Abstract
Elicitation of CD4 IFN-gamma T cell responses to Mycobacterium tuberculosis (MTB) is a rational vaccine strategy to prevent clinical tuberculosis. Diagnosis of MTB infection is based on T-cell immune memory to MTB antigens. The MTB proteome contains over four thousand open reading frames (ORFs). We conducted a pilot antigen identification study using 164 MTB proteins and MTB-specific T-cells expanded in vitro from 12 persons with latent MTB infection. Enrichment of MTB-reactive T-cells from PBMC used cell sorting or an alternate system compatible with limited resources. MTB proteins were used as single antigens or combinatorial matrices in proliferation and cytokine secretion readouts. Overall, our study found that 44 MTB proteins were antigenic, including 27 not previously characterized as CD4 T-cell antigens. Antigen truncation, peptide, NTM homology, and HLA class II tetramer studies confirmed malate synthase G (encoded by gene Rv1837) as a CD4 T-cell antigen. This simple, scalable system has potential utility for the identification of candidate MTB vaccine and biomarker antigens.
Keywords: Tuberculosis, CD4, T-cell, antigen, CD137, malate synthase, tetramer
1. Introduction
MTB causes 107 active tuberculosis infections (ATBI) and kills about 1.3 × 106 persons annually (27). Globally, 1 in 3 people have latent tuberculosis infection (LTBI) (48). The licensed BCG vaccine is poorly active against adult disease and there is a global effort to improve vaccines (37). The immune response to MTB infection includes CD4 T-cells with rearranged T-cell receptor (TCR) alpha beta receptors that recognize peptides derived from MTB-encoded proteins bound to human leukocyte antigen (HLA) class II. Notwithstanding disappointing results from a recent vaccine clinical trial of one MTB antigen (74) the elicitation or boosting of CD4 T-cells remains a valid proposed mechanism of action for candidate vaccines (77). It has been proposed that antigen and epitope choice may be important in the context of bacterial gene expression during different phases of MTB pathogenesis. The IFN-gamma axis is vital for host defense against MTB and CD4 T-cell decline in HIV infection is associated with severe MTB outcomes (18). Memory T-cells also form the basis for tests for MTB infection. The tuberculosis skin test (TST) measures in vivo leukocyte infiltration in response to a filtrate of MTB cultures, while licensed interferon-gamma release assays (IGRA) measure IFN-gamma production in response to MTB peptides from three or fewer MTB open reading frames (ORFs) (7, 50). Neither test discriminates between latent and active TB infection or predicts risk of progression from a latent to active state. There is an unmet need for biomarkers in this area.
MTB T-cell antigen discovery is thus relevant to vaccines and diagnostics. The complexity of the MTB proteome, encoded by 4,000 annotated genes, has hindered systematic screening of potential antigens in MTB. Approaches, as recently reviewed (25) have included expression libraries of MTB DNA fragments (52), prediction of HLA-binding peptides MTB ORFs (81), and expression of targeted subsets of MTB ORFs based on criteria such as phase- or nutrient-dependent gene expression (10, 26) or sequence motifs associated with protein secretion (9).
Advances in high throughput in vivo recombination and E. coli lysate-based in vitro transcription/translation (IVTT) allow expression of essentially the entire translated proteome of large-genome pathogens (6, 17, 21, 34). The proteins are useful for probing humoral responses (53). Our lab adapted these protein collections for CD4 T-cell research for viruses encoding up to 240 proteins (32, 34). IVTT proteins are suitable for CD4 T-cell studies because these immune cells typically detect microbial proteins after they are digested to linear peptides of 8 to about 20 amino acids. The peptides are not post-translationally modified, with recognized exceptions (55). Here, we report a novel approach to MTB CD4 T-cell antigen discovery that uses a proteome set (45, 46) originally created for antibody studies to probe the reactivity of polyclonal MTB-specific CD4 T-cell lines. We further developed modifications of the workflow to adapt to a resource-constrained, MTB-endemic region, obtaining adequate assay performance to confirm and extend MTB antigen discovery.
2. Materials and Methods
2.1 Subjects and specimens
Persons requiring LTBI evaluation for employee health in the US were screened with Quantiferon™ Gold In-Tube (QFT) (Qiagen, Germantown, MD) and participated in an institutional ethics committee-approved protocol and gave informed consent. Heparinized peripheral blood was obtained. For studies in India, institutional ethics committee approval was obtained to recover leukocyte buffy coats from blood donated by anonymous healthy donors at a blood bank, with no medical, demographic, or personal identifying information available. PBMC isolated by Ficoll-Hypaque density gradient centrifugation from blood or buffy coat were cryopreserved at 1–3 × 107 cells/vial. When QFT could not be done, thawed PBMC, with living cells re-isolated in some instances by Ficoll-Hypaque centrifugation, were assayed for LTBI using enzyme-linked spot assay (ELISPOT) (31). Plates were coated with monoclonal antibody (mAb) 1D1K (mAbTech, Mariemont, OH) specific for IFN-gamma. After washing, 3 × 105 cells/well were added in a final volume of 100 microliters T-cell medium (TCM). Stimuli included media negative and 1.6 μg/ml phytohemagglutinin (PHA) positive control. Peptides covering the sequence of MTB proteins early secretory antigen target (ESAT)-6 and cultured filtrate protein (CFP)-10 from BEI Resources (Manassas, VA) were added to 1 μg/ml final concentration each in 0.2% DMSO final as pools of 20 or 25 peptides. After 24 hours, cells were removed and IFN-gamma detected with biotinylated mAb 7-B6-1 (mAbTech), avidin-peroxidase, and AEC substrate with intermediate washes. Samples with > 10 spots/well for ESAT-6 and/or CFP-10 minus DMSO control were considered positive for LTBI (56). HLA typing was performed at the Puget Sound Blood Center, Seattle, Washington, USA. Procedures were approved by the relevant Institutional Review Board.
2.2 Expansion of MT-reactive cell populations from PBMC
Initial experiments adopted a strategy of sorting MTB-reactive cells using a surface activation marker, followed by non-specific polyclonal expansion. PBMC were plated at 4 × 106/well in 24-well plates in 2 ml T-cell medium TCM (41) with 1:400 native whole MTB antigen. After 18 hours, cells were stained with anti-CD3-phycoerythrin (PE), anti-CD4-fluorescien isothiocyanate, anti-CD137-allophycocyanin (Becton Dickinson, San Jose, CA), and 1 μg/ml 7-aminoactinomycin D (Life Technologies, Grand Island, NY). Viable CD3+/CD4+ lymphocytes, either CD137high or CD137negative, were sorted (FACSAria III, Becton Dickinson) (35). At least 1,000 sorted cells were expanded with PHA, feeder cells and 32 units/ml natural human (hn) IL-2 (41). A portion of the resultant bulk 1st generation (termed B1) cell lines were tested for whole MTB reactivity after 14 days. Remaining cells were expanded further using anti-CD3 mAb as mitogen, feeder cells and human recombinant IL-2 (42) for bulk 2nd generation (termed B2) cell lines that were cryopreserved in aliquots for testing. To make CD4 T-cell clones, 1.5 × 106 B2 cells from subject US3 were stimulated with an equal number of autologous PBMC as APC and 1 microgram/ml peptide Rv1837 278–292 (AVDAADKVLGYRNWL) in 2 ml TCM in a 24-well plate. At 18 hours, live CD3+ CD4+ CD137+ cells were sorted and cloned at 1 cell/well (39). Candidates were screened for 3H thymidine incorporation responses to 1 microgram /ml Rv1837 278–292 or whole 1:200 MTB antigen in separate wells using autologous gamma-irradiated PBMC as APC. Clones reactive to both peptide and MTB were further expanded (39, 47). To adapt these assays without a requirement for cell sorting or radioisotope use in a resource-limited setting in India, we modified the protocol in such a way that 5 days after initial plating of PBMC and MTB antigen, hnIL-2 was added at 32 units/ml. Cells were fed at least every other day with half-volume TCM/IL-2 and expanded as necessary. After 14–16 days, cells were expanded for a second 14–16 day cycle with PHA, IL-2, and irradiated random allogeneic PBMC (41). Epstein-Barr virus (EBV)-transformed lymphocyte continuous lines (LCL) were cultured as described (39).
2.3 T-cell assays
For direct ex vivo PBMC cytokine responses, thawed cells were incubated with MTB antigens or staphylococcal enterotoxin B (SEB) control and evaluated by intracellular cytokine cytometry (ICC). The protocol included Violet live/dead stain (Invitrogen, Carlsbad, CA), co-stimulatory antibodies and Brefeldin A as published (35). Alternatively, PBMC were stimulated without co-stimulatory antibodies/ Brefeldin A and the up-regulation of the activation/co-stimulatory molecule CD137 was assessed after 20 hours by flow cytometry (35). To measure B1 or B2 responder reactivity, we used a published (35) ICC assay. When used as APC, autologous PBMC were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) to allow exclusions from analysis of responder cells.
To determine individual antigen specificity, 2–4 × 104 bulk or cloned responder cells were plated/well in duplicate 96 U-bottoms with 5–10 × 104 autologous irradiated (3300 rad gamma irradiation) PBMC or EBV-LCL (8000 rad) and IVTT antigens or defined peptides in 200 microliters TCM (41). After three days, proliferation was assessed with 0.5–1.0 microcuries/well 3H thymidine (41). For selected assays, supernatant (100 microliters) cytokine concentrations were measured by bead ELISA at the Fred Hutchinson Cancer Research Center core facility, or by plate IFN-gamma ELISA as described (40). For the later assays, standard curves were done in most assays ranging from 2400 to 9.4 picograms/ml of recombinant IFN-gamma (Endogen) in two–fold dilutions, with a lower limit of detection of 18.7 to 37.5 picograms/ml. To determine HLA restricting loci, mAb that block all HLA DP, DR, or DQ allelic variants were included during assay setup (41). First-stage screens of the 164 selected MTB proteins used either individual proteins at 1:1000, or protein set arranged in a 12 × 16 matrix of row and column pools with final concentrations of each at 1:1200 to 1:1600. Screens were done in duplicate. For matrixes, each pool was scored as positive or negative and the intersections of each positive and negative pool was then scored as a candidate MTB antigen, as described (36). In second-stage assays, each candidate was checked in duplicate using IFN-gamma release.
For tetramer staining, cloned or bulk T-cells were stained with 1 microliter PE-labeled tetramer of Rv1837 284–298 and HLA DRB1*1501, prepared as described (60), for 30 minutes at room temperature followed by anti-CD4 FITC for 30 minutes at 4°C, washing and fixation. Control was DRB1*1501 tetramer with influenza B peptide GKTGTIVYQRGVLLPQK (Kwok et al. unpublished).
2.4 Antigens
Non-denatured whole MTB antigen was prepared by growth of MTB strain H37Rv (51) in 7H9 broth rotated at 37 °C in room air. After collection of stationary phase cells from 50 ml broth and two PBS washes, ~4 × 1010 cells were re-suspended in 4 ml PBS and disrupted with a Bead-beater™ (Biospec, Bartlesville, OK) 4 times, setting 6, 30 seconds with 5 seconds on ice preceding each. Lysate was centrifuged at 13,000 rpm in a tabletop microcentrifuge for 5 minutes at room temperature and supernatant filtered twice (0.2 micron Acrodisc™, Pall, Ann Arbor, MI). Denatured preparations had 1% final sodium dodecyl sulfate added after PBS washes, followed by 105 °C for 10 minutes, Bead-beater treatment and clarification. Antigens frozen at −80 °C were used after Lowenstein medium cultures were negative for growth at 3 weeks. PPD (Tubersol™, Aventis Pasteur, Toronto, Canada, 5 tuberculin units/0.1 ml) was used at 1:10 to 1:20 final dilution. Positive control was SEB (58).
