Summary
Here we show that bivalent domains and chromosome architecture for bivalent genes are dynamically regulated during the cell cycle in human pluripotent cells. Central to this is the transient increase in H3K4-trimethylation at developmental genes during G1, thereby creating a “window of opportunity” for cell-fate specification. This mechanism is controlled by CDK2-dependent phosphorylation of the MLL2 (KMT2B) histone methyl-transferase, which facilitates its recruitment to developmental genes in G1. MLL2 binding is required for changes in chromosome architecture around developmental genes and establishes promoter-enhancer looping interactions in a cell-cycle-dependent manner. These cell-cycle-regulated loops are shown to be essential for activation of bivalent genes and pluripotency exit. These findings demonstrate that bivalent domains are established to control the cell-cycle-dependent activation of developmental genes so that differentiation initiates from the G1 phase.
Highlights
-
•
Bivalent domains are unstable, dynamic, and cell-cycle regulated
-
•
CDK2 phosphorylates MLL2 and establishes bivalent domains in G1
-
•
Chromosome remodeling in G1 is required for the “poised” pluripotent state
In this report, Dalton and colleagues show that developmental genes are primed for activation in G1 phase of the cell cycle by a mechanism requiring convergence of the cell-cycle machinery with cell signaling pathways. This priming mechanism involves the establishment of bivalent epigenetic domains and dynamic changes in chromosome architecture around developmental genes.
Introduction
Transcription start sites (TSSs) of developmentally regulated genes are frequently marked by overlapping domains of active (H3K4me3) and repressive (H3K27me3) histone marks. These “bivalent” domains are generally believed to be stable in self-renewing pluripotent stem cells (PSCs) and serve to establish a “poised” transcriptional state (Bernstein et al., 2006; Mikkelsen et al., 2007). During lineage specification, the bivalent state is resolved, allowing developmental genes to be activated or more stably repressed, depending on the lineage being specified. The molecular mechanisms underpinning these epigenetic changes are poorly understood but are ultimately regulated through the concerted action of histone methyl-transferases (HMTs) and histone de-methylases (Voigt et al., 2013). In PSCs, H3K4me3 is established through the activity of trxG complexes containing MLL or SET enzymes (Bledau et al., 2014; Denissov et al., 2014; Hu et al., 2013), while the PRC2 complex establishes domains of H3K27 trimethylation (Boyer et al., 2006; Lee et al., 2006). JARID1 is thought to be important for the erasure of H3K4me3 (Christensen et al., 2007), while de-methylation of H3K27me3 is controlled by the activity of JMJD3 and UTX complexes (Agger et al., 2007). Although significant effort has been placed on understanding the biochemical role of these HMT complexes, only limited information is available on how this network of epigenetic modifiers is controlled in the pluripotent state and how they poise cells during the initial stages of differentiation.
A growing amount of evidence indicates that PSCs initiate their differentiation program from the G1 phase of the cell cycle where they are most susceptible to specification cues (Chetty et al., 2013; Jonk et al., 1992; Mummery et al., 1987; Pauklin and Vallier, 2013; Sela et al., 2012; Singh et al., 2013; Singh and Dalton, 2009). Weak transcriptional activation of developmental regulators has also been described in the G1 phase of self-renewing PSCs, accounting for population heterogeneity, but importantly also indicates that developmental genes are susceptible to transient activation during a narrow window of time during the cell cycle (Singh et al., 2013). These studies therefore suggest that the G1 phase represents a “window of opportunity” that places PSCs in a poised state. Despite the link between cell-cycle position and responsiveness to differentiation signals, the molecular mechanism underpinning this is not well understood.
In this report, we show that the bivalent state is not stable in PSCs but highly dynamic and cell-cycle regulated, thereby establishing “lineage-poised” and “lineage-restricted” states in PSCs. The cell-cycle machinery directs recruitment of the MLL2 (KMT2B) HMT complex to developmental genes, allowing for bivalent domains to be transiently established during a narrow window of time during G1, thus explaining the true nature of the poised state and a rationale for why cells commit to differentiation from G1. A key aspect of developmental gene activation is the reorganization of chromatin following establishment of the bivalent domain and recruitment of functionally important enhancers. These findings presented here have broad implications for other multipotent cell populations.
Results
The Bivalent State Is Unstable, Dynamic, and Cell-Cycle Regulated
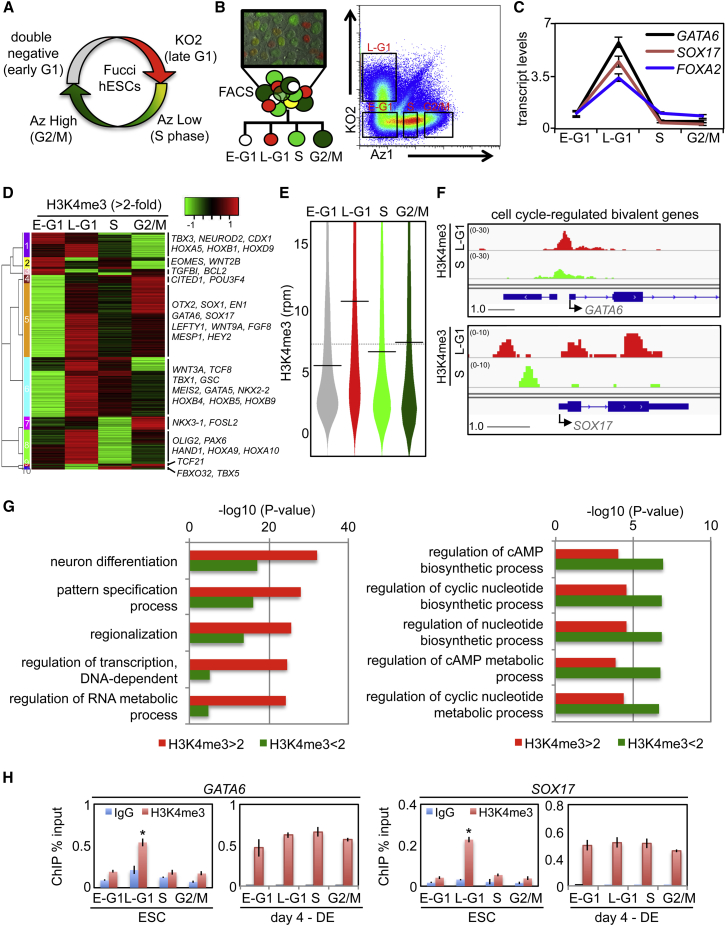
