Abstract
ModelDB (modeldb.yale.edu), a searchable repository of source code of more than 900 published computational neuroscience models, seeks to promote model reuse and reproducibility. Code sharing is a first step; however, model source code is often large and not easily understood. To aid users, we have developed ModelView, a web application for ModelDB that presents a graphical view of model structure augmented with contextual information for NEURON and NEURON-runnable (e.g. NeuroML, PyNN) models. Web presentation provides a rich, simulator-independent environment for interacting with graphs. The necessary data is generated by combining manual curation, text-mining the source code, querying ModelDB, and simulator introspection. Key features of the user interface along with the data analysis, storage, and visualization algorithms are explained. With this tool, researchers can examine and assess the structure of hundreds of models in ModelDB in a standardized presentation without installing any software, downloading the model, or reading model source code.
Keywords: ModelDB, repository, visualization, code analysis
1 Introduction
The principles of the scientific method require that research results be reproducible. In vivo and in vitro research have to account for noisy instrumentation and biological variation that do not affect in silico studies. Nonetheless, in silico models are nontrivial to reproduce correctly. The main challenge to reproducibility lies in the complexity of fully describing the model: for computational models every parameter and every set of dynamics must be explicitly specified by the researcher; no parameters or behaviors come “for free” with the specification of the model organism. To address this challenge, model repositories such as ModelDB (modeldb.yale.edu) (Migliore et al. 2003), BioModels (www.ebi.ack.uk/biomodels) (Le Novere et al. 2005), and Visiome (visiome.neuroinf.jp) (Usui 2003), tools such as Sumatra (Davison et al. 2014), and web portals for collaborative research (e.g. opensourcebrain.org) were created to assist researchers in sharing source code.
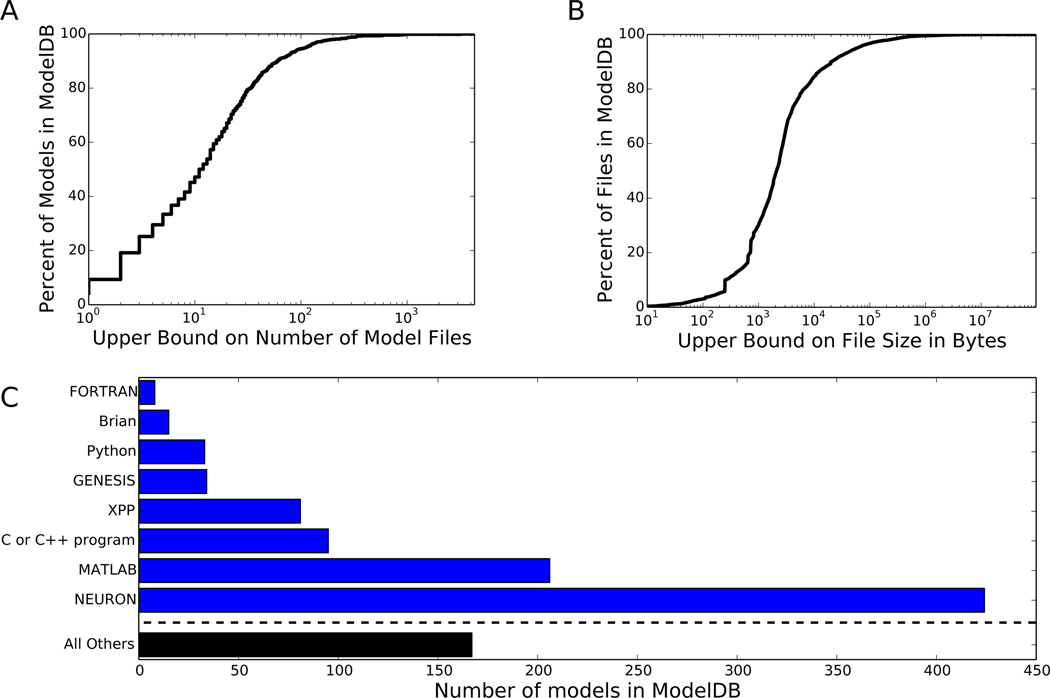
Computational models also present the significant challenge of “understandability”. Modern models are complex and their hierarchical structure and component properties are often obscured by their representation as an executable program text. On ModelDB, over half of the approximately 900 models are distributed across ten or more files (Fig. 1A). Over half of the files are more than two kilobytes in length, and over one-seventh are more than ten kilobytes in length (Fig. 1B).
Figure 1.
A view of the contents of ModelDB as of July 16, 2014, when ModelDB had 901 entries. (A) Cumulative distribution of the number of files per model. (B) Cumulative distribution of file sizes in bytes. (C) The distribution of simulator types in ModelDB. The total exceeds 901 because some models work with multiple simulators.
We have developed a two-phase process to facilitate the understanding of the runtime structure of models in ModelDB. In the analysis phase, data on the simulator state (parameter values, morphology, etc) after partly running a model is combined with information from other sources and stored in a file; this phase is run only once per model. The analysis process is necessarily simulator-specific. Our initial release focuses on support for NEURON (Hines and Carnevale 2001) models because this is the most commonly used simulator in ModelDB (Fig. 1C) and because NEURON can run some non-native models with the aid of standard formats like NeuroML (Gleeson et al 2010) and tools like neuroConstruct (Gleeson et al 2007) and PyNN (Davison et al 2008). In the visualization phase – which is completely simulator independent – a custom HTML5 web app for displaying hierarchically structured data loads the file generated by the analysis phase, and interactively presents it to the user. From the user perspective, this view into a model’s structure only requires pressing a button; the web app requires JavaScript support (enabled by default on most modern browsers), but does not require any browser plugins.
This paper begins with a description of how our scripts collect information about each model and ends with an explanation of ModelView’s user experience.
2 Methods
2.1 Overview
Our ModelView software consists of two key parts: (1) a backend, run in advance, that collects data (described in 2.2 – 2.5), and (2) a frontend web app, run on user demand, that displays the data (described in 2.6).
