Abstract
Mitochondrial electron transport drives ATP synthesis but also generates reactive oxygen species (ROS), which are both cellular signals and damaging oxidants. Superoxide production by respiratory complex III is implicated in diverse signaling events and pathologies but its role remains controversial. Using high-throughput screening we identified compounds that selectively eliminate superoxide production by complex III without altering oxidative phosphorylation; they modulate retrograde signaling including cellular responses to hypoxic and oxidative stress.
Mitochondria make ATP, but also leak electrons to produce superoxide and H2O2, reactive oxygen species (ROS) that have signaling functions and cause oxidative damage and pathology1,2. Demonstrating direct links between phenotypes and ROS remains difficult because methods for altering superoxide or H2O2 production also change energy metabolism. The mechanisms of electron leak from each of the ten or more superoxide and H2O2-producing mitochondrial sites have therefore attracted attention2 and have been shown to be thermodynamically similar but mechanistically unique. Importantly, the absolute and relative contribution from each site changes with metabolic context3.
Individual sites of ROS production are implicated in specific pathologies. Parkinson’s disease and longevity are linked to superoxide production from the flavin- and ubiquinone (Q)-binding sites of respiratory complex I (sites IF and IQ), respectively4,5; ROS from the complex II flavin (site IIF) is linked to Huntington’s disease and cancer6-8, and ROS from complexes I, II, and III, mitochondrial glycerol phosphate dehydrogenase (mGPDH) and matrix dehydrogenases are all invoked in ischemia/reperfusion injury9-12.
The outer Q-binding site of complex III (site IIIQo) is implicated in the broadest range of ROS-mediated signaling and pathologies1,13, partly because its capacity is large and it generates superoxide towards the cytosol, poising it to influence cellular events. Investigations of the cellular hypoxic response provided the first evidence of direct involvement of site IIIQo in cellular signaling1. During hypoxia, myxothiazol, which inhibits site IIIQo, decreased ROS production and blocked HIF-1α induction whereas antimycin A, which induces superoxide production from site IIIQo, increased ROS production and HIF-1α. Genetic manipulation of respiratory complexes provided further support and site IIIQo superoxide production was subsequently linked to H2O2-induced ROS production, AMPK, JNK and TGF-β signaling, K-ras- and ERK-mediated tumorigenicity, cellular differentiation, and T-cell activation1,14-17. However, these conclusions are not universally supported because other sites of ROS production and broad changes in metabolism are each implicated in mitochondrial control of these pathways18-21. Pharmacological support came from terpestacin, a fungal compound that inhibited site IIIQo ROS production and hypoxic signaling without altering basal respiration22. However, terpestacin depolarizes mitochondria22, shows evidence of uncoupling and inhibition of oxidative phosphorylation, and is not selective for site IIIQo (Online Methods and Supplementary Results, Supplementary Fig. 1). Ultimately, the ambiguity associated with pharmacological or genetic inhibition makes it problematic to assign ROS production to any particular mitochondrial site in cells.
Here, using high-throughput chemical screening and extensive validation, we introduce compounds that are selective Suppressors of site IIIQo Electron Leak (S3QELs, pronounced “sequels”) without otherwise altering energy metabolism. We identify multiple structural classes with similar effects on both superoxide production from complex III and downstream cellular signaling. By enabling experimental dissociation of energy metabolism from mitochondrial ROS production, S3QELs address a longstanding problem in redox biology and hold wide-ranging promise for studies of ROS production, cellular redox signaling and therapeutic intervention.
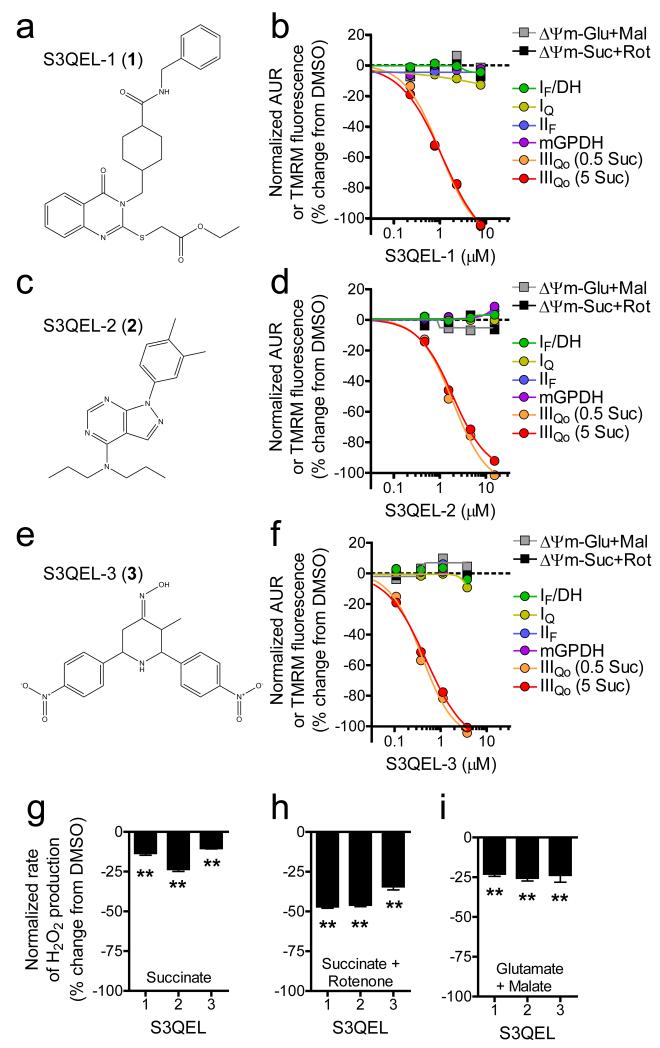
To identify S3QELs, we used an Amplex UltraRed-based detection system to screen 635,000 small molecules against H2O2 production caused by electron leak at sites IIIQo, IQ, and IIF in isolated muscle mitochondria and then rigorously eliminated compounds that were unselective for site IIIQo or inhibited energy metabolism (Supplementary Fig. 2, Online Methods, Supplementary Tables 1-2)23. S3QELs 1-3 (Fig. 1a-f) consistently met our strict criteria: they potently and selectively suppressed site IIIQo superoxide production without impairing any tested measure of bioenergetic function including mitochondrial membrane potential (ΔΨm).
