Abstract
Protein synthesis rates can affect gene expression and the folding and activity of the translation product. Interactions between the nascent polypeptide and the ribosome exit tunnel represent one mode of regulating synthesis rates. The SecM protein arrests its own translation, and release of arrest at the translocon has been proposed to occur by mechanical force. Using optical tweezers, we demonstrate that arrest of SecM-stalled ribosomes can indeed be rescued by force alone and that the force needed to release stalling can be generated in vivo by a nascent chain folding near the ribosome tunnel exit. We formulate a kinetic model describing how a protein can regulate its own synthesis by the force generated during folding, tuning ribosome activity to structure acquisition by a nascent polypeptide.
The ribosome translates mRNA into amino acid sequences that contain the information needed for the polypeptide to attain its native structure. Differential usage of synonymous codons and structural elements in the mRNA modulate polypeptide elongation rates. Such rate variations may be required for proper folding and processing of nascent proteins (1). Moreover, interactions of specific nascent chain sequences (2, 3) with the ribosome exit tunnel (4) result in reduced rates of elongation. The bacterial SecM protein represents an example of a stalling sequence that interacts with the ribosome exit tunnel and allosterically represses the peptidyl transferase activity of the ribosome (4–7). Translation of SecM regulates expression of SecA, the motor component of the bacterial Sec translocon (2). Release of stalling in vivo requires interactions between nascent SecM and the translocon machinery (8, 9). It has been suggested that mechanical force exerted by the translocon relieves elongation arrest and leads to translation restart (10).
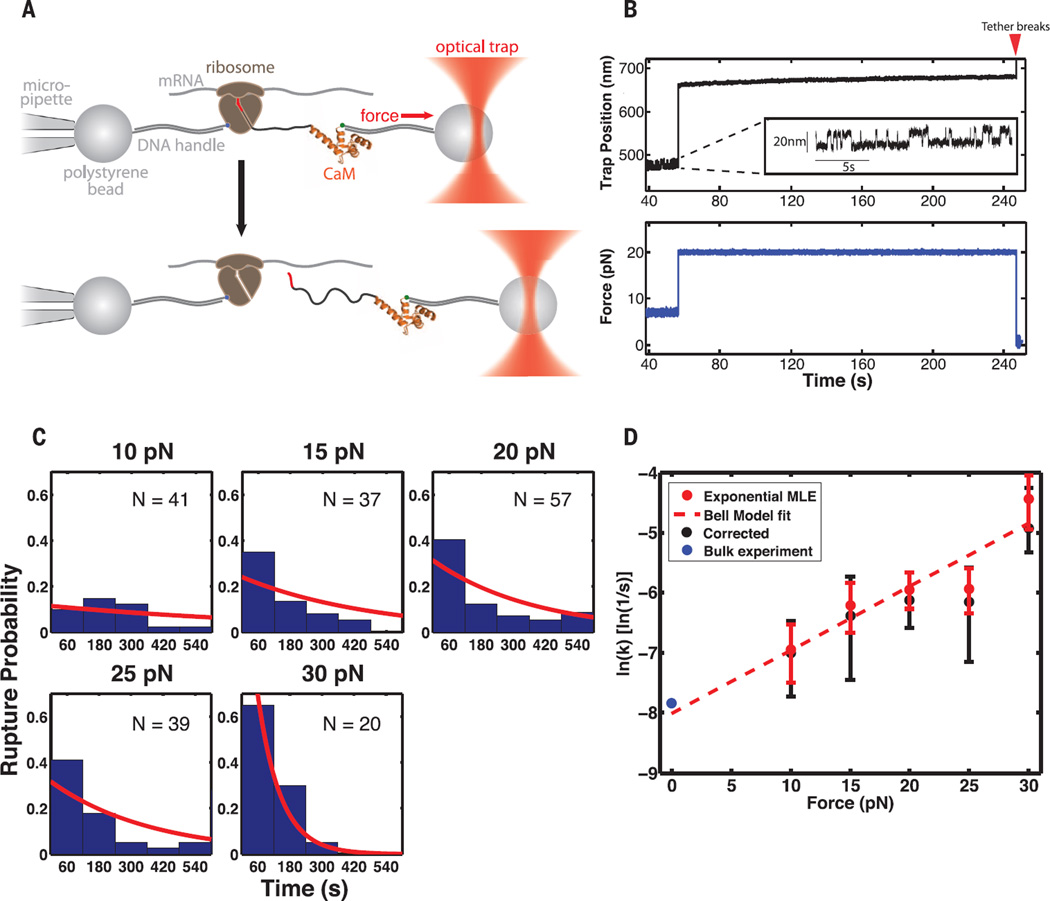
To investigate the effect of force on the release of SecM-stalled ribosome–nascent chains (RNCs), we adapted a single-molecule optical tweezers assay (11) (Fig. 1A), enabling the application of defined forces to single ribosome-associated nascent polypeptides. We generated stalled RNCs that contained the C-terminal domain of human calmodulin (CaM) (figs. S1 and S2). CaM provides a mechanical fingerprint (12) in our experiments by exhibiting equilibrium folding and unfolding (“hopping”) at ~7 pN (Fig. 1B and supplementary materials). To detect release of stalled ribosomes, we used the antibiotic puromycin. Puromycin binds to the ribosomal A site