Abstract
Acanthamoeba spp. are free‐living amoebae that are ubiquitous in natural environments. They can cause cutaneous, nasopharyngeal, and disseminated infection, leading to granulomatous amebic encephalitis (GAE) in immunocompromised individuals. In addition, they can cause amoebic keratitis in contact lens wearers. Acanthamoeba GAE is almost always fatal because of difficulty and delay in diagnosis and lack of optimal antimicrobial therapy. Here, we report the description of an unusual strain isolated from skin and brain of a GAE patient. The amoebae displayed large trophozoites and star‐shaped cysts, characteristics for acanthamoebas belonging to morphology Group 1. However, its unique morphology and growth characteristics differentiated this new strain from other Group 1 species. DNA sequence analysis, secondary structure prediction, and phylogenetic analysis of the 18S rRNA gene confirmed that this new strain belonged to Group 1, but that it was distinct from the other sequence types within that group. Thus, we hereby propose the establishment of a new species, Acanthamoeba byersi n. sp. as well as a new sequence type, T18, for this new strain. To our knowledge, this is the first report of a Group 1 Acanthamoeba that is indisputably pathogenic in humans.
Keywords: Group 1 Acanthamoeba; human isolate, nuclear small subunit ribosomal RNA gene; ribosomal secondary structure; sequence type T18; 18S rRNA gene
ACANTHAMOEBA spp. are free‐living amoebae that are ubiquitous in natural environments, such as soil and freshwaters, but are also found in a wide range of environments including dust particles in the air, bottled water, chlorinated pools, water taps and sink drains, flowerpots, aquariums, sewage, and brackish and marine waters. They have also been isolated from hydrotherapy baths, dental irrigation equipment, humidifiers, cooling systems, ventilators, and intensive care units (Marciano‐Cabral and Cabral 2003; Schuster and Visvesvara 2004). Acanthamoeba may enter the body via a break in the skin or inhalation of wind‐blown cysts and may cause cutaneous, nasopharyngeal, and disseminated infection and subsequently spread hematogenously to the central nervous system (CNS), where it can lead to granulomatous amebic encephalitis (GAE). Acanthamoeba GAE most often occurs in immunocompromised individuals, including HIV/AIDS patients, organ transplant recipients and debilitated persons. Acanthamoeba GAE has a worldwide distribution and often begins as a subclinical infection without any specific symptoms, making it difficult to diagnose. Symptoms are vague and may mimic neurocysticercosis, tuberculoma, or brain tumor. It is frequently identified in biopsy specimens of brain lesions during late stage of the disease process or at autopsy. Although a few cases have survived, Acanthamoeba GAE is almost always fatal because of difficulty and delay in diagnosis and lack of optimal antimicrobial therapy. Acanthamoeba also causes a sight‐threatening infection, Acanthamoeba keratitis (AK), in contact lens wearers or eye trauma patients (Visvesvara et al. 2011).
Acanthamoebas are classified into three main groups based on distinct morphological differences in the cysts (Pussard and Pons 1977; Visvesvara 1991). Group 1 acanthamoebas are characterized by large trophozoites measuring 25–35 μm and cysts greater than 18 μm in size. Group 1 acanthamoebas are mostly environmental and have not convincingly been associated with infections in humans or animals. Groups 2 and 3 acanthamoebas have smaller trophozoites compared to Group 1 and cysts smaller than 18 μm in size. Amoebae belonging to Group 2 cause the majority of reported human infections (both AK and Acanthamoeba GAE) and also constitute the majority of the strains isolated from the environment. Several of the Group 3 species have been shown to be pathogenic as well (Booton et al. 2005; Maciver et al. 2013).
Cyst morphology has also been used to define species within the three main groups. However, identification at the species level is problematic as species‐specific morphologic features are ambiguous and may vary with culture conditions (Visvesvara 1991). Group 1 is generally considered to include five species, Group 2 as many as 10 different species, and Group 3 up to five species, but several of them may be synonyms and some strains are likely to have been assigned to the wrong species due to ambiguous identification. As a complement to the morphologic classification, molecular analyses using nuclear small subunit ribosomal RNA (18S rRNA) gene sequences have divided Acanthamoeba into 17 sequence types (designated T1–T17, also called ribotypes or genotypes) (Corsaro and Venditti 2010; Gast 2001; Gast et al. 1996; Hewett et al. 2003; Horn et al. 1999; Nuprasert et al. 2010; Stothard et al. 1998). In some cases, the ribosomal sequence types correlate very well with the morphologically derived species designations (e.g. the association of sequence type T5 with Acanthamoeba lenticulata) while other species, especially the ones belonging to Group 2, are polyphyletic (Stothard et al. 1998).
