Abstract
The nuclear receptor corepressor 1 (NCoR1) is a transcriptional co-regulator that has wide-ranging effects on gene expression patterns. In the liver, NCoR1 represses lipid synthesis in the fasting state, whereas it inhibits the activation of PPARα upon feeding, thereby blunting ketogenesis. Here, we show that insulin via the activation of PKB/Akt induces the phosphorylation of NCoR1 on serine 1460, which selectively favors its interaction with PPARα and ERRα over LXRα. Phosphorylation of NCoR1 on S1460 selectively derepresses LXRα target genes, resulting in increased lipogenesis, while at the same time it inhibits PPARα and ERRα targets, thereby attenuating oxidative metabolism in the liver. The phosphorylation-gated differential recruitment of NCoR1 to different nuclear receptors explains the apparent paradox that liver-specific deletion of NCoR1 concurrently induces both lipogenesis and oxidative metabolism, due to a global derepression of LXRα, PPARα and ERRα activity. This phosphorylation-mediated recruitment switch of NCoR1 between nuclear receptor subsets hence provides a mechanism by which corepressors can selectively modulate liver energy metabolism during the fasting-feeding transition.
Keywords: Lipogenesis, Ketogenesis, Oxidative Phosphorylation, Insulin Resistance, Fatty Liver
INTRODUCTION
The nuclear receptor corepressor 1 (NCoR1) and the related corepressor, silencing mediator for retinoid or thyroid-hormone receptors (SMRT, aka NCoR2), serve as scaffolds that facilitate the interaction of several docking proteins to fine-tune transcription rates (1–3). The interaction between NCoR1 with nuclear receptors and histone deacetylases has been particularly well studied and shown to be vital for nuclear receptor-mediated down-regulation of gene expression (4–8). Consistent with this paradigm, mice with impaired HDAC3 activity, either because they lack the functional deacetylase-activating domains (DADs) in both NCoR1 and SMRT (NS-DADm) or because they carry a liver-specific mutation in HDAC3 (HDAC3hep−/− mice), suffer from hepatic steatosis as a consequence of the induction of lipogenic genes (9, 10). Collectively, these data suggest that HDAC3 and NCoR1 repress the expression of lipogenic genes.
Paradoxically, NCoR1 was also reported to be critical for the inhibition of PPARα and its downstream targets that control hepatic fatty oxidation and ketogenesis (11–13). Concordant with this suppressive role, NCoR1 also suppresses oxidative metabolism in the muscle (14, 15). Based on these at first sight contradictory observations, we postulated that NCoR1 might be able to select its repressor targets in a context-dependent manner to orchestrate liver energy metabolism during the feeding-fasting transition.
Insulin is a central regulator of carbohydrate and fat metabolism (16). Binding of insulin to its receptor results in the activation of a variety of downstream signaling pathways, such as those involving phosphoinositide 3-kinase (PI3K)/Akt and mechanistic target of rapamycin (mTOR) signaling, which mediate its pleiotropic metabolic effects on glucose uptake, hepatic glucose production, and hepatic lipid accumulation (16–18). Interestingly, previous work suggested that the repressive activity of NCoR1 is enhanced by high glucose levels, insulin and mTORC1 activation, all signals induced by feeding, which are tightly associated with Akt phosphorylation (13, 15). We therefore hypothesized that the insulin/Akt pathway differentially modulates the co-repressor activity of NCoR1 at lipogenic and oxidative phosphorylation (OxPhos)/ketogenic genes.
EXPERIMENTAL PROCEDURES
Generation of NCoR1 liver specific knockout mice
NCoR1 floxed (NCoR1L2/L2) mice were generated by a classical gene targeting strategy as described previously (15). Briefly, offspring that transmitted the mutated allele, in which the selection marker was excised, and that lost the Flp transgene (NCoR1L2/WT mice) were selected, mated with mouse albumin (Alb)-Cre mice (AlbcreTg/0), and then further intercrossed to generate mutant AlbcreTg/0/NCoR1L2/L2 mice, which were termed NCoR1hep−/− mice. Only congenic C57BL/6J NCoR1hep−/− mice were used in experiments (backcrossed for over 10 generations). Phenotyping experiments were performed using validated Eumorphia / EMPReSS standard operating protocols (www.eumorphia.org) (19, 20). Animal experiments were approved by the ethics committee of the canton of Vaud, Switzerland with the Permit ID #2474. Detailed description for animal procedures and biochemical measurements are available in Supporting Experimental Procedures (15).
In vitro Akt1 kinase assays
To generate fusion protein harboring the 1st (1453–1466 AA) and 2nd (2401–2415 AA) peptides of NCoR1, we used the glutathione S-transferase (GST) gene fusion system (Supporting Experimental Procedures). The kinase reaction was started by the addition 5 ng of recombinant Akt1 (Millipore, Billerica, MA) and 5 µg each GST peptide (GST-1st or GST-2nd) to kinase assay buffer [25 mM Tris pH 7.5, 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2 and a serine/threonine phosphatase inhibitor cocktail (Calbiochem, Darmstadt, Germany)]. The reaction was incubated for 75 min at 30 °C and terminated by the addition of the SDS sample buffer. The end products were resolved on SDS-PAGE (12–15%) and detected by immunoblotting with phospho-(Ser/Thr) Akt Substrate Antibody (Cell Signaling Technology, MA, #9614).
Cell Culture and Adenoviral Infection
HEK293T, Hep G2 and NCoR1L2/L2 MEF cells were cultured under normal culture condition with Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA). Mouse embryonic fibroblasts (MEFs) from NCoR1L2/L2 floxed mice were prepared and immortalized. Then, either LacZ or Cre recombinase-expressing adenovirus (LacZ or Cre) were infected at a MOI = 20 to generate NCoR1+/+ or −/− MEFs, respectively. A non-tumorigenic mouse hepatocyte cell line, i.e. alpha mouse liver 12 (AML12) was maintained in DMEM/F-12 medium (Invitrogen) as previously described (21). The plasmids and the method of site-directed mutagenesis are described in Supporting Experimental Procedures.
Oxygen Consumption Rate
The mitochondrial oxygen consumption rate (OCR) was measured using a Seahorse XF-96 extracellular flux analyzer as described (Seahorse Bioscience Inc., North Billerica, MA) (22). On the day before the experiment, the sensor cartridge was placed into the calibration buffer supplied by Seahorse Bioscience and incubated at 37°C in a non-CO2 incubator. MEFs were cultured on Seahorse XF-24 plates at a density of 20,000 cells per well. Cells were washed and incubated with assay medium (DMEM without bicarbonate) at 37°C in a non-CO2 incubator for 1-hr. All media and reagents were adjusted to pH 7.4 on the day of the assay. OCR was automatically calculated by the Seahorse XF-96 analyzer and four baseline measurements of OCR were taken.
RNA analysis
RNA was extracted using the TRIzol® reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from total RNA with the SuperScript First-Strand Synthesis System (Invitrogen) and random hexamer primers. The real-time PCR measurement of individual cDNAs was performed using SYBR green dye to measure duplex DNA formation with the LightCycler System (Roche Diagnostics, Meylan, France) (23). Primer details are listed in Supporting Table 1. Data analysis was performed using ABI Prism 7900HT SDS 2.0 software (Applied Biosystem). 2ΔCtn = 2CtGAPDH-Cttarget calculation was used for relative expression value. Affymetrix mouse 430_2 microarray analysis was performed according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA). Data were analyzed by Affymetrix MAS 5.0 software and GSEA (http://www.broad.mit.edu/gsea) (24).