Static analysis of a model is carried out by a Python program that performs a sequence of steps. First it downloads and uncompresses the model zip file from ModelDB. It then compiles and runs the model according to a model-specific manually-curated run protocol. After the first time step, the run is paused. A separate script is run that combines data from ModelDB, static analysis of the source code, and simulator introspection to generate a JavaScript Object Notation (JSON) file (Crockford 2006) which stores the data. Some models that are not originally designed for NEURON but that either are NeuroML models (e.g. Vervaeke et al 2010; modeldb.yale.edu/127996) or can be converted to NeuroML, can be supported by manually running neuroConstruct (Gleeson et al 2007) to first convert the NeuroML to NEURON and then employing the same scripts used for NEURON models.
Upon user request, the ModelView web app loads the JSON file, combines it with JSONP (a JSON variant that allows reading data from multiple servers) obtained from a ModelDB web service that identifies duplicate files across ModelDB, and displays it in the browser using NeuronWeb (McDougal, Flokos, Hines in preparation); NeuronWeb is a front-end under development in our lab to jQuery UI (jqueryui.com) – a Javascript library providing advanced GUI tools for the web – and other tools to facilitate the development of scientific web apps.
ModelView’s source code is available on ModelDB (modeldb.yale.edu/154872).
2.2 Generating the static data
The static data was generated from four sources: ModelDB, text-mining the source code, manual curation of run protocols, and simulator introspection. We consider each in turn.
2.2.1 ModelDB
The analysis script queries ModelDB to obtain the metadata associated with each model. This metadata is obtained from the main model page on ModelDB (ShowModel.asp); this page shows the model’s source code and metadata. To facilitate automatic parsing of the page, we modified this page to surround each piece of metadata with an HTML span tag labeled with an id attribute. The analysis script parses the HTML with BeautifulSoup (www.crummy.com/software/BeautifulSoup/) to extract the model title and zero or more paper Digital Object Identifiers (DOIs). Although every model in ModelDB is associated with at least one paper, some of the older papers do not have a DOI associated with them in our database. If no DOI is available, ModelView uses ModelDB’s link to the article on the journal’s website, and the analysis script displays a suggestion recommending additional curation.
2.2.2 Text Mining
The analysis script also downloads all of the model files and text mines the source code to identify information about NEURON mechanisms (channels, pumps, current clamps, etc…) defined in NMODL (Kohn et al. 1994). The model source tree is recursively searched for NMODL files, identified by their .mod extension. As such files are identified, they are scanned for POINT_PROCESS, SUFFIX, and USEION lines. The first two are used to determine the name of the mechanism, which immediately follows those tokens. USEION lines – there may be zero or more – are parsed with regular expression search patterns to locate the ionic attributes that they read or write (e.g. concentrations, currents, reversal potentials).
2.2.3 Manual Curation of Run Protocols
We manually curated one or more run protocols for each supported model. A run protocol corresponds to the set of instructions necessary to compile a model, launch NEURON, run a specific simulation within the model (some offer push buttons allowing the user to select one of many simulations), and perform any cleanup. Our analysis scripts run on a Linux machine using Python, so the compilation and cleanup instructions are written for the Linux shell, NEURON is launched in Python mode, and the run commands are in Python. For some models, the curation was accelerated by a script that scanned the source code to locate GUI control elements that potentially distinguish between different use cases.
2.2.4 Introspection
A script initializes and advances each model for one time step and then queries the simulator for information about the model’s structure. Compilation of mechanisms (ion channels, etc) is performed as specified in the run protocol. The NEURON libraries are imported inside a new Python process. The Python exec function is used to execute the commands one-at-a-time in the run part of the protocol, except that right before the last command is executed, a callback is passed to h.CVode().extra scatter gather. This method, originally developed for NEURON’s recent reaction-diffusion extension (McDougal et al. 2013) triggers the callback after every time step. In our case, the callback analyzes model structure and then exits the simulation instead of allowing it to advance to the next time step.
NEURON possesses introspection capabilities, that is, the ability to report what sections are present, what mechanisms are in those sections, and each mechanism’s parameters. In NEURON parlance, a “section” is an unbranched stretch of neurite (e.g. axon or dendrite); it is composed of “segments” which are the discrete compartments used for the numerical simulation. Indeed, NEURON’s own ModelView tool (Hines et al. 2007) uses these capabilities to display its structural analysis. For many parts of our analysis tree, such as the sections with homogeneous parameters, we began with the data from NEURON’s built-in ModelView; other parts, like the parameters for the mechanisms, were constructed de novo. The text from the tree was augmented with associated data from the source code or ModelDB: for example, the file name for each mechanism is identified from the text mining result and a link to the source code for that mechanism is provided by locating the file in ModelDB.
Graphics in ModelDB’s ModelView are generated independently of NEURON’s ModelView graphics. For those portions of the structure tree where the text is from NEURON’s ModelView, the analysis script parses the text to identify what information is to be presented.
Every graph involving a given cell had its segments processed in a consistent order. This consistency allows the segments to be identified once and have their names available to all graphs, and it allows interaction between multiple simultaneously presented graphs of the same phenomenon, as is available for mechanism parameter analysis (see section 3.2). The segments are followed by each section’s 0 end point and then by each section’s 1 end point. Generally, the 0 end of each child branch connects to the 1 end of the parent. In this case, the parent sections’ ends are the last points to be drawn; they are therefore on top and able to receive mouse events.
2.3 Encoding the static data
After the static data is collected, it is encoded in JSON (Crockford 2006) and stored on ModelDB’s server. The JSON object contains a number of attributes: neuron, title, short_title, neuronviewer, tree, colorbars, and modelview_version.
The JSON is structured to allow partial interoperability between versions of the viewer that are both newer and older than the analysis scripts. A modelview_version attribute allows newer viewers to identify old data files and respond appropriately; older viewers will ignore unknown attributes added in later versions of the data files.
The neuron attribute is a list of objects describing each neuron containing an identifier for each neuron (e.g. “root: soma”), the morphology (lists of lists of (x,y, z;d) points with a separate list for each NEURON segment), and a list of names for each segment. The section end points are included last in the morphology list to give them priority for mouse over events – events, such as displaying additional information, triggered when the mouse passes over an item, in this case the end point of a NEURON section. The neuronviewer attribute is a list of indices in the neuron list that are to get their own dialog; neuron indices may appear 0 or more times in this list.