Combined with fluorescent-activated cell sorting (FACS), the Fucci system (Sakaue-Sawano et al., 2008) allows cell-cycle events to be monitored without physiological perturbation induced by synchronizing drugs (Figures 1A and 1B). By RNA sequencing (RNA-seq) analysis of FACS-isolated human embryonic stem cell (hESC) Fucci cell-cycle fractions, we previously showed that transcription of developmental genes representative of the three embryonic germ layers (ectoderm, definitive endoderm [DE], and mesoderm) peaks during late G1 (Singh et al., 2013). This peak of transcription continues during the early stages of DNA replication but declines as cells transition through S phase and remains low for the remainder of the cell cycle (Figure 1C) (Singh et al., 2013). Since bivalent domains of histone modifications are known to regulate developmental genes in PSCs by placing them in a poised state (Bernstein et al., 2006; Mikkelsen et al., 2007), we reasoned that they could be subject to cell-cycle control, thus explaining why many of these genes display transcriptional periodicity. To assess this possibility, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis of H3K4me3 and H3K27me3 on Fucci hESC cell-cycle fractions (Tables S1 and S2). Approximately half of all bivalent genes (Sharov and Ko, 2007) show cell-cycle-linked periodicity for H3K4 trimethylation (1,029 of 2,095). Approximately 90% of these genes show increased H3K4me3 as cells transition through late G1 and perform roles primarily associated with developmental processes (Figure S1A). Consistent with RNA-seq data (Singh et al., 2013), H3K4me3 levels for bivalent genes that associated with specification of all three embryonic germ layers peaked in late G1 and declined during S phase (Figures 1D–1F and S1B). The residual group of bivalent genes displays no transcriptional periodicity or cell-cycle-dependent H3K4me3 and performs roles in non-developmental processes such as metabolism (Figures 1G, S1B, and S2A). Pluripotency regulators (OCT4, NANOG) and house-keeping genes (ADCY4, ESPN, GRM2) are constitutively H3K4 trimethylated in regions proximal to their transcription initiation sites (Figure S1B), and their transcripts show no periodicity during the cell cycle (Figure S1C) (Singh et al., 2013). ChIP-seq of H3K27me3 showed that this mark is not cell-cycle regulated (Figures S2B and S2C; Table S2). We validated the ChIP-seq data using quantitative ChIP-PCR (qChIP) for GATA6 and SOX17 (Figures 1H and S2D). Finally, we decided to examine how rapidly the loss of developmental transcripts and H3K4me3 occurs upon exit from G1 (Figures S2E and S2F). By evaluating early S-phase cells (i.e., those cells double positive for Az1 and KO2), we find that the loss of H3K4me3 precedes the loss of transcript levels, indicating a role for transcript degradation as cells exit the poised state. In summary, bivalent genes associated with developmental processes tend to be transcribed during the late G1 phase in hPSCs—this is correlated with increased H3K4 trimethylation in the domain proximal to TSSs. Bivalent domains are therefore primarily established in G1 and far more dynamic than previously believed. This challenges the conventional view of what bivalent modifications represent, how they are established and maintained, and what the poised pluripotent state is.
Figure 1.
H3K4 Trimethylation Is Cell-Cycle Regulated at Bivalent Domains of Developmentally Regulated Genes
(A) Diagram of the Fucci reporter system established in WA09 hESCs.
(B) The Fucci system can be used to isolate cell-cycle fractions in live hESC cells by FACS.
(C) qRT-PCR of Fucci-sorted cells exhibit periodicity of transcription for developmental genes. Data are the average of technical triplicates and representative of more than ten independent experiments.
(D) ChIP-seq of bivalent genes with cell-cycle-regulated H3K4me3 (>2-fold change between any two fractions) represented as a heatmap. Representative, developmental genes belonging to different cell-cycle-regulated clusters are shown. ChIP-seq data are from the pool of three independent experiments.
(E) Bean-plot diagram showing distribution of H3K4me3 levels for cell-cycle-regulated bivalent genes. The black horizontal line represents median values. Late G1 was significantly higher than S (p < 1 × 10−14). Statistical analyses are described in ChIP-seq data and are from the pool of three independent experiments.
(F) H3K4me3 ChIP-seq profiles of hESC Fucci fractions for GATA6 and SOX17. Scale bar indicates genomic distance in kb and value range given in RPM. ChIP-seq data are from the pool of three independent experiments.
(G) GO analysis comparing cell-cycle-regulated bivalent genes and non-cell-cycle-regulated bivalent genes.
(H) H3K4me3 qChIP using Fucci-isolated cell-cycle fractions from ESCs or DE at the GATA6 or SOX17 promoters. Data are the average of three independent replicates.
∗p < 0.05. Error bars in this figure represent the SEM. See also Figures S1 and S2 and Tables S1 and S2.
MLL2 Activity Is Cell-Cycle Regulated and Dynamically Regulates Bivalent Domains
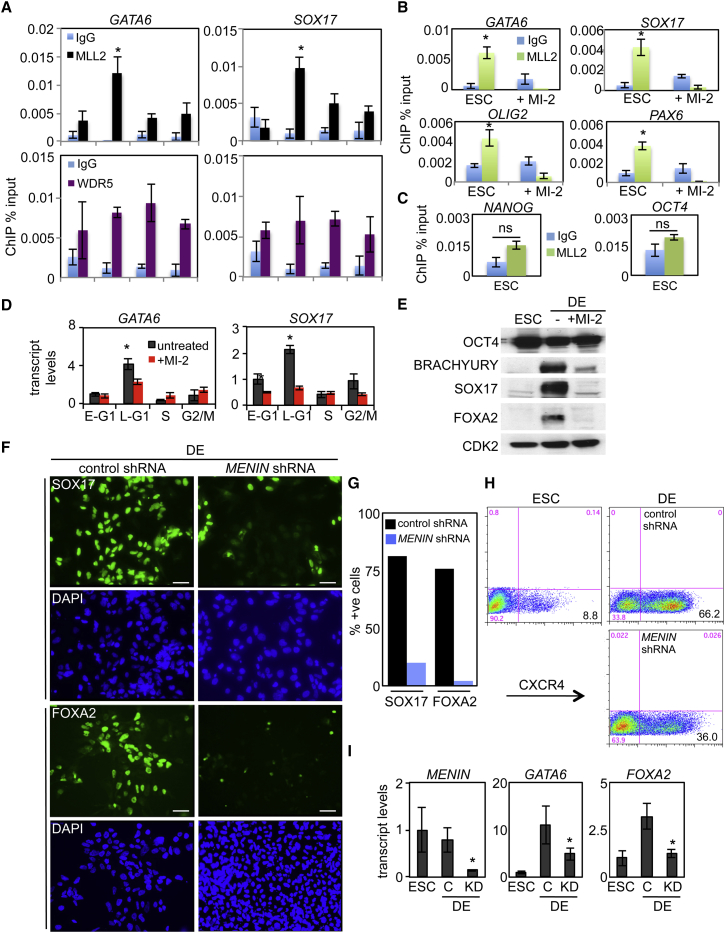
MLL2-containing HMT complexes are primarily responsible for H3K4me3 deposition at bivalent genes in ESCs (Denissov et al., 2014; Hu et al., 2013). MLL2 assembles into complexes with MENIN, which functions by recruitment of MLL1/2-specific complexes to chromatin and with other subunits such as WDR5 that are shared by other SET-family HMT complexes (Schuettengruber et al., 2011). To establish whether cell-cycle-dependent MLL2 activity could account for transcriptional periodicity and deposition of H3K4me3 during G1, we tested whether its recruitment to developmental genes was cell-cycle regulated. By qChIP, we found that MLL2 was significantly enriched at GATA6 and SOX17 promoters in late G1 (Figure 2A), coinciding with increased H3K4me3 and transcript levels (Figures 1C–1F), but showed no periodicity in binding to non-cell-cycle-regulated bivalent genes (Figure S1D). Other components of the MLL2-HMT complex such as WDR5 display no periodicity in binding activity (Figure 2A).