Figure 1. Chemical screening using isolated mitochondria identifies suppressors of site IIIQo superoxide production.
(a – f) Structures of S3QELs 1-3 and dose-response curves against two ΔΨm and six H2O2 endpoint screening assays (n = 1). Mean IC50 values against site IIIQo superoxide production were 0.75, 1.7, and 0.35 μM for S3QELs 1-3, respectively. (g – i) Effects of S3QELs 1-3 on the steady state rate of H2O2 production measured using the Amplex UltraRed assay (normalized mean ± SE, n = 3 biological replicates). **p < 0.01 versus DMSO in each condition; one-way ANOVA with Dunnett’s posttest. Glu, glutamate; Mal, malate; Suc, succinate; Rot, rotenone; IF/DH, site IF plus NADH-linked matrix dehydrogenases.
The screen used antimycin to induce strong superoxide production from site IIIQo. To determine if S3QELs required antimycin for their action, we tested them against H2O2 production and three independent bioenergetic assays in mitochondria respiring on different substrates in the absence of antimycin. S3QELs 1-3 suppressed H2O2 production independently of either antimycin or respiratory substrate (Fig. 1g-i and Supplementary Fig. 3a). The absolute and relative contributions of site IIIQo to total H2O2 production change depending on the reduction state of the Q-pool and the activity of other sites3. Importantly, each S3QEL suppressed more strongly when the predicted contribution3 from site IIIQo was higher; S3QELs lowered overall H2O2 production by an average of 16% with succinate alone (when it is dominated by superoxide from site IQ) but by 43% if site IQ production was eliminated by rotenone (Fig. 1g,h). Similarly, S3QELs eliminated the H2O2 production attributable to site IIIQo (~25% of the total) during oxidation of glutamate plus malate (Fig. 1i). Further agreement was found using glycerol phosphate or palmitoylcarnitine as substrates (Supplementary Fig. 3b). These condition-dependent effects on H2O2 production were not caused by changes in reduction state of the Q-pool, matrix NAD(P)H redox state, or respiration with any substrate (Supplementary Fig. 3c-e). Thus, following rigorous screening and validation, we conclude that S3QELs 1-3 selectively suppress site IIIQo superoxide production by the inhibited or uninhibited complex without affecting normal electron flux.
ROS produced by site IIIQo in cells are implicated in signaling, particularly the induction of HIF-1α in hypoxia1. Therefore, we tested S3QELs during hypoxic challenge of human embryonic kidney (HEK-293) cells.
Prolonged exposure to S3QELs was not toxic even when cells relied on mitochondrial metabolism (Supplementary Fig. 2e and Supplementary Fig. 4a). HEK-293 cells were exposed to high (20x IC50) levels of S3QELs and respiration driven by pyruvate and glutamine was measured for 3 h. Basal and uncoupled respiration were unaffected (Supplementary Fig. 4b).
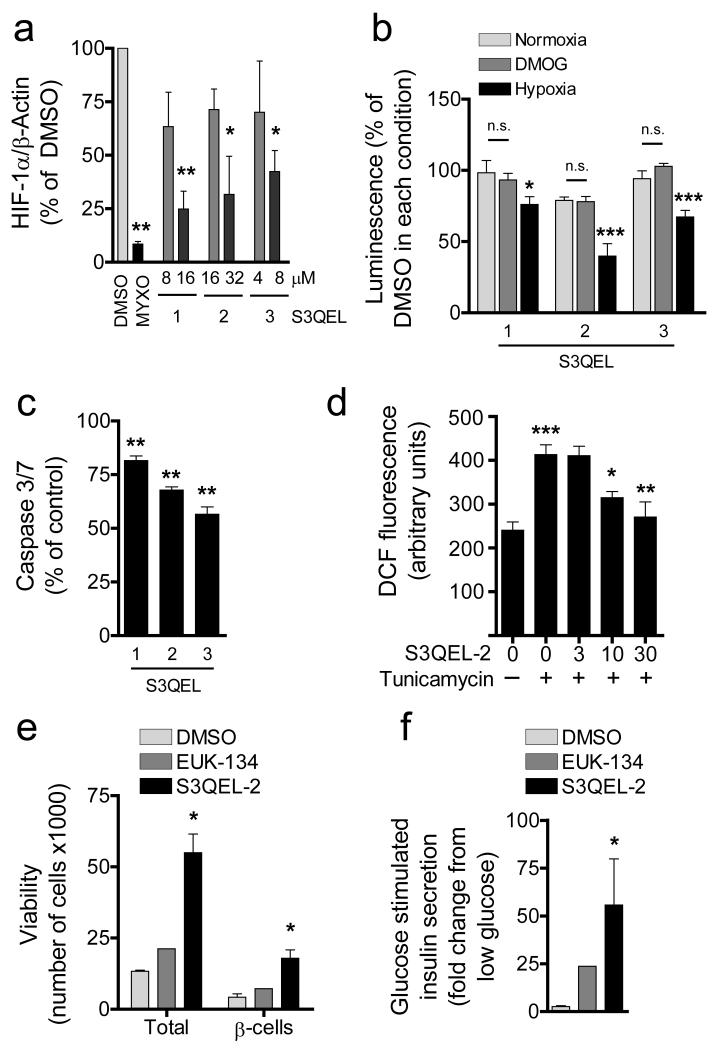
We confirmed standard regulation of HIF-1α by chemical induction at normoxia with CoCl2 and by hypoxia for 3.5 h (Supplementary Fig. 5). As reported1, the complex III inhibitor myxothiazol prevented HIF-1α stabilization (Fig. 2a). Remarkably, all three S3QELs inhibited HIF-1α accumulation (Fig. 2a) and a reporter of downstream transcriptional activity (Fig. 2b) without affecting cellular oxidative phosphorylation (Supplementary Fig. 4b) or hypoxia-independent stimulation of the HIF-1α pathway (Fig. 2b). Given the structural dissimilarity between the S3QELs, it is improbable that a common off-target mechanism explains their shared effect on HIF-1α regulation.
Figure 2. S3QELs modulate ROS-mediated signaling in cells.