Western Blot Analysis, Co-IP and ChIP Experiments
Western blot analysis was performed with commercially available antibodies listed in Supporting Experimental Procedures. But, polyclonal rabbit antibody to detect NCoR1, which is specifically phosphorylated on 1460 serine (Anti-pS1460 NCoR1 Ab) was generated in Young In Frontier Co., Ltd (Seoul, South Korea). For Co-IP studies, HEK293T cells were transfected with indicated plasmids. Immunoprecipitation was performed with 40 µL of anti-FLAG M2 Affinity Gel suspension (A2220, Sigma-Aldrich, St. Louis, Missouri) and the resulting immunoprecipitates was used for Western blot analysis. To evaluate the recruitment of NCoR1 to the LXRE, PPRE, ERRE of the mouse HA-LXRα, HA-PPARα or HA-ERRα promoters, AML12 cells (10×106 cells) were transfected with the indicated plasmids using lipofectamine 2000 according to the manufacturer’s protocol (Life Technologies Corporation, Carlsbad, CA). Two days after the transfection, the cells were used for ChIP experiments according to standardized protocols (EZ-ChIP™, Millipore). The primer sequences used for the ChIP experiments were: Srebp1c LXRE primers, 5’-AGG CTC TTT TCG GGG ATG G-3’ and 5’- TGG GGT TAC TGG CGG TCA C-3’; Cpt1a PPRE primers, 5’-CTT TCC TAC TGA GGC CCA GAT AG-3’ and 5’-TAC AGC CTA GAA CCC TGA CTG C-3’; Sdhb ERRE primers, 5’- CTT CCT GTA CAT TGG CTC GGA GAA ACC-3’ and 5’-CTT CAA GGA GAC CCC GAC CGT CGC CGC-3’. GW3965, Wy14643 and LY294002 were purchased from Sigma (Sigma-Aldrich).
Isolation of Mitochondria and Blue native-PAGE
Cells or tissues were homogenized in isolation buffer B (210 mM Mannitol, 70 mM sucrose, 1 mM EGTA, 5 mM HEPES, pH 7.2) with a Teflon-glass homogenizer. The homogenate was then centrifuged at 600 × g for 10 min at 4°C. The resulting supernatant was re-centrifuged at 17,000 × g for 10 min at 4°C. The mitochondrial fractions, which were recovered in the pellet, were washed with buffer B and resuspended in the same buffer. The mitochondria were either used immediately or stored at −80°C for later use. BN-PAGE was performed as described previously using the NativePAGE™ Novex® Bis-Tris Gel system (Invitrogen) (25). To detect OXPHOS complex, the Mitoprofile Total OXPHOS Rodent WB Antibody Cocktail (Mitosciences/Abcam) was used. After incubation in the primary antibody dilution, the membrane was washed and detected using the WesternBreeze® Chromogenic Western Blot Immunodetection Kit (Invitrogen).
Statistical Analyses
Statistical analysis was carried out by SPSS version 18.0 for Windows (SPSS Inc., Chicago, IL, USA) or GraphPad Prism (GraphPad Software, Inc., San Diego, CA, USA). Comparisons of average means were performed with Mann-Whitney U-test. Data are mean ± SEM and P < 0.05 was considered statistically significant (* P < 0.05, ** P < 0.01, *** P < 0.001). All reported P-values are two sided.
RESULTS
Insulin induces the phosphorylation of NCoR1 through Akt in vitro
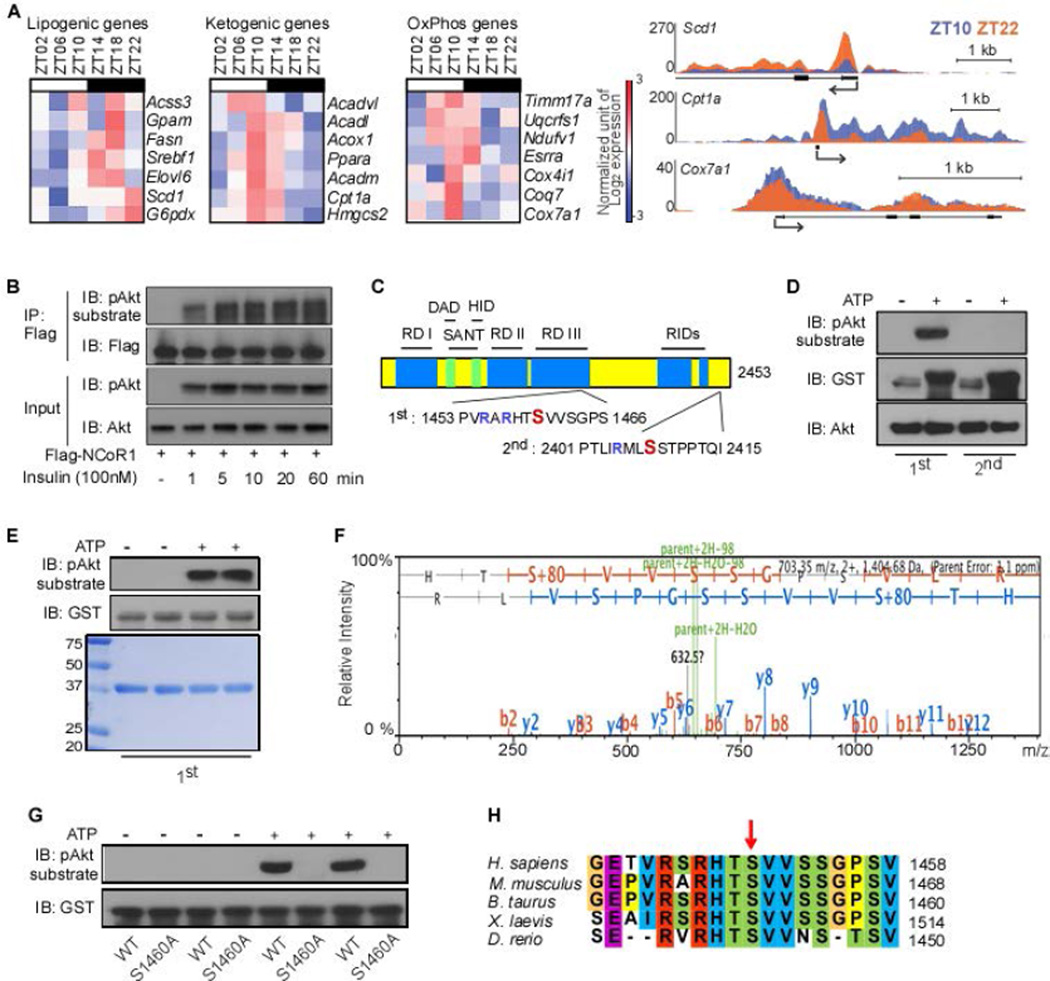
As a first step to test our rationale, we analyzed the expression of lipogenic versus ketogenic and OxPhos genes within the context of a normal diurnal cycle, using microarray gene expression data and ChIP-seq analysis (26). In line with our hypothesis, RNA Polymerase II (Pol II) occupancy on lipogenic, ketogenic and OxPhos genes in the liver reflected the diurnal expression pattern of these genes (Fig. 1A). Incited by these findings, we next investigated whether insulin could mediate this effect by triggering NCoR1 phosphorylation (27). To test this hypothesis, we analyzed the phosphorylation status of a Flag-tagged mouse NCoR1 construct that was transfected into HEK293T cells. Immunoprecipitation with a Flag antibody followed by immunoblotting with a phospho-Akt substrate antibody indicated that Flag-mNCoR1 was already phosphorylated 1 minute after insulin treatment (Fig. 1B). Bioinformatic analysis (http://scansite.mit.edu) for putative Akt phosphorylation sites within the mouse NCoR1 amino acid (AA) sequence identified serine 1460 as a candidate phosphorylation site (Fig. 1C, Supporting Fig. 1). The AA sequence harboring serine 1460 in NCoR1 (defined as the 1st sequence) was robustly phosphorylated by recombinant Akt1 in vitro, whereas no phosphorylation was observed using a downstream AA sequence harboring serine 2408 (2nd sequence), another potential phosphorylation site (Fig. 1D). Subsequent mass-spectrometry based on titanium dioxide (TiO2) phospho-peptide enrichment confirmed neutral loss of 98 Da (H3PO4) on serine 1460 when the 1st sequence was incubated with Akt (Fig. 1E,F). Mutation of serine 1460 to alanine (S1460A) resulted in the disappearance of a detectable phosphorylation signal (Fig. 1G). Moreover, this Akt site was conserved among all vertebrate NCoR1 sequences queried (Fig. 1H), further substantiating the evidence for this residue as a bona fide Akt phosphorylation site. Taken together, these data indicate that insulin induces the phosphorylation of serine 1460 within NCoR1 (pS1460 NCoR1) through the activation of the Akt kinase with very fast kinetics (1 min after insulin treatment).