Here, we describe the characterization of a novel species of Acanthamoeba, isolated from the skin and brain of an immunosuppressed patient that died of meningoencephalitis (D'Auria et al. 2012). Morphologic features and ribosomal sequence typing placed this new species within Group 1 Acanthamoeba.
Material and Methods
Case report
A detailed case report has been published elsewhere (D'Auria et al. 2012). Briefly, a 62‐yr‐old male received a double lung transplant in October 2009. In mid‐December, he developed skin lesions on his chest wall and posterior right lower extremity. At the end of January 2010, he was admitted to the hospital because of suspected rejection. A punch biopsy of the right chest lesion showed large mononuclear cells with prominent, centrally located nucleoli, consistent with amoebae. One week into his hospitalization, the patient became increasingly altered and agitated. CT and MRI scans revealed brain lesions consistent with meningoencephalitis. Granulomatous amebic encephalitis caused either by Balamuthia mandrillaris or Acanthamoeba species was suspected and the patient was started on a broad range of antimicrobials. Despite intensive treatment, the patient deteriorated and was declared brain dead in early February, 2010. Samples of the punch biopsy of the skin, a CSF sample collected 1 wk before death, and skin and brain tissue samples obtained at autopsy were forwarded to CDC and processed for culture, immunofluorescent antibody (IFA) staining, and real‐time PCR. Immunofluorescent antibody staining of the punch biopsy was positive for Acanthamoeba. DNA extracted from all types of specimens tested positive for Acanthamoeba in a triplex real‐time PCR assay that detects Acanthamoeba spp., B. mandrillaris, and Naegleria fowleri.
Culture isolation
Fresh unfixed skin and brain tissues were separately minced with about 0.5 ml of amoeba saline and inoculated on non‐nutrient agar plates coated with Escherichia coli and into human lung fibroblast (HLF) cultures along with 100 μg/ml gentamicin and incubated at 30 °C, 37 °C, and 40 °C. Amoebae that grew on agar plates were allowed to encyst, and those that grew in HLF cell culture were inoculated into proteose peptone yeast extract glucose medium containing 10% fetal bovine serum (PYGS) as described previously (Schuster 2002; Visvesvara and Balamuth 1975).
Ribosomal sequence typing
Cultured Acanthamoeba trophozoites were harvested from agar plates and DNA was extracted using the DNeasy tissue and blood kit (QIAGEN, Valencia, CA) following the instructions for nucleated cells. Three sets of PCR primers were used to amplify the full‐length or partial 18S rRNA gene. PCR primers JDP1 plus JDP2 (Schroeder et al. 2001) amplified a 450 base pair fragment; JDP1 plus B (Gast 2001) amplified a 1,456 base pair fragment; and CRN5 (Schroeder et al. 2001) plus B amplified the full‐length gene. All PCR reactions were performed in a 50 μM total volume using 0.2 μM of each primer, 1 μl of the extracted DNA, and the Platinum blue PCR supermix (Life Technologies, Grand Island, NY). PCR was performed in a GeneAmp 9700 thermocycler (Life Technologies) with a cycling structure of 2 min initial incubation at 95 °C followed by 40 cycles of 95 °C for 30 s, 53 °C for 30 s, and 72 °C for 2 min and 30 s. PCR products were separated on 1% agarose gel electrophoresis and purified using the QIAquick PCR purification kit (QIAGEN). The PCR products were cloned into pCR2.1 TOPO vectors using the topoisomerase method as described in the kit manual (Life Technologies). Plasmid preparations of 10 recombinant clones were purified from bacterial cultures using the QIAGEN miniprep method. Sequencing was performed using the BigDye V3.1 chemistry on an ABI Prism 3100 sequence analyzer (Life Technologies) using the amplification primers and internal 18S rRNA gene sequencing primers described elsewhere (Schroeder et al. 2001; Weekers et al. 1994).