and is incorporated into the nascent polypeptide, leading to its release from the ribosome (13). SecM-arrested ribosomes, containing a prolyl-tRNApro stably bound in the A site, are refractory to treatment with puromycin, but become sensitive after arrest release, proline incorporation, and translocation (14) (figs. S3 and S4). In the presence of puromycin and EF-G, arrest release will become apparent as a rupture of the tether (Fig. 1B and fig. S4).
Fig. 1. A direct applied force catalyzes release of SecM-mediated arrest.
(A) Experimental setup for optical tweezers experiments. When the nascent chain is transferred to puromycin, the assembly breaks. The structure of CaM was obtained from Protein Data Bank (PDB) ID 1CLL. (B) Example trace for restart experiment. After the “hopping” signature of CaM is observed (inset) at 7 pN, the force is raised to 20 pN. Red arrow: The tether breaks after ~3 min at 20 pN. (C) Restart lifetimes at each force. Red lines: Distributions returned by the right-censoring MLE. (D) Force-dependent rates for restart of SecM-stalled RNCs in the optical tweezers. Rates are determined as shown in (C), with error bars representing 95% CIs returned by the MLE. Red dotted line: Fit of Bell’s model to optical tweezers data. Δx‡: 0.4 nm (95% CI: 0.1 nm, 0.8 nm) and k0: 3.3 × 10−4 s−1 (95% CI: 0.5 × 10−4 s−1, 20 × 10−4 s−1). Black points: Rates determined with a method to account for nonspecific tether rupture (fig. S6). Error determined by bootstrapping. Blue dot: Lifetime obtained from bulk experiment (fig. S3).
We applied a defined, constant force to the molecule in the range of 10 to 30 pN and measured the time required to restart translation, as measured by tether rupture. The mean restart times decreased with increasing force (Fig. 1C). We calculated the rate of stalling rescue as a function of the applied force (Fig. 1, C and D, and figs. S5 and S6). By fitting the force-dependent rates to Bell’s model (15), we estimated a distance to the transition state (Δx‡) of 0.4 nm [95% confidence interval (CI): 0.1 nm, 0.8 nm] and a zero-force rupture rate (k0) of 3 × 10−4 s−1 (95% CI: 0.5×10−4 s−1, 20 × 10−4 s−1). This rate is in agreement with biochemical ensemble experiments, in which no force was applied (Fig. 1D, blue dot and fig. S3). In the force range of our experiments, release of SecM-mediated arrest is accelerated by more than an order of magnitude (Fig. 1D), supporting the hypothesis that SecM arrest is relieved by the mechanical force generated by the SecA adenosine triphosphatase (ATPase).