Figure 1. Serine 1460 of NCoR1 can be phosphorylated by Akt.
(A) Heat maps displaying expression values of each gene at the indicated Zeitgeber time and Pol II-density profiles of Scd1, Cpt1a, and Cox7a1 showing diurnal expression of lipogenic, ketogenic and OxPhos genes in liver (GSE35790). (B) NCoR1 phosphorylation by insulin treatment in HEK293T cells. Cells were transfected with Flag-tagged mouse NCoR1 (1 µg/well of 6-well plate). After 48 hours, cells were treated with insulin (100nM) for the indicated times. (C) Schematic presentation of putative mouse NCoR1 phosphorylation sites by Akt. Positions are numbered according to NCBI Reference Sequence: NP_001239242.1. RD, repression domain; SANT, the SANT-like domains; DAD, deacetylase activation domain; HID, histone interaction domain; RIDs, nuclear receptor interacting domains. AA sequences indicates (Arg-Xaa-)Arg-Xaa-Xaa-Ser/Thr motifs that have been reported to be phosphorylated by Akt/PKB in sequence 1 and 2. (D) In vitro kinase activity of Akt1 toward two short synthetic GST-tagged peptides, each containing 1 of the 2 putative Akt phosphorylation sites of NCoR1, was analyzed by autoradiography. (E) In vitro kinase activity of Akt1 toward a synthetic GST-tagged peptide containing 1460 serine residue of NCoR1. Coomassie stain was performed to visualize the short synthetic peptides, which were used for subsequent Mass spectrometry. (F) Diagrams showing the phosphorylation of 1460 serine residue of NCoR1 by LC-MS/MS. (G) In vitro kinase activity of Akt1 toward two short synthetic GST-tagged peptides, containing either the wild-type 1460 serine residue or mutant 1460 alanine residue of NCoR1. (H) Sequence alignment of the NCoR1 phosphorylation site in various vertebrate species. All immunoblots are representative of at least three independent experiments.
The phosphorylation of NCoR1 switches its recruitment to fatty acid catabolizing nuclear receptors
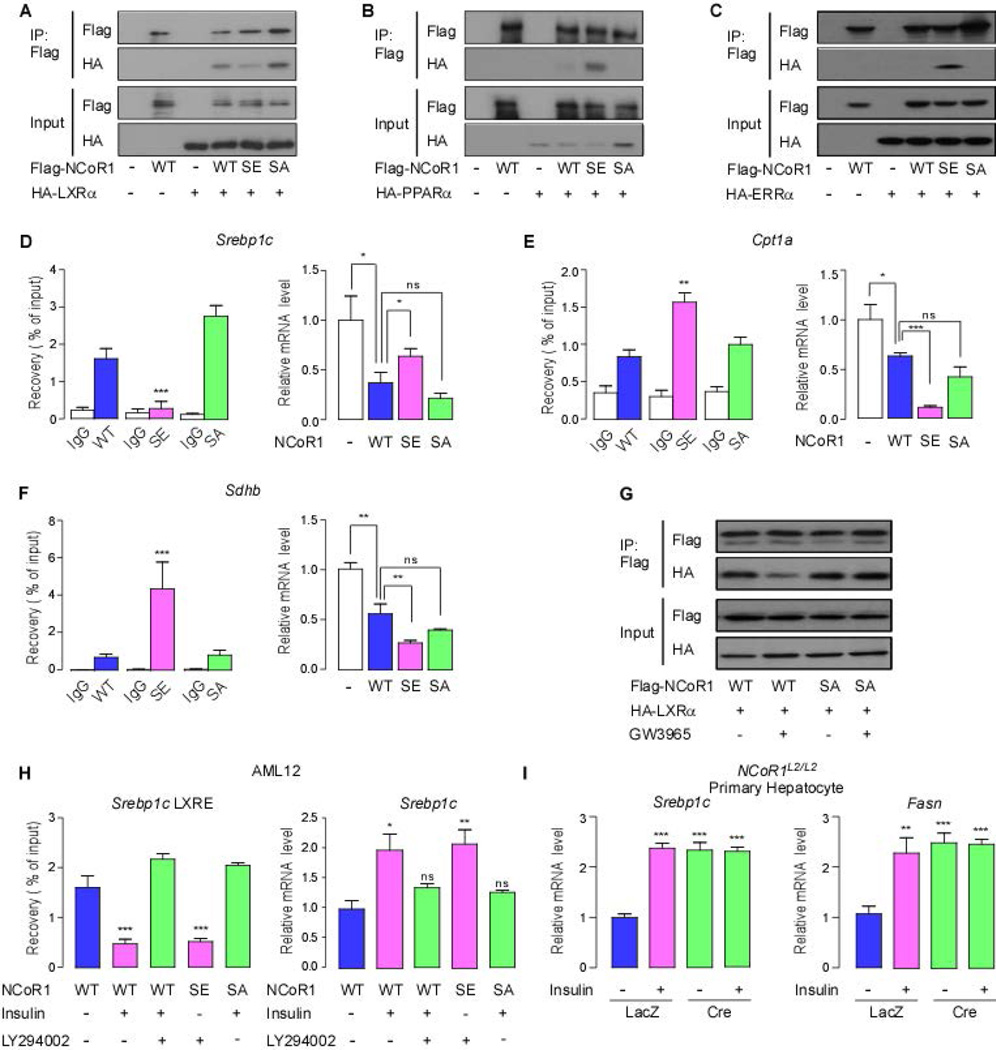
NCoR1 has been reported to interact with the nuclear receptors LXRα, PPARα and ERRα to repress the expression of genes involved in lipogenesis, ketogenesis and oxidative metabolism, respectively (7, 15, 28, 29). To investigate whether pS1460 NCoR1 can affect the binding with these nuclear receptors, we performed co-immunoprecipitation assays using two mutant constructs; S1460E NCoR1, simulating a constitutively phosphorylated form, and S1460A, a non-phosphorylatable form. Because the full-length cDNA of NCoR1 (7362bp) is too large to be incorporated into viral vectors, we used transient transfections of pCMX vectors expressing wild-type or mutant NCoR1. In IP experiments, S1460E NCoR1 showed decreased interaction with LXRα, whereas wild-type and S1460A NCoR1 avidly interacted with LXRα (Fig. 2A). Conversely, the phospho-mimetic S1460E NCoR1 interacted better with PPARα and ERRα, whereas S1460A was unable to interact (Fig. 2B,C). In chromatin IP (ChIP) experiments, we observed that S1460E NCoR1 resulted in reduced NCoR1 recruitment to the LXRE within the Srebp1c promoter (Fig. 2D). Conversely, the same S1460E NCoR1 mutant was more effectively recruited to the respective PPRE and ERRE within the Cpt1 and Sdhb promoters, compared with wild-type and S1460A NCoR1 (Fig. 2E,F). Consistent with co-IP and ChIP data, transfection of mouse AML12 (alpha mouse liver 12) cells with wild-type and S1460A NCoR1 reduced the mRNA expression of LXRα target genes such as Srebp1a, Srebp1c, and Me1, whereas S1460E NCoR1, which displayed reduced binding to LXRα, permitted LXRα target gene expression (Fig. 2D, Supporting Fig. 2A). In contrast to the observed permissive effect of S1460E NCoR1 on lipogenic gene expression, the phospho-mimetic mutation enhanced NCoR1-induced repression of genes controlled by ERRα and PPARα, including Cpt1a, mCad, Sdhb, Esrra and Ndufb3 (Fig. 2E,F, Supporting Fig. 2B). Furthermore, these specific gene-selective permissive actions of the different NCoR1 mutations were also consistently observed in the human hepatocarcinoma cell line, Hep G2 (Supporting Fig. 2C). To investigate the effect of LXRα or PPARα ligands on the selection of NCoR1 by the nuclear receptors, we treated AML12 cells with GW3965 (an LXRα agonist) or Wy14643 (a PPARα agonist) after transfection with either wild-type or mutant NCoR1. Interestingly, GW3965 promoted the dissociation of LXRα and wild-type NCoR1, whereas this LXRα agonist did not have any effect on the interaction between S1460A NCoR1 and LXRα, indicating that non-phosphorylatable NCoR1 can repress LXRα transactivation regardless of the presence of an LXRα agonist (Fig. 2G). Compatible with our findings using the LXRα agonist, the PPARα agonist Wy14643 also did not abrogate the interaction between the phospho-mimetic (SE) NCoR1 and PPARα (Supporting Fig. 2D). Collectively, our data support that the phosphorylation of NCoR1 at serine 1460 has a pivotal role in directing NCoR1 to select nuclear receptor sites.