Phylogenetic analysis
Full‐length 18S rRNA gene sequences from the new isolate were aligned with full‐length ribosomal gene sequences from other Acanthamoeba strains and isolates using ClustalW v.1.83 (Thompson et al. 1994) in the GeneStudio Pro v.2.1.2.4 software package (GeneStudio Inc., Suwanee, GA). Included sequences were (with GenBank accession numbers): Acanthamoeba astronyxis ATCC 30137 (AF019064), A. astronyxis ETW5 (DQ992178), Acanthamoeba tubiashi ATCC 30867 (AF019065), Acanthamoeba comandoni ATCC 30135 (AF019066), A. comandoni PJ (DQ185605), A. comandoni PSH (DQ185607), Acanthamoeba sp. E2 (GU808280–GU808282), Acanthamoeba sp. E1 (GU808277–GU808279), Acanthamoeba sp. E9 (GU808301–GU808302), and Acanthamoeba castellanii ATCC 50374(U07413). After flush trimming the ends and removing alignment columns containing ambiguous nucleotides, a phylogenetic analysis was performed on the alignment using the TREE‐PUZZLE program v.5.2 (Schmidt et al. 2002) with A. castellanii as an outgroup. The resulting phylogenetic tree was visualized with TreeView v.1.6.6 (Page 1996). The phylogeny program package PHYLIP v.3.68 (Felsenstein 1989) was used to confirm the tree topology using parsimony and distance methods.
18S rRNA secondary structure prediction
The secondary structure of the 18S rRNA gene was based upon the model proposed by Wuyts et al. (2000) with slight modifications. The program Mfold v.3.4 was used to determine optimal foldings of stems and loops (Zuker 2003). The sequences from the 10 clones were aligned using the program BioLign v.4.0.6 (Ibis Biosciences, Carlsbad, CA) and the base positions where variation occurred were noted. The final structure was drawn using the program Adobe Illustrator CS4.
Results
Morphological characterization
Both skin and brain specimens were positive for large Acanthamoeba trophozoites when inoculated onto agar plates coated with E. coli as well as in HLF cell cultures. Amoebae grew on the agar plate, ingested bacteria, and multiplied. In about a week, when all the bacteria were gone, the amoebae differentiated into double‐walled cysts. The amoebae grew well on the HLF cell culture and destroyed the monolayer within a week. It produced cytopathic effect (CPE) beginning on day 2 of inoculation. The CPE consisted of shrinkage of cell cytoplasm, nuclear pycnosis, and discontinuity of cell sheet resulting in the complete destruction of the cell sheet in about a week. The amoebae grew well at 30 °C and 37 °C in the PYGS medium. The amoebae could also grow at 40 °C on agar plate coated with bacteria, indicating that they are thermotolerant and have the ability to survive high fever that may occur in patients with GAE. The amoebae when examined under a phase contrast microscope measured 50–125 μm and exhibited large blunt thorn‐like processes, acanthopodia (Fig. 1), from the surface of the body, a feature that is characteristic of Acanthamoeba. The cysts measured 20–35 μm with a smooth, almost round outer wall, the ectocyst, and a thicker, inner cyst wall, the endocyst. The endocysts were mostly star‐shaped with five to eight arms or rays, although polygonal endocysts were occasionally seen (Fig. 2). The endocyst was widely separated from the ectocyst except at points where the rays contacted the ectocyst at different planes. The tips of the rays at the contact point appeared to be thick and crescent‐shaped and seemed to plug a pore, the ostiole. The crescent‐shaped plug, the operculum, was removed when the amoeba exited the cyst (Fig. 2). On the basis of these features, the amoebae were identified as Acanthamoeba sp. Group 1 and designated as CDC:V621.
Figure 1.
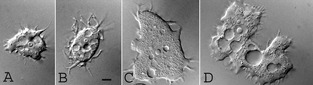
Acanthamoeba CDC:V621 trophozoites exhibiting varying sizes measuring from 50 to 110 μm. The trophozoites feature single (A–C) or double (D) nuclei or donut‐shaped nucleus (A, B). All trophozoites feature acanthopodia that are characteristically blunt at the tip, contractile vacuole, food vacuole, and a contractile vacuole. The large trophozoite is binucleate, indicating that it has completed karyokinesis and is awaiting cytokinesis. Bar = 10 μm.
Figure 2.
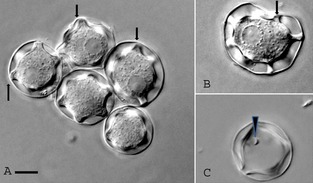
Cysts of CDC:V621 featuring the unique morphology of a nearly round ectocyst and stellate endocyst with five to eight arms or rays at different planes (A, B). Pores are present at the junction points of the ecto‐ and endocysts. The pores are plugged with the operculum (arrows), which is removed at the time of excystation (arrowhead in C). Bar = 10 μm.
