Abstract
Methods to introduce targeted double-strand breaks (DSBs) into DNA enable precise genome editing by increasing the rate at which externally supplied DNA fragments are incorporated into the genome through homologous recombination. The efficiency of these methods is limited by non-homologous end joining (NHEJ), an alternative DNA repair pathway that competes with homology-directed repair (HDR). To promote HDR at the expense of NHEJ, we targeted DNA ligase IV, a key enzyme in the NHEJ pathway, using the inhibitor Scr7. Scr7 treatment increased the efficiency of HDR-mediated genome editing using Cas9 in mammalian cell lines and in mice for all four genes examined up to 19-fold. This approach should be applicable to other customizable endonucleases, such as zinc finger nucleases and transcription activator like effector nucleases, and to non-mammalian cells with sufficiently conserved mechanisms of NHEJ and HDR.
The type II bacterial clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system is an efficient tool for the targeted introduction of mutations into eukaryotic genomes 1, 2. Properly designed single guide (sg) RNAs efficiently induce Cas9-mediated DSBs at desired target sites while minimizing off-target effects3, 4. The DSBs stimulate DNA repair by at least two distinct mechanisms—NHEJ and HDR—both of which are active in nearly all cell types and organisms 2. Cas9-mediated modification of the murine genome through NHEJ can reach efficiencies of 20% to 60% 5, 6. Because NHEJ is error-prone and introduces unpredictable patterns of insertions and deletions (indels) 6-8, it is suitable for introducing small random mutations. However, it does not enable precise genome editing by HDR-mediated incorporation of an exogenous DNA fragment. This has been achieved by co-injection of a targeted endonuclease and a single-stranded or double-stranded DNA template homologous to the sequences flanking the cleavage site 5, 6, 9. Because the frequency of HDR is inherently low 5, 6, 9, the efficiency of insertional mutagenesis using this strategy is only 0.5-20% 5, 6, 9. This poses a challenge for many applications, such as generating sufficient numbers of genome-edited animals.
HDR is less frequent than NHEJ and occurs only during S and G2 phase 5, 6, 9, whereas NHEJ occurs throughout the cell cycle 10. HDR occurs not sequentially but rather concurrently with NHEJ, and is increased in NHEJ-deficient cells, eg. Ku70-, XRCC4-, and DNA-PKcs-deficient cells11. We hypothesized that inhibition of NHEJ should improve the frequency of HDR. NHEJ is a broad term comprising two pathways: Ligase IV-dependent NHEJ [so-called canonical NHEJ (C-NHEJ)], which mainly functions in end-joining, and alternative NHEJ (alt-NHEJ/A-NHEJ/AEJ) pathways, which require DNA Ligase I/III and are induced by C-NHEJ pathway deficiencies12. In most cases, DNA repair after DSBs is mediated by the C-NHEJ pathway 13. We thus focused on DNA Ligase IV, a key enzyme for C-NHEJ, as a potential target to improve HDR efficiency. Indeed, the frequency of HDR stimulated by zinc finger nucleases is increased in Drosophila melanogaster strains lacking Ligase IV14. Homologous recombination is promoted in ligase IV-deficient NALM6 cells15. However, deletion of DNA Ligase IV in mice is late embryonic lethal 16, preventing the use of zygotes genetically deficient in DNA ligase IV as a strategy for making mutant mice.
To transiently target DNA Ligase IV, we used a DNA Ligase IV inhibitor, Scr7, initially identified as an anti-cancer agent 17. Scr7 targets the DNA binding domain of DNA Ligase IV, reducing its affinity for DSBs and inhibiting its function 17. Scr7 also inhibits DNA Ligase III (but not DNA Ligase I), albeit less efficiently 17. Treatment of mice with Scr7 affects lymphocyte development, as DNA Ligase IV plays a key role in the joining of coding ends during V(D)J recombination via C-NHEJ 16. The defects in lymphocyte development upon Scr7 treatment are transient and reversible, due to the non-covalent mode of binding of Scr7 17. We hypothesized that co-injection of CRISPR/Cas9 constructs with Scr7 into fertilized zygotes would allow mouse embryos to progress through development normally and show improved efficiency in obtaining defined DNA insertions. Accordingly, we used Scr7 to enhance the frequency of HDR by transiently blocking NHEJ (with the exception of DNA Ligase I-dependent alt-NHEJ), resulting in precise genome editing by CRISPR/Cas9 in both cultured cells and in mice.