(a) HIF-1α levels in HEK-293 cells treated with DMSO, S3QELs 1-3 (10 and 20x IC50) or 2 μM myxothiazol following 3.5 h hypoxic challenge (mean ± SE, n = 3 biological replicates). (b) Effect of 33 μM S3QELs on a HIF-1α-responsive luciferase reporter in HEK-293T cells following 4 h hypoxia, or normoxia ± 1 mM dimethyloxallyl glycine (DMOG), a ROS-independent stabilizer of HIF-1α. S3QELs significantly reduced HIF-1α transcription in hypoxia but not ± DMOG (mean ± SE, n = 5 or 6 biological replicates). (c) Effect of 10 μM S3QELs on tunicamycin-induced caspase 3/7 activation in INS1 insulinoma cells (mean ± SE, n = 3 or 6 biological replicates). (d) Effect of S3QEL-2 (0 – 30 μM) on total ROS in INS1 cells during tunicamycin-induced ER stress (mean ± SE, n = 4 or 6 biological replicates). (e - f) Effect of 30 μM S3QEL-2 or 30 μM EUK-134 on (e) viability of total and insulin-positive β-cells during isolation and culture of rat islets and (f) glucose-stimulated insulin secretion in islets isolated and cultured in DMSO or either compound (mean ± SE, n = 3 biological replicates for DMSO and S3QEL-2; mean for two biological replicates for EUK-134). Myxo, myxothiazol; DCF, 6-carboxy-2′,7′-dichlorofluorescein. *p < 0.05; **p < 0.01; ***p < 0.001; one-way ANOVA with Dunnett’s posttest versus DMSO (a-d) (versus tunicamycin for effects of S3QELs in c,d) or Student’s t-test with Welch’s correction versus DMSO (e,f).
To investigate the wider effects of S3QELs, we tested each for the ability to suppress toxicity in pancreatic β-cells, which are sensitive to oxidative stress due to poor antioxidant defense. In one model, INS1 β-cells were exposed to tunicamycin to induce oxidative stress through activation of an endoplasmic reticulum-mitochondrial ROS-JNK pathway24. S3QELs protected against caspase 3/7 activation (Fig. 2c) consistent with S3QEL-2 decreasing cellular ROS level (Fig. 2d). In a second model, S3QEL-2 enhanced both survival and function of primary pancreatic islets compared with equimolar amounts of the ROS scavenger, EUK-134 (Fig. 2e,f). We conclude that S3QELs 1-3 are cell-permeant, potent inhibitors of retrograde signaling mediated by superoxide from site IIIQo.
A major challenge in understanding the physiological roles of specific mitochondrial sites of ROS production has been the inability to manipulate ROS production without altering ATP synthesis and other metabolic processes. Recently, we identified CN-POBS, a compound that suppresses electron leak to O2 specifically at site IQ without affecting energy metabolism23. Here, we identify S3QELs as the first suppressors of superoxide production at site IIIQo that are selective, do not affect oxidative phosphorylation, and exert effects in cells and isolated tissues.
Conventional methods for assigning ROS production to individual sites are inherently ambiguous because they alter metabolism. For example, when myxothiazol is added to inhibit site IIIQo, upstream redox centers become more reduced, and their electron leak to oxygen increases3. S3QELs provide excellent tools to determine the contribution of site IIIQo when several sites operate simultaneously and provide the first means to eliminate IIIQo superoxide production under physiologically relevant conditions without altering underlying metabolism.
Stabilization of the semiquinone in site IIIQo depends on the reduction states of the b-cytochromes, which influence subunit interactions within complex III and modulate superoxide production13,25. Since S3QELs do not alter complex III electron transport rate, nor alter b-cytochrome reduction level, we propose that they do not change the concentration of the semiquinone but modify protein conformation to decrease the rate constant for electron transfer from the semiquinone to oxygen. Perhaps each structural group of S3QELs binds differently to the complex but affect this rate constant similarly. We can rule out direct, antioxidant-like quenching because of the low IC50 for S3QELs (the amount of superoxide suppressed exceeds the amount of added S3QEL), but not a site-specific catalytic antioxidant mechanism, although this would not easily explain the equivalent suppression seen at different rates and different reduction states of the b-cytochromes with low and high succinate concentrations.
S3QELs confirm the diverse cellular signaling effects of IIIQo superoxide production. S3QELs significantly, but incompletely, lowered HIF-1α induction. The greater effectiveness of myxothiazol likely resulted from its potent inhibition of site IIIQo combined with its inhibition of electron flow and consequent disruption of metabolism. The unique ability of S3QELs to modulate HIF-1α activation without directly impacting metabolism more clearly dissects the cellular hypoxic response.
S3QELs 1-3 protected against ROS-induced, JNK-mediated cell stress in pancreatic β-cells and S3QEL-2 strongly mitigated the oxidative stress-induced apoptosis that limits the yield of functional β-cells from intact islets. Both lines of evidence specifically indicate a role for superoxide from site IIIQo in β-cell survival and function. In conclusion, S3QELs provide exciting new ways to identify and target ROS-mediated signaling events in diverse biological systems.
ONLINE METHODS
Animal use
Studies involving isolated mitochondria utilized 8-week-old female Wistar (Harlan Laboratories) or male Sprague Dawley (Taconic Biosciences) rats. Studies involving the isolated pancreatic islets utilized 22-week-old male Sprague Dawley rats (Taconic Biosciences). All studies involving animals were conducted according to the guidelines of the relevant Institutional Animal Care and Use Committee. Studies involving tissues isolated from animals were not blinded or randomized.
Reagents and screening compounds
All reagents were from Sigma-Aldrich unless stated otherwise. The GNF Academic Screening Collection was composed from multiple sources and designed to select for optimal compound properties and eliminate undesirable functional groups (Supplementary Table 2). S3QELs were obtained from commercial sources and purity of powder stocks was confirmed by HPLC MS (Supplementary Note). S3QELs were obtained from Chemdiv unless specified otherwise: S3QEL-1 (catalog ID K284-4710), S3QEL-1.1 (K284-4711), S3QEL-1.2 (K284-4767), S3QEL-1.3 (K284-4794), S3QEL-2 (K405-3102), S3QEL-2.1 (Life Chemicals, F1886-0120), S3QEL-2.2 (Life Chemicals, F1886-0426), S3QEL-2.3 (K405-3096), S3QEL-2.4 (K405-3741), S3QEL-2.5 (K402-1025), S3QEL-2.6 (K402-0937), S3QEL-2.7 (K402-0893), S3QEL-2.8 (K402-0508), S3QEL-3 (Maybridge, JFD03367), S3QEL-4 (3377-0061), S3QEL-5 (3389-0595), S3QEL-6 (3786-1206), S3QEL-7 (8010-6022). Terpestacin was sourced from the internal Novartis compound library.