Figure 2. The NCoR1 phosphorylation determines nuclear receptor recruitment.
(A) Co-immunoprecipitation assay using wild-type Flag-NCoR1, Flag-S1460E NCoR1 (SE), Flag-S1460A NCoR1 (SA) and HA-LXRα in HEK293T cells. Cells were transfected with indicated plasmids (1 µg/well per 6-well plate). After 48 hours, cell lysates were immunoprecipitated with anti-Flag antibody. (B and C) Similar co-immunoprecipitation assays using the same NCoR1 constructs as in (A) and HA-PPARα (B) and HA-ERRα (C). (D-F) NCoR1 recruitment to the LXRE (D), PPRE (E) and ERRE (F) sites of the mouse Srebp1c, Cpt1a and Sdhb promoters determined by ChIP in AML12 cells and quantitative RT-PCR (qRT-PCR) analysis of the mRNA levels of Srebp1c (D), Cpt1a (E) and Sdhb (F) according to the sequence composition of NCoR1 phosphorylation site (each group, n>5). mRNA levels were analyzed in AML12 cells transfected with indicated NCoR1 plasmids (each group, n>7) (G) Co-immunoprecipitation assay using wild-type Flag-NCoR1, Flag-S1460A NCoR1 (SA) and HA-LXRα (1 µg/well per 6-well plate) in AML12 cells with or without GW3965 (1 µM) as indicated. (H) NCoR1 recruitment to the LXRE of the mouse Srebp1c, promoters determined by ChIP and quantitative RT-PCR (qRT-PCR) analysis of the mRNA levels of Srebp1c in AML12 cells according to the sequence composition of the S1460 NCoR1 phosphorylation site in the absence or presence of insulin (100 nM) for 6 h with or without LY294002 (30 µM) given 30 min before insulin treatment (each group, n>5). (I) qRT-PCR analysis to evaluate mRNA levels of Srebp1c and Fasn in primary hepatocytes from NCoR1L2/L2 mice infected with either an Ad-LacZ or Ad-Cre adenovirus grown in high glucose (25 mM) medium with or without insulin treatment (100nM) for 6 h. All immunoblots are representative of at least three independent experiments. Data are mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001.
In order to determine the physiologic relevance of NCoR1 phosphorylation by Akt/PKB, we performed ChIP experiments after insulin treatment with or without pre-treatment with the PI3K inhibitor, LY294002. NCoR1 recruitment on the LXRE within the Srebp1c promoter was significantly reduced by insulin treatment, which could be reversed by the PI3K inhibitor (Fig. 2H). However, the recruitment of the phospho-mimetic (SE) and non-phosphorylatable (SA) NCoR1 were not affected by PI3K inhibitor or insulin, respectively (Fig. 2H). In addition, qRT-PCR analysis indicated that the repressive action of NCoR1 on Srebp1c expression was removed by insulin/Akt signaling, consistent with our ChIP experiments (Fig. 2H). To verify our observation, we deleted NCoR1 in primary hepatocytes derived from NCoR1L2/L2 mice using an Adenovirus expressing the Cre-recombinase (Supporting Fig. 2E). Insulin treatment or NCoR1 deletion markedly increased mRNA expression of Srebp1c and Fasn, but insulin treatment did not lead to further increases of these LXRα target genes expression in NCoR1-deficient primary hepatocytes (Fig. 2I). Tail vein pDNA delivery into NCoR1hep−/− mice also induced changes in mRNA levels of Me1, Elovl6, mCad and Ndufb5 according to the sequence composition of NCoR1 phosphorylation site (Supporting Fig. 2F) (30, 31). Taken together, these ChIP and qRT-PCR data suggested that the NCoR1 phosphorylation by Akt/PKB may be operational in vivo at the period of fasting to feeding transition by insulin stimulation.
NCoR1 deletion suggests a dual repressor function for NCoR1
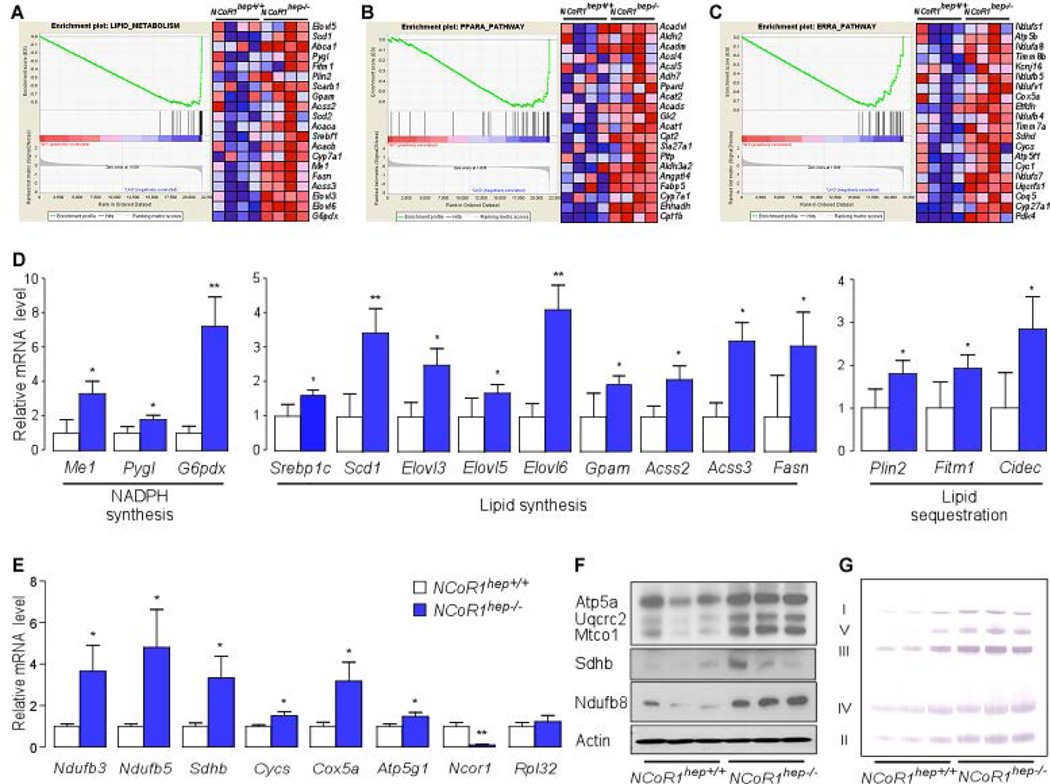
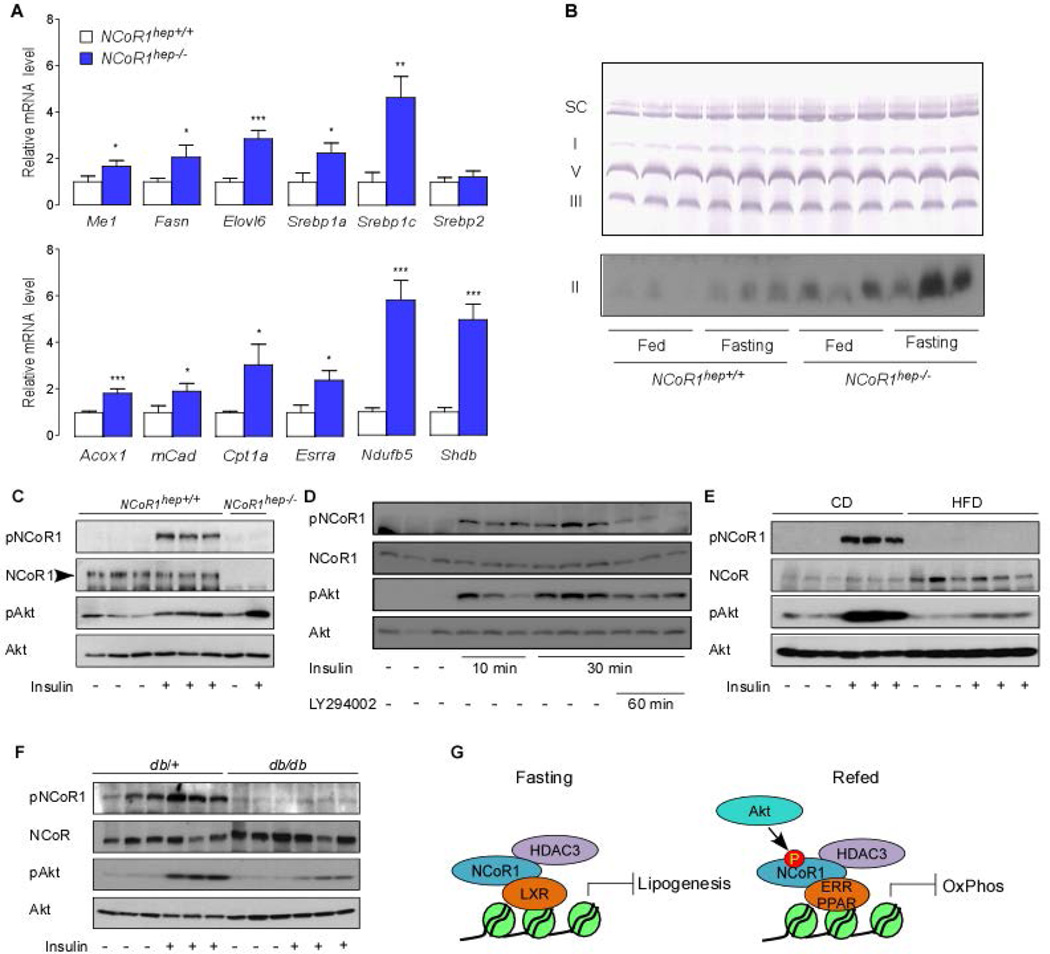