Figure 2.
The MENIN/MLL2 H3K4 Methyl Transferase Is Cell-Cycle Regulated and Controls the Activation of Bivalent Genes in G1
(A) qChIP of Fucci hESC cell-cycle fractions examining levels of MLL2 and WDR5 on the GATA6 and SOX17 promoters. Data are the average of three independent replicates.
(B) qChIP assays for MLL2 in untreated or MI-2 (25 μM for 24 hr)-treated WA09 ESCs at the indicated promoters. Data are the average of three independent replicates.
(C) qChIP assays for MLL2 in ESCs of pluripotency genes. Data are the average of three independent replicates.
(D) qRT-PCR transcript analysis of Fucci-sorted hESCs after treatment with the MLL/MENIN inhibitor, MI-2 (25 μM for 24 hr). Data are the average of three independent replicates.
(E) Immunoblot analysis of WA09 hESC and DE (2 days) lysates (20 μg per lane) with or without MI-2 (25 μM).
(F) Immunostaining for SOX17 (top) and FOXA2 (bottom) following the infection of GFP-control or MENIN shRNA lentivirus in WA09 hESCs or cells differentiated to DE for 3 days. Cells are co-stained with DAPI to visualize nuclei. Micron bar represents 50 μm.
(G) Quantitation of immunostaining represented in (E) for three independent fields, n > 1,000. Data are representative of three independent experiments.
(H) CXCR4 flow cytometry analysis of WA09 hESCs and DE (3 days differentiation) transduced with MENIN or GFP shRNA lentivirus. The percentage of CXCR4+ cells in each condition is indicated.
(I) qRT-PCR transcript analysis of WA09 hESCs and DE (3 days differentiation) following lentiviral infections with MENIN or GFP (control [C]) shRNA. Data are the average of three independent replicates.
∗p < 0.05. Error bars in this figure represent the SEM. See also Figure S3.
The small molecule MI-2 (Grembecka et al., 2012) blocks the MENIN-MLL2 interaction and as anticipated reduced recruitment of MLL2 to developmental genes such as GATA6, SOX17, PAX6, and OLIG2 (Figure 2B). MLL2 did not bind non-bivalent genes such as NANOG or OCT4 (Figure 2C). Next, we evaluated the role of MLL2/MENIN in ESCs and during DE differentiation. Here, MI-2 blocked the cell-cycle regulation of GATA6 and SOX17 (Figure 2D), along with the upregulation of developmental genes (GATA6, GATA4, FOXA2, SOX17), whereas non-endoderm genes such as NANOG, SOX2, FOXD3, CD9, and REX1 were unchanged or slightly upregulated (Figure S3A). Inhibition of the MENIN-MLL2 complex also blocked the upregulation of BRACHYURY, SOX17, and FOXA2 protein (Figure 2E). Recent studies indicate that loss of MLL2 may be compensated for by MLL1 (Denissov et al., 2014), so to circumvent this potential problem, we blocked MLL1/2 function by reducing levels of their common HMT-binding partner, MENIN. As anticipated, shRNA knockdown of MENIN leads to a loss of H3K4me3 on developmental genes (Figure S3B). Reduction of MENIN activity prevented the normal upregulation of protein levels for CXCR4, SOX17, and FOXA2, along with endoderm transcripts (GATA6 and FOXA2) (Figures 2F–2I). Collectively, these data indicate that MENIN-MLL2 HMT complexes are required for lineage priming through regulation of bivalent domains in PSCs and then for activation of the same developmental genes under differentiation conditions.
The Cell Cycle Controls MLL2 Recruitment to Bivalent Genes
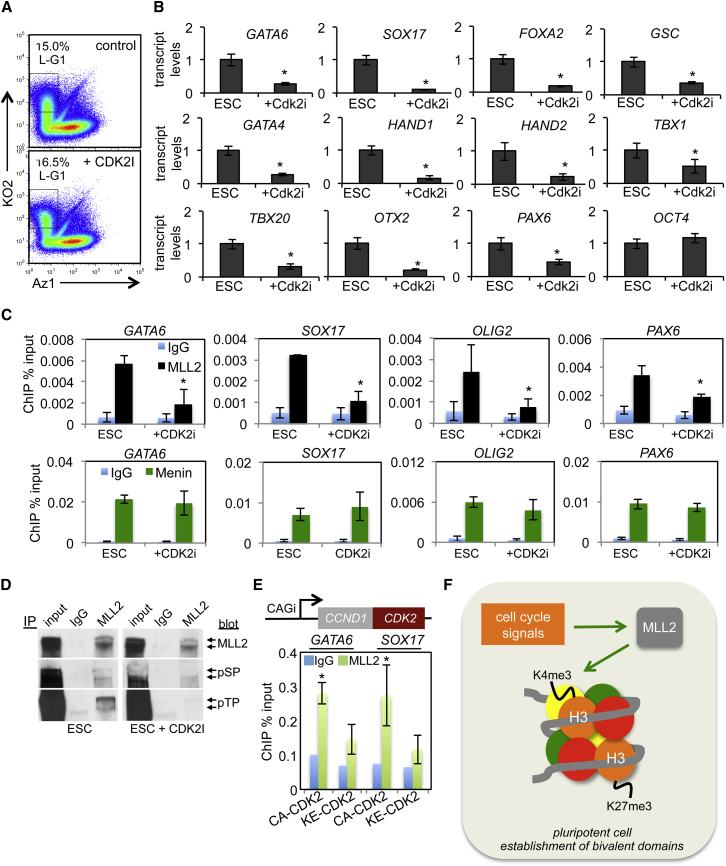
The accumulation of H3K4 trimethylation at bivalent domains and a parallel increase in transcription of these genes suggest a role for cyclin-dependent protein kinase (CDK) activities during the late G1 to early S-phase period. The most likely candidate was CDK2 because its activity is most closely associated with transition through late G1 phase and early S phase in cycling cells (Bertoli et al., 2013). To determine whether CDK2 activity was required for MLL2 recruitment to bivalent genes, we added the inhibitor CVT-313 (Brooks et al., 1997) for a brief period (4 hr) so as not to impose a cell-cycle block, and no major perturbations in cell-cycle progression were observed during this time (Figure 3A). The addition of CVT-313 suppressed developmental transcripts had no effect on OCT4 transcripts or transcripts for non-cell-cycle-regulated bivalent genes, and significantly reduced MLL2 recruitment to developmental genes (Figures 3B, 3C, and S3C). MENIN binding, however, was not affected by CDK2 inhibitor treatment (Figure 3C).