MTB ORFs (Supplementary Table 1) were cloned from strain H37Rv (Genbank NC_000962.2) as described (46) in-frame between N-terminal 6-histidine (HIS) and C-terminal influenza hemagglutinin (HA) sequences controlled by a T7 bacteriophage promoter. Constructs were sequence-verified. Genes with a delta (Δ) H have no C-terminal influenza hemagglutinin (HA) tag; genes designated delta SH have the HA tag and bacterial signal sequence deleted. For in vitro transcription/translation (IVTT) E. coli-based Expressway™ (Invitrogen) or RTS kits (5 Prime, Gaithersburg, MD) were used per the manufacturers. For inclusion bodies (IB), plasmids were transformed into BL21 E. coli, and master cell bank (MCB) stocks frozen in 25% glycerol. For protein expression, we modified a previous method (30). Flat-bottomed 96-well blocks (Qiagen) containing 1ml Luria broth with 100 micrograms/ml kanamycin were inoculated from the MCB and incubated 18–24 hours at 37 °C with 650 rpm shaking. These cultures were passaged to 1ml MagicMedia™ (Invitrogen)/kanamycin in Qiagen blocks incubated as above. The following day, cells harvested by centrifugation for 10 minutes at 6,000 × g at 4 °C were vortex re-suspended in 200 microliters BugBuster (BB) (EMD Millipore, Billerica, MA) supplemented with 1microliter/ml benzonase (Novagen, Madison WI, #70746), 1000 U/ml lysozyme (Novagen #71110), and one protease inhibitor pill/50 ml (Roche, Indianapolis, IN, #04693132001). The cells were incubated at 18 °C for 20 minutes with occasional vortexing. IBs were pelleted at 5000 × g for 15 minutes at 4 °C, re-suspended in 200microliters BB, and incubated 20 minutes with periodic vortexing. An additional 200 microliters of 0.1× BB was added and the IB’s pelleted by centrifugation, washed in 400 microliters 0.1× BB, incubated 5 minutes at room temperature, and pelleted. IBs were solubilized in 200 microliters of 0.2% sodium dodecyl sulfate (SDS) in PBS and placed at 4°C until clear (usually overnight). SDS was raised to 0.3–0.4% if necessary to solubilize proteins. Free SDS was precipitated by adding 1M KCl to 40 mM and cooling to 4 °C and centrifugation for 20 minutes at 6,000 × g at 4°C. 150 microliters of solubilized IB protein supernatant was decanted into 96-well plates containing 30 microliters of 50% glycerol and stored at −80°C. Protein expression was monitored as described (13).
For n=51 negative control antigens, ORFs or regions of ORFs from Francisella tularensis (FT) strain SchuS4 or Plasmodium falciparum (PF) strain 3D7, PCR-cloned with nomenclature as described (17, 19, 72, 73), were expressed in both IVTT and IB formats. The FT ORFs were 43, 284, 355, 362n, 443, 468, 476, 535, 552, 577, 701, 846, 920, 1052, 1188, 1254, 1306, 1396, 1510, 1609, 1631, 1695, 1729, 1776, 1844, 1903, 1923, 1952, 1957, 1973, and 2036. The PF ORFs (5) were PF3D7_1341300e2s1, PF3D7_1353900e1s1, PF3D7_0802900e1s1, PF3D7_1003500e2s1, PF3D7_1021700e4s1, PF3D7_1130100e1s1, PF3D7_1351400e2s1, PF3D7_1360100e3s1, PF3D7_1432100e6s1, PF3D7_1444100e1s1, PF3D7_0115000e2s1, PF3D7_0205500e3s1, PF3D7_0320900e2s1, PF3D7_0515600e10s1, PF3D7_0518700e3s1, PF3D7_0918300e1s1, PF3D7_0932200e3s1, PF3D7_1211400e4s1, and PF3D7_1218800e2s1, where e indicates exon number, and s indicates a segment number within a long exon. Instructions to access the PF primer sequences are published (17). As additional negative controls for IVTT only, 32 synthesis reactions were run with no added DNA giving a total of 83 negative control antigens.
Fragments of MTB ORF Rv1837 were amplified by PCR using full-length plasmid template (primers, Supplementary Table 2). Amplicons were cloned via pENTR221 (Invitrogen) into pDEST203 (35) with Gateway™ reagents, plasmids sequence-confirmed, and protein fragments expressed (Expressway™). Synthetic peptides covering Rv3875 (ESAT-6) (15-mers overlapping by 9–11 amino acids), portions of Rv1837 (15-mers overlapping by 11 amino acids and shorter peptides), and from bacterial homologs of Rv1837 (Sigma) were identity-confirmed by mass spectrometry and used as 10 mg/ml stocks in DMSO.
2.5 Statistical analysis
Cutoffs for positive responses to MTB antigens were calculated as previously described (33). Briefly, for 3H thymidine assays, the mean plus 3.09 times the standard deviation of the negative controls was calculated and MTB ORFs with CPM values above this were considered positive for a false discovery rate of 0.1%. For antigen determinations in ELISA-based work in India, we were able to run fewer negative controls due to limited cell number. Cutoffs were generally set using the mean plus 3 times the standard deviation of the negative controls.
3. Results
3.1 Subjects and specimens
PBMC were studied from 14 persons, comprising three US persons with LTBI, two US controls without LTBI (Table 1), and nine blood donors in India with LTBI. The US persons with LTBI were from endemic areas (East Asia, Eastern Europe), reported a history of positive tuberculin skin tests (TST), had positive Quantiferon™ IGRA tests, and negative evaluations for current or past ATBI. The US-born controls had negative TST and/or IGRA tests and no clinical history of MTB infection or exposure. In India, blood bank materials from a total of 47 persons were screened with an ESAT-6/CFP-10 peptide ELISPOT (56). Overall, 17 persons (36%) had ELISPOT results consistent with LTBI. We selected 9 subjects based on PBMC availability.
Table 1.
Subjects studied in this report.
| subject | age | national origin | BCGa | TSTb | IGRAc |
|---|---|---|---|---|---|
| US1 | 40 | China | yes | pos | pos |
| US2 | 50 | Poland | no | pos | pos |
| US3 | 45 | China | yes | pos | pos |
| US4 | 52 | US | no | neg | neg |
| US5 | 28 | US | no | neg | neg |
| Indiad | unknown | unknown | unknown | unknown | 17 of 47 (+) |
History of receipt of bacille Calmette-Guerin vaccine.
History of a positive tuberculosis skin test.
Results of a Quantiferon™ interferon gamma release assay for US subjects and ESAT-6/CFP-10 ELISPOT for India subjects.
No information is available from 47 anonymous blood donors studied in India.
3.2 Enrichment of MTB-specific CD4 T-cells
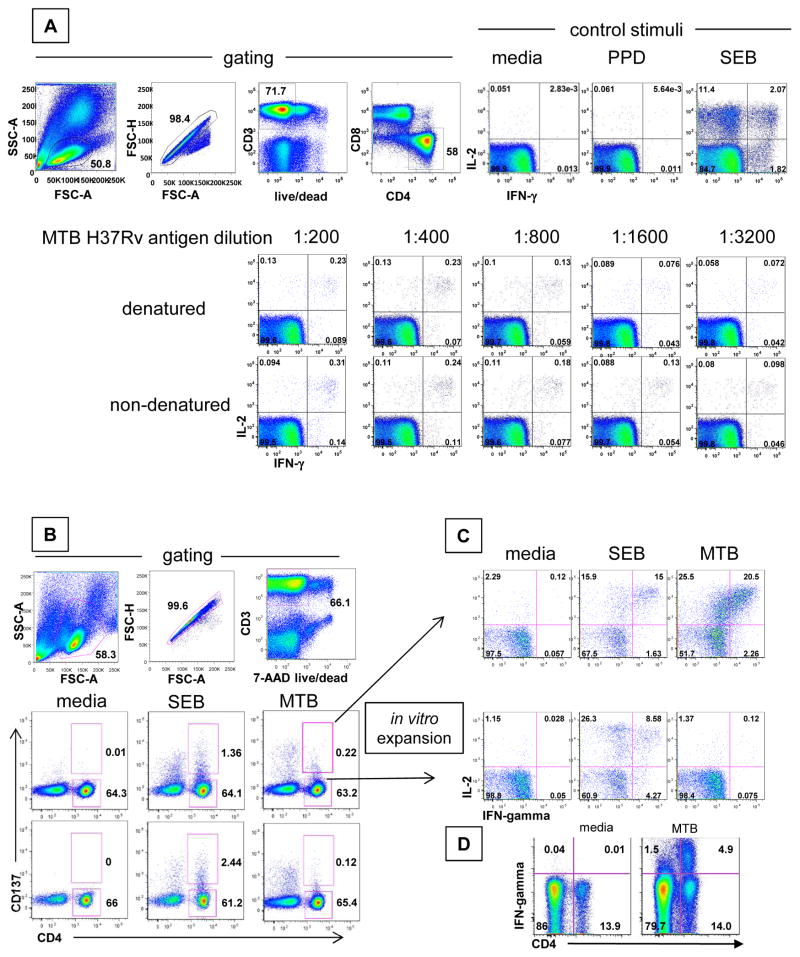
PBMC exposed ex vivo to either non-denatured or denatured MTB antigen showed populations of CD3+, CD4+ cells accumulating IL-2 and/or IFN-gamma. The fraction of reactive cells decreased with antigen dilution (Fig. 1A). More cells responded to whole MTB than to PPD. We chose a 1:400 dilution of non-denatured whole MTB for subsequent work. PBMC from one of two non-LTBI control persons were also positive for reactivity to whole MTB antigen by ICC (not shown) although the subject was negative for Quantiferon™ IGRA testing. Stimulation of PBMC with whole MTB antigen for 20 hours also resulted in up-regulation of surface expression of the co-stimulatory molecule CD137 (representative subject, Fig. 1B). PBMC from persons with LTBI, as well as the IGRA-negative person mentioned above, showed CD137 up-regulation on a small fraction of CD3+, CD4+ lymphocytes.
Figure 1.
Detection, enrichment and expansion of MTB-specific CD4 T cells from PBMC. A: Reactivity of PBMC for interferon-gamma (IFN-gamma) and interleukin 2 (IL-2) accumulation. Top row left: Gating for single, live, CD3+ CD4+ CD8- cells; right: reactivity to indicated substances. Next two rows: reactivity to dilutions of whole MTB antigens B: Ex vivo detection of MTB-reactive cells using CD137. Top row: Gating for single, live, CD3+ lymphocytes from an LTBI (+) subject. Middle row: surface expression of CD137 at 20 hours in response to the indicated stimuli from an LTBI (+) subject. Cells in the indicated gates after MTB stimulation were sorted and expanded. Bottom row: surface expression of CD137 at 20 hours in response to the indicated stimuli from an LTBI (-) subject. C: Reactivity of expanded CD4+CD137-high or CD4+CD137-negative cells from an LTBI (+) subject to the indicated stimuli. Numbers in quadrants or next to gates are cell percentages. D: Reactivity of bulk polyclonal MTB-IL-2-reactive responder lymphocytes from an Indian LTBI subject to media or whole killed MTB antigen. Numbers in quadrants are percent of gated singlet, live, CD3+ lymphocytes staining with the indicated antibodies.