Cotranslational insertion of transmembrane helices via the translocon can release SecM-mediated stalling, presumably by generating force (16). We wondered whether folding of a nascent globular protein domain could generate a force capable of modulating elongation by acting on peptide-tunnel interactions. Such interactions could serve to tune elongation rates to folding transitions (3, 4). The exit tunnel is too narrow to accommodate folded protein domains (17); therefore, as a nascent polypeptide emerges from the exit tunnel and folds in close proximity to the ribosome, it will be sterically excluded from the tunnel. This steric exclusion might generate a force that pulls on the nascent chain within the exit tunnel, which could modulate ribosome activity.
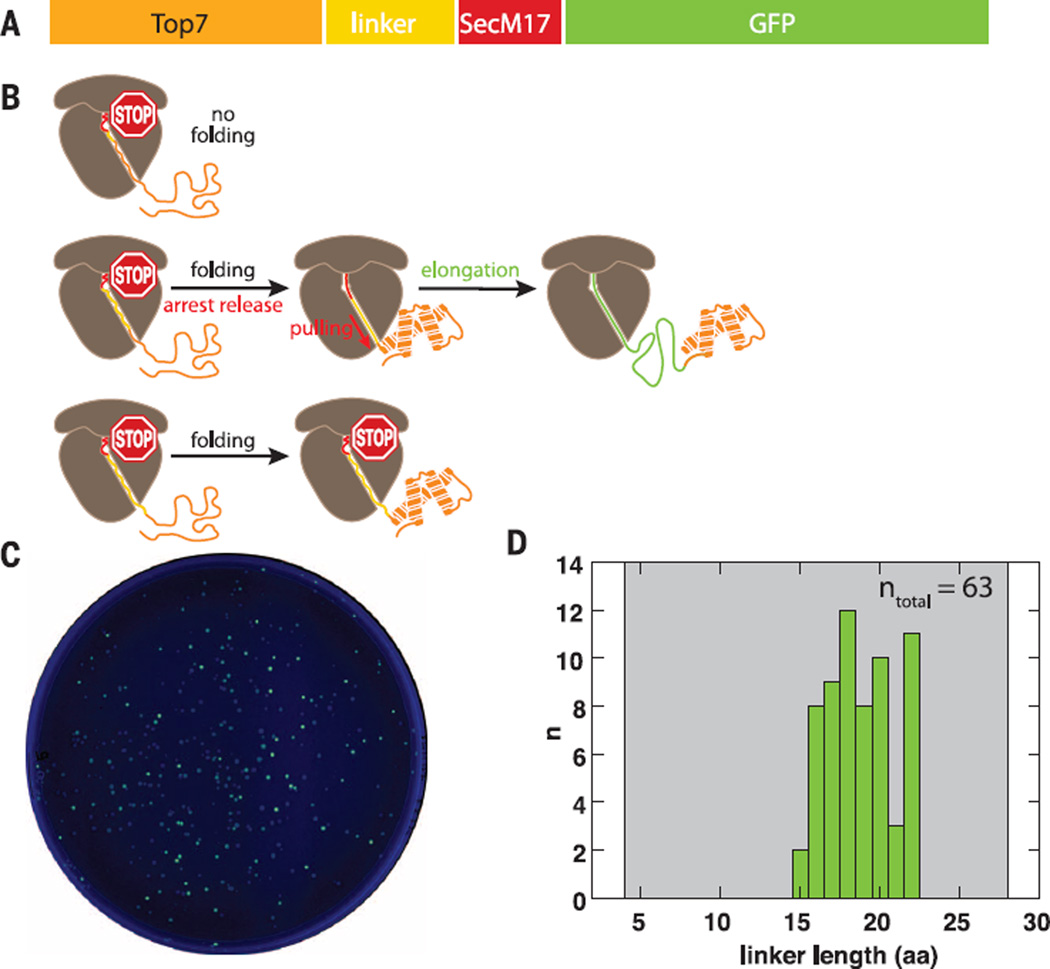
Having established that force accelerates SecM arrest release, we used SecM as a sensor to detect if nascent protein folding outside the ribosome can release the arrest. We constructed a library of plasmids encoding fusion proteins in which the stalling sequence (SecM17) is separated from the de novo–designed protein Top7 (18) by flexible linker sequences of various lengths, followed by a reporter green fluorescent protein (GFP) (Fig. 2A, fig. S7, and supplementary materials). Top 7 folds rapidly against an applied force in close proximity to the ribosome (fig. S8). The GFP coding sequence is translated only upon successful release of the SecM17-mediated translation arrest. Variations in the length of the linker separating Top7 and SecM17 would affect the translation outcome of these constructs (Fig. 2B). Short linker sequences will not allow folding of Top7 because the C terminus of the protein will be sequestered in the exit tunnel (Fig. 2B, top). Intermediate-length linkers will allow more of the Top7 sequence to emerge from the ribosome tunnel and for the protein to fold and produce the steric exclusion folding force (Fig. 2B, middle). And while longer linker sequences would also allow Top7 to fold, increased separation between the folding domain and the ribosome should abolish the proposed force-generating steric exclusion release mechanism (Fig. 2B, bottom).