The tree is a recursive data structure that defines a hierarchical view of the model structure. Each item in the tree list contains a text attribute and 0 or more optional fields: children, action, and noop. The children attribute stores a (sub)tree list. The action attribute lists objects identifying things that should be displayed when the row is selected. For example, action objects may call for displaying a neuron with a certain highlighting or rendering a 2D plot. An optional noop attribute indicates a non-selectable row.
2.4 Detecting stochastic models
The model analysis is run twice for each run protocol to detect models with stochastic features (e.g. random morphologies, random connections, random parameters). If the encoded data is not identical, a flag (“stochastic”) is set to true in the JSON. This ModelView web app (described in 2.6) displays an alert about stochastic features if this flag is set.
2.5 Generating the dynamic duplicate file data
To help place each model in context (see section 3.4), we added an automated step to ModelDB curation to allow the rapid identification of files reused elsewhere in the database and the locations of reuse. Our introduction of file duplicate checking was inspired by Channelpedia’s work-in-progress to mathematically classify ion channel models (Podlaski et al. 2014). Whenever a model is made public, currently about twice a week, a script runs that generates an MD5 (Rivest 1992) hash of each of the over 20,000 files in the database. i.e. Each file is reduced to a 128-bit number (the hash) in such a way that the same file will always generate the same number but that even slightly different files are likely to be associated with significantly different hashes. As they are generated, these hashes are stored in a Python dict, itself based on a hash table, where the hashes are keys and the values are lists of file paths with that hash. A JavaScript file is then output containing code to construct two associative arrays: one whose keys are file paths and values are hashes, and one whose keys are hashes and values are lists of files. These two associative arrays redundantly encode the same information, which allows O(1) lookup of both hashes and files at the cost of approximately doubling memory usage. This duplicate detection technique is independent of file type, and we have found it useful elsewhere in ModelDB, so we run it across all files for all models for all simulators in the database.
When ModelDB is updated, file reuse information can change. To avoid the complexities of modifying the JSON, file reuse data is loaded dynamically via a web service. Once loaded, this data is inserted into the tree. An include attribute on a given data tree row specifies the URL for dynamic data.
2.6 Visualizing the data
We wrote a custom web app to visualize hierarchically structured data that combines the strengths of many publicly available JavaScript libraries. This web app makes no assumptions about the simulator used to run the model. The ModelView page displays a loading message while it reads the location hash from the URL to determine what data to present. The ModelView scripts then request the model’s data (morphology, information tree, etc) from the server. One or more dialogs are created to display each neuron, a dialog is created to contain two-dimensional plots, and a dialog is created to contain the information tree. When a row is selected on the tree, the corresponding action is executed. Each action hides all of the non-tree dialogs and optionally displays one or more after updating their content. Mouseover events are captured and used to select associated hover text to display, if any.
ModelView generates its entire interface using JavaScript calls to an early version of NeuronWeb, a project from our lab to facilitate displaying scientific content in a web browser. NeuronWeb will be described in detail in a subsequent paper, but as used by ModelView, it provides a JavaScript API for dynamically constructing jQuery UI widgets (www.jqueryui.com), Flot charts (www.flotcharts.org), and jQuery Treeviews (www.github.com/jzaefferer/jquery-treeview). NeuronWeb provides a Neuron Viewer object that takes lists of segments of (x,y, z;d) information, projects it to a selectable two dimensional plane, and renders it with a Flot chart. Support for touch events for repositioning the dialogs is provided by jQuery UI Touch Punch (touchpunch.furf.com).
3 Usage
3.1 Getting started
To access ModelView, a user first locates a supported model of interest in ModelDB. At the time of this writing, 323 model entries support ModelView. A user who is only interested in supported models may indicate this on the search page.
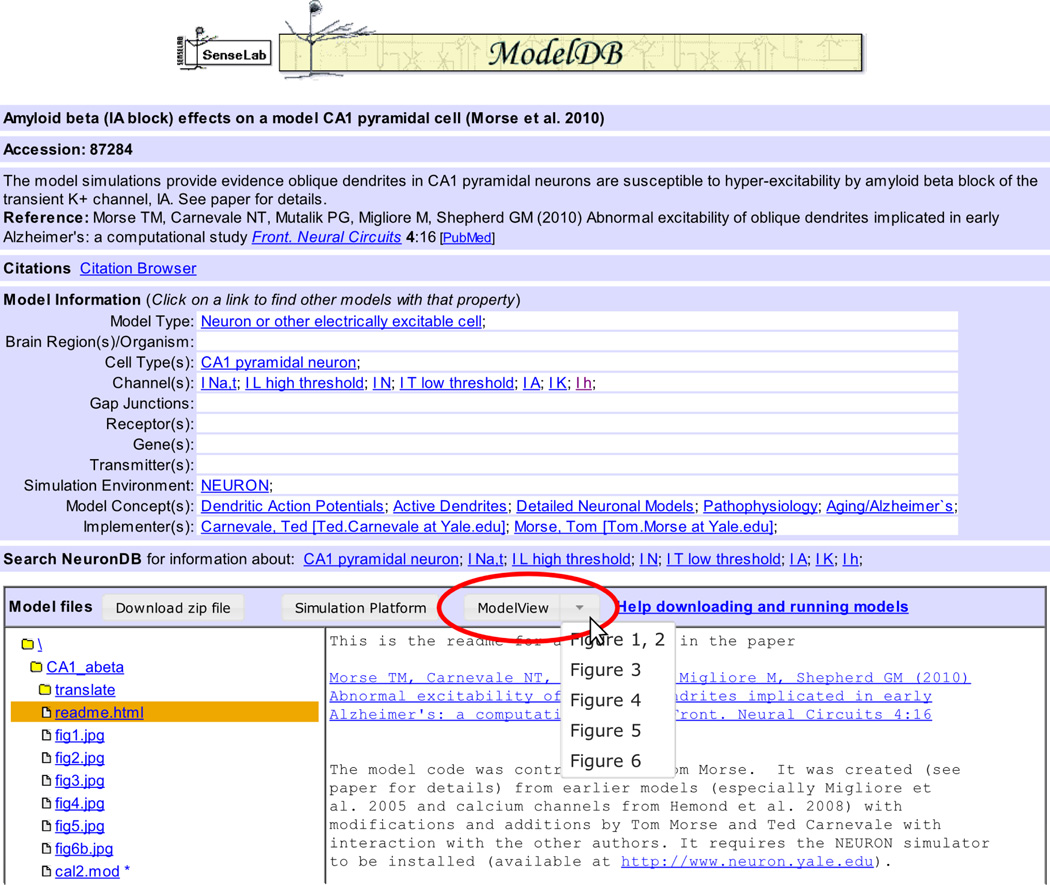
Opening a model displays the ShowModel page (Fig. 2). The top of the page provides metadata about the model as a whole. Below that section is a box labeled “Model files” which provides a file browser and displays the contents of the currently selected file. For ModelView supported models, a button labeled “ModelView” is provided. Clicking on that button launches the default view. The button is a stylized link, so right-clicking and selecting “open link in new tab” or browser-specific-equivalent also work. For those models with multiple associated run protocols, a down arrow appears next to the ModelView button (Fig. 2). Clicking this down arrow displays a drop-down list of associated run protocols; clicking on one of those launches Model-View for the corresponding protocol.