We next sorted CD137high or CD137negative CD3+, CD4+ cell populations from whole MTB-stimulated cultures. Both subsets expand briskly using non-specific mitogenic stimulation to yield polyclonal cell lines that were >95% CD3+, CD4+ (not shown). The CD137high- but not CD137negative-origin cells were enriched for IFN-gamma and IL-2 production when re-stimulated with whole MTB (representative subject, Fig. 1C). Reactivity to PPD was again much lower (not shown). CD137 selection enriched the frequencies of MTB-reactive CD4 T-cells on the order of 100-fold compared to the starting PBMC (Fig. 1B, 1C). The LTBI-negative subject with ex vivo cytokine and CD137 reactivity to MTB also had MTB reactivity in CD137high- origin B2 cells (not shown). CD4 T-cells expanded using MTB antigen and IL-2 rather than CD137 sorting generally showed up to one-third of the CD4 T-cells were MTB-specific using IFN-gamma responses (whole MTB antigen minus media control) as the readout. Each cell line reacted to SEB positive control (not shown).
3.3 Screen of the partial MTB proteome
We screened the polyclonal MTB-reactive CD4 T-cell lines for responses to 164 MTB proteins from strain H37Rv (46) (Supplementary Table 1). The criteria for selecting MTB ORFs included well-documented human CD4 T-cell responses, for example Rv3875 encoding ESAT-6 (14) or a predicted proline-glutamate (PE) or proline-proline-glutamate (PPE) motif associated with extracellular secretion (71). We also mined data from previous serological studies and picked proteins with differential serum IgG reactivity comparing persons with ATBI to MTB culture negative, or persons from MTB-endemic compared to non-endemic regions (45, 46). We selected optimal dilutions for IVTT proteins based on preliminary titration experiments and our prior experience using both expression formats (33, 35, 42–44).
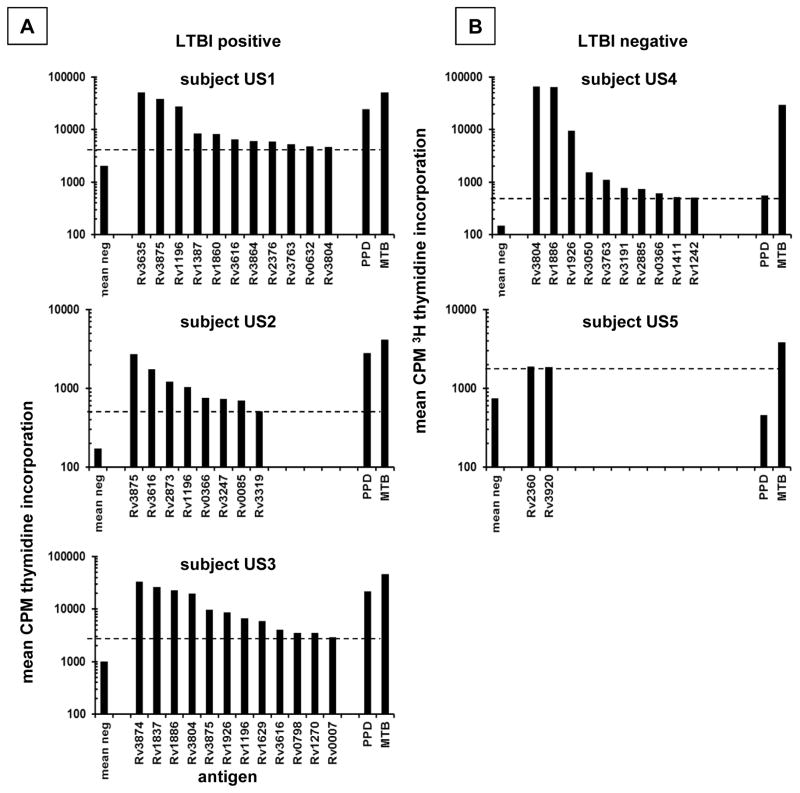
Using two different MTB protein expression formats, we observed a limited number of clearly positive protein-specific responses per person in the CD137high-origin cultures (Supplementary Fig. 1, Fig. 2). Comparison of MTB proteins expressed as IVTT proteins and solubilized inclusion bodies showed reasonable concordance between these formats (Supplementary Fig. 1). IVTT proteins were used for the bulk of the present report. Based on an objective cutoff using negative control antigens, we observed that TST (+), IGRA (+) persons born in MTB-endemic countries had 1 to 12 protein-specific responses per person (0.6% to 7.5% of the proteins tested) (Table 2). Interestingly, expanded CD137high origin B2 cells from two TST (-), IGRA (-) persons at low epidemiologic risk for MTB exposure also displayed proliferative responses to whole MTB and selected MTB proteins. Subject US5 had only weak responses to 2 MTB proteins just above the statistically defined cutoff, but US4 exhibited strong responses to several MTB proteins and whole MTB. We confirmed that US4 was IGRA negative with the Quantiferon™ test.
Figure 2.
A: Reactivity of CD137-high origin PBMC CD4 polyclonal T cell lines from US LTBI subjects to MTB proteins. MTB gene names for IVTT-expressed antigens and control antigens indicated on the X axis. Cell activation detected by 3H thymidine proliferative responses on Y axes; note axes limits vary for clarity. Only proteins with reactivity above the cutoff are shown in descending order. Each cell line was tested with each protein. Each bar is the mean of duplicates. Horizontal dashed lines indicate positivity cutoffs. B: Similar data for two US LTBI-negative persons.
Table 2.
MTB antigens scoring as positive for one or more subjects.
| ORF | subjecta | protein | novel | ||
|---|---|---|---|---|---|
| LTBI US | LTBI India | non-LTBI US | |||
| Rv0007 | 3 | conserved hypothetical membrane protein | yes | ||
| Rv0085 | 2 | hydrogenase C called hypC | yes | ||
| Rv0212 | 3 | asnC-family transcriptional regulator nadR | yes | ||
| Rv0366 | 2 | 4 | hypothetical conserved | yes | |
| Rv0632 | 1 | enoyl-CoA hydratase echA3 | yes | ||
| Rv0798 | 3 | 1 | CFP29 | yes | |
| Rv0984 | 4,5 | pterin-4-alpha-carbinolamine dehydratase moaB2 | yes | ||
| Rv1196 | 1, 2, 3 | 1,2,4,6,8 | PPE18 | ||
| Rv1270 | 3 | lipoprotein LprA | |||
| Rv1387 | 1 | 1,4,5,6,7 | PPE20 | ||
| Rv1411 | 4 | lipoporotein LprG | yes | ||
| Rv1629 | 3 | 3 | DNA polymerase I polA | yes | |
| Rv1728 | 1 | hypothetical conserved | yes | ||
| Rv1837 | 3 | 1 | malate synthase G glcB | yes | |
| Rv1860 | 1 | ala-pro rich secreted | |||
| Rv1886 | 3 | 4 | 4 | ||
| Rv1915 | 1 | isocitrate lyase aceAa | yes | ||
| Rv1926 | 3 | 4 | immunogenic protein mpt63 | yes | |
| Rv1980 | 5 | immunogenic protein mpt64 | |||
| Rv2031 | 2,5 | heat shock protein X, alpha-crystallin homolog | |||
| Rv2151 | 3 | cell division protein ftsQ | yes | ||
| Rv2360 | 5 | hypothetical protein | yes | ||
| Rv2376 | 1 | LMW antigen CFP2 | |||
| Rv2837 | 1 | hypothetical | yes | ||
| Rv2873 | 2 | 1 | lipoprotein MPT83 | ||
| Rv2875 | 1 | immunogenic secreted protein MPT70 | |||
| Rv2885 | 4 | transposase | yes | ||
| Rv3050 | 4 | transcriptional regulator asnC-family | yes | ||
| Rv3191 | 4 | transposase | yes | ||
| Rv3241 | 5 | Conserved protein, similar to many hypothetical proteins and to some putative ribosomal proteins | yes | ||
| Rv3243 | 1 | hypothetical protein | yes | ||
| Rv3247 | 2 | thymidylate synthase | |||
| Rv3319 | 2 | succinate dehydrogenase subunit sdhB | yes | ||
| Rv3345 | 5 | PE-PGRS family protein | yes | ||
| Rv3362 | 1 | ATP/GTP binding protein | yes | ||
| Rv3616 | 1, 2, 3 | 1,4,6 | ala-gly rich protein | ||
| Rv3635 | 1 | 1 | conserved membrane protein | yes | |
| Rv3763 | 1 | 4 | 19 kda lipoprotein antigen precursor lpqH | ||
| Rv3804 | 1, 3 | 1,4 | 4 | secreted fibronectin-binding protein antigen fbpA | |
| Rv3864 | 1 | 4,5,6 | hypothetical protein | ||
| Rv3874 | 3 | 1,2,4,5,6,7,9 | CFP-10, EsxB | ||
| Rv3875 | 1, 2, 3 | 1,4,5 | ESAT-6, EsxA | ||
| Rv3881 | 4,5 | espB, conserved alanine and glycine rich protein | yes | ||
| Rv3920 | 5 | hypothetical protein | yes | ||
We enrolled 3 subjects in the US listed as LTBI US 1–3, 2 subjects in the US who were IGRA-negative and at low risk for LTBI, listed as non-LTBI US 4–5, and 9 subjects in India listed as LTBI India 1–9. MTB-specific T-cells were enriched from US subjects using CD137 selection and from India subjects using MTB-IL-2.
3.4 ORFeome screens in a resource- and cell-limited setting
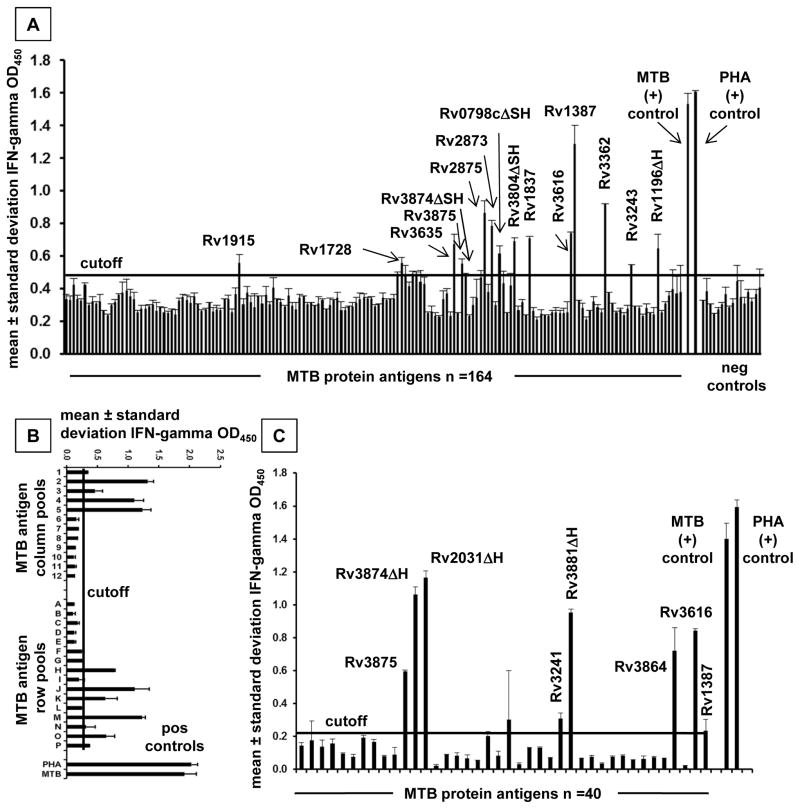
The workflow was modified as detailed in Materials and Methods to require neither cell sorting nor radioactive isotopes. We first compared proliferative and IFN-gamma and IL-17A secreted cytokine responses to 72 MTB ORFs for subject US3. Positivity thresholds were set for each assay using negative control protein antigens and mean plus 3.09 times standard deviation criteria used above. For proteins driving strong proliferation, all three readouts were above threshold (Supplementary Fig. 2). Some MTB proteins with weak positive proliferative responses were negative one or both cytokine ELISAs. IL-17A did not add sensitivity, as there were no IFN-gamma-negative/IL-17A-positive proteins. We concluded that IFN-gamma ELISA is a reasonable surrogate for 3H thymidine proliferation assays.