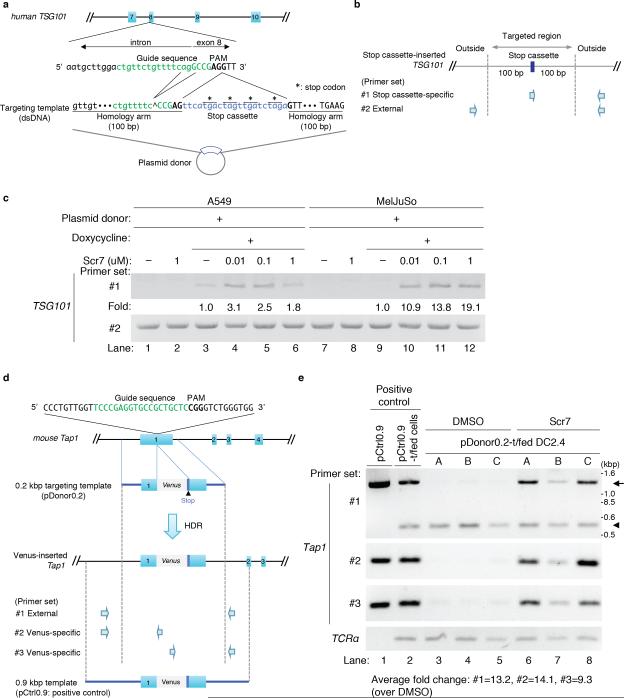
To investigate whether Scr7 improves the insertion efficiency of short DNA fragments at a given locus, we first tested whether Scr7 treatment enhances the rate of HDR in cultured human cell lines (Fig. 1). We generated epithelial (A549) and melanoma (MelJuSo) cell line derivatives expressing Cas9 and the TSG101 sgRNA, which targets exon 8 of the tumor susceptibility gene 10118, under the control of a doxycycline-inducible promoter (Fig. 1a and supplementary Fig. 1a and b, see also supplementary text ‘HDR-mediated targeting’). The targeting template comprises the intended insert (stop cassette) sandwiched between 100 base pair (bp) arms homologous to the sequence flanking the DSBs (Fig. 1a). In the targeting template, one adenine and two guanines in the guide sequence, in addition to the PAM motif, were deleted to avoid DSBs on the plasmid donor (Fig. 1a, targeting template). After 24 hours of plasmid donor transfection, cells were treated with doxycycline to induce Cas9 expression, and with various concentrations of Scr7, for 24 hours. The cells were then washed and incubated for an additional 24 hours in fresh media. We chose a range of 0 to 1 μM Scr7 to maintain cells capable of entering S/G2 phase, which is necessary for HDR (Supplementary Figs. 2a-c, and see Supplementary text ‘Cell viability upon Scr7 treatment’). For the A549 cells, which are relatively sensitive to Scr7, the efficiency of insertion at the target site in the presence of 0.01 μM Scr7 was improved ~3-fold relative to the untreated control. Efficiency was decreased at higher concentrations of Scr7 (0.1-1μM), but HDR frequencies at these concentrations were still higher than those in those in the untreated cells (Fig. 1b and c the lanes of A549). Similarly, the insertion efficiency was also enhanced in Scr7 treated MelJuSo cells in a dose-dependent manner up to 19-fold (Fig. 1c, the lanes labeled MelJuSo). To confirm whether precise insertion indeed increased upon Scr7 treatment, we performed both chip-based capillary electrophoresis and deep sequencing experiments (Supplementary Fig. 3a-e). Scr7 treatment increased both the frequency of all insertions, including imprecise insertions (Supplementary Fig. 3 b and e, ‘insertion’), and precise insertions, which increased from 0.02% to 12.8% upon 1 μM Scr7 treatment in MelJuSo cells (Supplementary Figs. 3e, ‘precise insertion’).