Overview of screening strategy
We adapted our recent chemical screen23 to 1536-well format and screened 635,000 small molecules at 10 μM against assays of superoxide and/or H2O2 production by sites IIIQo, IQ, and IIF in isolated rat skeletal muscle mitochondria (Supplementary Fig. 2a and Step 1 in Supplementary Table 1). Superoxide production from site IIIQo was defined as H2O2 production (after dismutation by endogenous or exogenous superoxide dismutases) in the presence of succinate plus antimycin A (to keep the ubiquinol-oxidizing site of complex III partly or fully reduced). H2O2 production was measured using the Amplex UltraRed assay (Life Technologies), in which horseradish peroxidase uses H2O2 to oxidize non-fluorescent Amplex UltraRed to a highly fluorescent resorufin product. Myxothiazol was included on each plate as a “positive” inhibitor of site IIIQo superoxide production and to monitor inter-plate and inter-day consistency of the assay. Over 8,000 compounds suppressed the site IIIQo signal by >40% and 6,674 of these were selective (Supplementary Fig. 2b). To validate their selectivity and eliminate any that altered energetics, we performed a series of secondary confirmation and stringency tests (Supplementary Table 1). First, hits were confirmed in triplicate for activity in the IIIQo, IQ, and IIF assays (Step 2 in Supplementary Table 1). In addition the hit compounds were tested for effects on ΔΨm and amount of cellular ATP to eliminate compounds that grossly altered mitochondrial energy metabolism or affected cell growth (Supplementary Fig. 2c-e and Steps 3 and 4 in Supplementary Table 1). We determined the IC50 values of the remaining 995 compounds against site IIIQo by generating dose-response profiles (Step 5 in Supplementary Table 1) and rescreened all with IC50 <3.2 μM (103 compounds) at 10x IC50 against a broader panel of two ΔΨm and six H2O2 production assays (Step 6 in Supplementary Table 1). Almost all compounds strongly suppressed site IIIQo superoxide production driven by either high or low succinate concentration (Supplementary Fig. 1f). However, many had off-target effects in this expanded set of assays; for example, several compounds influenced ΔΨm with NADH-linked substrates and/or ROS production from site IF plus matrix dehydrogenases. Notably, the compounds that remained selective were from numerous structural groups (Fig. 1 and Supplementary Figs. 6, 7 and 8). Because a major criterion was lack of interference with energy metabolism, we measured ADP-stimulated respiration as a more stringent test of ATP production (Step 7 in Supplementary Table 1). Of the remaining 71 hits, 63 had no unwanted effect (Supplementary Table 1). New stocks were procured for 59 of these and dose-response titrations were repeated against the expanded panel of ΔΨm and H2O2 production assays (Step 8 in Supplementary Table 1). Twenty compounds retained selectivity for site IIIQo without altering any other assay more than ± 20% (Supplementary Table 1). Of these, four were unavailable in sufficient quantity for further study while nine had markedly different potencies on retesting. Ultimately, seven compounds (S3QELs 1-7) representing different structural groups were found to have IC50 similar to the original stocks while retaining selectivity for site IIIQo over a broad concentration range (Fig. 1 and Supplementary Fig. 8). Four were subsequently eliminated due to potential off-target effects as described below. Additionally, structural analogs of S3QEL-1 and S3QEL-2 were identified and demonstrated to have similar activity profiles to the parent compounds (Supplementary Figs. 6 and 7).
Ultra high-throughput primary screen: sites IIIQo, IQ and IIF
The rationale and general design of the primary assays for production of H2O2 from sites IIIQo, IQ, and IIF (Step 1 in Supplementary Table 1) are described in detail elsewhere23. To enable screening of a 635,000 compound collection the protocols were modified. Briefly, freshly isolated rat muscle mitochondria were assayed in KHEB medium containing 120 mM KCl, 5 mM HEPES, 1 mM EGTA, and 0.3% (w/v) bovine serum albumin. We used individual media to assay H2O2 production from the three separate sites. Each medium contained Amplex UltraRed (50 μM), superoxide dismutase (5 U • ml−1), and horseradish peroxidase (1 U • ml−1) as well as one of the following components to drive H2O2 production predominantly from a single mitochondrial site (μM final concentrations): site IQ assay, 5000 succinate alone; site IIIQo assay, 5000 succinate with 2.5 antimycin A and 4 rotenone; site IIF assay, 1000 succinate with 2.5 antimycin A, 4 rotenone and 2 myxothiazol. Each assay was run on a different 1536-well microplate. To each well, 4 μl of assay medium was added followed by 50 nl of 1 mM test compound in DMSO. Assays were initiated by the addition of 1 μl freshly isolated rat skeletal muscle mitochondria (final concentration 0.2 mg protein • ml−1). The final screening concentration of the test compounds was 10 μM. Plates were incubated for 15 min at room temperature before fluorescence of the resorufin product of Amplex UltraRed oxidation was read on a BMG Labtech PHERAstar Plus microplate reader (λex = 540 nm, λem = 590 nm). Fluorescence intensity was normalized to the intra-plate median signal observed for each assay plate.
Hits in the IIIQo assay were first selected based on >40% inhibition (Supplementary Fig. 2c). Compounds that also had <50% effect in the IQ and <45% inhibition in the IIF assay were chosen as selective suppressors of superoxide production by site IIIQo and tested further.
High-throughput secondary screens: ΔΨm and cell viability
Hit compounds identified by the primary screen were then tested in confirmation and counterscreen assays. Compounds were tested for their ability to inhibit IIIQo, IQ and IIF assays at 10 μM as described for the primary screen, but with triplicate determinations of activity (Step 2 in Supplementary Table 1). Compounds that displayed a median inhibition of >35% in the IIIQo assay, but <30% inhibition in signal (approximately 3 standard deviations) from sites IQ or IIF were chosen for further testing.