Scan of the 164 individual MT proteins for subject D1 revealed a discrete pattern of positive and negative responses (Fig. 3A). In other subjects, we studied the MTB proteins initially as combinatorial matrix row and column pools to reduce the PBMC required as APC (representative subject D5, Fig. 3B). Secondary assays of individual proteins at the intersection of positive matrix row and column pools (Fig. 3C) confirmed hits and allowed discrete single protein positive calls to be made.
Figure 3.
A: Reactivity of bulk polyclonal MTB-reactive responder lymphocytes from an Indian LTBI subject to MTB proteins. Antigens are identified on the X-axis, and IFN-gamma responses on the Y-axis. B: Reactivity of bulk polyclonal MTB reactive responder lymphocytes from a Delhi subject to MTB proteins pooled from rows and columns of a combinatorial matrix. C: Breakdown of candidate MTB single protein antigens from Figure 3B in a follow-up assay. Positive responses are labeled. For each assay, bars are means of duplicate and error bars are standard deviations. The Δ symbol specifies omission of C-terminal influenza hemagglutinin (H) tag and in some cases N-terminal histidine (S) tag from selected MTB proteins as per Materials and Methods.
3.5 Confirmation of positive CD4 T cell responses and evaluation of cross-reactivity
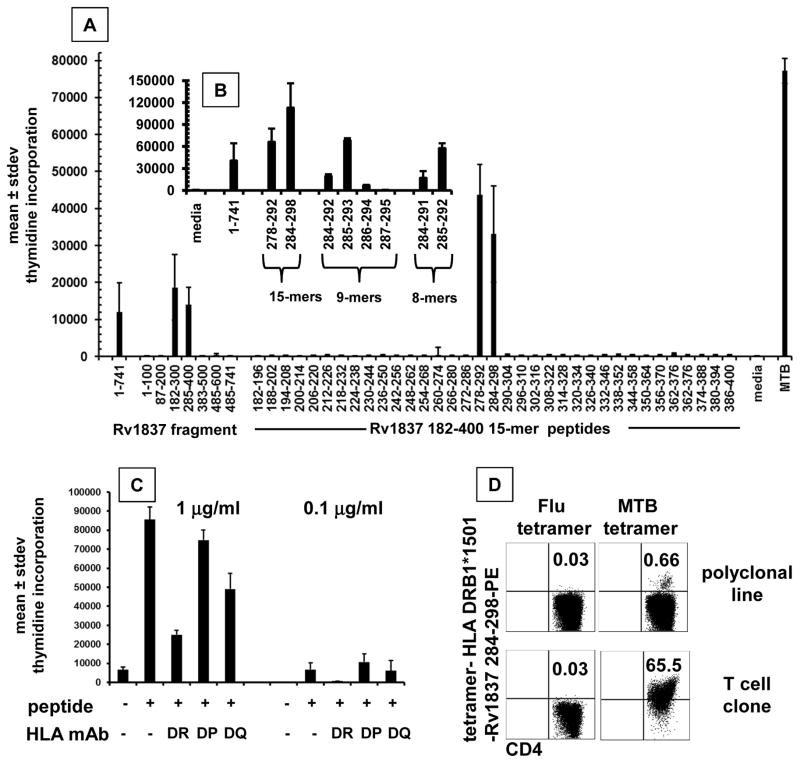
Reactivity with full-length proteins typically implies recognition of one or more internal linear peptides. To test this, we showed that polyclonal CD4 CD137-high origin MTB-reactive T-cells from subject US3 reacted to several 15-mer peptides in ESAT-6 (Fig. 4). Recognition of multiple epitopes is consistent with literature (4, 75). We then explored epitope-level recognition of the candidate T-cell antigen malate synthase G (glcB), encoded by Rv1837. This protein was chosen for validation studies because it has not previously been described as a T cell antigen, because enzymes are less frequently described as MTB T-cell antigens, because it has homologs in all prokaryotes but eukaryotes, and because abundant responder cells were available. Protein fragments of the 741 amino acid-long protein were expressed in pDEST203 (35) and tested with responder cells from US3. Regions 182–300 and 285–400 were positive (Fig. 5A). Peptide 15-mers 278–292 AVDAADKVLGYRNWL and 284–298 KVLGYRNWLGLNKGD were also reactive, consistent with an epitope near 284–292, KVLGYRNWL. In the 285–400 fragment expression in pDEST203, the vector contributes an initial phenylalanine at position 284 in lieu of K284 in MTB, implying that K284 may not be essential for T-cell recognition. Truncation analysis showed that peptides as short as the 8-mer 285–292 were active (Fig. 5B).
Figure 4.
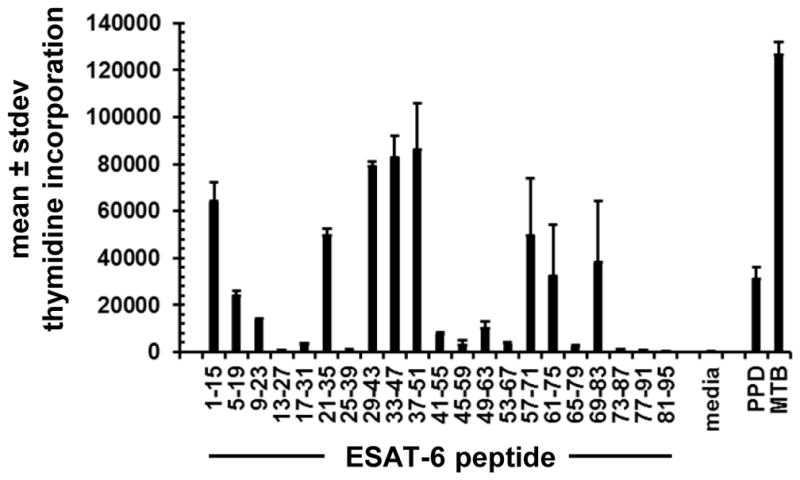
Recognition of peptides from MTB protein ESAT-6 encoded by gene Rv3875 by CD137-high origin polyclonal PBMC CD4 T-cells selected from subject US3. Amino acids of 15-mer peptides are on the X-axis. Control antigens are at right. Bars are mean of duplicate and error bars are standard deviations. Proliferative responses are indicated on the Y-axis.
Figure 5.
A: Recognition of full length, truncated fragments, and peptides from MTB RV1837 by bulk MTB-reactive CD4 T-cells. Antigens indicated on the X-axis include IVTT-expressed proteins at 1:1000, peptides at 1 microgram/ml, and whole MTB antigen. B: responses to selected peptides at 1 microgram/ml in a follow-up experiment. C: Inhibition of proliferation of CD4+ T-cell clone specific for MTB Rv1837 in response to peptide Rv1837 284–298 at 1.0 or 0.1 microgram/ml in the absence or presence of mAb that bind to and inhibit all allelic variants HLA DR, DP, or DQ. For A, B, and C, Y-axis is mean and standard deviation of duplicate. D: Tetrameric complexes of HLA DRB1*1501 and MTB Rv1837 peptide specifically bind to MTB-reactive T-cells. Fluorescent tetramers were assembled with Rv1837 284–298 or control influenza B peptide. Cultured T-cell line (top) or T-cell clone (bottom) reactive with this peptide were incubated with tetramer followed by anti-CD4 and analyzed by flow cytometry. The percentage of cells in the right upper quadrant is indicated for each dot-plot.
CD4 clone US3-TCC2 with strong reactivity to 284–292 and whole MTB (not shown) was selected for detailed study. Reactivity to both 278–292 and 284–298 recapitulated the parent bulk culture (Fig. 5B, Table 3, supplementary Fig. 3). The shortest active peptide was octamer 285–292 with sequence VLGYRNWL, confirming that K284 was not required. While nonamer 285–293 was also fully active, nonamer 286–294 was less active, implying that V285 was required for strong T-cell activation. Similarly, nonamer 283–291 was inactive, such that L292 was required (not shown). HLA restriction was determined with 15-mer 284–298. Only anti-HLA-DR blocked proliferation (Fig. 5C). The subject was heterozygous for DRB1*1501 and *0402 and also had the DRB4*0401 and DRB5*0501 alleles. The peptide had a predicted (76) high binding efficiency to only HLA DRB1*1501 (Table 3). We made fluorescent tetrameric complexes of DRB1*1501 with 284–298. The reagent specifically bound TCC2 and 0.66% of CD4 T-cells in the parental MTB-reactive polyclonal cell line (Fig. 5D).
Table 3.
Reactivity of cloned MTB-reactive CD4 T-cells to peptides from MTB Rv1837 and selected malate synthase G homologs from other bacterial species.
| bacteria | T-cell response a | sequence b | bindingc | accession d |
|---|---|---|---|---|
|
| ||||
| media | 444 ± 70 | |||
| MTB 278–292 | 29,067 ± 3,449 | AVDAADKVLGYRNWL | ||
| MTB 284–298 | 47,773 ± 5,152 | KVLGYRNWLGLNKGD | 13.6 nM | |
| MTB 284–292 | 54,297 ± 1,987 | KVLGYRNWL | ||
| Mycobacterium ulcerans | 45,914 ± 4,343 | KALGYRNWL | 45.8 nM | ABL05304.1 |
| Mycobacterium gilvum | 44,368 ± 18,553 | KVLAYRNWL | 8.3 nM | YP_001134631.1 |
| Mycobacterium leprae | 18,819 ± 7,387 | KVRGYRNWL | 18.4 nM | YP_002504009.1 |
| Rhodococcus equi | 50,665 ± 3,741 | KVVGYRNWL | 19.7 nM | WP_022596098.1 |
| Mycobacterium xenopi | 1,599 ± 479 | KTRAYRNWL | 80.8 nM | WP_003921805.1 |
| Pseudomonas putida | 957 ± 11 | KVIVYRNWL | 9.6 nM | WP_020192754.1 |
| Rhodococcus fascians | 42,814 ± 2,243 | AVDADDKVLGYRNWLGLMKGD | Q9AE55.1 | |
| Mycobacterium marinum | 44,066 ± 4,457 | AVDADDKVLGYRNWLGLNRGD | WP_020728966.1 | |
| Mycobacterium abscesses | 25,802 ± 16,682 | AVDAEDKVLGYRNWLGLNKGD | WP_005091776.1 | |
| Mycobacterium neoaurum | 29,558 ± 15,103 | AVDAEDKVLGYRNWLGLNRGD | YP_008907621.1 | |
Mean and standard deviation of duplicate 3H thymidine incorporation proliferation assays with 1 μg/ml of the indicated peptide. Results showed an identical trend for each peptide in a separate experiment.
