1073/pnas.1219047110. This work demonstrates that RIDD occurs also in plants, and a connection between the UPR and the cytosolic protein response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Chen Y, Brandizzi F. AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 2012;69:266–277. doi: 10.1111/j.1365-313X.2011.04788.x. This work highlights that AtIRE1A and AtIRE1B have overlapping functions that are essential for the plant UPR, uncovers a genetic interaction between IRE1 proteins and a G protein-signaling component (i.e. AGB1) in the UPR and shows that AtIRE1A/AtIRE1B-dependent and AGB1-dependent signaling pathways have a critical role not only in stress tolerance, but also in normal vegetative growth (i.e. root growth and specifically in the maintenance of optimal root cell elongation) [DOI] [PubMed] [Google Scholar]
- 18••.Deng Y, Srivastava R, Howell SH. Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110:19633–19638. doi: 10.1073/pnas.1314749110. This work explores the role of the protein kinase and RNAse domains of AtIRE1 in plant growth, reproduction and response to ER stress and demonstrates that AtRE1 controls male gametophyte development independently from AtbZIP60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader? Biochem Soc Trans. 2014;42:1135–1140. doi: 10.1042/BST20140142. [DOI] [PubMed] [Google Scholar]
- 21.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 2001;127:949–962. [PMC free article] [PubMed] [Google Scholar]
- 23.Noh SJ, Kwon CS, Chung WI. Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim Biophys Acta. 2002;1575:130–134. doi: 10.1016/s0167-4781(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 24.Nagashima Y, Iwata Y, Ashida M, Mishiba K, Koizumi N. Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 2014;55:1772–1778. doi: 10.1093/pcp/pcu108. [DOI] [PubMed] [Google Scholar]
- 25••.Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–4651. doi: 10.1105/tpc.112.101535. This work provides evidence in support of a strong connection between ER stress and autophagy in plants and its dependence on AtIRE1b but independence from AtbZIP60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 2013;76:274–286. doi: 10.1111/tpj.12287. This work identifies a novel ER stress cis-element (UPRE-III) in the promoter of NAC103 to which AtbZIP60 binds inducing the expression of NAC103 under ER stress. In turn, NAC103-TF is translated and modulates the expression of the UPR downstream genes. [DOI] [PubMed] [Google Scholar]
- 27.Henriquez-Valencia C, Moreno AA, Sandoval-Ibanez O, Mitina I, Blanco-Herrera F, Cifuentes-Esquivel N, Orellana A. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J Cell Biochem. 2015 doi: 10.1002/jcb.25121. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Chen H, Brandizzi F, Verchot J, Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015;11:e1005164. doi: 10.1371/journal.pgen.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell. 2013;25:1416–1429. doi: 10.1105/tpc.113.110684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 32•.Lai YS, Stefano G, Brandizzi F. ER stress signaling requires RHD3, a functionally conserved ER-shaping GTPase. J Cell Sci. 2014;127:3227–3232. doi: 10.1242/jcs.147447. This work demonstrates that the ER shaping RHD3 has a role in modulating the IRE1 arm of the plant UPR and establishes a connection between ER network integrity and AtIRE1 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefano G, Renna L, Moss T, McNew JA, Brandizzi F. In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 2012;69:957–966. doi: 10.1111/j.1365-313X.2011.04846.x. [DOI] [PubMed] [Google Scholar]
- 34.Maneta-Peyret L, Lai YS, Stefano G, Fouillen L, Brandizzi F, Moreau P. Phospholipid biosynthesis increases in RHD3-defective mutants. Plant Signal Behav. 2014;9 doi: 10.4161/psb.29657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woehlbier U, Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem Sci. 2011;36:329–337. doi: 10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37••.Chen Y, Aung K, Rolcik J, Walicki K, Friml J, Brandizzi F. Inter-regulation of the unfolded protein response and auxin signaling. Plant J. 2014;77:97–107. doi: 10.1111/tpj.12373. This work demonstrates link between the UPR and the auxin, a critical phytohormone for plant growth and physiology and shows that AtIRE1 is required for auxin homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Smith SM. Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci Signal. 2010;3:ra69. doi: 10.1126/scisignal.2001140. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi S, Wakasa Y, Takaiwa F. Functional integration between defence and IRE1-mediated ER stress response in rice. Sci Rep. 2012;2:670. doi: 10.1038/srep00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakasa Y, Hayashi S, Takaiwa F. Effect of overexpression of kinase- or RNase-deficient OsIRE1 on the endoplasmic reticulum stress response in transgenic rice plants. Plant Signal Behav. 2013;8 doi: 10.4161/psb.25343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leborgne-Castel N, Jelitto-Van Dooren EP, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell. 1999;11:459–470. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho HH, Silva PA, Mendes GC, Brustolini OJ, Pimenta MR, Gouveia BC, Valente MA, Ramos HJ, Soares-Ramos JR, Fontes EP. The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 2014;164:654–670. doi: 10.1104/pp.113.231928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao H, Brandizzi F, Benning C, Larkin RM. A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:16398–16403. doi: 10.1073/pnas.0808463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi H, Kawakatsu T, Wakasa Y, Hayashi S, Takaiwa F. A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 2012;53:144–153. doi: 10.1093/pcp/pcr157. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava R, Chen Y, Deng Y, Brandizzi F, Howell SH. Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 2012;70:1033–1042. doi: 10.1111/j.1365-313X.2012.04943.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Zhang SS, Lu SJ, Liu JX. Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Sci China Life Sci. 2015 doi: 10.1007/s11427-015-4807-6. [DOI] [PubMed] [Google Scholar]
- 49.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 50.Basu D, Le J, Zakharova T, Mallery EL, Szymanski DB. A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc Natl Acad Sci U S A. 2008;105:4044–4050. doi: 10.1073/pnas.0710294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 53.Wang S, Narendra S, Fedoroff N. Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc Natl Acad Sci U S A. 2007;104:3817–3822. doi: 10.1073/pnas.0611735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Yang ZT, Lu SJ, Wang MJ, Bi DL, Sun L, Zhou SF, Song ZT, Liu JX. A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 2014;79:1033–1043. doi: 10.1111/tpj.12604. This work shows that the plant-specific plasma membrane-tethered transcription factor NAC062 is upregulated by ER stress through AtbZIP60-TF and in response to ER stress it relocated from the plasma membrane to the nucleus, where it plays important roles in regulating UPR downstream gene expression. [DOI] [PubMed] [Google Scholar]
- 55•.Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014;10:e1004243. doi: 10.1371/journal.pgen.1004243. This work demonstrates that NAC089 has a pro-apoptotic role in the plant UPR. Thus this work contributes to the knowledge of the identity of proteins that execute death in the plant UPR. [DOI] [PMC free article] [PubMed] [Google Scholar]