Fig. 2. Nascent protein folding near the ribosome tunnel exit can rescue SecM-mediated stalling.
(A) Primary sequence of the construct used in the GFP reporter assay. (B) Schematic illustrating the translation outcome for a short (top), intermediate (middle), and long (bottom) linker. (C) Ultraviolet-illuminated image of colonies transformed with the linker library and grown under inducing conditions. (D) Histogramof linker lengths recovered by sequencing of fluorescent colonies. Gray shaded area: library range.
We transformed Escherichia coli with the plasmid library containing linker lengths varying from 4 to 28 amino acids. When grown under inducing conditions, a fraction of the colonies exhibited green fluorescence, indicating accumulation of GFP (Fig. 2C) and suggesting that SecM17-mediated stalling had been rescued in some of the transformants. We isolated and sequenced plasmid DNA from 63 fluorescent colonies. Plasmids isolated from fluorescent colonies contained linker sequences between 15 and 22 amino acids in length (Fig. 2D and fig. S9). Given that the SecM17 sequence contributes 16 amino acids to the polypeptide and the ribosome tunnel can accommodate 30 to 35 residues (17), a linker length of 15 to 22 amino acids corresponds to having the protein sequence barely outside the tunnel exit. These results suggest that nascent chain folding near the ribosome tunnel exit can result in release of SecM arrest by stretching the polypeptide in the tunnel.
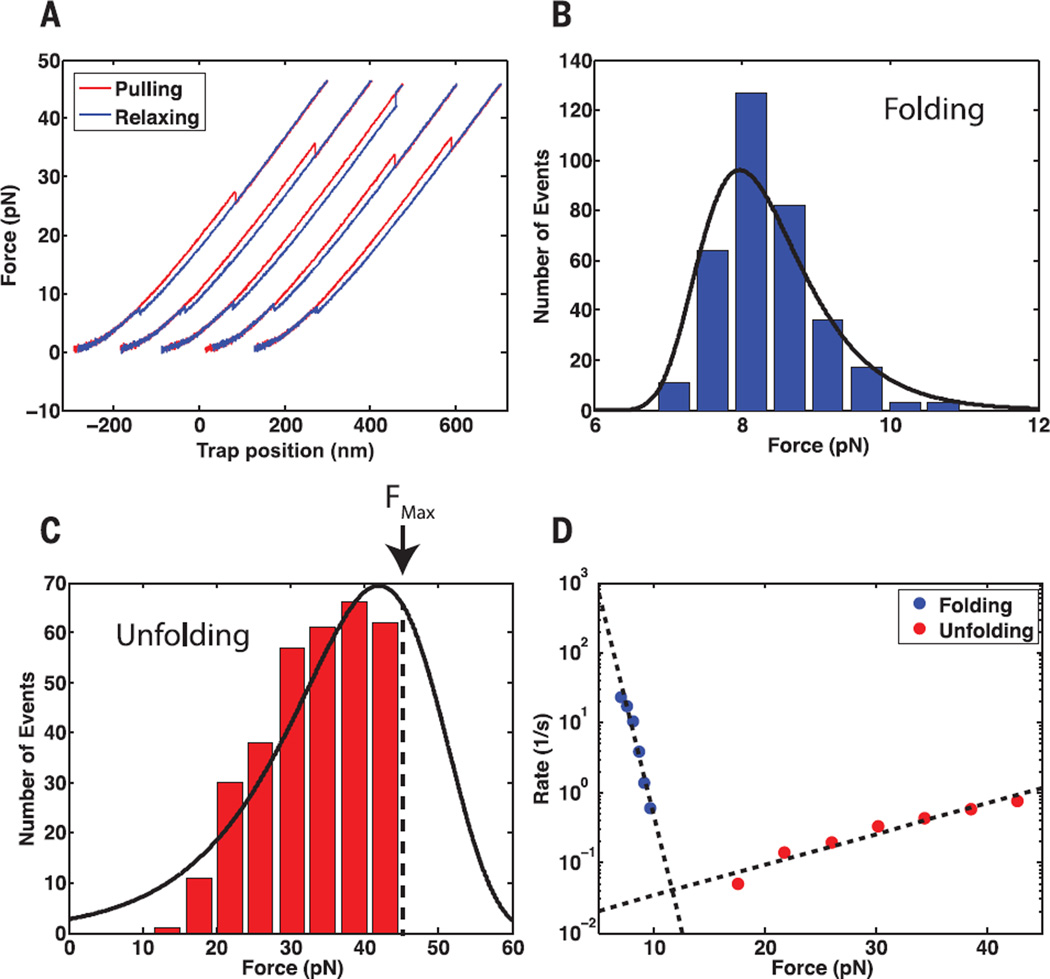
When Top 7 folds near the tunnel exit, it does so against the steric exclusion force that it generates in the process. The protein must be able to fold against this force and remain folded for a sufficiently long period of time to release stalling by SecM. To estimate the forces generated by the protein, we performed optical tweezers force spectroscopy measurements with single Top7 molecules