Differences from Mycobacterium tuberculosis are indicated in bold underline.
Predicted binding to HLA DRB1*15:01 of nonamer region of peptide from valine 285 to glycine 293 upon input of genomic 15-mer corresponding to MTB 284–298 (lysine to aspartic acid) to IEDB (76) binding predictor using nn_align_IC50 algorithm.
Accession number for representative gene or genome containing the indicated sequence in malate synthase G.
MTB is phylogenetically related to non-tuberculous mycobacteria (NTM) and other bacteria. Some NTM are pathogens, and NTM infection may interfere with diagnostic tests for T-cell memory to MTB (56). To evaluate cross-reactivity we obtained peptides from homologs of Rv1837 (Table 3) with variant amino acids within or flanking 284–292. Among variants within 284–292, peptides from Mycobacterium ulcerans, Mycobacterium gilvum, Mycobacterium leprae, and Rhodococcus equi were cross-recognized (Table 2). These differed from MTB at a single amino acid. Cross-reactivity suggested that conservative single amino acid changes at positions 2, 3, or 4, and even a non-conservative change at position 3 were tolerated by this T-cell clone. Peptides from Mycobacterium xenopi, Pseudomonas aeruginosa, Pseudomonas putida, and Nocaria farcinica, which differed at multiple positions within amino acids 2 through 4, were not cross-recognized. Thus, this region contributes to HLA and/or TCR binding. It is controversial whether changes in amino acids lateral to core CD4 T-cell epitopes alter antigen recognition (78). We examined 21-mer peptides with variations in amino acids that flank the 284–292 epitope and observed no interference with T-cell reactivity for Rhodococcus fascians, Mycobacterium neoaurum, or Mycobacterium abscesses (Table 3). For Mycobacterium marinum, responses appeared to be enhanced by flanking mutations. Dose-response curves showed a general pattern of lower EC50 values for longer peptides, as reported by others (38), with some showing good activity at 10−9 M or lower (Supplementary Fig. 3).
3.6 Characteristics of the MTB proteins eliciting CD4 T-cell responses
Overall, we detected responses to 44 distinct MTB proteins (28% of the 164 studied) (Table 3). Among these, 33 were immunogenic in just LTBI (+) subjects, 6 solely in non-LTBI control subjects, and 5 in persons in each group. The most frequently recognized MTB proteins overall included CFP-10 (encoded by Rv3874, 8 of 12 LTBI subjects) and ESAT-6 (encoded by Rv3875, 6 of 12 LTBI subjects). These are well known MTB T-cell antigens (56). Other prevalent antigens included PPE18 (encoded by Rv1196, 8 of 12 LTBI subjects), also known as MTB39a, which is known to drive human T-cell responses and is a vaccine candidate (16), and PPE20 (encoded by Rv1387, 6 of 12 LTBI subjects). An alanine-glycine rich protein (encoded by Rv3616) was recognized by 6 of 12 subjects. Rv3616 is both a virulence factor and a CD8 antigen in mice with characteristics of the Esx family of bacterial proteins.
4. Discussion
MTB infections have devastating clinical impact. While there has been considerable recent progress in diagnosis and therapy (56, 79), broad gains in TB control will likely require an effective vaccine. Recently, an efficacy trial based on boosting T-cell responses to a single MTB antigen reported negative results (74). It is not fully understood if the conceptual underpinning of CD4-based vaccines is flawed, or if this candidate was insufficiently potent. MTB research vaccine priorities are undergoing a re-evaluation in the wake of this trial (15). Some data indicate that MTB-specific T-cell responses may target conserved epitopes and thus might benefit the pathogen (62). Regardless, persons with defects in IFN-gamma pathways are susceptible to Mycobacterial infections, and IFN-gamma can activate macrophages to eliminate MTB (18). In recognition of the possible value of T-cell antigens for designing vaccines and diagnostics, many labs are continuing to screen the MTB ORFeome for novel antigens and epitopes.
Recently, Lendestam Alrehamn et al. reported the results of a large-scale peptide-based survey of PBMC CD4 responses amongst 28 persons with LTBI (54). A total of 20,160 15-mer peptides were selected on the basis of predicted binding to population-prevalent HLA DR, DP, and DQ heterodimers. The selection process included peptide variants found in an expanded set of MTB genomes. The readout was direct PBMC IFN-gamma secretion via ELISPOT. The principal conclusions included the detection of a mean of 24 epitopes per person, a focus on antigens encoded in discrete pathogenicity islands in the MTB genome including secreted and secretory apparatus proteins as well as PE/PPE proteins, pronounced immunodominance hierarchies within-subject and within-population, and the presence of the chemokine receptors CXCR3 and CCR6 on most MTB-reactive CD4 T-cells.
In the present report, we present alternative technologies for dissecting the CD4 T-cell response to MTB. The two methods used to enrich MTB-reactive T-cells from PBMC were not directly compared. Both yielded discrete “hits” with some overlap in both known and novel CD4 antigens. The CD137-based method tended to yield bulk cultures that were more highly enriched for MTB-reactive cells, targeted just CD3+ CD4+ T-cells, and could be adapted for other markers such as chemokine receptors. The whole MTB co-culture method was methodologically simpler and did not require cell sorting. Enrichment of MTB-reactive cells has previously been reported using CD154, a TNF receptor family member distinct from CD137, and other methods (12, 24, 59).
We explored readout methods using a radioactive tracer to measure cell proliferation, and cytokine secretion. Direct comparison showed slightly greater sensitivity for the proliferation assay. Each requires autologous PBMC as APC, but we were able to reduce the overall PBMC requirement in a two-step IFN-gamma workflow progressing from a pool matrix to single candidate T-cell antigens. The average number of antigens identified per person was higher in the US, but the reasons for this are unclear and the number of subjects studied in each site was small. In addition to methodological differences, the biological characteristics of the test subjects may also have differed between groups. For example, repeated exposure to MTB may be more common in India. We also compared two expression systems for MTB proteins that are compatible with high throughput. The results were largely concordant for inclusion bodies and IVTT-expressed proteins. The reasons for discordant results are not clear but could include variation in partition of some MTB proteins to inclusion bodies, or differential cleavage of some MTB in one or the other expression system that interrupts CD4 T-cell epitopes.
The number of subjects and portion of the MTB proteome investigated in this report do not yet allow broad comparison with the data presented by Lendestam Alrehamn et al. (54). Both studies observed prevalent responses to secreted and secretory proteins, such as ESAT-6 and CFP-10, as well as to PPE and PE proteins. The latter families contain regions of shared protein sequence (64), and murine T-cells can react with several discrete gene products (65). Investigation of this hypothesis will require determination of reactive peptides. The peptide survey found an average of 24 antigenic ORFs per person, while the average breadth we observed amongst 12 persons with LTBI, across the two technology platforms in two countries, was 7 ORFs per person. PPD was historically used as a starting point to identify MTB CD4 T cell antigens, given its’ utility in the TST. We observed a much higher abundance of CD4 T cells in PBMC responding to whole MTB compared to PPD (Fig. 1), and both the peptide survey (54) and this present report noted prominent antigenicity for proteins that have not been detected in PPD (11). While Lendestam Alrehamn et al. (54) included peptides from variant MTB genomes, our protein set was derived from a single MTB strain, H37Rv. However, the cloning and expression pipeline we used can incorporate variant proteins. Other groups have made impressive efforts to purify large numbers of MTB and other microbial proteins for T cell research using affinity tags, including automated and micro-scaled methods (10, 66).
Challenges remain with such systems; for example, not all proteins with polyhistidine tags can be purified with such methods. Another method of surveying the antigens within MTB has been to use microarrays printed with plasmids encoding each gene of interest, followed by in situ protein expression and then probing with human sera to detect binding antibodies (63). Thus far, this system has not been adapted for querying the T-cell response. Direct ex vivo methods generally require quite pure proteins, while our general workflow, using in vitro expanded cells, is able to use non-purified proteins made in high throughput.
The sensitivity of our method to requires further characterization. We began with 5–10 × 106 PBMC per subject to either sort CD137-reactive cells, or re-stimulate memory cells with whole antigen. Very rare responder T-cell clonotypes might be missed, but this is larger than the 106 or fewer cells typically interrogated by ELISPOT or ICC per reaction ex vivo. Although memory T-cells can be triggered by as few as 1 to 10 peptide-MHC ligands per APC (61) implying that antigenic proteins can be present in relatively small amounts, or as elements of complex mixtures, it is possible that low peptide abundance in our antigen preparation could also have been limiting. Study of whole MTB antigens prepared under distinct oxygen, pH, and carbon source conditions will be required to determine if antigen abundance is an important variable. We readily detected responses to secreted MTB proteins such as ESAT-6 and CFP-10, indicating that these proteins were not excluded from our whole antigen. Regarding the antigen set, we were able to observe responses in pools of up to 24 IVTT proteins, as previously reported (36) for HSV.
It is difficult to definitively prove that responses to MTB proteins are driven by MTB infection. It is now recognized that the human naïve CD4 T-cell repertoire contains peptide-reactive T-cells that are readily detectable in PBMC if one increases sensitivity enough (70). The balance of priming and tolerance to sequence-related proteins expressed by NTM and other, non-Mycobacterial bacteria may further contribute to the reactivities noted in this study. CD4 reactivity to selected MTB proteins was noted in our limited survey of LTBI-negative persons, and similar results have been reported previously (54). While reactivity to some specific MTB proteins was only noted for LTBI-negative subjects, study of larger panels of LTBI-positive subjects is required to determine the consistency of this pattern. Study of larger numbers of LTBI-negative persons, such as the ELISPOT-negative subjects from India, could further test the discriminatory power of T-cell responses to individual proteins. We encountered difficulty in raising polyclonal T-cell lines from such IGRA-negative donors using whole MTB antigen, consistent with a low abundance of MTB-reactive T-cells. Direct ex vivo methods may be required to address the sensitivity and specificity of reactivity to individual MTB proteins and peptides with regards to LTBI.
Among the 164 MTB proteins studied, we detected T-cell responses to 44 proteins in just 12 persons with LTBI. In addition to validating reactivity using ESAT-6 peptides, we also studied a novel hit in malate synthase G encoded by Rv1837. Malate synthase G is also called GlcB (68), MS (1), MSG (3), and 81(88)-kDa protein (67). GlcB synthesizes malate from glyoxylate and acetyl-coenzyme A in the glyoxylate cycle used for gluconeogenesis from lipids (68). The glyxoylate cycle is absent in humans and is a proposed anti-tuberculous drug target (20). It is important for pathogenesis in animals (57) and is modulated when MTB is grown in specific cells (28) or nutrient conditions (68). GlcB has previously been shown (1) to be an antibody target in MTB-infected humans. IgG responses to recombinant GLcB are present in persons with ATBI and may be useful in differentiating ATBI from other respiratory diseases (2) or identifying incipient ATBI (67) or tuberculous meningitis (29). This is the first report of GlcB T-cell immunogenicity. GlcB was reported to be absent in several PPD preparations (11), highlighting the importance of including metabolic enzymes and other non-secreted proteins in study of immune responses to bacteria.