Figure 1. The NHEJ inhibitor Scr7 enhances the efficiency of insertional mutagenesis in cell lines.
(a) Schematic of sgRNA/ targeting template-targeting site at TSG101 exon 8. The guide sequence of human TSG101 sgRNA is indicated in green. The protospacer-adjacent motif (PAM) is in bold typeface. The exon sequence is capitalized. The targeting template contains 100 base pairs (bp) homology arms that the double strand breaks (DSBs) on both sides. To avoid CRISPR/Cas9-based DSBs in the plasmid donor containing the targeting template, the ‘agg’ nucleotides in the guide sequence were deleted. The stop cassette is labeled in blue. (*: stop codon).
(b) Primer sets to detect the insertion of the stop cassette. The presence of the stop cassette in the TSG101 locus was analyzed using an internal forward primer specific for the stop cassette and an external reverse primer specific for a sequence outside of the flanking arm (Fig. 1b primer set #1, reverse primer: 419 bp downstream of the PAM motif). The genomic region is targeted by external primers as an internal control (Fig. 1b, primer set #2, forward primer: 229 bp upstream of the PAM motif, reverse primer: identical to the reverse primer of set #1).
(c) Increased frequency of HDR in the presence of Scr7 in A549 and MelJuSo cells. The Cas9-inducible A549 and MelJuSo cell lines stably expressing TSG101 sgRNA were transfected with the plasmid donor. Twenty-four hours after transfection, cells were treated with doxycycline and Scr7 at the indicated concentration for another 24 hours. Doxycycline and Scr7 were removed by washing, and the cells were cultured for another 24 hours in fresh media, and finally lysed to collect genomic DNA. Analyses were performed using primer sets #1 and #2. Data are representative of 3 experiments. The HDR frequency -fold change was calculated by determining the ratio of band intensities at the different concentrations of Scr7 (lane 4-6 and lane 10-12) over the intensities observed in the absence of Scr7 (lane 3 and 9). See Online Methods for the detailed method for quantification of band intensities.
(d) Strategy for insertion of a Venus reporter gene into the mouse Tap1 exon1. The guide sequence of mouse Tap1 sgRNA is indicated in green. The PAM is designated by bold-typeface. The targeting template was introduced into a plasmid (plasmid donor). A Venus reporter gene (813 bp) is shown in gray and contains the stop codon. We prepared two constructs: pDonor0.2 as a template donor and pCtrl0.9 as a positive control for a PCR. pDonor0.2 contains 213 base pair (bp) flanking arms on either side of a Venus reporter gene to induce HDR in front of the PAM motif (GFP mutant Venus size: 813 bp); pCtrl0.9 has longer homology arms (Fig. 1f, bottom; the length of upstream homology arm: 914 bps; downstream length: 660 bp), and was used to provide a comparative PCR product of the Venus-inserted Tap1 locus in lane 1 and 2 of Figure 1g.
Primer sets to detect the insertion of the Venus gene and positive control plasmid. External primers were designed outside of the homology arms of the 200 bp targeting template. Primer set #1: a pair of external forward and reverse primers designed outside of the homology arms on the 0.2 kbp targeting template (forward primer: 258 bp upstream of the PAM motif, reverse primer: 252 bp downstream of the PAM motif), primer set #2: the forward primer from primer set #1 and a Venus-specific reverse primer, primer set #3: a Venus-specific forward primer and the external reverse primer from primer set #1. As a positive control for PCR, Vector pCtrl0.9 containing longer homology arms flanking both sides was prepared (length of the left homology arm: 914 bps, length of the right homology arm: 660 bp).