tethered by their termini (fig. S10). We measured the distributions of lifetimes of both the unfolded and folded states (Fig. 3, A to C). From these distributions, we extracted the force-dependent rates of folding and unfolding events (Fig. 3D and supplementary materials) (19). Folding rates decrease with increasing force applied to the protein, and unfolding rates increase. The intersection of the folding and unfolding distributions occurs at ~12 pN and represents the force at which the protein has equal probability of being folded or unfolded, a mean lifetime of 28 s for both states. Thus, as it emerges on the surface of the ribosome and folds, Top7 can exert a force of at least 12 pN for many seconds on the nascent chain still in the tunnel, before it unfolds. On the basis of our single-molecule results, we propose a kinetic scheme that describes how folding can modulate arrest release rates (Fig. 4A):
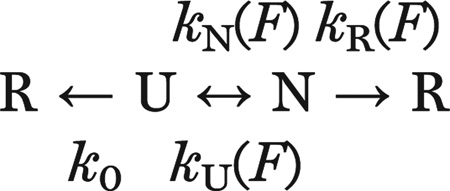 |
Fig. 3. Top7 refolds against an applied mechanical load.
(A) Example force ramp cycles for a single Top7 molecule. Pulling is shown in red, relaxing in blue. Successive cycles are offset along the x axis for display purposes. (B and C) Folding and unfolding force distributions, respectively, for Top7 at a pulling speed of 100 nm/s. Black line: Distributions reconstructed from the force-dependent rates in (D). The unfolding-force distribution in (C) is right-censored because the maximum force in pulling experiments was set at 45 pN to avoid tether rupture. (D) Force-dependent rates of folding and unfolding extracted from the distributions in (B and C). Dashed lines: fit of Bell’s model to the force-dependent rates. For folding, Δx‡: 6 nm (95% CI: 4 nm, 8 nm) and k0: 1 × 106 s−1 (95% CI: 0.04 × 106 s−1, 30 × 106 s−1). For unfolding, Δx‡: 0.4 nm (95% CI: 0.3 nm, 0.6 nm) and k0: 0.01 s−1 (95% CI: 0.003 s−1, 0.03 s−1) (supplementary materials).