The bulk MTB-reactive CD4 T-cells studied reacted to two overlapping fragments of GlcB. After making CD4 T-cell clones, specificity was further probed using synthetic peptides. Based on data from inhibition of proliferation using anti-HLA class II framework monoclonal antibodies (Fig. 5C), and from positive binding to a tetramer made from peptide and recombinant HLA DRB1*15:01 (Fig. 5D), supported by the high predicted peptide binding affinity for the subject’s HLA DRB1*15:01 allele (Table 3) but not their other HLA DR alleles (DRB1*04:02, DRB4*01:10, DRB5*01:01), we can define HLA DRB1*15:01 as the restricting HLA molecule. Gaseitsiwe et al. (23) found that 674 of 7,466 peptides selected from 61 ORFs in MTB H37Rv bound DRB1*15:01, but did not include Rv1837 or T-cell readouts. The observed reactivity octamer GlcB 285–292, is consistent with classical CD4 T-cell TCR alpha-beta recognition of HLA-peptide. The nonamer 285–293 is predicted to have very high affinity for HLA DRB1*15:01, with our data indicating that 285–292 also must bind adequately to provide a ligand for the TCR expressed by our indicator T-cell clone.
The minimal epitope 285–292 is identical in several NTM and the region is highly conserved in many bacteria. Indeed, we observed cross-reactivity with both NTM and non-NTM homolog peptides. Of interest, the variant Mycobacterium xenopi and Pseudomonas putida peptides that failed to elicit CD4 T-cell proliferation (Table 3) retained predicted high affinity binding to HLA DRB1*15:01. This is consistent with the established model (69) in which specific amino acid residues, separate from those involved in HLA binding, provide TCR contacts that are critical for controlling T-cell activation. GlcB is a relatively conserved protein, such that many additional bacterial species have GlcB amino acid sequences related to those tested. For example, the KVLAYRNWL variant in Mycobacterium gilvum, differing by one amino acid from the canonical KVLGYRNWL in MTB strain H37Rv, is present in many strains of Mycobacterium tuberculosis, and also in members of the Polaromonas, Rhodobacteraceae, Ideonella, Mesorhizobium, Leptothrix, Cheloativorans, Pseudomonas, Marinobacter, Lutibaclum, Azoarcus, Loktanella, Alcaligenes, Pusillimonas, Ketogulonicigenium, and Azosprillum genera. Exposure to this type of related sequence from non-MTB bacteria could account for the detection of responses to MTB proteins in LTBI-negative persons, noted in this and previous reports (54). Several reports have linked DRB1*1501 with susceptibility to ATBI, especially in Asia (reviewed in (80)). This is unlikely to be related to responses to a single epitope. However, the recent demonstration of antigen-specific regulatory T-cells in MTB infection (49) and the possibility that T-cell responses to commensal bacteria may promote tolerance rather than TH1 immunity (8) illustrate the potential for complexity in the response to bacterial communities with highly overlapping antigen proteins.
In addition to confirm previously known population-prevalent T-cell antigens, we uncovered evidence of human CD4 T cell reactivity to myriad other MTB proteins. Several factors make definitive assignment of these proteins as novel or known T-cell antigens very difficult. These include the volume and pace of MTB T-cell research, overlapping systems of gene and protein nomenclature, access challenges using search engines for data in figures or tables, and incomplete nature of MTB and epitope database annotations. Lindestam Arlehamn et al. (54) in 2013 synthesized 20,610 peptides predicted to be encoded by MTB genes and to be likely CD4 epitopes, and tested these against PBMC from persons with LTBI. The resulting publication both greatly expanded the set of known MTB antigens, and reviewed prior knowledge. We used this source, surveyed the Tuberculosis database (22), queried IEDB (76) using Mycobacterium tuberculosis and T-cell reactivity as search terms, and manually searched Pubmed.
Synthesizing these sources, we preliminarily assigned 27 of 44 (61%) of the MTB ORFs we found to be T-cell antigens as novel (Table 2).
In summary, the detection and ranking of candidate vaccine and diagnostic antigens amongst the large MTB proteome remains an important challenge. In the present report, we describe workflows that allow the use of unpurified MTB proteins, available from proteome-wide antibody research, for the discovery of MTB-specific CD4 T-cell responses. By making adjustments to our protocols, we were able to adapt to antigen discovery to a setting with challenging availability of cell sorting and radioisotopes. Our long-germ goal is to scale up to the entire proteome by leveraging existing ORFeome expression libraries (45, 46) and perform immune-epidemiologic studies in proximity to MTB-endemic populations. A representative novel antigen was confirmed at the peptide level, with reactivity to NTM homologs illustrating the complexity of T-cell responses to bacteria. Several novel MTB proteins recognized by human T-cells have been discovered and are rational candidates for future research concerning biomarkers for MTB disease status, or as possible vaccine antigens. Finally, the methods developed herein may also be applicable to dissection of the memory T-cell response to other large-genome pathogens.
Supplementary Material
Supplementary Table 1. MTB ORFs studied in this report.
Supplementary Figure 1. Responses of bulk polyclonal MTB-reactive, CD137high-origin CD4 T-cell lines from 3 donors to the set of 164 MTB proteins expressed either by IVTT (X-axis) or inclusion bodies (Y-axis). Each dot represents a protein run in singlicate in both formats. The identity of proteins reactive in both formats is provided. Abbreviations within protein names are discussed in Materials and Methods.
Supplementary Figure 2. Comparison of cytokine and proliferation assay readouts for bulk polyclonal MTB-reactive, CD137high-origin CD4 T-cell line from donor US3. A subset of 72 IVTT proteins were assayed in duplicate and 72 hour supernatants removed before addition of 3H thymidine for proliferation assays. IL-17A and IFN-gamma were assayed by bead-based ELISA. A: Cutoffs (three colored horizontal lines) were set using the mean plus 3.09 times the standard deviation of the negative control protein set for each assay. Y axis is mean values in cpm for proliferation, of picograms/ml for cytokines, for each readout for each MTB protein scored as positive in the proliferation format (blue vertical lines). There were no MTB proteins that were positive by either cytokine assay and negative proliferation. Antigen identity is on the X-axis; IVTT proteins were used at 1:1000 and PPD and MTB as indicated in the text. B: Distribution of cytokine results for each MTB protein scored as positive in the proliferation assay.
Supplementary Figure 3. Dose-response curves for MTB-reactive CD4 T-cell clone responder cells, autologous EBV-LCL antigen presenting cells, and defined peptides as indicated in the legend. Y axis is mean of duplicate proliferative responses, while X axis is concentration of peptides. Changes in amino acid sequence from the MTB H37Rv strain are indicated in red in the legend.
Supplementary Table 2. Regions of primers from the MTB strain H37Rv genome used to generate MTB RV1837c fragments by PCR. Each is written in 5′ to 3′ direction. The 5′ primers in addition, at their 5′ end, each have the att homology sequence GGGGACAAGTTTGTACAAAAAAGCAGGCTTC. Similarly the 3′ primers in addition, at their 5′ end, each have the att homology sequence GGGGACCACTTTGTACAAGAAAGCTGGGTC.
Acknowledgments
We wish to thank Vu Huynh and Andy Teng (Antigen Discovery, Inc.) for help with protein quality control by microarray printing, and Rick Lawlor for cytokine ELISAs at Fred Hutchinson Cancer Research Center. Cloning of the ORFS in the Mycobacterium tuberculosis genome was supported by the Foundation of Innovative New Diagnostics (FIND) and by the Bill and Melinda Gates Foundation. The National Institute of Immunology, Delhi, India, kindly allowed use of their cell irradiator. Mycobacterium tuberculosis peptides were kindly provided by BEI Resources, funded by the NIAID at the U.S. National Institutes of Health. This work was partially supported by the U.S. Public Health Services, National Institutes of Health RO1094019 (D.M.K.), R44 AI58365 (D.H.D.), and HHSN272200900043C (W.W.K.).
Abbreviations (currently most are also spelled out in text at first use)
- MTB
Mycobacterium tuberculosis
- ORF
open reading frame
- IVTT
in vitro transcription translation
- ATBI
active Mycobacterium tuberculosis infection
- LTBI
latent Mycobacterium tuberculosis infection
- TST
tuberculin skin test
- PPD
purified protein derivative
- IGRA
interferon gamma release assay
- ICC
intracellular cytokine cytometry
- IFN-gamma
interferon gamma
- IL-2
interleukin 2
- PBMC
peripheral blood mononuclear cell
- CFSE
carboxyfluorescein succinimidyl ester
- TCR
T cell receptor
- PCR
polymerase chain reaction
- RTS
rapid translation system
- DMSO
dimethylsulfoxide
- CFP
cultured filtrate protein
- ESAT
early secretory antigen target
- PHA
phytohemagglutinin
- PBMC
peripheral blood mononuclear cells
- SEB
Staphylococcal enterotoxin B
- IB
inclusion body
- mAb
monoclonal antibody
- TCM
T-cell medium
- MCB
master cell bank
- SDS
sodium dodecyl sulfate
- PE
phycoerythrin
- EBV
Epstein-Barr virus
- LCL
lymphocyte continuous line
Footnotes
Conflict of Interest Statement.
D.H.D. and X.L. declare a financial interest in Antigen Discovery, Inc. D.H.D and X.L. and the University of California, Irvine may financially benefit from this interest if the company is successful in marketing its products that are related to this research. The terms of this arrangement have been reviewed and approved by the University of California, Irvine in accordance with its conflict of interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kaustuv Nayak, Email: kaustuvnayak@gmail.com.
Lichen Jing, Email: lchjing@u.washington.edu.
Ronnie M. Russell, Email: ronnie53@u.washington.edu.
D. Huw Davies, Email: ddavies@uci.edu.
Gary Hermanson, Email: ghermason@antigendiscovery.com.
Douglas M. Molina, Email: dmolina@antigendiscovery.com.
Xiaowu Liang, Email: xliang@immport-inc.com.
David R. Sherman, Email: david.sherman@seattlebiomed.org.
William W. Kwok, Email: bkwok@benaroyaresearch.org.
Junbao Yang, Email: jyang@benaroyaresearch.org.
John Kenneth, Email: Johnkennet@gmail.com.
Syed F. Ahamed, Email: mail2fazil@gmail.com.
Anmol Chandele, Email: chandeleanmol@gmail.com.
Murali-Krishna Kaja, Email: murali.kaja@emory.edu.
David M. Koelle, Email: viralimm@u.washington.edu.