(e) Improvement of HDR-mediated insertion efficiency by inclusion of Scr7 in DC2.4 cells. Lane 1 serves as a positive control, using the pCtrl0.9 as a PCR template to provide the comparative PCR products of a Venus-inserted Tap1 locus. The Cas9-inducible DC2.4 cell line stably expressing Tap1 sgRNA was transfected with the pCtrl0.9 as an additional positive control (lane 2), or with pDonor0.2 (lane 3-8). After 24 hours of transfection, cells were treated with doxycycline and 1 μM of Scr7 for another 24 hours, after which Scr7 and doxycycline were removed by washing. The cells were cultured for another 24 hours, and lysed to collect genomic DNA. The genomic DNA was analyzed using the primer sets as shown in Fig. 1g. (arrow: the amplicon of the Venus-inserted Tap1 locus, arrowhead: the amplicon of original Tap1 locus) The samples analyzed were derived from 3 independent experiments (A-C), i.e. the sample in lane 3 is matched with the sample in lane 6, 4 with 5, and 7 with 8. The fold change in HDR frequency was calculated by determining the ratio of band intensities in the presence of Scr7 (lane 3-5) over the intensities observed in the absence of Scr7 (lane 6-8). For the panel of primer set #1, the ratio of the inserted band intensities (arrow) over total band intensities (arrow + arrowhead) was calculated, and then the -fold changes were determined. We observed the appearance of an ~800 bp larger DNA fragment (arrow) upon HDR-based Venus insertion as compared to the original Tap1 DNA fragment (arrowhead) (Fig. 1g, upper panel, individual experiment: A-C), as evidence for integration of the Venus gene into the target locus. Sequence analysis confirmed that the new, longer DNA fragment (Fig. 1g, bands indicated by arrow) amplified by primer set #1 corresponded to precise insertion of the Venus reporter gene (Supplementary Fig. 4a).
To assess the effect of Scr7 on insertion of longer DNA fragments, we also generated a Cas9-inducible murine bone marrow-derived dendritic cell line (DC2.4 cells) stably expressing an sgRNA that targeted the Transporter, ATP-binding cassette, major histocompatibility complex 1 (Tap1)19 locus at exon 1, and inserted the Venus fluorescent gene using pDonor0.2 (Figs. 1d, and Supplementary Fig. 1c sgRNA targeting efficiency). TAP1-knockout mice are deficient in antigen presentation, but otherwise healthy. Because Scr7 was only slightly toxic even at concentrations >10 μM (Supplementary Fig. 2d), the DC2.4 cells were treated with 1μM of Scr7, but otherwise underwent the same procedure as outlined for the A549 and MelJuSo cells. After treatment, we performed genotyping using two Venus-specific primer sets (#2 and #3), in addition to an external primer set (#1) (Fig. 1d). Scr7 treatment improved the efficiency of insertion at the target site ~ 13-fold. The -fold change of inserted alleles amplified by the Venus-specific internal primer sets #2 and #3 was similar to that using the primer set #1 (Figs. 1d and e, middle panels, average fold change: #2; ~14-fold, #3; ~9-fold). Furthermore, expression of Venus was observed by immunoblot analysis and fluorescence microscopy (Supplementary Figs. 1c, 4a-c, Scr7(-): Venus positive cells = 4.58%; Scr7(+):Venus positive cells = 58.3%). Scr7 thus enhances the HDR frequency in cell lines for insertions as large as ~800 bp.
Given the increase in HDR frequency in cell lines exposed to Scr7, we next tested whether Scr7 would improve precise genome editing in mouse zygotes. First, we determined a baseline efficiency of deletion and insertion mutagenesis in zygotes when targeting multiple genes in the absence of Scr7. These included Osteosarcoma amplified 9 (Os9, three types of sgRNAs) 20 and Leucine rich repeat containing 8 family, member D (Lrrc8d) 21 as targets of NHEJ-mediated deletion mutagenesis; and Mesothelin 22 and Immunoglobulin heavy constant gamma 3 23 as targets for insertional mutagenesis through HDR. CRISPR/Cas9 constructs were microinjected into the cytoplasm of fertilized zygotes. Manipulated embryos were cultured to the two-cell stage and transferred into pseudopregnant females. The tails of pups carried to term were harvested for genotyping. Supplementary Table 1 shows the ratio of insertion- and/or deletion-mutated alleles for each gene over the total number of alleles present in the embryos obtained. The frequencies of NHEJ and HDR were calculated as the ratio of deletion and insertion alleles relative to wild-type alleles (Supplementary Table 1, NHEJ [%], HDR [%]). We confirmed that deletional mutagenesis consistently occurred at ~ 20% to ~60%, as previously reported (OS9: deletionsgRNA#1 = 28.6%; deletionsgRNA#2 = 45.8%; deletionsgRNA#3 = 20.0%, LRRC8D: deletion = 66.7%) 6. The efficiency of insertional mutagenesis in the same experiment was lower than that of deletional mutagenesis (mesothelin: deletion = 19.4%; insertion = 11.1%, IgG3: deletion = 44.4%; insertion = 16.7%), again confirming previous reports 5, 6 (Supplementary Table 1).