Figure 3.
CDK2 Phosphorylates MLL2 to Regulate Its Binding
(A) Cell-cycle Fucci profiles of untreated and CDK2 inhibitor (CDK2I, CVT-313, 20 μM) treated cells for 4 hr.
(B) qRT-PCR analysis of transcript levels in ESCs treated with CVT-313 for 4 hr. Data are the average of three independent replicates.
(C) qChIP for MLL2 and MENIN in ESCs treated with CVT-313 for 4 hr. Data are the average of three independent replicates.
(D) MLL2 immunoprecipitations (IPs) from ESC lysates (200 μg protein; ±CVT-313) were probed with MLL2, phospho-threonine (pTP), or phospho-serine (pSP) antibodies. IgG was used as an IP control, and whole cell lysate was immunoblotted (input) alongside IPs.
(E) ESCs were transfected with a construct expressing constitutively active (CA) or inactive (KE mutant) CDK2-CCND1 gene fusion driven by CAGi promoter, and then qChIP assays for MLL2 were performed 36 hr later for GATA6 and SOX17. Data are the average of three independent replicates.
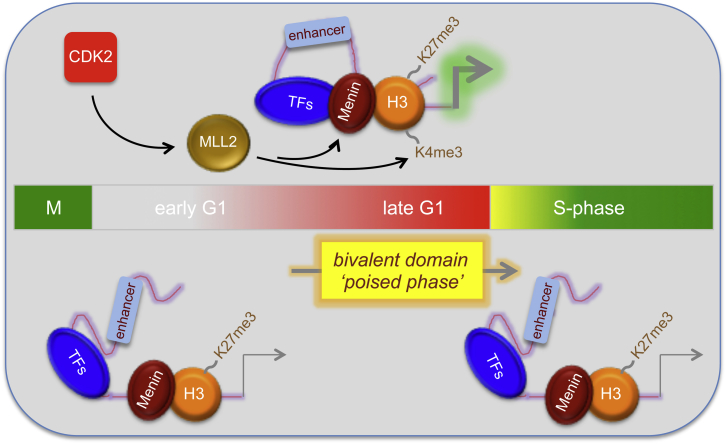
(F) Model depicting the cell-cycle control of bivalent domains. All data are representative of biological replicates.
Error bars in this figure represent the SEM. ∗p < 0.05.
Previous studies seeking to identify CDK2 substrates discovered that MLL2 is phosphorylated by CDK2 on threonine-5099 (Badeaux and Shi, 2013; Chi et al., 2008). Immunoprecipitation of MLL2, followed by immunoblotting for phospho-serine and phospho-threonine residues, in the presence or absence of the CDK2 inhibitor, CVT-313, confirmed that MLL2 is a bona fide CDK2 target (Figure 3D). Since MLL2 binding is stable on non-cell-cycle-regulated bivalent genes (Figure S1D), we reasoned that CDK2 may phosphorylate MLL2 to facilitate its binding only for cell-cycle-regulated bivalent genes. In support of this, we find that CDK2 specifically binds to developmental gene promoters (Figure S3D). Finally, a constitutively active version of CDK2 was expressed in hESCs and its effects compared with that of an inactive mutant (Chytil et al., 2004). Here, increased CDK2 activity promoted recruitment of MLL2 to GATA6 and SOX17 promoters compared with the control (Figure 3E). We conclude that CDK2 signals through MLL2 to establish H3K4me3-marked bivalent domains in G1 (Figure 3F). This explains the small pulse of G1-specific transcription of developmental genes described previously (Singh et al., 2013).
Bivalent Genes Undergo Chromatin Remodeling during the Cell Cycle to Establish Promoter-Enhancer Interactions
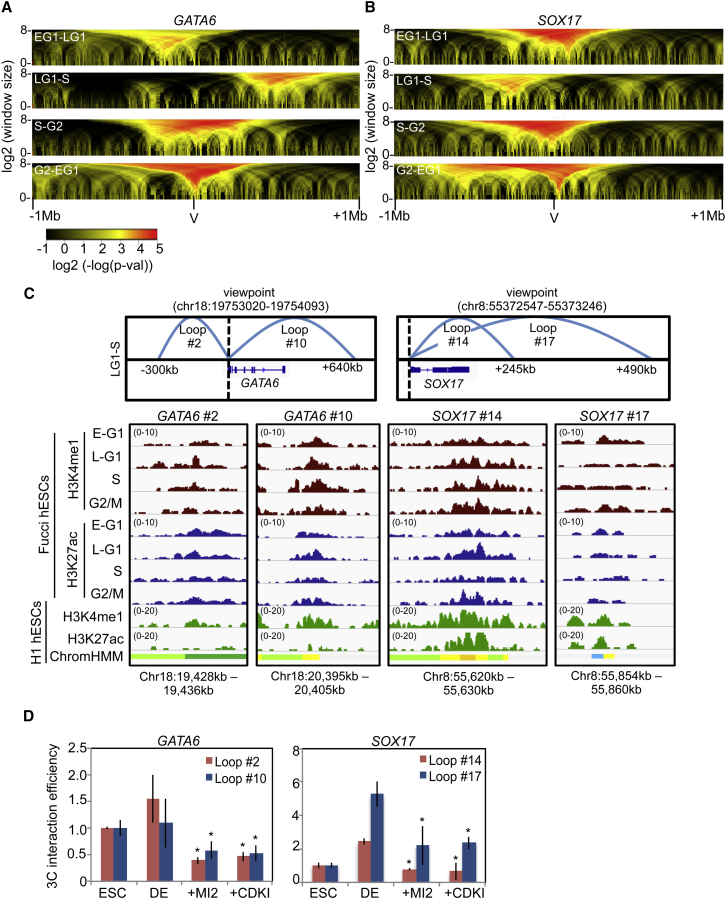
Since the epigenetic status of a gene is often closely correlated with its chromatin structure, we asked whether chromosome dynamics around developmental genes change in the cell cycle and whether this is regulated by changes in H3K4 trimethylation status at bivalent domains. This was approached by circular chromatin conformation capture sequencing (4C-seq) assays (Gheldof et al., 2012; Splinter et al., 2012) in the four Fucci cell-cycle fractions. Focus was directed toward GATA6 and SOX17 because these two genes have cell-cycle-regulated transcription and H3K4me3 patterns. For this analysis, we used TSSs for GATA6 and SOX17 as “viewpoints” and analyzed regions spanning 1 Mb upstream and downstream. This generated “domainograms” to provide a visual representation of regional chromosome architecture (Figures 4A and 4B). Although most chromatin structures around these genes remain intact during the cell cycle, several changes were identified by subtractive analysis of 4C data plots (Figures 4A, 4B, and S4A–S4D). Interestingly, several chromatin structures were established as cells transitioned from early G1 to late G1 and then lost as cells transition through S phase. These cell-cycle-regulated chromatin loop structures were validated by chromatin conformation capture qPCR (3C-qPCR) in separate biological samples (Figure S4E).