Molecular characterization
Genetic characterization of the Acanthamoeba isolate (CDC:V621) was attempted using the established PCR primers JDP1 and JDP2, as well as by amplifying larger fragments of the 18S rRNA gene. However, the resulting electropherograms were of bad quality with overlapping peaks resulting in ambiguous base calling. The full‐length 18S rRNA PCR product was therefore cloned to obtain interpretable DNA sequences. Ten recombinant clones were analyzed, each one displaying a unique sequence. The sequence differences between the clones consisted mostly of insertions/deletions and transitions; only three transversions were detected. The length of the 18S rRNA gene was between 2,586 and 2,597 base pairs (primer sequences not included). Eight of the clones were very similar with just two to nine differences scattered throughout the sequence, whereas clones 1 and 6 exhibited more divergence with on average 14 and 42 sequence differences as compared to the other clones. The difference in sequences between the 10 clones is noted in Fig. 3. Despite the sequence variability among the clones, similarity searches via the GenBank server produced the same results for all the clones: the highest similarity (approximately 95%) to Acanthamoeba strains E1 and E9, belonging to the newly described T17 sequence type, followed by A. tubiashi strain ATCC 30867 (89% similarity) and A. comandoni isolates (86–88% similarity).
Figure 3.
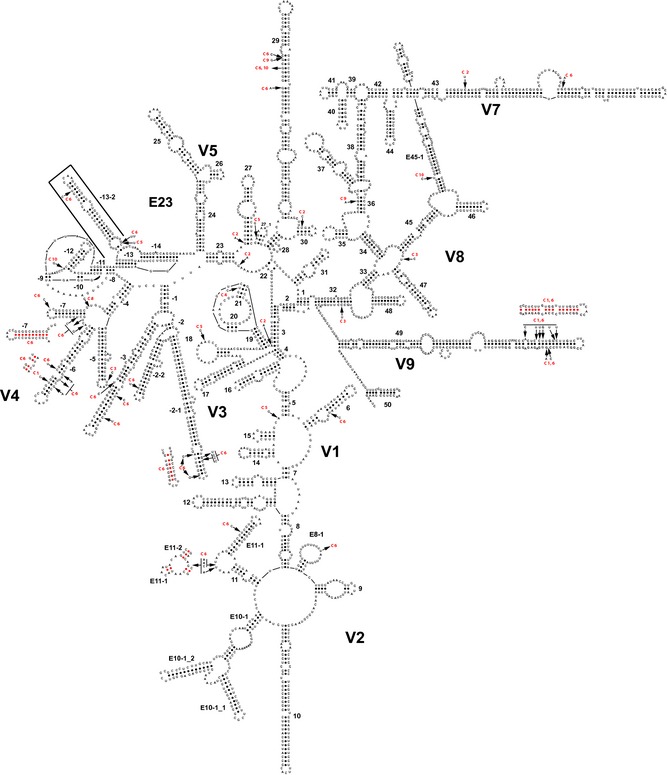
A predicted secondary structure for clone 4 of the 18S rRNA gene of CDC:V621 with areas of micro‐heterogeneity among the 10 clones indicated by arrows and the clone where the variation occurred designated with the clone number. The long inserted stem, E23‐13‐2, is outlined by a thick three‐sided box. Insertions are indicated by an arrow that does not point to a specific base. Deletions are indicated by an arrow pointing away from the specific base that has been deleted. Transitions and transversions are indicated where a base at the shaft end of the arrow is different from the base at the arrowhead point.
Phylogenetic analysis using full‐length 18S rRNA sequences unambiguously placed CDC:V621 among the other Group 1 species A. tubiashi, A. astronyxis, and A. comandoni. The 10 clones of CDC:V621 formed a single clade separated from the other Group 1 Acanthamoeba sequences (Fig. 4). The T17 strains formed a sister group to CDC:V621, illustrating that CDC:V621 and the T17 strains are closely related but genetically distinct. The DNA sequence difference in pair‐wise alignments between the T17 strains and CDC:V621 ranged between 4.2% and 4.6%.
Figure 4.
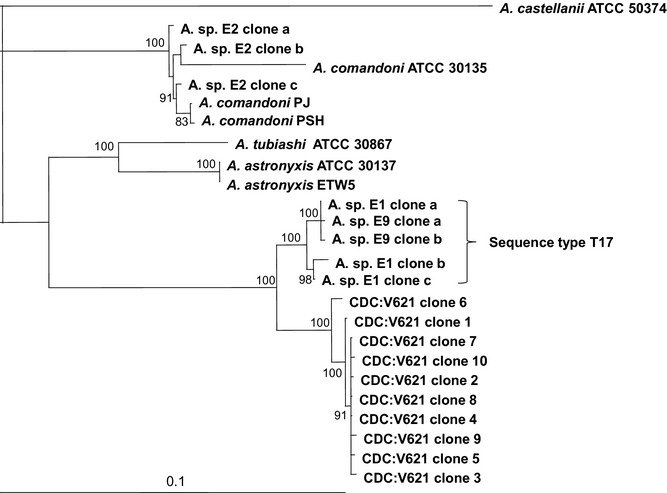
Phylogram based on 18S rRNA gene sequences from Acanthamoeba Group 1 strains. A representative from Group 2 (i.e., the type strain of A. castellanii) was added as the outgroup. All 10 sequences obtained from CDC:V621 formed a single clade, separate from other Acanthamoeba sequences. The phylogenetic tree was created with the quartet puzzling maximum likelihood method using the program TREE‐PUZZLE. The scale bar estimates evolutionary distance (nucleotide substitutions per position).