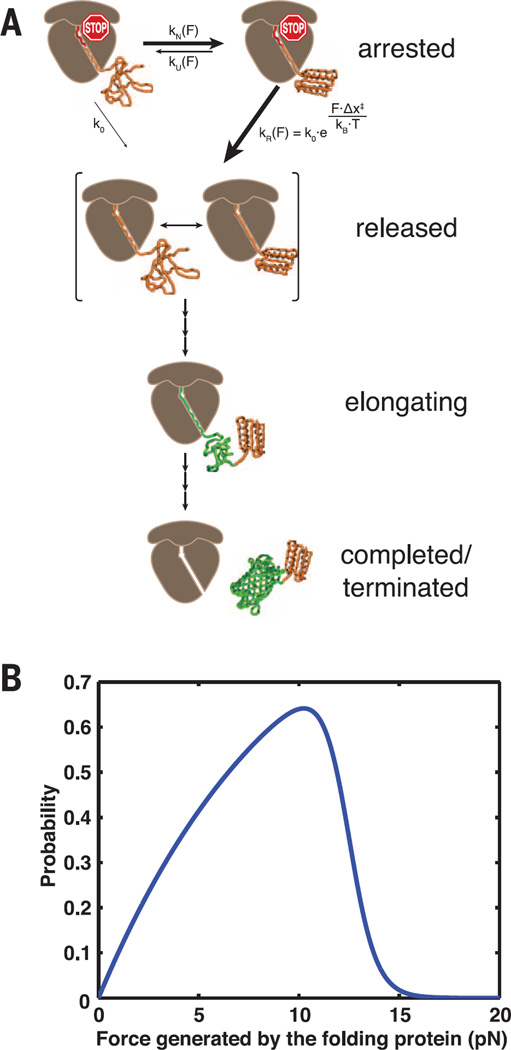
Fig. 4. Kinetic model for folding-induced release of stalled ribosomes.
(A) Kinetic scheme illustrating the pathway to release of translation arrest. The nascent polypeptide can transit reversibly between the native and unfolded states, with rate constants kN(F) and kU(F). Once folded, the nascent protein both generates and experiences a force, “F,” which can drive it either irreversibly to the “released” state, with rate constant kR(F), or back to the unfolded state. In addition, the stall can be released via the irreversible spontaneous process from the unfolded state, with rate constant k0, which is independent of force. PDB: 1QYS and 1EMA. (B) The probability of force-catalyzed stall release is plotted as a function of the folding force.
U and N are the unfolded and natively folded states of Top7; F is the force, and R the stall-released ribosome state, which can be accessed from the folded (N) state [at a rate accelerated by the exclusion steric force, kR(F)] or from the unfolded (U) state (at the basal, force-independent rate, k0). kN(F) and kU(F) represent the force-dependent folding and unfolding rates of Top7 obtained from single-molecule experiments (fig. S11).
The effectiveness with which the pulling force catalyzes stall release depends on the force generated upon folding, the probability that the protein folds at that force, and the lifetime of the folded state. To determine how translation stall release rates depend on these factors, we solved the kinetic scheme above for the effective stall release rate, kReff(F) (supplementary materials). An approximate solution that assumes equilibrium between N and U (good for kU >> kR) yields (see supplementary materials for the exact solution):
where f0 is the fraction of natively folded protein assuming equilibrium with the unfolded state. f0 decreases with force as the equilibrium is tilted toward the unfolded state; in contrast, the term in parentheses increases with force as the release rate increases and the protein is biased toward the unfolded state (fig. S12). Owing mainly to the difference in experimental geometry between the dual-tethered Top7 in the optical tweezers and the singly tethered Top7 in the in vivo experiments, the force-dependent rates likely represent an underestimate to the true rates (supplementary materials). Although the experimentally observed quantity, kReff (F), represents the composite kinetics of both the spontaneous and force-dependent release rates, we calculated the probability that release proceeds by the mechanical, as opposed to the spontaneous, process. This function provides the most likely force at which folding of the nascent protein leads to the release of the ribosome stall and can be expressed as:
A plot of P(F) versus force shows that the probability of force-catalyzed stall release is a maximum near 10 pN (Fig. 4B). At forces below 12 pN, Top7 is mostly folded (N), so release proceeds largely through the mechanical path, whose rate, kR, increases with increasing force; at forces higher than 12 pN, the protein spends shorter and shorter times in the folded state, so release occurs more and more via the slower, spontaneous path, k0, from the unfolded state.