Compounds were also tested for effects on ΔΨm (Step 3 in Supplementary Table 1). The assay for ΔΨm was essentially as previously described23 but adapted to high-throughput screening. 4 μl of KHEB medium containing 2.5 μM of the potentiometric dye tetramethylrhodamine methyl ester (TMRM, Life Technologies), 5 mM glutamate and 5 mM malate was added to each well of a 1536 microtiter plate. Glutamate and malate are metabolized by matrix enzymes to generate NADH, which is oxidized by the respiratory chain leading to production of ΔΨm. As above, 50 nl of 1 mM test compound in DMSO was added to the medium and the assay was initiated by addition of 1 μl freshly isolated rat skeletal muscle mitochondria (0.2 mg protein • ml−1). The final screening concentration of the test compounds was 10 μM. The plates were incubated for 10 min at room temperature then resorufin fluorescence was read on a BMG LabTech PHERAstar Plus microplate reader (λex = 540 nm, λem = 590 nm). Fluorescence intensity was normalized to the intra-plate median signal observed for each assay plate. Compounds that altered the fluorescent signal beyond ± 30% (approximately 3 standard deviations, Supplementary Fig. 2d) were removed from further testing.
The effect of compounds (10 μM) on viability and growth of HEK-293T cells (ATCC) cultured in glucose-free medium (DMEM with 10% v/v fetal bovine serum) containing 2 mM pyruvate with 2 mM glutamine and 20 mM galactose was assessed after 72 h exposure (GalacTox, Step 4 in Supplementary Table 1) by measuring total ATP with standard procedures (Cell Titer Glo, Promega). Replacing glucose with pyruvate, glutamine and galactose enforces reliance on mitochondrial over glycolytic ATP production and is particularly useful for uncovering mitochondrial toxicities of candidate drugs26. The average effect of compounds was normalized to the intraplate median signal. Compounds that lowered viability >20% were removed from further testing (Supplementary Fig. 2e). HEK-293T cells were from ATCC and regularly tested for mycoplasma contamination.
Initial dose-response screens: sites IIIQo and IQ
Using the protocol described above for the ultra high-throughput primary screen, the remaining compounds were tested in a dose response for inhibition of H2O2 production by sites IIIQo and IQ (Step 5 in Supplementary Table 1). Each compound was tested in duplicate at eight doses between 5 nM and 10 μM. Endpoint fluorescence values were normalized to intraplate DMSO wells.
Expanded rescreens: five sites of H2O2 production and two assays of ΔΨm
An expanded rescreen at 10x IC50 (determined versus site IIIQo superoxide production), and a dose-response expanded rescreen with newly-sourced compounds (Steps 6 and 8 in Supplementary Table 1; Supplementary Fig. 2f) were performed as described previously23. Compound concentrations and criteria for selectivity for these steps are outlined in Supplementary Table 1.
The expanded rescreens were in 96-well format in a total volume of 100 μL and included two assays for ΔΨm driven by 5 mM glutamate plus 5 mM malate or 5 mM succinate plus 4 μM rotenone. These different substrates are metabolized by different enzymes and their reducing equivalents enter the electron transport chain primarily via complex I (glutamate plus malate) or complex II (succinate). Therefore, the two ΔΨm assays identify inhibitors of most complexes and several upstream enzymes or transporters, and all uncouplers and other membrane-permeabilizing agents. Additionally, six site-specific assays for H2O2 production included the following additions: site IF plus matrix dehydrogenases, 5 mM glutamate plus 5 mM malate and 4 μM rotenone; site IQ, 5 mM succinate; site IIIQo, 0.5 mM or 5 mM succinate plus 4 μM rotenone and 2.5 μM antimycin A; site IIF, 0.1 mM succinate plus 4 μM rotenone, 2.5 μM antimycin A, and 2 μM myxothiazol; site mGPDH, 25 mM glycerol phosphate plus 4 μM rotenone, 1 mM malonate, 2.5 μM antimycin A, and 2 μM myxothiazol. Endpoint fluorescence readings after 30 min were normalized to vehicle (DMSO) and known inhibitor controls as described23 and expressed as % change from DMSO. Inhibitor controls for sites IF/matrix DH, IQ, IIIQo and IIF, were respectively as follows: 20 mM aspartate, 1 μM FCCP, 2 μM myxothiazol, 10 mM malonate. At the time of the screen, selective inhibitors of mGPDH were not known27. Therefore, the FCCP-inhibited signal in the site IQ assay was used as background normalization factor for the mGPDH assay23. The relationship between site IIIQo superoxide production and the reduction state of the Q-pool is bell-shaped in the presence of antimycin (i.e. superoxide production is highest when the Q-pool is only partially reduced by substrate)25. Two assays differing only in the concentration of succinate were used to evaluate effects on site IIIQo superoxide production. Targeting this site with either subsaturating (0.5 mM) or saturating (5 mM) succinate helped eliminate compounds that subtly influenced succinate oxidation or otherwise partially oxidized the Q-pool. Such unwanted compounds were found to decrease superoxide production from site IIIQo with subsaturating succinate (peak superoxide production) but increase it at saturating succinate (moderate superoxide production). More potent non-selective inhibitors presumably were eliminated in other counterscreens that relied on robust succinate oxidation or Q-pool reduction (e.g. H2O2 production related to site IQ, or ΔΨm). Terpestacin was tested against four assays of H2O2 production at concentrations at which it was previously shown to be active against mitochondrial ROS production22. Terpestacin showed no sign of inhibition of H2O2 production related to site IIIQo and was not selective.