Figure 4.
Functional Chromosome Architectural Changes Occur around Bivalent Genes in G1
(A and B) Domainograms from 4C-seq analysis of GATA6 (A) and SOX17 (B) depicting subtractions between cell-cycle fractions as indicated, with 1 Mb regions upstream and downstream of the viewpoints (V) (GATA6 chr18:19,753,020-19,754,093; SOX17, chr8:55372547-55373246).
(C) Spider-plot diagrams are shown for two interaction loops (with approximate distances from the viewpoint) that map to putative enhancers at GATA6 or SOX17 loci (top). ChIP-seq profiles for H3K4me1 and H3K27ac in Fucci cell-cycle fractions and from ENCODE data in WA01 (H1) hESCs for putative enhancer loops identified (bottom). The ChromHMM track identifies chromatin features based on the local epigenetic landscape (green, transcribed region; yellow, weak enhancer; orange, strong enhancer; and blue, insulator). Range is given in RPM.
(D) 3C-qPCR examining cell-cycle-regulated interaction loops in WA09 ESCs and DE (3-days differentiation) following treatment with MI-2 (25 μM) for the final 24 hr or CVT-313 (20 μM) for the final 4 hr. Data are normalized to ESC levels. Data are the average of three independent replicates.
∗p < 0.05. Error bars in this figure represent the SEM. See also Figure S4 and Table S3.
To characterize the functional significance of G1-specific chromosome structures, we asked whether loop contact regions overlap with known gene regulatory elements using data provided by the NIH Roadmap Epigenomics Project (http://www.roadmapepigenomics.org), along with chromatin state dynamics analysis in hESCs (Ernst et al., 2011). Several interaction sites were identified that mapped to enhancer elements, based on histone modification profiles (Table S3). ChIP-seq for H3K27ac and H3K4me1 enhancer marks was then performed to independently evaluate the epigenetic landscape around G1-specific structures in the four Fucci cell-cycle fractions (Figure 4C). Specific focus was on loops that form in G1 phase at the GATA6 and SOX17 loci (Figures 4C and S5C–S5E). These data show that cell-cycle-regulated architectural changes facilitate interactions between bivalent promoters with active enhancers in late G1. Enhancer histone marks do not appear to vary with cell-cycle position at enhancers or promoters (Figures 4C and S4F), indicating that they are not responsible for architectural changes around bivalent genes in G1.
Studies so far have focused on the bivalent, poised pluripotent state where developmental genes are transiently activated and domains of H3K4 trimethylation are temporarily established in late G1. We then asked whether these loops that showed enrichment of histone marks associated with enhancers are relevant under conditions when developmental genes are activated during lineage-specific differentiation. To investigate whether cell-cycle-regulated loops at GATA6 and SOX17 loci also form during DE differentiation, we performed 3C-qPCR analysis. This analysis showed that cell-cycle-regulated loops identified in PSCs were maintained at these genes as they become fully activated during DE differentiation (Figure 4D). Continued MENIN/MLL2 activity and CDK2 activity are required for maintenance of these loops as brief exposure to MI-2 and CDK inhibitor erased these architectural structures.
Cell-Cycle-Dependent Recruitment of Enhancers Is Required for Activation of Bivalent Genes
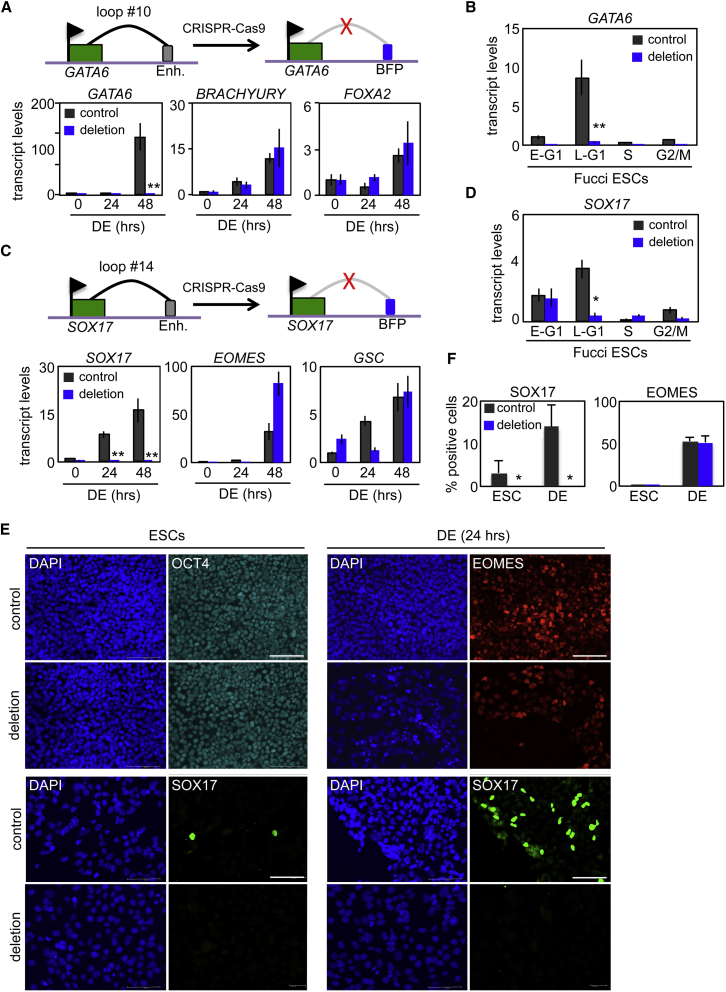
4C-seq identified enhancers that interact in a cell-cycle-regulated manner with GATA6 and SOX17 bivalent promoters. To demonstrate that these enhancers are important for gene activation and differentiation, we utilized CRISPR technology to delete two of these elements. By using two CRISPR guide RNAs, along with a targeting vector consisting of 1 kb homology arms and a reporter cassette containing a CAGi promoter driving BFP-IRES-Zeocin, we deleted the enhancer for GATA6 that corresponded to interaction from loop #10 in Fucci hESCs (Figure S5). Deletion of this enhancer blocked the upregulation of GATA6 transcription under differentiation conditions, while other genes such as BRACHYURY and FOXA2 were activated as expected (Figure 5A). Using FACS-sorted hESC Fucci fractions, the specific loss of GATA6 transcripts in late G1 was confirmed (Figure 5B).
Figure 5.