Our results provide evidence that the translocon must generate at least 10 pN of force to relieve SecM-induced ribosomal arrest. It appears that another translocase, ClpX, operates in a similar force regime (20, 21). Given that a number of polypeptide sequences are known to stall the ribosome (3, 22–24), our results suggest that force can play a generally important role as a regulator of elongation. In instances where ribosome–nascent chain interactions are less robust than the SecM system, folding could play an important role in modulating translation elongation and vice versa. For example, if such interactions occur near the C terminus of a newly synthesized domain, elongation would slow down when the polypeptide segment just outside the ribosome begins to acquire stable structure, permitting the folding of this segment to be completed before more of the polypeptide is synthesized. Likewise, folding of the domain, and the force so generated, may provide the signal for speeding up elongation through and beyond the regulatory signal. A force could be generated not only by nascent chain folding, but also by the binding of partners to the nascent chain outside the ribosome. In eukaryotic cells, ribosome profiling experiments have shown that the chaperone Hsp70 relieves global stalling of ribosomes near the beginning of genes (25, 26), perhaps by generating a pulling force similar to the scenario of Hsp70 binding to protein aggregates (27). Thus, force generated by either nascent chain folding or chaperone binding could constitute an important feedback mechanism to tune elongation to folding.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Chistol and L. Alexander for guidance regarding data analysis and the kinetic model, and members of the Bustamante and Tinoco laboratories for helpful discussions on the manuscript. D.H.G acknowledges the NSF’s Graduate Research Fellowship; C.M.K acknowledges support from the NIH K99 Award 5K99GM086516 and the QB3 Institute, Berkeley (Distinguished Postdoctoral Fellowship); M.R. acknowledges support from the Damon Runyon Cancer Research Foundation (DRG-2096-11). C.B. acknowledges support from NIH grant 5R01GM32543; I.T. acknowledges support from NIH grant GM10840.
Footnotes
www.sciencemag.org/content/348/6233/457/suppl/DC1
Materials and Methods
Figs. S1 to S12
References (28–30)
REFERENCES AND NOTES
- 1.Nissley DA, O’Brien EP. JACS. 2014;136:17892–17898. doi: 10.1021/ja510082j. [DOI] [PubMed] [Google Scholar]
- 2.Ito K, Chiba S. Annu. Rev. Biochem. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 3.Deutsch . Regulatory Nascent Polypeptides. Tokyo: Springer; 2014. pp. 61–86. [Google Scholar]
- 4.Wilson DN, Beckmann R. Curr. Opin. Struct. Biol. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Tsai A, Kornberg G, Johansson M, Chen J, Puglisi JD. Cell Rep. 2014;7:1521–1533. doi: 10.1016/j.celrep.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa H, Ito K. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 7.Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. Biophys. J. 2012;103:331–341. doi: 10.1016/j.bpj.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap MN, Bernstein HD. Mol. Microbiol. 2011;81:540–553. doi: 10.1111/j.1365-2958.2011.07713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamori K, Chiba S, Ito K. FEBS Lett. 2014;588:3098–3103. doi: 10.1016/j.febslet.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 10.Butkus ME, Prundeanu LB, Oliver DB. J. Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Jr, Bustamante C. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junker JP, Ziegler F, Rief M. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 13.Nathans P. Natl. Acad. Sci. U.S.A. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muto H, Nakatogawa H, Ito K. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Bell GI. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 16.Ismail N, Hedman R, Schiller N, von Heijne G. Nat. Struct. Mol. Biol. 2012;19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voss NR, Gerstein M, Steitz TA, Moore PB. J. Mol. Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Kuhlman B, et al. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 19.Dudko OK, Hummer G, Szabo A. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maillard RA, et al. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolstenhulme CJ, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ude S, et al. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 24.Doerfel LK, et al. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 25.Shalgi R, et al. Mol. Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Han Y, Qian SB. Mol. Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.