Steady-state measurements of rates of H2O2 production and levels of NAD(P)H and cytochrome b566 reduction in respiring mitochondria
The effect of S3QELs on rates of H2O2 production and the reduction states of matrix NAD(P)H and cytochrome b566 (Fig. 1g-i and Supplementary Fig. 3a-d) were determined at 37°C in KHEB medium as previously described2,3 with minor modifications. Measurements of rates of H2O2 production using Amplex UltraRed oxidation to resorufin (λex = 540 nm, λem = 590 nm) and measurements of endogenous NAD(P)H (λex = 340 nm, λem = 460 nm) were performed on a BMG LabTech PHERAstar Plus microplate reader. All components except skeletal muscle mitochondria (0.2 mg protein • ml−1) were pre-warmed. Briefly, endogenous substrates were depleted during a 5 min baseline prior to the addition of compounds. Exogenous substrates were then added to induce H2O2 production and reduction of matrix NAD(P)H and steady-state rates or reduction levels were measured over the following 2 – 8 min. At the end of the NAD(P)H measurements, 5 mM glutamate plus 5 mM malate and 4 μM rotenone were added to all wells to obtain internal calibrations of 100% reduction level. Rates of H2O2 production were normalized to intraplate vehicle control wells (DMSO) and the effects of S3QELs were expressed as % change from DMSO. Cytochrome b566 reduction level was measured on an Olis DW-2 dual wavelength spectrophotometer as previously described2,3 except 0% reduction was taken after a total of 10 min depletion of endogenous substrates and 5 min after the addition of compound. Sequential additions of exogenous substrate and antimycin A (100% reduction) were used to determine relative effects of S3QELs on reduction state of the Q-pool during substrate oxidation.
Mitochondrial respiration
Mitochondrial respiration (Supplementary Fig. 1b; Supplementary Fig. 3e-g; Step 7 in Supplementary Table 1) was measured as described previously on a Seahorse XF24 (or, for terpestacin, XF96) instrument27. Briefly, mitochondria were assayed in a mannitol- and sucrose-based medium (Seahorse MAS buffer28) containing 0.3% (w/v) bovine serum albumin. Baseline measurements of substrate-only (state 2) respiration were made in the absence and presence of compound, followed by injection of 5 mM ADP (phosphorylating state 3) and then 1 μg • ml−1 oligomycin (non-phosphorylating state 4o). The following substrates were used: 5 mM glutamate plus 5 mM malate; 5 mM succinate plus 4 μM rotenone; and 27 mM glycerol phosphate plus 4 μM rotenone23. In addition, 15 – 60 μM palmitoylcarnitine plus 1 mM malate was tested to verify that S3QELs did not inhibit fatty acid oxidation. In this condition, the rate of respiration is strongly dependent upon palmitoylcarnitine concentration (Supplementary Fig. 3f). Because high levels of palmitoylcarnitine can uncouple mitochondria, extra additions of 15 μM palmitoylcarnitine were made with each port injection. Pilot tests indicated that these cumulative amounts of palmitoylcarnitine resulted in the highest state 3 rates and state 3/state 4o coupling ratios. Wells were loaded with 0.5 – 6 μg rat muscle mitochondrial protein depending on the condition tested. Each substrate condition was tested on different plates and each S3QEL was tested at least three times against each substrate. S3QELs were injected to a final concentration up to 10x IC50 against site IIIQo superoxide production. Myxothiazol was tested at 5x IC50 against site IIIQo superoxide production (0.08 μM), to demonstrate the potent effect of this inhibitor on mitochondrial respiration (Supplementary Fig. 3g). Terpestacin at 10 – 30 μM showed evidence of uncoupling and inhibition of oxidative phosphorylation and was not tested further (Supplementary Fig. 1b). Criteria for selectivity for the preliminary screening are outlined in Supplementary Table 1. In follow-up screening with freshly-acquired S3QELs, selectivity for inhibition of H2O2 production and not electron flux was evaluated statistically by one-way ANOVA with post-hoc analysis. S3QEL-5 showed a small but significant inhibition of state 4o respiration with succinate oxidation and was not tested further.
Cellular respiration
Basal and uncoupled respiration rates of HEK-293 cells were measured in the presence of either 25 mM glucose or 4 mM pyruvate plus 2 mM glutamine and 20 mM galactose. 15,000 cells were seeded in each well of Seahorse XF24 assay plates and grown for 24 h in DMEM with 25 mM glucose, 10% (v/v) fetal bovine serum, 1 U • ml−1 penicillin, 100 μg • ml−1 streptomycin, and 2 mM glutamine. Subsequently, medium was changed to include 20 mM galactose plus 4 mM sodium pyruvate in place of glucose as this was found to maximize mitochondrial respiration compared to cells maintained continually in glucose (Supplementary Fig. 4a). After a further 24 h, cells were rinsed in prewarmed assay medium (120 mM NaCl, 7 mM KCl, 0.8 mM KH2PO4, 10 mM NaHCO3, 2.4 mM Na2SO4, 20 mM galactose, 4 mM sodium pyruvate, 2 mM glutamine, 1.8 mM CaCl2, 0.8 mM MgCl2, 0.3% (w/v) bovine serum albumin, 40 mM TES pH 7.4) and equilibrated for ~30 min at 37°C prior to measurement of respiration (Supplementary Fig. 4b). S3QELs or vehicle control (DMSO) were injected via port A to a final concentration up to 20x IC50 against site IIIQo superoxide production and repeated measurements were taken every 30 – 60 min for 3 h after compound addition. Subsequently, uncoupled respiration was driven by the addition 10 μM FCCP with 2 μg • ml−1 oligomycin. To control for well-to-well variation in cell density, all rates were normalized to the initial basal rate prior to the addition of compound. S3QEL-4 and S3QEL-6 progressively inhibited basal respiration probably due to cell detachment. The pre-post uncoupler transition was proportionally similar to other S3QELs, indicating that the cells that remained attached were still metabolically active. Regardless, these two S3QELs were not tested further.
Hypoxia-induced HIF-1α activation
The effect of S3QELs on HIF-1α accumulation during hypoxic challenge was tested in HEK-293 cells exposed for 3.5 h to 1% (v/v) O2 in an incubator equipped with control of N2, CO2 and O2. Cells were seeded at 15,000 cells per well and incubated for 48 h in standard culture medium prior to replacement with medium pre-equilibrated overnight at either 1% or atmospheric O2. S3QELs (at 10x and 20x IC50 against site IIIQo superoxide production), 100 μM CoCl2, or 2 μM myxothiazol were pre-equilibrated for 1 h in the same media and then added at the same time as the media switch. Cells were returned to incubators held at 1% or atmospheric O2 for 3.5 h. After hypoxic challenge, cells were rapidly rinsed once with ice-cold 1x PBS before addition of 2x Laemmli Lysis Buffer (BioRad), then the plate was sealed with adhesive foil, and frozen at −80°C. Frozen samples were thawed and collected on ice and then pulse-sonicated and boiled for 5 min prior to blotting 2 μl (~1/8th total) for levels of HIF-1α (R & D Systems, MAB1536, 1:500) and β-actin (Sigma-Aldrich, A3853, 1:5000) (Fig. 2a and Supplementary Fig. 5). S3QELs were compared to vehicle control wells (DMSO). Statistical significance was determined by one-way ANOVA with posthoc analysis. S3QEL-7 did not alter HIF-1α induction at 20x IC50 against site IIIQo superoxide production, presumably because it was not sufficiently cell permeant at this concentration, and was not tested further.