Figure 2.
To launch the ModelView web app from the file browser of a supported model, click the ModelView button (indicated by the oval). For models like this one with multiple run protocols, a down arrow appears next to the ModelView button. Clicking on the down arrow shows a list of the available protocols.
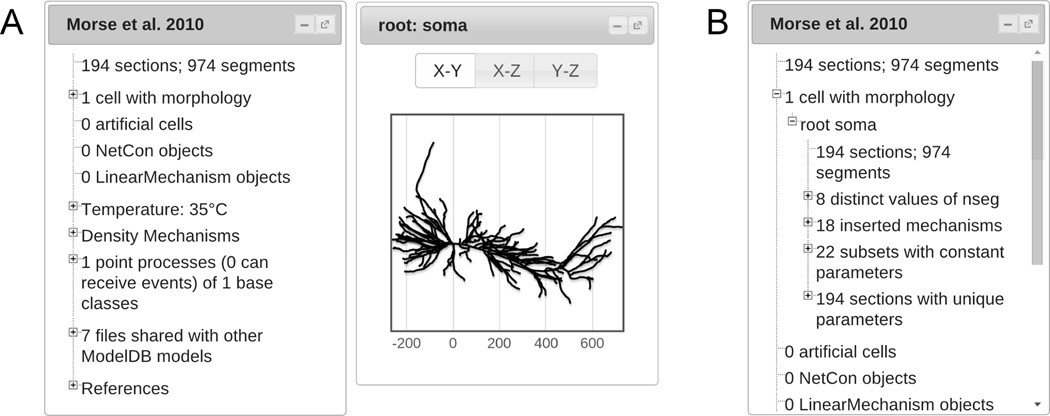
The ModelView web app opens to display a tree dialog inside the web browser and – if the number of neurons in the model is small – dialogs for each of the neurons (Fig. 3A). All dialogs displayed by the web app may be resized, moved, minimized or maximized within the browser. Selecting entries in the tree dialog controls what is displayed in the rest of the web app. Some rows open, close, or change other dialogs that display corresponding information about the model. At all times, the row, if any, corresponding to the information displayed in other open dialogs is highlighted. Although the details of the rows are model-dependent, the overall structure of the tree is independent of the specific model implementation; in particular, ModelView is unaffected by coding style or programming language (HOC or Python).
Figure 3.
(A) The initial ModelView window for the first run protocol for ModelDB entry 87284. (B) The information tree with “1 cell with morphology” and “root soma” expanded.
Cell-specific views of model structure are available by selecting the “1 cell with morphology” or – if the model contains more than one cell – “n cells with morphology” line. To select, simply click on the text. This action also opens a dialog for each cell, displaying cell morphology. Each morphology may be zoomed, panned, or viewed from a different angle. Hovering over a cell with the mouse pointer displays NEURON segment names (e.g. basal[25](0.5)) and (x,y, z) coordinates. To expand a section of the tree without changing the current view, click on the “+” icon to the left of the text. Once expanded, the contents may be collapsed by clicking on the parent header text or by clicking on the “−” icon to the left of the text; as before, the second approach does not affect any other dialogs currently displayed.
3.2 View by cell
Inside “n cell(s) with morphology”, there is a tree for each cell in the model, labeled by the cell “root” – typically the name of the cell’s soma (Fig. 3B). The first row lists the number of sections and segments in the cell.
The second row is a tree with information about the distribution of the number of segments (numerical compartments) per section. Selecting the first row inside that tree highlights the section with the greatest distance between electrical nodes and augments the mouse over text – text displayed when the mouse passes over a section in the tree – with information about the local value of dx. Other rows provide information about the number of segments relative to dlambda, the local electrical space constant at 100 Hz where capacitive current dominates the membrane current (Hines and Carnevale 2001).