Secondary structure predictions
The secondary structure of the complete 18S rRNA of CDC:V621 is depicted in Fig. 3. When analyzing this structure, it became evident that this amoeba had hyper development of the E23‐1 through E23‐7 region with a bifurcated E23‐2 stem not previously identified in Groups 2 and 3 acanthamoebas. It also displayed a unique fairly long stem, E23‐13‐2, inserted in the loop between the E23‐13 and E23‐14 helices. The size of the insert varied from 54 to 56 bases in the various clones of CDC:V621, with the stem containing 20 base pairs. In addition, the E10‐1 stem in CDC:V621 was bifurcated. Partial reconstruction of the secondary structures of these regions in other acanthamoebas revealed that the unique features found in CDC:V621 were characteristics for Group 1 acanthamoebas.
Discussion
The genus Acanthamoeba consists of several species of free‐living amoebae that are ubiquitous in the environment. The amoeba described in this study initially caused skin lesions unresponsive to empiric treatment and subsequently disseminated to the CNS, causing a fatal GAE. Balamuthia mandrillaris, another free‐living amoeba, can also cause skin infections and GAE. Acanthamoeba and Balamuthia appear similar in tissue sections and both produce cysts in the tissues. To distinguish these two amoebae in clinical specimens, electron microscopy, immunostaining of tissue sections, or molecular analysis have to be performed. The Acanthamoeba infection in our patient was identified by immunofluorescence testing using rabbit antibodies to Acanthamoeba as well as real‐time PCR performed on tissues (D'Auria et al. 2012). Culture isolation of the amoeba from skin and brain tissues was successful and yielded an Acanthamoeba strain designated CDC:V621. This new strain exhibited large trophozoites and cysts characteristic for the Group 1 Acanthamoeba. Molecular and structural analysis of the 18S rRNA gene concluded that it is genetically most similar to the other Group 1 species within the Acanthamoeba genus, confirming the description of CDC:V621 as a new Acanthamoeba Group I strain.
Acanthamoeba‐specific PCR primers were used to amplify the 18S rRNA gene from CDC:V621; although single bands of the expected size could be seen on agarose gels following amplification, direct DNA sequencing of obtained PCR products failed due to overlapping peaks in the sequence electropherogram. This is a typical result when there is a mixture of two or more distinct PCR products in the amplified reaction. As discussed in a previous publication, the most likely explanation for this is the sequence variability among the many copies of the ribosomal gene within individual amoebae (Stothard et al. 1998). Acanthamoeba castellanii Neff strain appears to have 700 copies of the 18S rRNA gene in its genome (Byers et al. 1990) and it is reasonable that other Acanthamoeba contain numerous copies as well. The presence of micro‐heterogeneity at the 18S rRNA gene level has been detected in strains from all three morphology groups, including Group 1 (Nuprasert et al. 2010; Stothard et al. 1998). An alternative explanation for the sequence difference between the clones, errors produced by the Taq polymerase, may have contributed to some of the point mutations seen in the clones but cannot account for the majority of sequence variation, especially in clones 1 and 6. Thus, we conclude that CDC:V621 has several distinct alleles of the 18S rRNA gene. The greatest sequence difference observed between the clones was 1.7%. Most of the differences were represented as deletions, insertions, or transitions; only three transversions occurred, of which two were in stem 49. Variations between alleles of ribosomal RNA genes in amoebae can be quite substantial. For example, in the amoeba Vannella epipetala the difference between 11 clones was as high as 3.8% (Amaral‐Zettler et al. 2006).