We had previously generated Kell-LPETG mice by the introduction of a linker and an in-frame sortase recognition motif (LPETG) at the C-terminus of the type II membrane protein Kell24 . To examine whether treatment with Scr7 enhances the efficiency of HDR-mediated insertional mutagenesis in mice, we therefore chose the Kell gene as a target. As a second target, we introduced the LPETG sortase motif at the 3’ end of the immunoglobulin kappa constant region. We designed sgRNAs targeting the last exon of the Kell gene and the last exon of the Immunoglobulin kappa (Igκ) constant region(Igkc) as well as single-stranded DNA oligonucleotides [targeting template (ssDNA)] to introduce the desired insertion in close proximity to the DSBs (Fig. 2a). Zygotes co-injected with CRISPR/Cas9 components (sgRNA, Cas9 mRNA and targeting template) and 1 μM Scr7 developed normally to the blastocyst stage with an ~80% survival rate, comparable to the condition without Scr7 (Fig. 2b, Supplementary Figs. 5a-c). The genotype of each blastocyst was then analyzed by PCR using an LPETG-specific primer. The insertion efficiency with Scr7-coinjection significantly increased (p = 0.0012) compared to blastocysts not injected with Scr7 (Fig. 2c, Table 1). Zygotes co-injected with Scr7 were also cultured to either the two-cell or blastocyst stage and then transferred into pseudo-pregnant mice (Fig. 2b). DNA from the resulting E10 embryos was analyzed by digestion with MboI, by direct sequencing and by PCR with an LPETG-specific primer set (Figs. 2d-f, Table 1). For the Kell locus, we detected both NHEJ-mediated deletion alleles (shown as Δ/Δ or +/Δ in Fig. 2d and Supplementary Fig. 5d) and LPETG-inserted alleles (shown as di/di or +/di in Fig. 2d) in the absence of Scr7. In the presence of Scr7, only LPETG-insertion alleles were observed (Fig. 2d and Table 1). After comparing the number of wild-type, deletion and insertion alleles, we found the insertion efficiency in Scr7-coinjected E10 embryos to be significantly enhanced compared to E10 embryos not injected with Scr7 (p = 0.003). We observed a similar effect of co-injection with Scr7 on the insertion efficiency of LPETG-encoding DNA fragment into the Igkc locus (Figs. 2e and f; LPETG-specific primer set #1 and external primer set #2, Supplementary Fig. 5e; RFLP by MboI digestion, and Table 1), suggesting that improvements in insertion efficiency upon inhibition of NHEJ are not limited to a particular locus.
Figure 2. Co-injection of Scr7 enhances the efficiency of precise genome editing in mouse embryos.
(a) Strategy for introduction of a sortase target motif at the stop codon of the mouse Kell locus and the Ig κ constant region. sgRNA guide sequences of the Kell 3’ exon and the Igκ constant region are indicated in green. The Kell 3’ exon and the Igκ constant region are capitalized. A PAM motif is indicated in bold. The targeting single-stranded (ss) DNA template [targeting template (ssDNA)] contains ~80 nt of homology flanking both sides of the DSB. In the case of Igκ, the PAM is deleted to avoid inducing DSBs on the targeting ssDNA template as shown by “^”. The insertion cassette, including sortase recognition motif and linker, is labeled in blue. (*: stop codons).
(b) A mixture of the CRISPR components (Cas9 mRNA, sgRNA and targeting template) was prepared, and 10 mM of Scr7 NHEJ inhibitor was then added to the mix (final concentration: 1mM). Fertilized zygotes were collected from oviducts of super-ovulated female mice, and the mixture was injected into the cytoplasm at the pronuclear stage. The injected zygotes were transferred at the 2-cell stage into the oviduct of pseudo-pregnant females.