Given that our cellular model of NCoR1 regulation may have important implications for the role of NCoR1 in controlling hepatic lipid metabolism, we crossed mice bearing floxed NCoR1 alleles (NCoR1L2/L2) with transgenic mice expressing the Cre-recombinase specifically in hepatocytes to yield NCoR1hep−/− mice (15, 24). To determine whether an a priori defined set of genes shows statistically significant, concordant differences between NCoR1hep−/− and NCoR1hep+/+ ad libitum fed mice, we performed Affymetrix Gene 1.0 ST arrays, followed by Gene Set Enrichment Analysis (GSEA) using LXRα (Lipid metabolism), PPARα and ERRα target gene sets (32–35). As expected from previous work on the NS-DADm and HDAC3hep−/− mice, several lipogenic LXR target genes were coordinately induced in NCoR1hep−/− livers (Fig. 3A) (9, 10). Furthermore, genes related to PPARα and ERRα signaling pathways were also strongly induced by the hepatic NCoR1 deletion (Fig. 3B,C) (36, 37). Both the PPARs and ERRs play a common role in control of mitochondrial physiology and fatty acid oxidation (37–40). GSEA using an electron transport system (OxPhos) gene set identified a coordinated induction of genes for OxPhos in NCoR1hep−/− livers (Supporting Fig. 3). To validate the GSEA data, we used qRT-PCR analysis. Transcript levels of genes involved in NADPH synthesis (Me1, Pygl, G6pdx), lipid synthesis (Srebp1c, Scd1, Elovl3, Elovl5, Elovl6, Gpam, Acss2, Acss3, Fasn) and lipid sequestration (Plin2, Fitm1, Cidec), were all increased (Fig. 3D). Interestingly, expression of all of these genes were also reported to be increased in livers from HDAC3hep−/− mice (9). Of note, most of these induced transcripts are LXRα dependent. In addition, the mRNA levels of OxPhos genes such as Ndufb3, Ndufb5, Sdhb, Cycs, Cox5a and Atp5a1 were also coordinately increased in NCoR1hep−/− livers (Fig. 3E). Western blot and BN-PAGE analysis furthermore indicated an increased abundance of OxPhos proteins and enhanced complex formation in livers from chow fed NCoR1hep−/− mice (Fig. 3F,G).
Figure 3. Liver specific NCoR1 deletion in ad libitum fed mice and NCoR1 deletion in MEFs results in a robust induction of lipogenic, ketogenic and OxPhos genes.
(A–C) Gene Set Enrichment Analysis of gene expression profiles in livers of chow fed NCoR1hep−/− mice at 25 weeks of age (n=4/group). The gene set encompassing lipid metabolism has a nominal p = 0.03 and False discovery rate (FDR) q = 0.052 (A), PPARα has nominal p < 0.001 and FDR q = 0.024 (B), and the ERRα has nominal p = 0.028 and FDR q = 0.056 (C). (D and E) qRT-PCR analysis of mRNA expression in the livers of male chow fed mice at 25 weeks of age. (n=7/group). mRNA expression for the indicated genes involved in NADPH synthesis, lipid synthesis, lipid sequestration (D) and OxPhos (E). (F) Western blot analysis showing Atp5a, Uqcrc2, Mtco1, Sdhb and Ndufb8 levels in NCoR1hep+/+ and NCoR1hep−/− livers of chow fed 25-wk old male NCoR1hep+/+ and NCoR1hep−/− mice. (G) BN-PAGE of OXPHOS complexes in mitochondria isolated from NCoR1hep+/+ and NCoR1hep−/− livers.
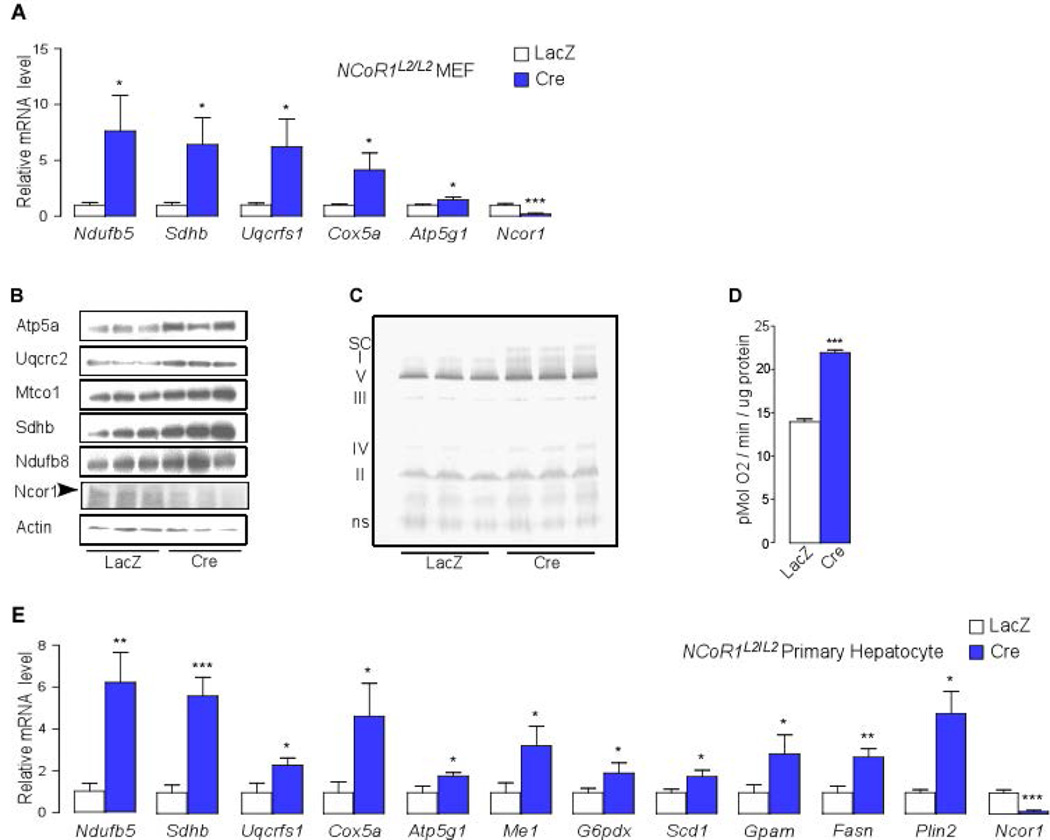
To investigate whether these effects of NCoR1 deletion could be induced acutely and cell-autonomously, we compared OxPhos components at the level of complex assembly and mRNA and subunit protein expression in NCoR1L2/L2 MEFs, infected with adenoviruses expressing either the Cre-recombinase (Ad-Cre), to delete NCoR1 gene, or LacZ (Ad-LacZ). In agreement with data from NCoR1hep−/− livers, transcripts of the OxPhos genes, OxPhos complexes, OxPhos supercomplexes, and individual subunit proteins were higher in NCoR1−/− MEFs, collectively contributing to significantly elevated oxygen consumption (Fig. 4A–D). Moreover, acute NCoR1 deletion in primary hepatocytes from NCoR1L2/L2 mice, infected with Ad-Cre or Ad-LacZ, also resulted in higher mRNA expression of genes involved in OxPhos (Ndufb5, Sdhb, Uqcrfs1, Cox5a, Atp5g1), NADPH synthesis (Me1, G6pdx), lipid synthesis (Scd1, Gpam, Fasn) and lipid sequestration (Plin2) (Fig. 4E).