Previous studies using DNA sequencing analysis of the 18S rRNA gene have identified 17 sequence types and these are designated T1–T17. Based on phylogenetic analysis of 18 strains belonging to morphology Groups 2 and 3, Gast et al. (1996) divided the strains into four sequence types (T1–T4) and defined a sequence type as sequences with at least 6% difference from members of the other sequence types. A subsequent study including more sequences from all three morphology groups established the presence of eight additional sequence types (T5–T12) and adjusted the sequence difference threshold between the sequence types to 5% (Stothard et al. 1998). This arbitrary number of 5% dissimilarity has since been used to create five additional sequence types (T13–T17) (Corsaro and Venditti 2010; Gast 2001; Hewett et al. 2003; Horn et al. 1999; Nuprasert et al. 2010). The vast majority of Acanthamoeba isolates from the environment as well as isolates associated with human infections belong to sequence type T4 (Booton et al. 2005; Maciver et al. 2013). In contrast, isolation of Acanthamoeba with Group 1 morphological features is rare. Three species of Group 1 Acanthamoeba, namely A. astronyxis, A. tubiashi, and A. comandoni, belong to sequence types T7, T8, and T9, respectively. The fourth species classified as Group 1, Acanthamoeba echinulata, has not yet been associated with a sequence type as no 18S rRNA gene sequence from that species is available in public databases. Recently, two environmental isolates were found to exhibit Group 1 morphology, but as their 18S rRNA genes differed more than 12% from all known Acanthamoeba sequences, a new sequence type, designated T17, was created for them and added to Group 1 (Nuprasert et al. 2010). The DNA sequences obtained from the amoebae CDC:V621 were approximately 4.3% dissimilar from the T17 strains. Although this is slightly lower than 5% sequence divergence, the phylogenetic analysis clearly distinguishes CDC:V621 as a separate lineage with an evolutionary distance of 2.7% from the T17 strains. Therefore, we consider CDC:V621 as a new and unique sequence type and identified it as sequence type T18.
The proposed secondary structure of the 18S rRNA molecule from CDC:V621 presents some unique features, especially the new stem identified between stems E23‐13 and E23‐14. An extra stem in this region has previously been identified in another amoeba, Corallomyxa tenera, and was given the designation E23‐13‐1 (Tekle et al. 2007). The new stem identified in CDC:V621, designated E23‐13‐2, is present and quite pronounced in all Group 1 Acanthamoeba strains examined to date; the size of the insert is 54–95 bases with stems containing 20–42 base pairs. In contrast, Acanthamoeba belonging to Groups 2 and 3 have either no or a considerably smaller E23‐13‐2 stem, containing less than 18 bases. In addition, the CDC:V621 amoeba had extra stems and bifurcated stems in other regions of the ribosomal RNA molecule similar to other Group 1 acanthamoebas and distinct from Groups 2 and 3 acanthamoebas. Thus, the secondary structure prediction of the 18S rRNA provides additional support for the placement of CDC:V621 in Group 1 Acanthamoeba.
To our knowledge, this is the first description of a Group 1 Acanthamoeba that is indisputably pathogenic in humans. Acanthamoeba astronyxis has been suggested to have caused human infections in one case where it was cultured from the CSF of a patient with encephalitic syndrome, but the patient recovered without any anti‐Acanthamoeba medication (Callicott 1968). It is believed that the isolation was probably due to contamination (Martinez 1980). The Acanthamoeba isolate described here, however, caused a devastating cutaneous and CNS infection leading to the death of the patient.
In conclusion, CDC:V621 is unusual for several reasons and differs from other Group 1 Acanthamoeba in its morphological, physiological, and genetic features: (1) its ability to grow on mammalian cell cultures at higher temperature (37 °C) with an optimum temperature of growth of 30 °C, (2) large size with measurements of 50–125 μm, and (3) substantial differences in its 18S rRNA gene from the other species included in Group 1 Acanthamoeba. Therefore, we establish CDC:V621 as a new species, and propose its diagnosis as follows.
Taxonomic Summary
Super Group: AMOEBOZOA Luhe, 1913, emend. Cavalier‐Smith, 1998 [Eumycetozoa Zopf 1884, emend Olive 1975]
Discosea Cavalier‐Smith et al., 2004 (R)
Longamoebia Cavalier‐Smith & Smirnov in Smirnov et al., 2011 (R)
Centramoebida Rogerson & Patterson 2002, emend. Cavalier‐Smith 2004, (R)
Genus: Acanthamoeba Volkonsky, 1931
Species: Acanthamoeba byersi n. sp.
Diagnosis. A new species of Acanthamoeba (CDC:V621) is described based upon morphologic, physiologic, and DNA sequencing data. The life cycle has amoeboid (50–125 μm) and cyst (20–35 μm) stages.
Description. The amoeboid stage is flattened with prominent subpseudopodia that are flexible and thorn like with blunt tips, acanthopodia. Trophozoite with or without a uroid, the cyst is large, bounded by a double wall with an outer smooth, almost round ectocyst and an inner mostly stellate endocyst with five to eight arms in different planes touching the ectocyst. Flagellate stage not seen even after suspending in distilled water for 24 h.