(c) Genotyping results of the Kell-targeted blastocysts. Co-injection was conducted as in (b). The co-injected zygotes were cultured up to the blastocyst stage. The blastocysts were subjected to PCR using an LPETG-specific forward primer and a Kell-specific reverse primer. The PCR-positive blastocysts were counted as insertion positive. P value was calculated by Fischer's test in Prism6 software. (p=0.0012)
(d) Restriction fragment length polymorphism (RFLP) analysis results of the Kell-targeted E10 embryos. Upper: the untreated, original amplicons using Kell external primer, lower: the digested amplicon using a unique enzyme MboI to LPETG cassette, including a linker sequence and the LPETG sortase-targeting motif. Genotypes and sequencing results are shown as ‘Genotype’(+: wild-type allele,Δ: deletion mutant allele, di: insertion mutant allele) and ‘Observed indel’ at the bottom (minus: deletion; plus: insertion).
(e) Primer sets to detect insertion of the sortase motif. External primers were designed outside of the homology arms flanking the DSBs. Primer set #1: a sortase motif-specific forward primer, and external reverse primer targeting the Igκ constant region > 250 bp downstream of the DSB, primer set #2: a pair of external forward and reverse primers targeting the genomic region > 250 bp up- and down-stream of the DSB.
(f) Genotyping results of the Ig κ constant region-targeted E10 embryos. Upper: genotyping results using primer set #1 in Fig. 2e. The amplicon obtained using primer set #1 in figure 2e is shown as an arrow. (*: non-specific product) Lower: the amplicon obtained using primer set #2 in figure 2e. Genotypes and sequencing results are shown as ‘Genotype’(+: wild-type allele,Δ: deletion mutant allele, di: insertion mutant allele) and ‘Observed indel’ at the bottom (minus: deletion; plus: insertion). RFLP analysis by digestion with MboI is shown in the Supplementary Figure 5e.
Table 1.
mice by the Scr7-based approach
| Sample number |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | Description | Background | Scr7 | Blastocyst | Non-ins | Insertion | HDR (%) |
| Kell | LPETG insertion | BDF | – | 41 | 30 | 11 | 26.8 |
| + | 59 | 24 | 35 | 59.3 | |||
| Allele number |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | Background | Scr7 | Embryo | Newborn | Wildtype | Deletion | Insertion | NHEJ (%) | HDR (%) |
| Kell | LPETG insertion | BDF | – | 14 | *** | 11 | 9 | 8 | 32.1 | 28.6 |
| + | 12 | 10 | 0 | 14 | 0.0 | 58.3 | ||||
| Igk constant | LPETG insertion | BDF | – | 10 | * | 14 | 5 | 1 | 25.0 | 5.0 |
| + | 11 | 17 | 0 | 5 | 0.0 | 22.7 | ||||
| Os9 | Stop codon | C67BL/6 | + | 6 | 7 | 0 | 5 | 0.0 | 25.0 | |
| Sgms2 | Stop cassette | BDF | + | 4 | 7 | 0 | 1 | 0.0 | 12.5 | |
Blastocysts, E10 embryos and newborn pups were prepared as in Fig. 2 and Supplementary Fig. 6 with Scr7. The genotypes of the samples were analyzed by insertion-specific primer genotyping, and/or RFLP. The genotypes were confirmed by direct sequencing. Percentages of the NHEJ and HDR were calculated by the number of the deletion and/or the insertion alleles against that of total alleles.
p-value < 0.05
p-value < 0.005 comparing the frequency of deletion, insertion, and wildtype allele in the mouse cohort generated with or without NHEJ inhibitor treatment.
(#1: p = 0.003, #2: p = 0.02) Non-ins: Non-insertion.