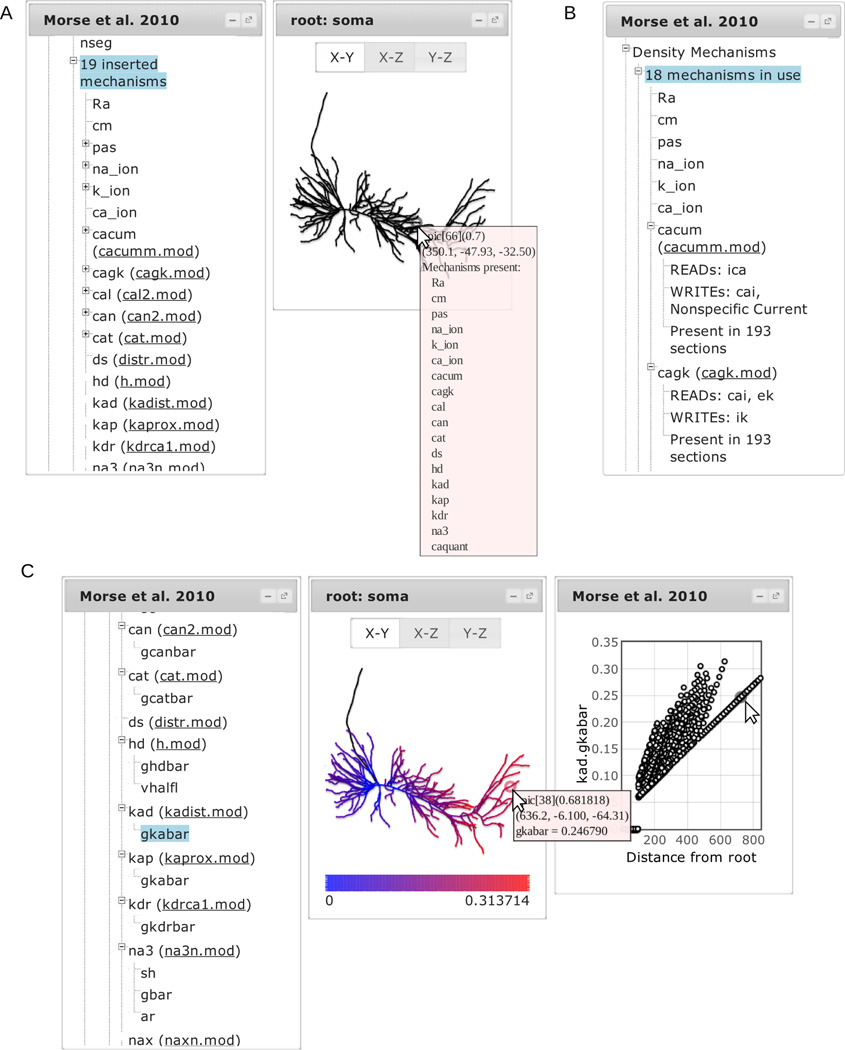
Selecting the third row of “n cell(s) with morphology” – “n inserted mechanisms” – lists each channel or ionic accumulation model inserted into the cell. Mouseover texts on the morphology viewer now include information on what mechanisms are present in the region under the mouse cursor (Fig. 4A). Selecting a mechanism highlights the portions of the cell containing that mechanism. When the mechanism is defined by a file, a link to the file appears next to the mechanism name in parentheses. Most mechanisms have associated subtrees listing their parameters. Selecting a parameter colorizes the morphology by parameter value and opens another dialog plotting the parameter value as a function of distance from the root. Mousing over a point in the value vs distance plot shows the name of the NEURON segment and (x,y, z) location. Mousing over the morphology plot shows this information along with the parameter’s value. The two graphs are synchronized; hovering the mouse pointer over a point on one graph highlights the corresponding point in the other (Fig. 4C).
Figure 4.
Exploring mechanisms. (A) Selecting a cell and then selecting inserted mechanisms lists all the mechanisms present within the cell. Mousing over the cell displays a popup listing the mechanisms that are located in the corresponding segment. (C) Selecting a mechanism displays its associated range variables; selecting a variable displays its values on the plots. (B) Selecting “Density Mechanisms” instead of a single cell lists all the density mechanisms in the model; selecting one shows what ionic states it reads or writes.
The fourth row, “n sections with constant parameters”, lists regions of the cell where one or more parameters is fixed for multiple sections. Selecting a region highlights it on the cell; its subtree lists what parameters are constant and what values they take. We found this analysis is useful to help identify logical units within the cell (e.g. basal dendrites).
The fifth row, “n sections with unique parameters”, conversely, lists sections that contain a parameter value or values not found elsewhere on the cell; selecting a section highlights it on the cell. A section’s subtree displays the unique parameter(s) and their value(s).
3.3 View by mechanism
NEURON has two main ways of representing ion channel and pump kinetics, and these are listed separately in ModelView.
3.3.1 Density mechanisms
NEURON uses “density mechanisms” to represent biological processes whose parameters or variables are best described in “density” units (e.g. current, ion flux, or number of ion channels per unit area). To explore the mechanisms, expand the “Density Mechanisms” tree.
The first subtree lists the number of unique density mechanisms in use across all cells. When a mechanism is associated with a file, a link to the file is provided in parentheses next to the mechanism name and a further subtree displays information about what, if any, concentrations and currents the mechanism reads or writes. The prevalence of the mechanism across the sections of the model is also provided (Fig. 4B).
A mechanism parameter may be either global (value independent of spatial location) or local (value may vary with position; also called “range variable”). These two possibilities are explored in the “Homogeneous Parameters” and “Heterogeneous Parameters” subtrees. Each row in the “Heterogeneous Parameters” subtree lists a parameter name and the number of unique values. If the number of unique values is small, each value is listed along with its frequency in a subtree. Global parameters are necessarily constant throughout the model and are listed by mechanism.
“KSChan definitions for density mechanisms” lists channels created with NEURON’s Channel Builder tool. A subtree for each of these channels lists the equations describing channel kinetics with further subtrees for gating variable dynamics. This method of specifying channel dynamics is not currently widely used in ModelDB models. For an example of how this information is presented, see the ModelView for modeldb.yale.edu/116740.