Growth in vitro. Grows well on agar plate coated with a Gram‐negative bacterium such as E. coli at 30 °C, 37 °C and 40 °C. Also grows well in axenic medium supplemented with fetal bovine serum, known to support growth of other Acanthamoeba strains, at 30 °C and 37 °C. Also grows well on mammalian tissue cultures (e.g., HLF cell monolayer) at 30 °C and 37 °C. The optimal growth temperature is 30 °C. The amoeba produces CPEs on monolayers of HLF cells. The normal habitat of the amoeba is not known although the amoeba was isolated from the brain and skin tissues of a patient with typical symptoms of granulomatous amoebic encephalitis.
Etymology. The new species of Acanthamoeba is named A. byersi in recognition of the extensive molecular work performed on Acanthamoeba spp. by the late Dr Thomas J. Byers (1935–2003), Professor of Molecular, Cellular and Developmental Biology Program at The Ohio State University, Columbus, Ohio. Dr Byers was an active member and a tireless worker for the International Society of Protozoologists (now International Society of Protistologists) and responsible for proposing a the new name of The Journal of Eukaryotic Microbiology, for the previously named Journal of Protozoology, to attract new interest and provide additional support for the flagging Journal of Protozoology. Dr Byers was responsible for organizing the first Acanthamoeba conference in 1978 at The Ohio State University in Columbus, Ohio as at that time the first few cases of AK were described. Now the conference is known as the International Meeting on the Biology and Pathogenicity of Free‐Living Amoebae and includes all pathogenic free‐living amoebae. Dr Byers was instrumental for using both nuclear and mitochondrial ribosomal genes as targets for taxonomic classification and phylogenetic relationships of the genus Acanthamoeba. Dr Byers was a gifted teacher and a tireless research worker, a generous collaborator and a helpful friend of one of the authors (G.S.V.).
Type species. Acanthamoeba byersi, n. sp.
Deposition of type material. Type culture has been deposited at the American Type Culture Collection.
Gene sequence data. The 18S rRNA gene sequences are deposited in GenBank under accession numbers KC822461–KC822470.
Acknowledgments
We are indebted to Drs Jamie Lin and Andrea D'Auria for providing us with the patient specimens and relevant history of the case. Dr Norman J. Pieniazek assisted with the phylogenetic analysis.
Literature Cited
- Amaral‐Zettler, L. A. , Cole, J. , Laatsch, A. D. , Nerad, T. A. , Anderson, O. R. & Reysenbach, A. L. 2006. Vannella epipetala n. sp. isolated from the leaf surface of Spondias mombin (Anacardiaceae) growing in the dry forest of Costa Rica. J. Eukaryot. Microbiol., 53:522–530. [DOI] [PubMed] [Google Scholar]
- Booton, G. C. , Visvesvara, G. S. , Byers, T. J. , Kelly, D. J. & Fuerst, P. A. 2005. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J. Clin. Microbiol., 43:1689–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, T. J. , Hugo, E. R. & Stewart, V. J. 1990. Genes of Acanthamoeba: DNA, RNA and protein sequences (a review). J. Protozool., 37:17S–25S. [DOI] [PubMed] [Google Scholar]
- Callicott Jr, J. H. 1968. Amebic meningoencephalitis due to free‐living amebas of the Hartmannella (Acanthamoeba)‐Naegleria group. Am. J. Clin. Pathol., 49:84–91. [DOI] [PubMed] [Google Scholar]
- Corsaro, D. & Venditti, D. 2010. Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida). Parasitol. Res., 107:233–238. [DOI] [PubMed] [Google Scholar]
- D'Auria, A. , Jaime, L. , Geiseler, P. , Qvarnstrom, Y. , Bandea, R. , Roy, S. L. , Sriram, R. , Paddock, C. , Zaki, S. , Kim, G. & Visvesvara, G. S. 2012. Cutaneous acanthamoebiasis with CNS involvement post‐transplantation: implication for differential diagnosis of skin lesions in immunocompromised patients. J. Neuroparasitol., 3:1–7. [Google Scholar]
- Felsenstein, J. 1989. PHYLIP: phylogenetic inference package (version 3.2). Cladistics, 5:164–166. [Google Scholar]
- Gast, R. J. 2001. Development of an Acanthamoeba‐specific reverse dot‐blot and the discovery of a new ribotype. J. Eukaryot. Microbiol., 48:609–615. [DOI] [PubMed] [Google Scholar]
- Gast, R. J. , Ledee, D. R. , Fuerst, P. A. & Byers, T. J. 1996. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J. Eukaryot. Microbiol., 43:498–504. [DOI] [PubMed] [Google Scholar]