Figure 4.
Acute NCoR1 deletion results in induction of lipogenic and OxPhos genes. (A) qRT-PCR analysis to evaluate mRNA levels of Ndufb5, Sdhb, Uqcrfs1, Cox5a, Atp5g1 and Ncor1 in NCoR1L2/L2 MEF cells infected with either an Ad-LacZ or Ad-Cre adenovirus grown in high glucose (25 mM) medium. (B) Western blot analysis showing Atp5a, Uqcrc2, Mtco1, Sdhb and Ndufb8 levels in NCoR1L2/L2 MEFs infected during 72-hr with either an Ad-LacZ or Ad-Cre adenovirus. (C) BN-PAGE of OXPHOS complexes in mitochondria isolated from NCoR1L2/L2 MEFs infected with either an Ad-LacZ or Ad-Cre adenovirus. (D) Oxygen consumption rate (OCR) was measured in NCoR1L2/L2 MEFs infected with either an Ad-LacZ or Ad-Cre adenovirus grown in high glucose (25 mM) medium (n=6/group). Data are mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001. (E) qRT-PCR analysis to evaluate mRNA levels of Ndufb5, Sdhb, Uqcrfs1, Cox5a, Atp5g1, Me1, G6pdx, Scd1, Gpam, Fasn, Plin2 and Ncor1 in primary hepatocytes from NCoR1L2/L2 mice infected with either an Ad-LacZ or Ad-Cre adenovirus grown in high glucose (25 mM) medium.
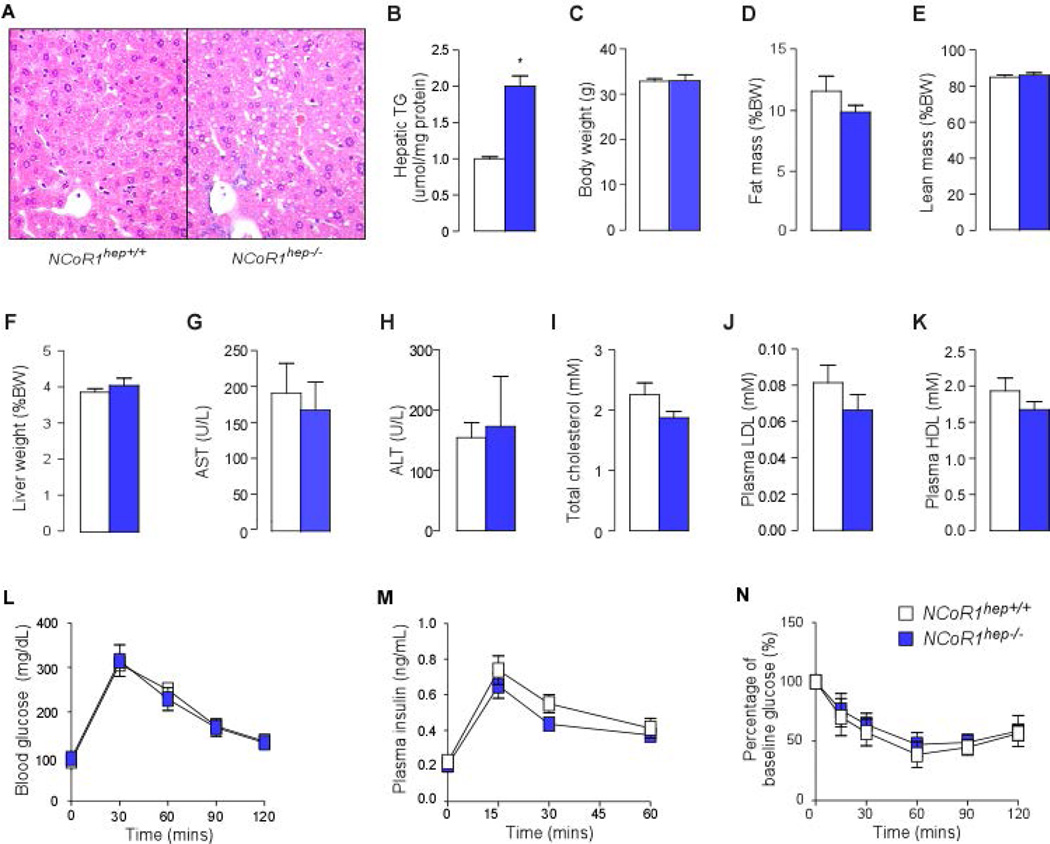
Given the concurrent induction of genes involved in lipogenesis and lipid oxidation upon NCoR1 loss-of-function in both mice and cell models, we next determined its phenotypic impact in NCoR1hep−/− mice. Hematoxylin and Eosin (H&E) staining indicates the accumulation of very small lipid-like droplets within the hepatocytes of NCoR1hep−/− mice fasted for 4-hr (Fig. 5A). Consistent with these histological findings, triglyceride content was two-fold elevated in livers from NCoR1hep−/− compared to control NCoR1hep+/+ mice (Fig. 5B). Despite the marked accumulation of triglycerides in the liver, NCoR1hep−/− mice displayed no differences in anthropometric and plasma biochemical parameters, fasting blood glucose, glucose tolerance and insulin tolerance (Fig. 5C–N).
Figure 5. NCoR1hep−/− mice have hepatic lipid accumulation.
(A) Hematoxylin and eosin staining of liver sections from 25-wk old male chow fed NCoR1hep+/+ and NCoR1hep−/− mice that were fasted for 4-hr. (B) Hepatic triglyceride measurements in livers from 25-wk old male chow fed NCoR1hep+/+ and NCoR1hep−/− mice that were fasted for 4-hr (n = 7/group). Anthropometric (C–F) and biochemical analysis (G–K) of NCoR1hep−/− mice compared to NCoR1hep+/+ mice. (L and M) Blood glucose and insulin levels before and during intraperitoneal glucose tolerance test (ipGTT) in 25-wk old chow fed male NCoR1hep+/+ and NCoR1hep−/− mice after a 4-hr fast (n > 7/group). (N) Percentage of baseline glucose levels during Insulin tolerance test (ITT) in 25-wk old chow fed male NCoR1hep+/+ and NCoR1hep−/− mice after overnight fasting (n > 7/group). All data shown as mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001.