Cell-Cycle-Dependent Recruitment of Enhancer Elements Are Required for the Activation of Bivalent Genes
(A) Organization of the GATA6 locus around a region displaying cell-cycle-dependent changes in chromosome architecture. The specific loop structure (loop #10), its associated enhancer region, and CRISPR-Cas9-directed disruption of a 1 kb region encompassing the enhancer and the resulting insertion of a BFP reporter cassette are indicated (top). qRT-PCR transcript analysis of WT (control) and genome-edited (deletion) ESCs was performed over the first 48 hr of differentiation toward DE for indicated genes (bottom).
(B) Analysis of GATA6 transcript levels during the cell cycle in Fucci ESC cell-cycle fractions from WT (control) and genome-edited (deletion; loop #10) cells. Data are the average of three independent replicates.
(C) Organization of the SOX17 locus around a region displaying cell-cycle-dependent changes in chromosome architecture. The specific lop structure (loop #14), its associated enhancer region, and CRISPR-Cas9-directed disruption of a 1 kb region encompassing the enhancer and the resulting insertion of a BFP reporter cassette are indicated (top). qRT-PCR transcript analysis of WT (control) and genome-edited (deletion) ESCs was performed over the first 48 hr of differentiation toward DE for indicated genes (bottom). Data are the average of three independent replicates.
(D) Analysis of SOX17 transcript levels during the cell cycle in Fucci ESC cell-cycle fractions from WT (control) and genome-edited (deletion; loop #14) cells. Data are the average of three independent replicates.
(E) Immunostaining of WT (control) and genome-edited (SOX17 locus; loop #14) cells under self-renewal (ESCs) or differentiation (DE, 24 hr) conditions.
(F) Quantitation of immunostaining in (E) from three separate fields of view for each antibody (n > 1000). Micron bar is 100 μm. Data are representative of three independent experiments.
∗p < 0.05, ∗∗p < 0.01. Error bars in this figure represent the SEM. See also Figure S5.
Using a similar approach, an enhancer that forms part of a cell-cycle-dependent loop at the SOX17 locus was also disrupted (Figure S6). Disrupting this enhancer blocked SOX17 transcription during endoderm differentiation, while other endoderm genes such as EOMES and GSC were activated with typical kinetics and magnitude (Figure 5C). In addition, we confirmed that the loss of SOX17 transcripts was specific to the late G1 phase by sorting Fucci fractions in hESCs (Figure 5D). Finally, we showed that cells lacking an intact, cell-cycle-regulated enhancer at the SOX17 locus failed to accumulate SOX17 protein during DE differentiation (Figures 5E and 5F). In contrast, EOMES expression was unaffected. These data demonstrate that cell-cycle-dependent chromosome architectural changes are critical for activation of developmentally regulated genes. Bivalent marks regulated by CDK-dependent signals are an absolute requirement for architectural changes and for normal transcriptional activation during differentiation. An overall model depicting our results is shown in Figure 6.
Figure 6.
Model Depicting How the Cell Cycle Regulates Bivalent Domains and Chromatin Structure during Self-Renewal and Differentiation of PSCs
Discussion
In 2006, Bernstein and colleagues showed that developmental genes are held in a poised chromatin state by domains of overlapping H3K4me3 and H3K27me3 histone marks proximal to TSSs. The basic assumption of this model was that in an H3K4me3-marked state, repressive domains serve to silence developmental genes. Following exposure to specification signals, these genes then become rapidly activated following erasure of the H3K27me3 domain. Recently, however, we showed that instead of being silent, bivalently marked genes are transcribed during the late G1 phase in human PSCs (Singh et al., 2013). In this report, we provide a mechanism for this by showing that H3K4me3 domains are only established at developmental genes in G1 phase and that this is due to cell-cycle-dependent recruitment of the HMT complex subunit MLL2. These observations establish that bivalent domains are highly unstable and form only during a narrow window of time during the cell cycle. Moreover, they show that PSCs alternate between the poised and lineage-restricted states.
The brief activation of developmental genes in late G1 phase explains why PSCs are prone to initiate differentiation from this period of the cell cycle (Chetty et al., 2013; Jonk et al., 1992; Mummery et al., 1987; Pauklin and Vallier, 2013; Sela et al., 2012; Singh et al., 2013), but the question of why G1 is special in terms of cell-fate decisions is not immediately obvious. The most likely explanation is that a “permissive” chromatin environment is established in G1 allowing for transcription factor loading and chromatin remodeling immediately after chromosome segregation but before replication (Egli et al., 2008). This is supported by observations showing that de novo chromatin domains are preferentially established during G1 phase of the cell cycle (Thomson et al., 2004). Data in this report provide the first detailed evidence at the molecular level to support this model.
While the majority developmental transcription factors display periodicity for H3K4me3 on their gene promoters, some such as BRACHYURY, do not. Similarly, BRACHYURY transcript does not oscillate during the cell cycle (Singh et al., 2013). Further studies will be needed to understand how this small subset of developmental genes and the remaining non-cell-cycle-regulated genes involved in metabolism are transcriptionally regulated and how this impacts upon lineage specification. Consistent with the stable H3K4me3 and transcript levels of non-cell-cycle-regulated bivalent genes, we find that MLL2 binding does not oscillate on their promoters during the cell cycle. Since the non-cell-cycle-regulated bivalent genes represent approximately 50% of all bivalent genes, this also explains why the total levels of MLL2 phosphorylation did not peak in late G1 (data not shown). Phosphoregulation of MLL2 is likely to be a complex process. We evaluated this issue in cell-cycle fractions, and although phosphorylated in a CDK2-dependent manner in G1, MLL2 is also phosphorylated in other cell-cycle phases when CDK2 is less active. This indicates that other CDKs and non-CDKs control MLL2 activity. It is possible, for example, that CDK2 controls MLL2 recruitment to bivalent genes in G1, while other phosphorylation events regulate its recruitment to other targets and possibly release from bivalent genes outside of G1.
A second major aspect of the poised, bivalent state involves changes in chromosome architecture around developmental genes in G1. Following recruitment of MLL2 and deposition of H3K4me3, distal enhancers are recruited to proximal sites by a looping mechanism. Recruitment of enhancer-containing structures to developmental genes is dependent on the establishment of bivalent domains in G1 and not by epigenetic changes at the enhancer itself. The cell-cycle-dependent structures and their associated enhancers persist during DE differentiation are critical for transcriptional activation and are important for the differentiation program itself. Results described in this report provide a foundational mechanism to explain the poised state originally described by Bernstein and colleagues several years ago. Our data show that rather than being in a stable poised state, hPSCs are only bivalent and poised for differentiation during G1 phase. This raises many questions about the regulation of H3K4 trimethylation that have not been addressed by this study. For example, how are H3K4me3 marks rapidly erased in S phase? Are histone de-methylases under the control of the cell-cycle machinery? Are bivalent domains in multipotent stem cells under the control of the cell cycle? Future studies are needed to address these important questions.