References
- 1.Achkar JM, Dong Y, Holzman RS, Belisle J, Kourbeti IS, Sherpa T, Condos R, Rom WN, Laal S. Mycobacterium tuberculosis malate synthase- and MPT51-based serodiagnostic assay as an adjunct to rapid identification of pulmonary tuberculosis. Clinical and vaccine immunology : CVI. 2006;13:1291–1293. doi: 10.1128/CVI.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achkar JM, Jenny-Avital E, Yu X, Burger S, Leibert E, Bilder PW, Almo SC, Casadevall A, Laal S. Antibodies against immunodominant antigens of Mycobacterium tuberculosis in subjects with suspected tuberculosis in the United States compared by HIV status. Clin Vaccine Immunol. 2010;17:384–392. doi: 10.1128/CVI.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anstrom DM, Remington SJ. The product complex of M. tuberculosis malate synthase revisited. Protein science : a publication of the Protein Society. 2006;15:2002–2007. doi: 10.1110/ps.062300206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arend SM, Geluk A, van Meijgaarden KE, van Dissel JT, Theisen M, Andersen P, Ottenhoff TH. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect Immun. 2000;68:3314–3321. doi: 10.1128/iai.68.6.3314-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic acids research. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, Felgner PL. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infection and immunity. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck JS, Morley SM, Lowe JG, Brown RA, Grange JM, Gibbs JH, Potts RC, Kardjito T. Diversity in migration of CD4 and CD8 lymphocytes in different microanatomical compartments of the skin in the tuberculin reaction in man. British journal of experimental pathology. 1988;69:771–780. [PMC free article] [PubMed] [Google Scholar]
- 8.Belkaid Y, Bouladoux N, Hand TW. Effector and memory T cell responses to commensal bacteria. Trends in immunology. 2013;34:299–306. doi: 10.1016/j.it.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertholet S, Ireton GC, Kahn M, Guderian J, Mohamath R, Stride N, Laughlin EM, Baldwin SL, Vedvick TS, Coler RN, Reed SG. Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis. Journal of immunology. 2008;181:7948–7957. doi: 10.4049/jimmunol.181.11.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chegou NN, Black GF, Loxton AG, Stanley K, Essone PN, Klein MR, Parida SK, Kaufmann SH, Doherty TM, Friggen AH, Franken KL, Ottenhoff TH, Walzl G. Potential of novel Mycobacterium tuberculosis infection phase-dependent antigens in the diagnosis of TB disease in a high burden setting. BMC infectious diseases. 2012;12:10. doi: 10.1186/1471-2334-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YS, Dobos KM, Prenni J, Yang H, Hess A, Rosenkrands I, Andersen P, Ryoo SW, Bai GH, Brennan MJ, Izzo A, Bielefeldt-Ohmann H, Belisle JT. Deciphering the proteome of the in vivo diagnostic reagent “purified protein derivative” from Mycobacterium tuberculosis. Proteomics. 2012;12:979–991. doi: 10.1002/pmic.201100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Commandeur S, Coppola M, Dijkman K, Friggen AH, van Meijgaarden KE, van den Eeden SJ, Wilson L, van der Ploeg-van Schip JJ, Franken KL, Geluk A, Ottenhoff TH. Clonal analysis of the T-cell response to in vivo expressed Mycobacterium tuberculosis protein Rv2034, using a CD154 expression based T-cell cloning method. PLoS One. 2014;9:e99203. doi: 10.1371/journal.pone.0099203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies DH, Chun S, Hermanson G, Tucker JA, Jain A, Nakajima R, Pablo J, Felgner PL, Liang X. T cell antigen discovery using soluble vaccinia proteome reveals recognition of antigens with both virion and nonvirion association. Journal of immunology. 2014;193:1812–1827. doi: 10.4049/jimmunol.1400663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day CL, Mkhwanazi N, Reddy S, Mncube Z, van der Stok M, Klenerman P, Walker BD. Detection of polyfunctional Mycobacterium tuberculosis-specific T cells and association with viral load in HIV-1-infected persons. J Infect Dis. 2008;197:990–999. doi: 10.1086/529048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delogu G, Manganelli R, Brennan MJ. Critical Research Concepts in TB Vaccine Development. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2013 doi: 10.1111/1469-0691.12460. [DOI] [PubMed] [Google Scholar]
- 16.Dillon DC, Alderson MR, Day CH, Lewinsohn DM, Coler R, Bement T, Campos-Neto A, Skeiky YA, Orme IM, Roberts A, Steen S, Dalemans W, Badaro R, Reed SG. Molecular characterization and human T-cell responses to a member of a novel Mycobacterium tuberculosis mtb39 gene family. Infection and immunity. 1999;67:2941–2950. doi: 10.1128/iai.67.6.2941-2950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, Randall A, Molina D, Liang X, Freilich DA, Oloo JA, Blair PL, Aguiar JC, Baldi P, Davies DH, Felgner PL. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorman SE, Picard C, Lammas D, Heyne K, van Dissel JT, Baretto R, Rosenzweig SD, Newport M, Levin M, Roesler J, Kumararatne D, Casanova JL, Holland SM. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 19.Eyles JE, Unal B, Hartley MG, Newstead SL, Flick-Smith H, Prior JL, Oyston PC, Randall A, Mu Y, Hirst S, Molina DM, Davies DH, Milne T, Griffin KF, Baldi P, Titball RW, Felgner PL. Immunodominant Francisella tularensis antigens identified using proteome microarray. ((c))Crown Copyright 2007 Dstl. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 20.Fang X, Wallqvist A, Reifman J. Modeling synergistic drug inhibition of Mycobacterium tuberculosis growth in murine macrophages. Mol Biosyst. 2011;7:2622–2636. doi: 10.1039/c1mb05106g. [DOI] [PubMed] [Google Scholar]
- 21.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, Molina DM, Hirst S, Chew JS, Wang D, Tan G, Duffield M, Yang R, Neel J, Chantratita N, Bancroft G, Lertmemongkolchai G, Davies DH, Baldi P, Peacock S, Titball RW. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13499–13504. doi: 10.1073/pnas.0812080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galagan JE, Sisk P, Stolte C, Weiner B, Koehrsen M, Wymore F, Reddy TB, Zucker JD, Engels R, Gellesch M, Hubble J, Jin H, Larson L, Mao M, Nitzberg M, White J, Zachariah ZK, Sherlock G, Ball CA, Schoolnik GK. TB database 2010: overview and update. Tuberculosis. 2010;90:225–235. doi: 10.1016/j.tube.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Gaseitsiwe S, Valentini D, Mahdavifar S, Reilly M, Ehrnst A, Maeurer M. Peptide microarray-based identification of Mycobacterium tuberculosis epitope binding to HLADRB1* 0101, DRB1*1501, and DRB1*0401. Clinical and vaccine immunology : CVI. 2010;17:168–175. doi: 10.1128/CVI.00208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geiger R, Duhen T, Lanzavecchia A, Sallusto F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. The Journal of experimental medicine. 2009;206:1525–1534. doi: 10.1084/jem.20090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geluk A, van Meijgaarden KE, Joosten SA, Commandeur S, Ottenhoff TH. Innovative Strategies to Identify M. tuberculosis Antigens and Epitopes Using Genome-Wide Analyses. Frontiers in immunology. 2014;5:256. doi: 10.3389/fimmu.2014.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gideon HP, Wilkinson KA, Rustad TR, Oni T, Guio H, Sherman DR, Vordermeier HM, Robertson BD, Young DB, Wilkinson RJ. Bioinformatic and Empirical Analysis of Novel Hypoxia-Inducible Targets of the Human Antituberculosis T Cell Response. J Immunol. 2012 doi: 10.4049/jimmunol.1202281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaziou P, Sismanidis C, Floyd K, Raviglione M. Global Epidemiology of Tuberculosis. Cold Spring Harbor perspectives in medicine. 2014 doi: 10.1101/cshperspect.a017798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci U S A. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haldar S, Sankhyan N, Sharma N, Bansal A, Jain V, Gupta VK, Juneja M, Mishra D, Kapil A, Singh UB, Gulati S, Kalra V, Tyagi JS. Detection of Mycobacterium tuberculosis GlcB or HspX Antigens or devR DNA impacts the rapid diagnosis of tuberculous meningitis in children. PLoS One. 2012;7:e44630. doi: 10.1371/journal.pone.0044630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermanson G, Chun S, Felgner J, Tan X, Pablo J, Nakajima-Sasaki R, Molina DM, Felgner PL, Liang X, Davies DH. Measurement of antibody responses to Modified Vaccinia virus Ankara (MVA) and Dryvax((R)) using proteome microarrays and development of recombinant protein ELISAs. Vaccine. 2012;30:614–625. doi: 10.1016/j.vaccine.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wegener F, Elliott M, Grabstein L, Posavad C, Corey L. Diversity of the CD8+ T cell response to herpes simplex virus type 2 proteins among persons with genital herpes. Journal of virology. 2006;80:5509–5515. doi: 10.1128/JVI.02659-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, Dunkley KM, Aldaz-Carroll L, Eisenberg RJ, Cohen GH, Kwok WW, Sette A, Koelle DM. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. Journal of immunology. 2007;178:6374–6386. doi: 10.4049/jimmunol.178.10.6374. [DOI] [PubMed] [Google Scholar]
- 33.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T cell profiling. Journal of virology. 2008 doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jing L, Haas J, Chong TM, Bruckner JJ, Dann GC, Dong L, Marshak JO, McClurkan CL, Yamamoto TN, Bailer SM, Laing KJ, Wald A, Verjans GMGM, Koelle DM. Herpes simplex virus type 1 T-cells antigens in humans revealed by cross-presentation and genome-wide screening. Journal of Clinical Investigation. 2012;122:654–673. doi: 10.1172/JCI60556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jing L, Schiffer JT, Chong TM, Bruckner JJ, Davies DH, Felgner PL, Haas J, Wald A, Verjans GM, Koelle DM. CD4 T-cell memory responses to viral infections of humans show pronounced immunodominance independent of duration or viral persistence. Journal of virology. 2013;87:2617–2627. doi: 10.1128/JVI.03047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann SH. Fact and fiction in tuberculosis vaccine research: 10 years later. Lancet Infect Dis. 2011;11:633–640. doi: 10.1016/S1473-3099(11)70146-3. [DOI] [PubMed] [Google Scholar]
- 38.Kiecker F, Streitz M, Ay B, Cherepnev G, Volk HD, Volkmer-Engert R, Kern F. Analysis of antigen-specific T-cell responses with synthetic peptides--what kind of peptide for which purpose? Human immunology. 2004;65:523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Koelle DM. Expression cloning for the discovery of viral antigens and epitopes recognized by T-cells. Methods. 2003;29:213–226. doi: 10.1016/s1046-2023(02)00344-4. [DOI] [PubMed] [Google Scholar]
- 40.Koelle DM, Chen H, Gavin MA, Wald A, Kwok WW, Corey L. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. Journal of immunology. 2001;166:4049–4058. doi: 10.4049/jimmunol.166.6.4049. [DOI] [PubMed] [Google Scholar]