Expression of PPARα in the liver is relatively low in the fed state (12, 13, 41). PPARα expression is, however, induced upon fasting and then plays a critical role in the regulation of hepatic gluconeogenesis and fatty acid oxidation (37). In addition, ERRα also takes part in induction of OxPhos genes upon fasting (37). To support our hypothesis of autonomous and constitutive activation of hepatic LXRα, PPARα and ERRα in livers from NCoR1hep−/− mice, we analyzed liver phenotypes in NCoR1hep−/− and NCoR1hep+/+ mice after a 24-hr fast. Consistent with our data in mice fed ad libitum, LXRα target genes, such as Me1, Fasn, Elovl6 and Srebp1, were also abundantly expressed in NCoR1hep−/− livers even under fasting condition (Fig. 6A). In addition, the mRNA expression of fasting-induced PPARα and ERRα target genes such as Acox1, mCAD, Cpt1a, Esrra, Ndufb5 and Sdhb were markedly increased in livers from fasted NCoR1hep−/− mice (Fig. 6A). In accordance with the mRNA expression of fasting-induced PPARα target genes, BN-PAGE also showed higher levels of OxPhos complexes in livers from NCoR1hep−/− mice under both fasting and fed conditions (Fig. 6B) (42). Collectively these data indicate that LXRα, PPARα and ERRα transcriptional activity is constitutively turned on in NCoR1hep−/− livers, which supports the dual repressor function of NCoR1.
Figure 6. Metabolic impact of NCoR1 deletion on livers of fasted mice and impaired phosphorylation of serine 1460 NCoR1 in mouse models of diabesity.
(A) mRNA levels of Me1, Fasn, Elovl6, Srebp1, Srebp2, Acox1, mCad, Cpt1a, Esrra, Ndufb5 and Sdhb were evaluated by qRT-PCR in livers of NCoR1hep+/+ and NCoR1hep−/− mice fasted for 24-hr (n > 7/group). All data shown as mean ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001. (B) BN-PAGE of OXPHOS complexes in mitochondria isolated from liver of 25-wk old male NCoR1hep+/+ and NCoR1hep−/− mice either fed or fasted for 24-hr. Upper panel is a representative result developed by chromogen-based detection technique to visualize supercomplexes (SC), complex I, V and III. The lower panel shows HRP-based detection to evaluate complex II amount. (C) Western blot analysis indicating that insulin increases pS1460 NCoR1 in livers from 25-wk old NCoR1hep+/+ and NCoR1hep−/− male chow fed mice. Animals were injected with either PBS or insulin (0.5 U/kg body weight) at 10 min before sacrificing. (D) Western blot analysis showing that PI3K inhibition attenuates the insulin-induced increases of pS1460 NCoR1 in livers from 25-wk old 4-hr fasted C57BL/6J male chow fed mice. After 4 hrs of fasting, animals were injected with either PBS or insulin (0.5 U/kg body weight) for 10 min or 30 min with or without LY294002 (40 mg/kg) which was given 60 min before sacrifice. (E) pS1460 NCoR1 levels in livers from 25-wk old C57BL/6J male mice fed with either chow or high fat diet for 12-wk. (F) pS1460 NCoR1 levels in livers from 12-wk old male db/db mice. Animals in (E) and (F) were fasted for 4-hr. The experimental protocol is similar to that used in (C). All immunoblots are representative of at least three independent experiments. (G) Working model, summarizing how NCoR1 may control the fasting-feeding transitions in liver in a phosphorylation dependent manner.
Insulin resistance attenuates NCoR1 phosphorylation by insulin in vivo
To directly address whether insulin induces NCoR1 phosphorylation at serine 1460 in vivo, we generated a polyclonal rabbit antibody to detect NCoR1 that is specifically phosphorylated on 1460 serine (Anti-pS1460 NCoR1 Ab). After initial validation with ELISA (data not shown) and western blot analysis (Supporting Fig. 4A), we applied this antibody to detect pS1460 NCoR1 in HEK293T cells transfected with Flag-mNCoR1 or Flag-S1460A mNCoR1 in the absence or presence of insulin in the medium. pS1460 NCoR1 was readily detected in Flag-mNCoR1 transfected cells and the intensity of the band was increased by insulin (Supporting Fig. 4B). However, no pS1460 NCoR1 signal was observed in Flag-S1460A mNCoR1 transfected cells treated with or without insulin. Notably, ten minutes after insulin injection the amount of pS1460 NCoR1 was increased in livers from chow fed 25-wk old NCoR1hep+/+ mice, an effect that was absent in NCoR1hep−/− livers (Fig. 6C). In addition, pS1460 NCoR1 was persistently detected 30 min after insulin in NCoR1hep+/+ mice and the pretreatment with the PI3K inhibitor LY294002 attenuated the phosphorylation of S1460 NCoR1 in parallel with Akt/PKB phosphorylation status (Fig. 6D). The robust NCoR1 phosphorylation by insulin incited us to analyze whether NCoR1 phosphorylation was deregulated in livers of mice with either a diet-induced (by feeding a high fat diet) or a genetic (which carry a mutation in the Lepr gene) form of insulin resistance (43). Consistent with the data from NCoR1hep+/+ mice liver, NCoR1 was hyperphosphorylated by insulin in chow fed C57BL/6J mice (Fig. 6E). However, pS1460 NCoR1 was barely detectable in C57BL/6J livers from mice that were fed with high fat diet, even though insulin still slightly increased Akt phosphorylation levels (Fig. 6E). In line with these data obtained in mice with diet-induced insulin resistance, pS1460 NCoR1 was also not induced by insulin in livers harvested from 12-wk old db/db mice (Fig. 6F).
DISCUSSION
Our work identifies a novel phosphorylation-dependent switch, which enables NCoR1 to select its repressive targets pending on the cellular energy status. The regulation of the phosphorylation status of serine 1460 in fact confers to NCoR1 the capacity to co-repress lipogenic nuclear receptors, such as LXRα, in its unphosphorylated state during fasting, or to co-repress oxidative nuclear receptors, such as PPARα and ERRα, when it is phosphorylated during feeding (Fig. 6G). Since the transcriptional activity of LXRα and PPARα is dependent on their binding with specific ligands, such as oxysterols and fatty acids, one may consider that the ligand-dependent dissociation of NCoR1 from LXRα in the fed state and from liganded PPARα in the fasted state might be sufficient to select NCoR1’s repressor targets (44, 45). The phosphorylation of NCoR1 by insulin-induced Akt/PKB activation was, however, robustly induced with very fast kinetics (i.e. 1 min in vitro and 10 min in vivo after insulin treatment), indicating that NCoR1 phosphorylation directly alters protein functions of NCoR1 most likely already before LXRα activation is initiated by ligand-binding. It is in fact improbable that the levels of LXRα ligands change within a time-frame of a few minutes. We hence postulate that NCoR1 phosphorylation by insulin/Akt is acting independently and in parallel to the action of LXR ligands and has a permissive role for LXRα transactivation. Supporting our hypothesis, LXRα or PPARα agonists did not induce the dissociation of non-phosphorylatable NCoR1 or phospho-mimetic NCoR1 from their ligand-activated nuclear receptors. In addition, livers of NCoR1hep−/− mice showed a supplemental increase of OxPhos in the fasting compared to the fed state, supporting a synergistic effect of de-repression, by the deletion of NCoR1, and PPARα/ERRα transactivation (Fig. 6B). Taken together, the phosphorylation-dependent regulation of NCoR1 adds another layer of control on top of the ligand-dependent interaction of NCoR1 with its client nuclear receptors. This makes NCoR1 an exquisite energy sensor, which allows cells to adapt their energy homeostasis, via the selective fine-tuning of appropriate gene expression programs.
Our data also explain the apparent paradox between previous reports that showed rather incompatible activities of NCoR1 on liver metabolism (9, 12, 13, 41). PPARα, the transcriptional activator of ketogenesis, is repressed in livers from aged mice by hyperactive mTOR signaling (13). Recent data have identified that S6K2 phosphorylated by mTORC1, enhances NCoR1’s repressor activity on PPARα transactivation (12). However, these observations of NCoR1 function seem to be in contradiction with its role established in early neural development and in glioma cells (3, 46, 47), suggesting that NCoR1 is exported out of nucleus by Akt phosphorylation. Thus, the fact that the transcriptional repressive activity of NCoR1 is enhanced in nutrient affluent conditions (15), which are usually associated with enhanced Akt/PKB phosphorylation, seems in conflict with the transcriptional de-repression mechanism of NCoR1 induced by Akt/PKB phosphorylation (7). In our model, NCoR1 can both decrease the expression of lipogenic genes—through LXRα repression—and the expression of ketogenic and OxPhos genes—via PPARα and ERRα—pending on the fasting and fed condition. As a result, phosphorylation of S1460 NCoR1 by Akt/PKB explains these apparent contradictory effects. This regulatory switch is, however, absent in NCoR1hep−/− livers, resulting in the concurrent activation of fat oxidation and lipogenic pathways. A similar de-repression of hepatic fat oxidation and lipogenic pathway, is also observed in the NS-DADm and HDAC3hep−/− mice (9, 10), in whom the function of HDAC3, which is responsible for NCoR1’s repressive activity, is impeded. Of note, like in the muscle, the NCoR1/HDAC3 tandem also in the liver coordinates the multiple hormonal and metabolic adaptations that govern hepatic lipid homeostasis via their wide-ranging impact on mitochondrial function (15).