3.3.2 Point processes
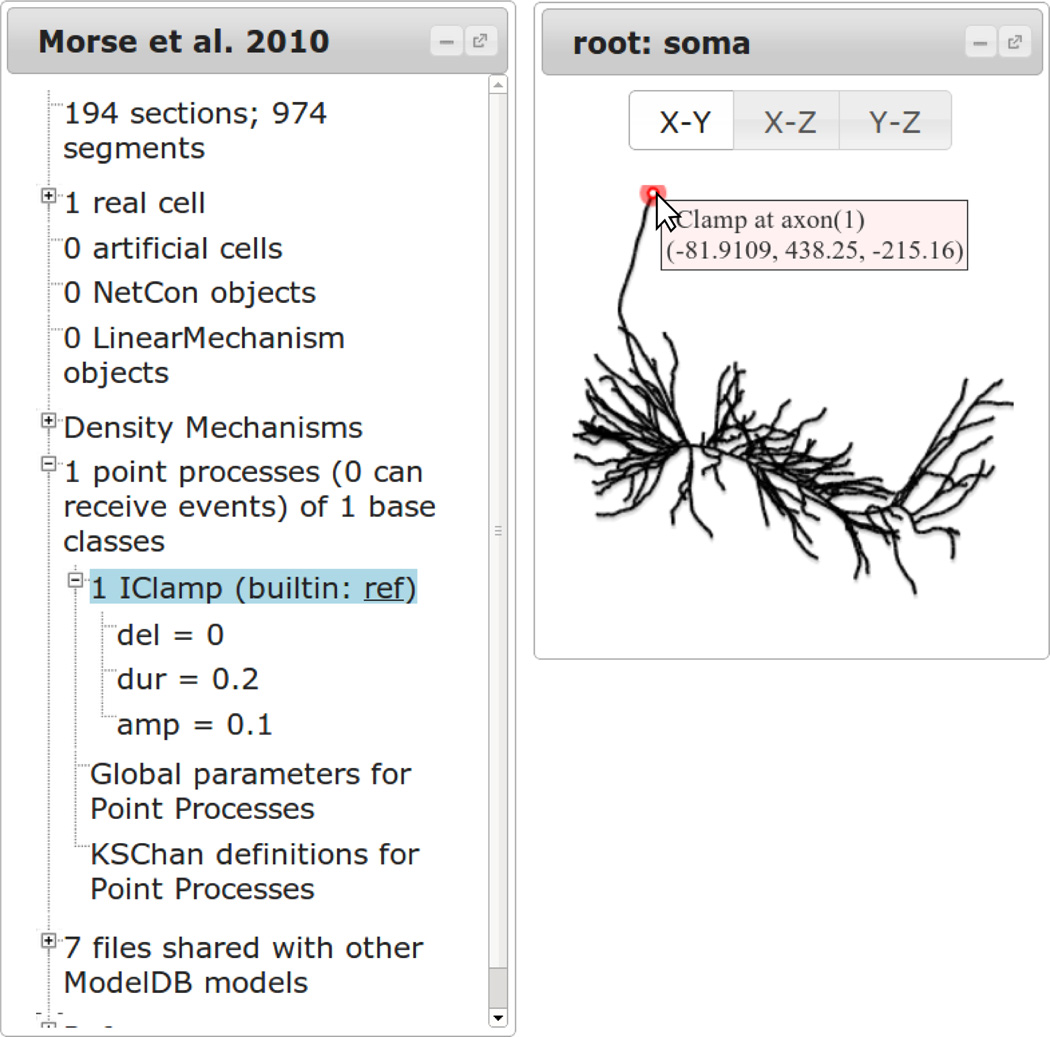
Point processes are mechanisms located at specific points on the neuron. These are used for modeling synapses and current clamps or for fine control of the placement of ion channels. These are listed under the “n point processes…” tree. If n > 0; then each class of point process has its own subtree, headed with an instance count, the name of the class, and a link to the source code for user-created point processes or to NEURON’s help documentation for built-in classes. The subtree lists the parameters used. Selecting the class row highlights the location(s) of the point process on the model’s neurons. Mousing over the location of the point process highlights the mouse over circle and displays the name of the point process and the location (Fig. 5). This allows clear identification of the point process locations while preserving the default mouse over behavior for each neuron.
Figure 5.
Selecting a type of point process highlights all of them on the neuron viewer. The exact location of a specific point process may be determined by hovering over the point (right). Parameter values are listed in the tree. For built-in point processes, a link is provided to the documentation; for user-created point processes, a link is provided to the source for that mechanism.
3.4 Context
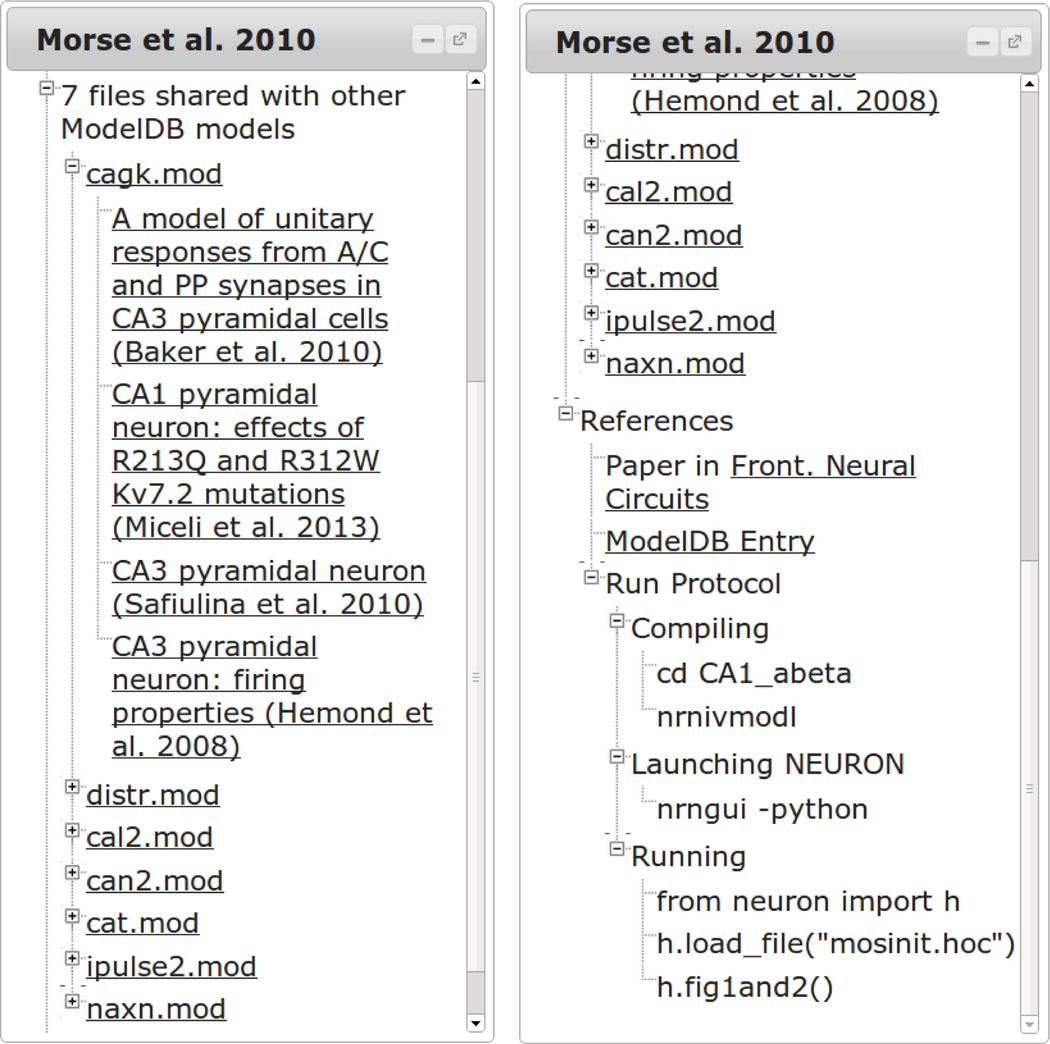
Expanding the “n files shared with other ModelDB models” (for n > 0) displays each file in the model that is used in some other model in the database. The filenames link to the file in the current model and head a subtree linking to the file in other models labeled by their ModelDB titles (left panel of Fig. 6). As of July 16, 2014, there were 2359 files repeated identically in the database. An A-type potassium channel model, kaprox.mod, is the most reused ion channel mechanism in the database, employed in 29 different published models. It first appeared in a model associated with Migliore et al 1999 (modeldb.yale.edu/2796).