In summary, this report defines a level of regulation that is of fundamental importance to pluripotency and to mechanisms of cell-fate commitment. By establishing links between the cell-cycle machinery, epigenetic regulators, and chromatin architecture, we now have a framework that can be used to further explore the fundamental aspects of pluripotency and cell-fate specification.
Experimental Procedures
hESC Culture and Differentiation
Fucci WA09 hESCs were generated and grown as described previously in media containing HEREGULIN β1 (10 ng/ml, Peprotech), ACTIVIN A (10 ng/ml, R&D Systems), and LONGR3 IGF-1 human (200 ng/ml, Sigma) (Singh et al., 2013; Singh et al., 2012). Endoderm differentiation was performed by removing HEREGULIN β1 and IGF-1, and increasing the concentration of ACTIVIN A to 100 ng/ml. SB431542 (20 μM, Tocris Bioscience), U0126 (20 μM, LC Laboratories), MI-2 (25 μM, Xcess Biosciences), and CVT-313 (20 μM, EMD Biosciences) were used as indicated. Expression constructs for CA-CDK2, a constitutively active and stabilized CYCLIN D1- CDK2 fusion protein, and KE-CDK2, a CYCLIN D1-CDK2 fusion protein carrying an inactivating K114E mutation, were kind gifts from Brian Law (Chytil et al., 2004).
Immunostaining, Immunoblotting, and qRT-PCR
qRT-PCR was performed using Taqman “Assays on Demand” (Invitrogen) on a Viia7 instrument (Life Technologies). All assays were performed in triplicate, normalized to GAPDH, and analyzed using the ΔΔCT method. Data are representative of multiple independent experiments. Immunoblotting, immunoprecipitations, and immunostaining were performed as previously described (Singh et al., 2012), with antibodies raised against MLL1 (A300-086A), MLL2 (A300-113A, Bethyl Laboratories); MENIN (ab2605), JMJD3 (ab85392, Abcam), WDR82 (kind gift from David Skalnik, IUPUI School of Science); pSMAD2 (3104S), pSerine-CDKs Substrate, P-S-100 (9477), pThreonine-Proline (9391) (Cell Signaling Technology); OCT4 (sc-8628), CDK2 (sc-163) (Santa Cruz Biotechnology); BRACHYURY (AF2085), SOX17 (AF1924) (R&D systems); FOXA2 (07-633, Millipore).
4C-Seq and 3C-qPCR
4C-seq analysis was performed as previously described (Splinter et al., 2012). Approximately 10 million cells from FACS-isolated Fucci hESC fractions were fixed in 1% formaldehyde and quenched with 1 M glycine. Following cell lysis, nuclei were digested with Hind III and ligated overnight under diluted conditions. Following the reversal of crosslinks, chromatin was digested with Nla III overnight. Next the chromatin was re-ligated and used to generate DNA libraries by PCR using the Expand Long Template Polymerase kit (#11759060001, Roche). PCR primer details are available upon request. PCR products were then purified using the High Pure PCR Product Purification Kit (#11732676001, Roche) and used for 3C-qPCR or 4C-seq. 3C-qPCR was performed using indicated primers (Table S4) with Kapa SYBR Fast Kit (KK4602, KAPA Biosystems) according to the manufacturer’s instructions on a ViiA7 Real-Time PCR instrument (Life Technologies). 4C-seq was performed using an Illumina HiSeq instrument to obtain 180–200 M, 50 bp paired-end reads (HudsonAlpha). Following sequencing, the resulting data were analyzed as follows. A reference genome was built for sites around Hind III sites. Reads were then trimmed to remove adaptor sequences, followed by mapping to the customized reference genome with Bowtie. p values were then calculated for various window sizes and plotted as domainographs (de Wit et al., 2008). For differential interaction site identification between cell-cycle fractions, counts were normalized and subtracted between samples, followed by p value calculations, as performed for single-sample analysis. Log-transformed p values were then summed at a window size of 16, and the resulting peaks indicated the cell-cycle-regulated chromatin interactions. Human genome hg19 is used for mapping.
ChIP-Seq and qChIP
Human Fucci ESCs were sorted into early G1 (DN, double negative), late G1 (KO2, Kusabira orange-2), S phase (AzL, Azami green low), and G2/M (AzH, Azami green high) fractions by FACS on a MoFlo instrument (Beckman Coulter). WA09 hESCs expressing no fluorescence, Cdt1-KO2 only, and Geminin-Az1 only, were used as FACS gating controls. Following FACS isolation, cells were fixed in 1% formaldehyde and quenched with 2.5 M glycine. Approximately 10 million cells per sample were sonicated with a Covaris S220 for 8 min at 200 cycles per burst. Chromatin was collected and then enriched by ChIP with H3K4me3, H3K27me3, H3K27ac, or H3K4me1 antibodies using Protein G Dynabeads (Invitrogen). Sequencing samples were prepared according to the manufacturer’s protocol and sequenced with an Illumina HiSeq instrument (Hudson Alpha). In addition to the ChIP-seq samples of histone marks, input samples were used as sequencing controls. For each sample, ∼40 M reads were obtained by single-end (50 bp) sequencing. Alignment of sequencing reads against human genome (hg19) was performed using the Burrows-Wheeler Aligner (BWA) tool (Li and Durbin, 2009) with the default parameters documented in the bwa-0.5.9 version. Uniquely mapped reads were chosen for the sequencing analysis. To eliminate noise and to take into account the unequal total numbers of reads for different sample, we identified the ChIP signal enriched regions: the 2.5 kb region from 1 kb upstream of the TSS to 1.5 kb downstream of the TSS as H3K4me3 signal-enriched region. For each bivalent gene obtained from Sharov and Ko (2007), we calculated the normalized RPM (reads per million) at ChIP signal enriched regions for H3K4me3 and H3K27me3, respectively. A list of bivalent genes was compiled that show a >2-fold RPM change between any of the adjacent cell-cycle time points and included these in heatmap analysis. For ChIP-seq with immunoprecipitations against H3K27ac and H3K4me1, reads were mapped using Bowtie (Langmead et al., 2009). HPeak (Qin et al., 2010) was used to call peaks from mapped reads. Gene Ontology (GO) analysis was conducted using R (http://www.r-project.org/). Differential binding sites were identified using a conditional binomial model based on counts falling in specific peak regions of each sample. Histone tracks in H1 hESCs (H3K4me1 and H3K27ac) from ENCODE and chromatin state dynamics (ChromHMM) tracks (Ernst et al., 2011) were downloaded for inclusion in ChIP-seq analysis. The categories considered as strong or weak enhancers had overlapping H3K27ac peaks. qChIP was performed as previously described using the Kapa SYBR Fast Kit (KK4602, KAPA Biosystems) according to the manufacturer’s instructions on a ViiA7 Real-Time PCR instrument (Life Technologies). Antibodies for ChIP were as follows: H3K4me3 (ab8580 and ab1012, Abcam), H3K27me3 (ab6002, Abcam), H3K4me1 (ab8895, Abcam), H3K27ac (ab4729, Abcam), MENIN (ab2605, Abcam), Wdr5 (ab56919, Abcam), MLL2/TRX2 (A300-113A, Bethyl Laboratories), and Cdk2 D12 antibody (sc-6248. Santa Cruz Biotechnology). See Table S4 for primer sequences.