How the S1460 residue in NCoR1 affects its interaction with nuclear receptors from a mechanistic and structural point of view is a puzzling question as the S1460 residue is distant from the known nuclear receptor interaction motifs in NCoR1. We speculate that NCoR1 binds in a constitutive manner with nuclear receptors and that its phosphorylation status does not determine whether it interacts, but only dictates its preference for certain receptors above others (see model in Fig. 6G). This function is not incompatible with the S1460 residue not being part of the NCoR1 receptor interaction motifs. This is, however, in contrast with the ligand-dependent interactions between nuclear receptors and cofactors, which are strictly governed by NCoR1’s nuclear receptor interaction domains. Future structural studies will help to elucidate these interesting questions.
Nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) are the most common liver disease in developed countries (48). To understand the pathogenic mechanisms of NAFLD and NASH, abnormalities in hepatocellular lipid metabolism, such as de novo lipogenesis have been extensively investigated. However, the role of mitochondrial oxidative metabolism governing the terminal disposal of fat via β-oxidation and ketogenesis in the pathogenesis of NAFLD and NASH has been less illuminated. In our experiment, the livers of mice with either a diet-induced or a genetic form of insulin resistance showed dysregulation of NCoR1 phosphorylation, implying activated hepatic ketogenesis through the release of NCoR1 repression on ERRα/PPARα. Loss of Akt/PKB-induced ERRα/PPARα repression in the insulin resistant state could be a potential compensatory mechanism to increase fat oxidation and alleviate the onset of NAFLD and NASH. Because ketogenesis eliminates two-thirds of the lipids entering the liver, NAFLD and NASH progression may depend on the dysregulation of this potential compensatory mechanism, which will be a fertile subject of future studies (49, 50).
In conclusion, we show that a phosphorylation-dependent switch enables NCoR1 to select the nuclear receptors that it targets for repression, pending on the cellular energy status. NCoR1 hence seems to act as a hormonally regulated nuclear scaffold protein that amplifies and controls insulin signaling, in a similar fashion as the scaffolding protein IRS does in the cytoplasm. We hope that these studies will incite further work to explore the role of NCoR1 as a key metabolic regulator with roles in health and disease.
Supplementary Material
Acknowledgments
Financial Support: J.A. is the Nestlé Chair in Energy Metabolism. This research was supported by the EPFL, NIH (R01-AG043930) the SNSF (31003A-140780 awarded to J.A. and 310030-143748 awarded to K.S.). YSJ was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2012R1A2A2A01014672). AM was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research.
Abbreviations
- NCoR1
nuclear receptor co-repressor 1
- PKB
protein kinase B
- PPARα
peroxisome proliferator-activated receptor alpha
- ERRα
estrogen-related receptor alpha
- LXRα
liver X receptor alpha
- SMRT
silencing mediator for retinoid or thyroid-hormone receptors
- DADs
deacetylase-activating domains
- HDAC3
histone deacetylase 3
- PI3K
phosphoinositide-3-kinase
- mTOR
mechanistic target of rapamycin
- mTORC1
mechanistic target of rapamycin complex 1
- OxPhos
oxidative phosphorylation
- GST
glutathione S-transferase
- MEF
mouse embryonic fibroblasts
- OCR
oxygen consumption rate
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- ChIP
chromatin immunoprecipitation
- LXRE
LXR response element
- PPRE
PPAR response element
- ERRE
ERR response element
- GSEA
gene set enrichment analysis
- IRS
insulin receptor substrate
Contributor Information
Young Suk Jo, Email: JOYS@yuhs.ac.
Dongryeol Ryu, Email: dongryeol.ryu@epfl.ch.
Adriano Maida, Email: a.maida@dkfz-heidelberg.de.
Xu Wang, Email: xu.wang@epfl.ch.
Ronald M. Evans, Email: evans@salk.edu.
Kristina Schoonjans, Email: kristina.schoonjans@epfl.ch.
Johan Auwerx, Email: admin.auwerx@epfl.ch.
References
- 1.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 2.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 3.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 4.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 5.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 7.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 9.Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 12.Kim K, Pyo S, Um SH. S6 kinase 2 deficiency enhances ketone body production and increases peroxisome proliferator-activated receptor alpha activity in the liver. Hepatology. 2012;55:1727–1737. doi: 10.1002/hep.25537. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Schindler J, Summermatter S, Salatino S, Zorzato F, Beer M, Balwierz PJ, van Nimwegen E, et al. The corepressor NCoR1 antagonizes PGC-1alpha and estrogen-related receptor alpha in the regulation of skeletal muscle function and oxidative metabolism. Mol Cell Biol. 2012;32:4913–4924. doi: 10.1128/MCB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, Heligon C, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 17.Brown MS, Goldstein JL. Selective versus total insulin resistance: a pathogenic paradox. Cell Metab. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Champy MF, Selloum M, Piard L, Zeitler V, Caradec C, Chambon P, Auwerx J. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm Genome. 2004;15:768–783. doi: 10.1007/s00335-004-2393-1. [DOI] [PubMed] [Google Scholar]
- 20.Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, et al. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome. 2008;19:318–331. doi: 10.1007/s00335-008-9107-z. [DOI] [PubMed] [Google Scholar]
- 21.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 25.Kim SJ, Kwon MC, Ryu MJ, Chung HK, Tadi S, Kim YK, Kim JM, et al. CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 2012;16:274–283. doi: 10.1016/j.cmet.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 26.Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, et al. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci U S A. 2008;105:19544–19549. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor alpha interacting protein. J Biol Chem. 1999;274:15901–15907. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- 30.Herweijer H, Wolff JA. Progress and prospects: naked DNA gene transfer and therapy. Gene Ther. 2003;10:453–458. doi: 10.1038/sj.gt.3301983. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen TH, Ferry N. Liver gene therapy: advances and hurdles. Gene Ther. 2004;11(Suppl 1):S76–S84. doi: 10.1038/sj.gt.3302373. [DOI] [PubMed] [Google Scholar]
- 32.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 37.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 39.Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 40.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 41.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soboll S, Oh MH, Brown GC. Control of oxidative phosphorylation, gluconeogenesis, ureagenesis and ATP turnover in isolated perfused rat liver analyzed by top-down metabolic control analysis. Eur J Biochem. 1998;254:194–201. doi: 10.1046/j.1432-1327.1998.2540194.x. [DOI] [PubMed] [Google Scholar]
- 43.Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, Friedman JM. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 44.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 45.Forman BM, Ruan B, Chen J, Schroepfer GJ, Jr, Evans RM. The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc Natl Acad Sci U S A. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 47.Park DM, Li J, Okamoto H, Akeju O, Kim SH, Lubensky I, Vortmeyer A, et al. N-CoR pathway targeting induces glioblastoma derived cancer stem cell differentiation. Cell Cycle. 2007;6:467–470. doi: 10.4161/cc.6.4.3856. [DOI] [PubMed] [Google Scholar]
- 48.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 49.Williamson JR, Scholz R, Browning ET. Control mechanisms of gluconeogenesis and ketogenesis. II. Interactions between fatty acid oxidation and the citric acid cycle in perfused rat liver. J Biol Chem. 1969;244:4617–4627. [PubMed] [Google Scholar]
- 50.Cotter DG, Ercal B, Huang X, Leid JM, d'Avignon DA, Graham MJ, Dietzen DJ, et al. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J Clin Invest. 2014;124:5175–5190. doi: 10.1172/JCI76388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.