- Hewett, M. K. , Robinson, B. S. , Monis, P. T. & Saint, C. P. 2003. Identification of a new Acanthamoeba 18S rRNA gene sequence type, corresponding to the species Acanthamoeba jacobsi Sawyer, Nerad and Visvesvara, 1992 (Lobosea: Acanthamoebidae). Acta Protozool., 42:325–329. [Google Scholar]
- Horn, M. , Fritsche, T. R. , Gautom, R. K. , Schleifer, K. H. & Wagner, M. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus . Environ. Microbiol., 1:357–367. [DOI] [PubMed] [Google Scholar]
- Maciver, S. K. , Asif, M. , Simmen, M. W. & Lorenzo‐Morales, J. 2013. A systematic analysis of Acanthamoeba genotype frequency correlated with source and pathogenicity: T4 is confirmed as a pathogen‐rich genotype. Eur. J. Protistol., 49:217–221. [DOI] [PubMed] [Google Scholar]
- Marciano‐Cabral, F. & Cabral, G. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev., 16:273–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, A. J. 1980. Is Acanthamoeba encephalitis an opportunistic infection? Neurology, 30:567–574. [DOI] [PubMed] [Google Scholar]
- Nuprasert, W. , Putaporntip, C. , Pariyakanok, L. & Jongwutiwes, S. 2010. Identification of a novel t17 genotype of acanthamoeba from environmental isolates and t10 genotype causing keratitis in Thailand. J. Clin. Microbiol., 48:4636–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12:357–358. [DOI] [PubMed] [Google Scholar]
- Pussard, M. & Pons, R. 1977. Morphologie de la paroi kystique et taxonomie du genre Acanthamoeba (Protozoa: Amoebida). Protistologica, 8:557–598. [Google Scholar]
- Schmidt, H. A. , Strimmer, K. , Vingron, M. & von Haeseler, A. 2002. TREE‐PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics, 18:502–504. [DOI] [PubMed] [Google Scholar]
- Schroeder, J. M. , Booton, G. C. , Hay, J. , Niszl, I. A. , Seal, D. V. , Markus, M. B. , Fuerst, P. A. & Byers, T. J. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol., 39:1903–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, F. L. 2002. Cultivation of pathogenic and opportunistic free‐living amebas. Clin. Microbiol. Rev., 15:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, F. L. & Visvesvara, G. S. 2004. Free‐living amoebae as opportunistic and non‐opportunistic pathogens of humans and animals. Int. J. Parasitol., 34:1001–1027. [DOI] [PubMed] [Google Scholar]
- Stothard, D. R. , Schroeder‐Diedrich, J. M. , Awwad, M. H. , Gast, R. J. , Ledee, D. R. , Rodriguez‐Zaragoza, S. , Dean, C. L. , Fuerst, P. A. & Byers, T. J. 1998. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J. Eukaryot. Microbiol., 45:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle, Y. I. , Grant, J. , Cole, J. C. , Nerad, T. A. , Anderson, O. R. , Patterson, D. J. & Katz, L. A. 2007. A multigene analysis of Corallomyxa tenera sp. nov. suggests its membership in a clade that includes Gromia, Haplosporidia and Foraminifera. Protist, 158:457–472. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. & Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res., 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvesvara, G. S. 1991. Classification of Acanthamoeba . Rev. Infect. Dis., 13(Suppl. 5):S369–S372. [DOI] [PubMed] [Google Scholar]
- Visvesvara, G. S. & Balamuth, W. 1975. Comparative studies on related free‐living and pathogenic amebae with special reference to Acanthamoeba . J. Protozool., 22:245–256. [DOI] [PubMed] [Google Scholar]
- Visvesvara, G. S. , Roy, S. L. & Maguire, J. H. 2011. Pathogenic and opportunistic free‐living amebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia pedata In: Guerrant R. L., Walker D. H. & Weller P. F. (ed.), Tropical Infectious Diseases – Principles, Pathogens & Practice, 3rd edn Elsevier, Churchill Livingstone, Philadelphia, PA: p. 707–713. [Google Scholar]
- Weekers, P. H. , Gast, R. J. , Fuerst, P. A. & Byers, T. J. 1994. Sequence variations in small‐subunit ribosomal RNAs of Hartmannella vermiformis and their phylogenetic implications. Mol. Biol. Evol., 11:684–690. [DOI] [PubMed] [Google Scholar]
- Wuyts, J. , De Rijk, P. , Van de Peer, Y. , Pison, G. , Rousseeuw, P. & De Wachter, R. 2000. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res., 28:4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]