- 41.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, Triezenberg SJ. Antigenic specificity of human CD4+ T cell clones recovered from recurrent genital HSV-2 lesions. Journal of virology. 1994;68:2803–2810. doi: 10.1128/jvi.68.5.2803-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koelle DM, Posavad CM, Barnum GR, Johnson ML, Frank JM, Corey L. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. Journal of Clinical Investigation. 1998;101:1500–1508. doi: 10.1172/JCI1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koelle DM, Reymond SN, Chen H, Kwok WW, McClurkan C, Gyaltsong T, Petersdorf EW, Rotkis W, Talley AR, Harrison DA. Tegument-specific, virus-reactive CD4 T cells localize to the cornea in herpes simplex virus interstitial keratitis in humans. Journal of virology. 2000;74:10930–10938. doi: 10.1128/jvi.74.23.10930-10938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koelle DM, Schomogyi M, McClurkan C, Reymond SN, Chen HB. CD4 T-cell responses to herpes simplex virus type 2 major capsid protein VP5: comparison with responses to tegument and envelope glycoproteins. J Virol. 2000;74:11422–11425. doi: 10.1128/jvi.74.23.11422-11425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunnath-Velayudhan S, Davidow AL, Wang HY, Molina DM, Huynh VT, Salamon H, Pine R, Michel G, Perkins MD, Xiaowu L, Felgner PL, Flynn JL, Catanzaro A, Gennaro ML. Proteome-scale antibody responses and outcome of Mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J Infect Dis. 2012;206:697–705. doi: 10.1093/infdis/jis421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunnath-Velayudhan S, Salamon H, Wang HY, Davidow AL, Molina DM, Huynh VT, Cirillo DM, Michel G, Talbot EA, Perkins MD, Felgner PL, Liang X, Gennaro ML. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwok WW, Liu AW, Novak EJ, Gebe JA, Reymond SN, Ettinger RA, Nepom GT, Koelle DM. HLA-DQ tetramers identify epitope-specific T-cells in peripheral blood of herpes simplex virus-2-infected individuals: direct detection of immunodominant antigen responsive cells. Journal of immunology. 2000;164:4244–4249. doi: 10.4049/jimmunol.164.8.4244. [DOI] [PubMed] [Google Scholar]
- 48.Lambert PH, Hawkridge T, Hanekom WA. New vaccines against tuberculosis. Clin Chest Med. 2009;30:811–826. x. doi: 10.1016/j.ccm.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 49.Larson RP, Shafiani S, Urdahl KB. Foxp3(+) regulatory T cells in tuberculosis. Advances in experimental medicine and biology. 2013;783:165–180. doi: 10.1007/978-1-4614-6111-1_9. [DOI] [PubMed] [Google Scholar]
- 50.Leung WL, Law KL, Leung VS, Yip CW, Leung CC, Tam CM, Kam KM. Comparison of intracellular cytokine flow cytometry and an enzyme immunoassay for evaluation of cellular immune response to active tuberculosis. Clin Vaccine Immunol. 2009;16:344–351. doi: 10.1128/CVI.00159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Lewinsohn DM, Zhu L, Madison VJ, Dillon DC, Fling SP, Reed SG, Grabstein KH, Alderson MR. Classically restricted human CD8+ T lymphocytes derived from Mycobacterium tuberculosis-infected cells: definition of antigenic specificity. Journal of immunology. 2001;166:439–446. doi: 10.4049/jimmunol.166.1.439. [DOI] [PubMed] [Google Scholar]
- 53.Liang L, Tan X, Juarez S, Villaverde H, Pablo J, Nakajima-Sasaki R, Gotuzzo E, Saito M, Hermanson G, Molina D, Felgner S, Morrow WJ, Liang X, Gilman RH, Davies DH, Tsolis RM, Vinetz JM, Felgner PL. Systems biology approach predicts antibody signature associated with Brucella melitensis infection in humans. Journal of proteome research. 2011;10:4813–4824. doi: 10.1021/pr200619r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS pathogens. 2013;9:e1003130. doi: 10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mannering SI, Harrison LC, Williamson NA, Morris JS, Thearle DJ, Jensen KP, Kay TW, Rossjohn J, Falk BA, Nepom GT, Purcell AW. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J Exp Med. 2005;202:1191–1197. doi: 10.1084/jem.20051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazurek GH, Jereb J, Vernon A, LoBue P, Goldberg S, Castro K, Committee IE Centers for Disease, C and Prevention. Updated guidelines for using Interferon Gamma Release Assays to detect Mycobacterium tuberculosis infection - United States, 2010. MMWR. Recommendations and reports : Morbidity and mortality weekly report. Recommendations and reports / Centers for Disease Control. 2010;59:1–25. [PubMed] [Google Scholar]
- 57.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 58.Moss NJ, Magaret A, Laing KJ, Kask AS, Wang M, Mark KE, Schiffer JT, Wald A, Koelle DM. Peripheral blood CD4 T-cell and plasmacytoid dendritic cell (pDC) reactivity to herpes simplex virus 2 and pDC number do not correlate with the clinical or virologic severity of recurrent genital herpes. J Virol. 2012;86:9952–9963. doi: 10.1128/JVI.00829-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mustafa AS, Oftung F, Amoudy HA, Madi NM, Abal AT, Shaban F, Rosen Krands I, Andersen P. Multiple epitopes from the Mycobacterium tuberculosis ESAT-6 antigen are recognized by antigen-specific human T cell lines. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2000;30(Suppl 3):S201–205. doi: 10.1086/313862. [DOI] [PubMed] [Google Scholar]
- 60.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. Journal of immunology. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 61.Pamer E, Cresswell P. Mechanisms of MHC class-I-restricted antigen processing. Annual review of immunology. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 62.Philips JA, Ernst JD. Tuberculosis Pathogenesis and Immunity. Annu Rev Pathol. 2012:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 63.Prados-Rosales R, Carreno LJ, Batista-Gonzalez A, Baena A, Venkataswamy MM, Xu J, Yu X, Wallstrom G, Magee DM, LaBaer J, Achkar JM, Jacobs WR, Jr, Chan J, Porcelli SA, Casadevall A. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio. 2014;5:e01921–01914. doi: 10.1128/mBio.01921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sayes F, Sun L, Di Luca M, Simeone R, Degaiffier N, Fiette L, Esin S, Brosch R, Bottai D, Leclerc C, Majlessi L. Strong immunogenicity and cross-reactivity of Mycobacterium tuberculosis ESX-5 type VII secretion: encoded PE-PPE proteins predicts vaccine potential. Cell host & microbe. 2012;11:352–363. doi: 10.1016/j.chom.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 66.Sheikh A, Khanam F, Sayeed MA, Rahman T, Pacek M, Hu Y, Rollins A, Bhuiyan MS, Rollins S, Kalsy A, Arifuzzaman M, Leung DT, Sarracino DA, Krastins B, Charles RC, Larocque RC, Cravioto A, Calderwood SB, Brooks WA, Harris JB, Labaer J, Qadri F, Ryan ET. Interferon-gamma and proliferation responses to Salmonella enterica Serotype Typhi proteins in patients with S. Typhi Bacteremia in Dhaka, Bangladesh. PLoS neglected tropical diseases. 2011;5:e1193. doi: 10.1371/journal.pntd.0001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh KK, Dong Y, Belisle JT, Harder J, Arora VK, Laal S. Antigens of Mycobacterium tuberculosis recognized by antibodies during incipient, subclinical tuberculosis. Clin Diagn Lab Immunol. 2005;12:354–358. doi: 10.1128/CDLI.12.2.354-358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith CV, Huang CC, Miczak A, Russell DG, Sacchettini JC, Honer zu Bentrup K. Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:1735–1743. doi: 10.1074/jbc.M209248200. [DOI] [PubMed] [Google Scholar]
- 69.Stern LJ, Calvo-Calle JM. HLA-DR: molecular insights and vaccine design. Current pharmaceutical design. 2009;15:3249–3261. doi: 10.2174/138161209789105171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memoryphenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sultana R, Tanneeru K, Guruprasad L. The PE-PPE domain in mycobacterium reveals a serine alpha/beta hydrolase fold and function: an in-silico analysis. PLoS One. 2011;6:e16745. doi: 10.1371/journal.pone.0016745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sundaresh S, Doolan DL, Hirst S, Mu Y, Unal B, Davies DH, Felgner PL, Baldi P. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics. 2006;22:1760–1766. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
- 73.Sundaresh S, Randall A, Unal B, Petersen JM, Belisle JT, Hartley MG, Duffield M, Titball RW, Davies DH, Felgner PL, Baldi P. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 74.Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, Mahomed H, McShane H, Team MATS. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013;381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Dissel JT, Soonawala D, Joosten SA, Prins C, Arend SM, Bang P, Tingskov PN, Lingnau K, Nouta J, Hoff ST, Rosenkrands I, Kromann I, Ottenhoff TH, Doherty TM, Andersen P. Ag85B-ESAT-6 adjuvanted with IC31(R) promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in volunteers with previous BCG vaccination or tuberculosis infection. Vaccine. 2011;29:2100–2109. doi: 10.1016/j.vaccine.2010.12.135. [DOI] [PubMed] [Google Scholar]
- 76.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic acids research. 2010;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walker KB, Brennan MJ, Ho MM, Eskola J, Thiry G, Sadoff J, Dobbelaer R, Grode L, Liu MA, Fruth U, Lambert PH. The second Geneva Consensus: Recommendations for novel live TB vaccines. Vaccine. 2010 doi: 10.1016/j.vaccine.2009.12.083. [DOI] [PubMed] [Google Scholar]
- 78.Weaver JM, Lazarski CA, Richards KA, Chaves FA, Jenks SA, Menges PR, Sant AJ. Immunodominance of CD4 T cells to foreign antigens is peptide intrinsic and independent of molecular context: implications for vaccine design. Journal of immunology. 2008;181:3039–3048. doi: 10.4049/jimmunol.181.5.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wong EB, Cohen KA, Bishai WR. Rising to the challenge: new therapies for tuberculosis. Trends in microbiology. 2013;21:493–501. doi: 10.1016/j.tim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yim JJ, Selvaraj P. Genetic susceptibility in tuberculosis. Respirology. 2010;15:241–256. doi: 10.1111/j.1440-1843.2009.01690.x. [DOI] [PubMed] [Google Scholar]
- 81.Zhu YH, Gao YF, Chen F, Liu W, Zhai MX, Zhai WJ, Qi YM, Ye Y. Identification of novel T cell epitopes from efflux pumps of Mycobacterium tuberculosis. Immunol Lett. 2011;140:68–73. doi: 10.1016/j.imlet.2011.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. MTB ORFs studied in this report.
Supplementary Figure 1. Responses of bulk polyclonal MTB-reactive, CD137high-origin CD4 T-cell lines from 3 donors to the set of 164 MTB proteins expressed either by IVTT (X-axis) or inclusion bodies (Y-axis). Each dot represents a protein run in singlicate in both formats. The identity of proteins reactive in both formats is provided. Abbreviations within protein names are discussed in Materials and Methods.
Supplementary Figure 2. Comparison of cytokine and proliferation assay readouts for bulk polyclonal MTB-reactive, CD137high-origin CD4 T-cell line from donor US3. A subset of 72 IVTT proteins were assayed in duplicate and 72 hour supernatants removed before addition of 3H thymidine for proliferation assays. IL-17A and IFN-gamma were assayed by bead-based ELISA. A: Cutoffs (three colored horizontal lines) were set using the mean plus 3.09 times the standard deviation of the negative control protein set for each assay. Y axis is mean values in cpm for proliferation, of picograms/ml for cytokines, for each readout for each MTB protein scored as positive in the proliferation format (blue vertical lines). There were no MTB proteins that were positive by either cytokine assay and negative proliferation. Antigen identity is on the X-axis; IVTT proteins were used at 1:1000 and PPD and MTB as indicated in the text. B: Distribution of cytokine results for each MTB protein scored as positive in the proliferation assay.
Supplementary Figure 3. Dose-response curves for MTB-reactive CD4 T-cell clone responder cells, autologous EBV-LCL antigen presenting cells, and defined peptides as indicated in the legend. Y axis is mean of duplicate proliferative responses, while X axis is concentration of peptides. Changes in amino acid sequence from the MTB H37Rv strain are indicated in red in the legend.
Supplementary Table 2. Regions of primers from the MTB strain H37Rv genome used to generate MTB RV1837c fragments by PCR. Each is written in 5′ to 3′ direction. The 5′ primers in addition, at their 5′ end, each have the att homology sequence GGGGACAAGTTTGTACAAAAAAGCAGGCTTC. Similarly the 3′ primers in addition, at their 5′ end, each have the att homology sequence GGGGACCACTTTGTACAAGAAAGCTGGGTC.