The effect of S3QELs on HIF-1α-dependent transcription was evaluated in HEK-293T cells with a NanoLuc luciferase-reporter construct (Promega) downstream of a minimal hypoxia response element (HRE) containing a tandem 4x repeat of the sequence CTGCACGTA that results in the promoter being dependent on HIF-1α accumulation in the nucleus for expression29. Cells were bulk transfected with Fugene transfection reagent (Promega) in Optimem serum-free medium and seeded at 16000 cells per well of a 384 well plate in 25 μl DMEM, 10% (v/v) fetal bovine serum (FBS, Omega Scientific) and 1% Pen-Strep/Fungizone and cultured for 24 h. Cells were subsequently treated for 1 h with 33 μM S3QELs followed by 4 h incubation in 1% O2 (Oxoid AnearoGen Compact System, Thermo Fisher Scientific). Luminescence due to HRE-driven luciferase expression was measured on a Perkin Elmer Envision, following addition 12.5 μl Nano-Glo reagent (Promega) and incubation at room temperature for 5 min (Fig. 2b). To determine if effects on transcription were hypoxia-dependent, parallel assays were performed under normoxia (20% O2) in the presence or absence of 1 mM dimethyloxallyl glycine (DMOG). DMOG chemically increases HIF-1α levels in a ROS- and hypoxia-independent manner by inhibiting proteins containing prolyl hydroxylase domains. The relative effect of S3QELs was normalized to the relative effect of DMSO under each condition. Hypoxia or DMOG each increased luciferase ~ 2.5 fold versus normoxia alone. Note that S3QEL-2 had a small (likely direct) effect in normoxia ± DMOG, but its effect was much greater in hypoxia. Statistical significance was determined by one-way ANOVA with posthoc analysis.
Endoplasmic reticular (ER) stress protection assay in pancreatic β-cells
To assess the ability of S3QELs to protect cells under ROS-induced cell stress, we induced ER stress in pancreatic INS1 β-cells using tunicamycin. ER stress is linked to elevated mitochondrial ROS production through the action of JNK14 and JNK activation has been linked to complex III superoxide production1,30,31. INS1 cells were plated at 8000 cells per well in a 384 well plate and incubated for 24 h in DMEM with 10% (v/v) FBS prior to the addition of 1 μg • ml−1 tunicamycin to induce ER stress and either DMSO or 10 μM S3QELs. After a further 24 h incubation, caspase 3/7 activation was measured on a luminometer to quantify relative levels of apoptotic cell death (Caspase Glo 3/7, Promega) (Fig. 2c). Caspase 3/7 levels in the presence of tunicamycin plus S3QELs were normalized to tunicamycin alone.
Cellular ROS production
Total cellular ROS levels were monitored using 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate, di(acetoxymethyl ester) (carboxy-H2DCFDA-AM, Life Technologies). INS1 cells were seeded in 5 μl of growth medium at a density of 2250 cells per well in 1536-well assay plates and incubated for 24 h. Cells were stained for DNA and ROS by incubating with 8.1 μM Hoechst 33342 (Life Technologies) and 5 μM carboxy-H2DCFDA-AM for 30 min. Cells were gently washed three times and further incubated for 6 h with 1 μg • ml−1 tunicamycin and 0, 3, 10 or 30 μM S3QEL-2 (all with a final concentration of 0.5% v/v DMSO). Parallel incubations in the absence of tunicamycin were used as controls. Cells were imaged on an ImageXpress Micro (Molecular Devices) using standard DAPI and FITC filter sets. Images were analyzed using a custom MetaXpress analysis to measure the average cellular fluorescence signal from oxidized 6-carboxy-2′,7′-dichlorofluorescein (DCF) after subtraction of the average background signal per well (Fig. 2d). Average cellular DCF fluorescence with S3QEL-2 and/or tunicamycin-treatment was compared to untreated cells by one-way ANOVA.
Intact islet protection assay
The ability of S3QEL-2 to protect pancreatic β-cells during isolation and culture was determined by measuring viable cell yield and function in cells 48 h post isolation.
Islets were isolated from 22 week old Sprague Dawley male rats intraductally perfused with 15 ml Hank’s Balanced Salt Solution (HBSS) containing 1 mg/ml collagenase P solution (Roche) supplemented with 30 μM EUK-134, 30 μM S3QEL-2 or DMSO vehicle control. Pancreata were further dissected and digested for 15 min at 37°C. Digested pancreata were then filtered and washed in HBSS with 10% (v/v) FBS before resuspending in histopaque. HBSS was layered and islets isolated by density gradient centrifugation (1500 rpm × 4 min at 4°C). Islets were collected from the mid-tube interface and washed in HBSS (1600 rpm × 3 min at 4°C) then allowed to gravity settle twice in HBSS with 10% (v/v) FBS, and then finally in RPMI-1640 medium supplemented with 2.05 mM L-glutamine, 5.5 mM glucose, 10% (v/v) FBS, 2% Penicillin-streptomycin and 55 μM β-mercaptoethanol to give complete RPMI medium (c.RPMI) for supplementation with 30 μM EUK-134, 30 μM S3QEL-2 or DMSO for routine culture at 37°C. During one of nine independent islet preparations, there was a technical problem causing incomplete perfusion from one of the preparations performed in the presence of EUK-134. This preparation was subsequently found to have abnormally low yield (<10% of the total cellular yield of the other two islet preparations perfused with EUK-134) and was excluded from further analysis.