Figure 6.
Understanding context. Links are provided to models reusing some of the same files (left). The References section provides links to the paper or papers that use a given model, the corresponding ModelDB entry, and step-by-step instructions for running the model in a way that produces the structure visualized in ModelView (right).
The “References” tree provides additional context. Links are provided to each paper known to use the current model. By policy, every model in ModelDB is associated with at least one publication. A link is also provided to the main ModelDB entry. The “Run Protocol” subtree lists steps for compiling the model, launching NEURON, and running the model in a way that produces the same structure visualized in Model-View. These steps are for NEURON compiled with Python support.
3.5 Additional information
The ModelView tool also provides information about additional NEURON features that are not specific to modeling individual neurons, including NetCon objects for network models, artificial spiking cells, and LinearMechanism objects for modeling gap junctions, electrical circuits, and more. If a model is identified as potentially being temperature dependent, the simulation temperature is reported in degrees celsius and any MOD files (mechanisms) that are potentially temperature dependent are listed.
4 Discussion
To facilitate the study of computational neuroscience models on ModelDB, we have developed a web-based analysis tool called ModelView. A single click transports the user from the default code presentation to a web app that allows a graphical exploration of the model’s structure. ModelView graphically presents the shape of the cell(s) involved, their ion channel distributions, the distribution of channel parameters and more.
By making this information more accessible, ModelView facilitates many types of research. Without installing a simulator or reading any code, users can see if a model incorporates features (e.g. shape, temperature, ion channels) that their research suggest are important. Knowing that information helps them to decide how much significance to give to the model’s results, to decide if the model is suitable as a base for further research. A model that clearly does not include effects that are believed to be significant presents a research opportunity, as the researchers could explore how incorporating their data would change the results. Those who prefer to work in a different simulator may use the ModelView summary to help them reimplement a model. We have developed a script, json_to_py.py, available on the ModelDB entry for ModelView (modeldb.yale.edu/154872) that can reconstruct many cells as NEURON Python implementations from their ModelView data. This script can be used as a template and a starting point for other automated model translators.
The json_to_py.py script also formed part of our strategy to address the question of validation. We took models from ModelDB and recreated models generated by the script, carefully applied the same experimental conditions, and compared the spike trains. Some care is necessary when setting up the experiments because many of the models on ModelDB do not have a clean separation in the code between describing the model and describing the experiment. Using the script and data obtained via the “Download Data” button at the bottom-right of the web app, interested users may conduct their own such validation tests.
Advanced users can interpret such downloaded data – it is encoded in JSON – as a form of a NoSQL database. It is encoded for presentation, but it is still structured, and the json_to_py.py script offers an example of how to extract quantitative data. We intend to eventually make much of the ModelView data searchable, but downloading the data necessarily offers at least as many exploration possibilities as we can offer. Alternatively, our code and in particular the list of model run protocols that we have curated can be used to explore the distribution of any discoverable aspect of the supported models, not just the ones included in ModelView.
The development of run protocols is an extension of ModelDB’s policy of including a mosinit.hoc file for each NEURON model to provide a standard entry point for running a simulation. Unlike mosinit.hoc, run protocols explicitly specify arbitrary shell-runnable compilation instructions. Furthermore, following the run protocol execution instructions requires no interaction with the NEURON GUI, which allows automated analysis of the supported subset of ModelDB.
The graphical exploration features of ModelView are made possible by an analysis script that is run when a model is first added to ModelDB. It initializes and runs the simulation for one time step to setup its structure. It then queries the simulator to identify morphology, mechanisms, parameters, etc, and augments this information with metadata from ModelDB and with the results of a static source code analysis.
ModelView introspects the “flat” and generally very large internal NEURON data that authoritatively defines a simulatable model and attempts to represent this data in as compact a form as feasible. Most variables are constants over large regions or even globally. For example, in a hundred compartment model, the hundred values for specific membrane capacitance are, much more often than not, a constant 1 µF/cm2 in all of the compartments. Parameters that vary with position on the cell are usually of particular interest and it is generally helpful to view these values as a function of distance from the soma. Generally, the compact ModelView analysis corresponds closely to the logical data hierarchies specified in the interpreter program and helps focus on “unusual” values whose causes can then be more easily determined from a targeted code inspection by normal text search. For example it is very common for a value to be specified early in a parameter file and then to be reset to a different value later in the code sequence to define a particular variant of a simulation run.