Genome Editing with CRISPR-Cas9
For genome editing at the GATA6 and SOX17 loci, target sequences for guide RNAs (gRNA) were designed using http://crispr.mit.edu/. Two gRNAs were used for each locus. The following sequences were used for gRNAs: GATA6, upper gRNA1: TGTACGAATTACAATTTGGCTGG, GTTATGTACGAATTACAATTTGG, lower gRNA2: AAATGTTAGGCTTAATCAAATGG, AATGGCATAGCCTCAATACTTGG; SOX17, upper gRNA1: GCAATGAAAGTCGCTGTCAGCGG; and lower gRNA2: TTCACCCATTGCAAGCTTCCCGG. The target gRNA expression constructs were ordered as individual 455 bp gBlocks (Genscript), which has U6 promotor, target sequences, guide RNA scaffold, and terminal signal, cloned into pUC57 vector. The specificity of the gRNAs was confirmed by transfecting the gRNA construct with an hCas9 expression plasmid (#41815, Addgene) into WA09 hESCs by lipofectamine 2000 for use in a SURVEYOR assay as previously described (Ran et al., 2013). For performance of gene knockouts in Fucci hESCs, a targeting plasmid consisting of a CAG promoter driving BFP-IRES-Zeo with 1 kb homology arms was used to delete a 1 kb region containing the indicated enhancer elements. Fucci hESCs were transfected with the hCas9 plasmid, gRNA1 and gRNA2 plasmids, and the targeting plasmid using the Neon transfection system (Invitrogen) according to manufacturer instructions. Following transfection, hESCs were selected with Zeocin for 2 weeks and further purified by FACS isolation of BFP to isolate cells containing genomic deletions. Targeted deletion of the enhancer elements was verified by PCR and agarose gel electrophoresis using primers within BFP and outside of the homology arm. See Table S4 for primer sequences.
Author Contributions
A.M.S. and Y.S. designed, performed, and analyzed the experiments and wrote the manuscript. L.L., W.Z., S.Z., and Z.Q. performed bioinformatics analyses. T.W. performed and analyzed experiments. S.D. oversaw the project, designed and analyzed the experiments, and wrote the manuscript.
Acknowledgments
This work was supported by grants to S.D. from the National Institute of Child Health and Human Development (HD049647) and the National Institute for General Medical Sciences (GM75334). We thank Julie Nelson for assistance with FACS.
Published: August 13, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.07.005.
Accession Numbers
The accession number for sequencing data described in this report is GEO: GSE61176.
Supplemental Information
To be included in this list, a >2-fold difference in H3K4me3 levels was required between any two Fucci cell-cycle fractions.
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Badeaux A.I., Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat. Rev. Mol. Cell Biol. 2013;14:211–224. [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J.M., de Bruin R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledau A.S., Schmidt K., Neumann K., Hill U., Ciotta G., Gupta A., Torres D.C., Fu J., Kranz A., Stewart A.F., Anastassiadis K. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–1035. doi: 10.1242/dev.098152. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brooks E.E., Gray N.S., Joly A., Kerwar S.S., Lum R., Mackman R.L., Norman T.C., Rosete J., Rowe M., Schow S.R. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J. Biol. Chem. 1997;272:29207–29211. doi: 10.1074/jbc.272.46.29207. [DOI] [PubMed] [Google Scholar]
- Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Welcker M., Hizli A.A., Posakony J.J., Aebersold R., Clurman B.E. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Chytil A., Waltner-Law M., West R., Friedman D., Aakre M., Barker D., Law B. Construction of a cyclin D1-Cdk2 fusion protein to model the biological functions of cyclin D1-Cdk2 complexes. J. Biol. Chem. 2004;279:47688–47698. doi: 10.1074/jbc.M405938200. [DOI] [PubMed] [Google Scholar]
- de Wit E., Braunschweig U., Greil F., Bussemaker H.J., van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4:e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S., Hofemeister H., Marks H., Kranz A., Ciotta G., Singh S., Anastassiadis K., Stunnenberg H.G., Stewart A.F. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Egli D., Birkhoff G., Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof N., Leleu M., Noordermeer D., Rougemont J., Reymond A. Detecting long-range chromatin interactions using the chromosome conformation capture sequencing (4C-seq) method. Methods Mol. Biol. 2012;786:211–225. doi: 10.1007/978-1-61779-292-2_13. [DOI] [PubMed] [Google Scholar]
- Grembecka J., He S., Shi A., Purohit T., Muntean A.G., Sorenson R.J., Showalter H.D., Murai M.J., Belcher A.M., Hartley T. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Garruss A.S., Gao X., Morgan M.A., Cook M., Smith E.R., Shilatifard A. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1093–1097. doi: 10.1038/nsmb.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonk L.J., de Jonge M.E., Kruyt F.A., Mummery C.L., van der Saag P.T., Kruijer W. Aggregation and cell cycle dependent retinoic acid receptor mRNA expression in P19 embryonal carcinoma cells. Mech. Dev. 1992;36:165–172. doi: 10.1016/0925-4773(92)90067-t. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery C.L., van Rooijen M.A., van den Brink S.E., de Laat S.W. Cell cycle analysis during retinoic acid induced differentiation of a human embryonal carcinoma-derived cell line. Cell Differ. 1987;20:153–160. doi: 10.1016/0045-6039(87)90429-5. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z.S., Yu J., Shen J., Maher C.A., Hu M., Kalyana-Sundaram S., Yu J., Chinnaiyan A.M. HPeak: an HMM-based algorithm for defining read-enriched regions in ChIP-Seq data. BMC Bioinformatics. 2010;11:369. doi: 10.1186/1471-2105-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Martinez A.M., Iovino N., Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Sela Y., Molotski N., Golan S., Itskovitz-Eldor J., Soen Y. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of pRB. Stem Cells. 2012;30:1097–1108. doi: 10.1002/stem.1078. [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Ko M.S. Human ES cell profiling broadens the reach of bivalent domains. Cell Stem Cell. 2007;1:237–238. doi: 10.1016/j.stem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Singh A.M., Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Reynolds D., Cliff T., Ohtsuka S., Mattheyses A.L., Sun Y., Menendez L., Kulik M., Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.M., Chappell J., Trost R., Lin L., Wang T., Tang J., Matlock B.K., Weller K.P., Wu H., Zhao S. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Reports. 2013;1:532–544. doi: 10.1016/j.stemcr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., de Wit E., van de Werken H.J., Klous P., de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Thomson I., Gilchrist S., Bickmore W.A., Chubb J.R. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To be included in this list, a >2-fold difference in H3K4me3 levels was required between any two Fucci cell-cycle fractions.