Viability of insulin expressing (β-cells) and non-expressing cells was determined by flow cytometry. Whole islets were cultured in a 6-well plate for 48 h at 37°C with 5% CO2 in c.RPMI in the presence of DMSO or 30 μM EUK-134 or 30 μM S3QEL-2 to test for viability. Islets were then dissociated by incubation with 0.05% (w/v) trypsin and EDTA (Gibco) for 5 min at 4°C. The cells were then resuspended in c.RPMI in the presence of compound or DMSO vehicle. Cells were stained using LIVE/DEAD® Fixable Blue Dead Cell Stain for UV excitation (Life Technologies), followed by fixation and permeabilization (Cytofix/Cytoperm, BD Biosciences), staining with anti-insulin-APC conjugated antibody (R&D) and purified rabbit anti-active caspase-3 (BD) or their respective isotype controls (Rat IgG2a APC, R&D; polyclonal rabbit IgG, R&D), and then labeling with goat anti-rabbit IgG PE secondary antibody. Flow cytometry was performed on an AutoLSR II (BD Bioscience) (Fig. 2e). Cells were gated according to their forward and side scatter signal to identify single cells and then scored for viability and insulin staining.
For GSIS determination, islets were selected based on similar size and level of border integrity from each rat at the time of isolation and picked into cRPMI medium in a 96 well tissue-culture treated plate at a density of 3 islets per well. After 48 h of culture in the presence of DMSO or either compound, the medium was refreshed to a volume of 100 μl and the islets were incubated for 1.5 h at 37°C. 20 μl of medium was removed to determine the baseline level of insulin in the medium. The islets were then incubated for a further 1 h before a second 20 μl sample was taken and 40 μl of RPMI medium containing 80 mM glucose was added to give a final glucose concentration of 35 mM. After 1 h incubation, 20 μl of medium was removed. Insulin concentration in the medium samples was assayed using a HTRF insulin assay kit (Cisbio) and the relative rates of insulin secretion in low and high glucose calculated (Fig. 2f).
Statistical analysis
Two-tailed Student’s t-test or one-way ANOVA with Dunnett’s posthoc analysis were used for statistical analysis where indicated. The D’Agostino and Pearson omnibus normality test was used to verify that data were normally distributed. For comparisons of two groups with unequal variances, Welch’s correction was used. In multiple comparisons with sufficient sample sizes, Bartlett’s test was used to test for equal variance. Variance could not be determined for comparisons of small sizes. Results of pilot studies and previously completed experiments were used to determine sample sizes.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants R01 AG033542 (MDB) and TL1 AG032116 (ALO), and The Ellison Medical Foundation grant AG-SS-2288-09 (MDB).
Footnotes
Author Contributions: ALO, EKA and MDB devised the project, designed and interpreted the screen and wrote the paper. ALO, LV, CNT, JEB, JTM, VJD, SJA, JL, RLSG, IVP and SLM carried out the experiments. MP advised on compound selection. All authors edited and approved the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
REFERENCES
- 1.Sena LA, Chandel NS. Mol. Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinlan CL, Perevoschikova IV, Goncalves RLS, Hey-Mogensen M, Brand MD. Meth. Enzymol. 2013;526:189–217. doi: 10.1016/B978-0-12-405883-5.00012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, Orr AL, Brand MD. Redox Biol. 2013;1:304–312. doi: 10.1016/j.redox.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherer TB, et al. J. Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert AJ, Buckingham JA, Boysen HM, Brand MD. Aging Cell. 2010;9:78–91. doi: 10.1111/j.1474-9726.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin MT, Beal MF. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 7.Ishii T, et al. Cancer Res. 2005;65:203–209. [PubMed] [Google Scholar]
- 8.Quinlan CL, et al. J. Biol. Chem. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heather LC, et al. Cardiovasc. Res. 2010;85:127–136. doi: 10.1093/cvr/cvp276. [DOI] [PubMed] [Google Scholar]
- 10.Adam-Vizi V, Tretter L. Neurochem. Int. 2013;62:757–763. doi: 10.1016/j.neuint.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Chouchani ET, et al. Nat. Med. 2013;19:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dröse S. Biochim. Biophys. Acta. 2013;1827:578–587. doi: 10.1016/j.bbabio.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Bleier L, Dröse S. Biochim. Biophys. Acta. 2013;1827:1320–1331. doi: 10.1016/j.bbabio.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Sena LA, et al. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M, et al. J. Biol. Chem. 2013;288:770–777. doi: 10.1074/jbc.M112.431973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg F, et al. Proc. Natl Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viola HM, Hool LC. J. Mol. Cell. Cardiol. 2010;49:875–885. doi: 10.1016/j.yjmcc.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Kaminski MM, et al. Cell Rep. 2012;2:1300–1315. doi: 10.1016/j.celrep.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Srinivas V, et al. J. Biol. Chem. 2001;276:21995–21998. doi: 10.1074/jbc.C100177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua YL, et al. J. Biol. Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S-J, Hwang AB, Kenyon C. Curr. Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung HJ, et al. J. Biol. Chem. 2010;285:11584–11595. doi: 10.1074/jbc.M109.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr AL, et al. Free Rad. Biol. Med. 2013;65:1047–1059. doi: 10.1016/j.freeradbiomed.2013.08.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Win S, Than TA, Fernandez-Checa JC, Kaplowitz N. Cell Death Dis. 2014;5:e989. doi: 10.1038/cddis.2013.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan CL, Gerencser AA, Treberg JR, Brand MD. J. Biol. Chem. 2011;286:31361–72. doi: 10.1074/jbc.M111.267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 26.Swiss R, Niles A, Cali JJ, Nadanaciva S, Will Y. Toxicol. in Vitro. 2013;27:1789–1797. doi: 10.1016/j.tiv.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Orr AL, et al. PLoS ONE. 2014;9:e89938. doi: 10.1371/journal.pone.0089938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers GW, et al. PLoS ONE. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaluz S, Kaluzová M, Stanbridge EJ. Biochem. Biophys. Res. Commun. 2008;370:613–8. doi: 10.1016/j.bbrc.2008.03.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soberanes S, et al. J. Biol. Chem. 2009;284:2176–2186. doi: 10.1074/jbc.M808844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers JW, LoGrasso PV. J. Biol. Chem. 2011;286:16052–16062. doi: 10.1074/jbc.M111.223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.