The analysis script is necessarily simulator specific, but as support for multi-simulator descriptions (e.g. NeuroML and PyNN) increases, this will become less of an issue. We initially focused on NEURON support because it is the most common simulator needed for models in ModelDB. We have since used NeuroConstruct to convert NeuroML to NEURON; this pathway allows us to support NeuroML. Tools that convert models from other simulators into NeuroML would provide a pathway for supporting those other simulators. Alternatively, we are open to collaborations with others to develop more simulator-specific analysis tools; advanced versions of json_to_py.py could be developed that would use ModelView as an intermediate step to converting to NeuroML.
Unlike many web apps, ModelView’s web presentation phase uses a non-fixed interface with dialogs that appear, disappear, and can be moved or resized. This design simplifies the interface as each dialog only needs to serve one function, allows future expandability as new features can be added without modifying the existing interface, and it allows the user to choose a layout which emphasizes the important parts for an analysis of interest. This flexibility is especially important for network models with many cells of only a few distinct types as the dialogs of the repeated cells can be minimized to make space to study the unique types.
We intend to continue developing ModelView. Increased interaction with this new tool will suggest new opportunities for introspection, for text-mining the source code, and for integration with databases. We currently have several specific plans for future enhancements: for example, we would like to improve support for network models. Each cell can currently be examined individually, but we intend to add support for aggregating them by type and for examining the network topology. We want to make the ModelView data searchable: for example, modelers interested only in point cell models have no way of restricting their searches to those models even though Model-View knows for each individual model if it only has point cells or not.
It is our hope that ModelDB’s ModelView will promote understanding and code reuse within the computational neuroscience community, and we envision that our approach can be adapted for other domains as well.
Information Sharing Statement.
The analysis, visualization, and inversion code for ModelView is available on ModelDB at: modeldb.yale.edu/154872. The JSON data is available from addresses of the form http://senselab.med.yale.edu/modeldb/getJsonFile.asp?file=VIEW_ID.json, where VIEW_ID is either the model accession number or the model accession number separated by an underscore and then an identifier (typically a small positive integer) distinguishing which of many simulations is to be viewed. The analyzed models are independently downloadable from modeldb.yale.edu. The set of all run protocols is at http://senselab.med.yale.edu/modeldb/run_protocols.json. These files may be downloaded anonymously; no login is required.
Acknowledgements
We thank the laboratory of GM Shepherd for valuable suggestions for improving ModelView’s usability, P. Miller, L. Marenco, and N.T. Carnevale for comments on the manuscript, and Nicole Flokos for her contributions to the NeuronWeb library. This research was supported by NIH T15 LM007056, NIH R01 NS11613, and NIH R01 DC009977.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Crockford, Douglas. The application/json media type for JavaScript Object Notation (JSON) 2006 [Google Scholar]
- 2.Davison AP, Brüderle D, Eppler J, Kremkow J, Muller E, Pecevski D, Perrinet L, Yger P. PyNN: a common interface for neuronal network simulators. Frontiers in neuroinformatics. 2008;2 doi: 10.3389/neuro.11.011.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davison AP, Mattioni M, Samarkanov D, Teleńczuk B. Sumatra: A Toolkit for Reproducible Research. In: Stodden V, Leisch F, Peng RD, editors. Implementing Reproducible Research. Boca Raton, Florida: Chapman & Hall/CRC; 2014. pp. 57–79. [Google Scholar]
- 4.Gleeson P, Steuber V, Silver RA. neuroConstruct: A Tool for Modeling Networks of Neurons in 3D Space. Neuron. 2007;54(2):219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gleeson P, Crook S, Cannon RC, Hines ML, Billings GO, Farinella M, Morse TM, Davison AP, Ray S, Bhalla US, Barnes SR, Dimitrova YD, Silver RA. NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail. PLoS computational biology. 2010;6(6):e1000815. doi: 10.1371/journal.pcbi.1000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hines ML, Carnevale NT. NEURON: a tool for neuroscientists. The Neuroscientist. 2001;7:123–135. doi: 10.1177/107385840100700207. [DOI] [PubMed] [Google Scholar]
- 7.Hines ML, Morse TM, Carnevale NT. Model Structural Analysis in NEURON. Methods Mol Biol. 2007;401:91–102. doi: 10.1007/978-1-59745-520-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohn MC, Hines ML, Kootsey JM, Feezor MD. A Block Organized Model Builder. Mathl and Comput Modeling. 1994;19:75–97. [Google Scholar]
- 9.McDougal RA, Hines ML, Lytton WW. Reaction-diffusion in the NEURON simulator. Front Neuroinf. 2013;7:28. doi: 10.3389/fninf.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migliore M, Hoffman DA, Magee JC, Johnston D. Role of an A-type K+ conductance in the back-propagation of action potentials in the dendrites of hippocampal pyramidal neurons. J Comp Neurosci. 1999;7:5–15. doi: 10.1023/a:1008906225285. [DOI] [PubMed] [Google Scholar]
- 11.Migliore M, Morse TM, Davison AP, Marenco L, Shepherd GM, Hines ML. ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics. 2003;1:135–139. doi: 10.1385/NI:1:1:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morse TM, Carnevale NT, Mutalik PG, Migliore M, Shepherd GM. Abnormal excitability of oblique dendrites implicated in early Alzheimer’s: a computational study. Front. Neural Circuits. 2010;4:16. doi: 10.3389/fncir.2010.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic acids research. 2006;34(suppl 1):D689–D691. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podlaski WF, Ranjan R, Seeholzer, Markram H, Gerstner W, Vogels T. Neuroscience 2013 Abstracts. San Diego, CA: Society for Neuroscience; 2013. Visualizing the similarity and pedigree of NEURON ion channel models available on ModelDB. Program No. 678.31. Online. [Google Scholar]
- 15.Rivest R. The MD5 Message-Digest Algorithm. RFC1321. Internet Engineering Task Force. 1992 [Google Scholar]
- 16.Usui S. Visiome: neuroinformatics research in vision project. Neural Networks. 2003;16(9):1293–1300. doi: 10.1016/j.neunet.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Vervaeke K, Lorincz A, Gleeson P, Farinella M, Nusser Z, Silver RA. Rapid Desynchronization of an Electrically Coupled Interneuron Network with Sparse Excitatory Synaptic Input. Neuron. 2010;67:435–451. doi: 10.1016/j.neuron.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]