Inclusion of the DNA ligase IV inhibitor, Scr7, improved insertion efficiency of both short and long DNA fragments at the TSG101 and Tap1 target loci, respectively (Fig. 1). In A549 cells, the use of higher concentrations of Scr7 attenuated HDR. This likely reflects cell type-dependent toxicity of Scr7 (Supplementary Fig. 2). Co-injection of zygotes with Scr7 significantly improved the efficiency of HDR-mediated insertional mutagenesis (Fig. 2c, p=0.0012). Modification of the Kell and the Igkc loci in the presence of Scr7 yielded only embryos with the desired LPETG sequence, and no deletion mutant embryos (Figs. 2d and f, Table 1). Because total indel-frequency did not change in either example (Supplementary Figs. 3b and e, the columns ‘indel’), increased efficiency of insertions upon treatment with Scr7 correlates with a decrease in deletions (Supplementary Figs. 3b and e, the columns of ‘insertion’ and ‘deletion’). Scr7 thus promotes the frequency of HDR through inhibition of NHEJ, and favors precise insertions over random deletions.
As inhibition of NHEJ can induce apoptosis 17, we considered the possibility that co-injection of Scr7 into zygotes reduces the number of deletion mutants without affecting the efficiency of insertional mutagenesis. However, treatment with Scr7 affected neither viability of the injected zygotes nor the number of live pups born from Scr7-injected embryos (Supplementary Fig. 5). We also addressed the possibility that Scr7 had a detrimental effect on mouse generation, eg. by failure of germline transmission and/or increase of off-target effects. The offspring of Scr7-generated pups carried the same desired mutation as that of the parental mice, suggesting successful germline transmission (Supplementary Fig. 6, see also Supplementary Text ‘Germline transmission) without off-target effects evaluated by the direct sequencing analysis (Supplementary Fig. 7, see also Supplementary Text ‘Effect on off-target by Scr7 treatment).
It is also possible that Ligase IV deficiency may induce alternative NHEJ pathways, which produce longer indels and are mediated by Ligase I/III25, 26. However, we looked in-depth at the Kell locus targeted in zygotes (Supplementary Fig. 8, see also Supplementary Text ‘Long indel analysis), and saw no obvious signs of alternative NHEJ. This may be due to Scr7 inhibition of Ligase III activity, or to alternative NHEJ pathways being less efficient in zygotes. For cell-based experiments, the effect of alternative NHEJ pathways warrants further investigation, because Ligase III-dependent chromosome translocations occur more frequently in cancer cells26.
Although in this study we used Scr7 to promote HDR frequency, we predict that other NHEJ inhibitors, for example compounds that target DNA-dependent protein kinases, might also promote HDR. From a practical standpoint, we observe an improvement in the efficiency of precise genome editing by CRISPR/Cas9 in fertilized zygotes, which facilitates the generation of gene-targeted mice with insertions at multiple loci. Given that the molecular mechanisms involved in NHEJ and HDR are conserved across eukaryotes, the method may be applicable to non-mammalian species as well.
Methods
NHEJ inhibitor treatment
The sgRNA stable Cas9-inducible cells were transfected with the plasmid donor, and treated with Scr7 (Excess Bioscience; #M60082-2s) at the indicated concentrations (A549 and MelJuSo: 0.01 ~ 1 μM and DC2.4: 1 μM for HDR-stimulation, A549, MelJuSo and DC2.4: 0.01 ~ 10 μM for MTT assay) and 1 μg/mL of doxycycline (Clontech; #631311). After 24 hours, cells were incubated without Scr7 for an additional 24 hours, and then genomic DNA was collected and analyzed. See the legend of Supplementary Fig. 2 for the time course of the cell viability assay.
Zygote co-injections with NHEJ inhibitor
All animal procedures were performed according to NIH guidelines and approved by the Committee on Animal Care at MIT. Intra-cytoplasmic injection of fertilized zygotes was performed as previously described6. Super-ovulated female B6D2F1 mice (7– 8 weeks of age) were mated to B6D2F1 stud males, and fertilized zygotes were collected from oviducts. Cas9 mRNA (100 ng/mL), sgRNA (50 ng/mL), template oligos (100 ng/ml) were mixed, and Scr7 (1mM, where indicated) were mixed and injected into zygotes at the pronuclear stage (24 hours post-HCG). The injected zygotes were cultured in KSOM with amino acids at 37°C and 6% CO2 to the two cell stage, and then were implanted into pseudo-pregnant ICR females.
Mouse and blastocyst genotyping
Mouse-tails were lysed at 55°C for 12 hours using tail lysis buffer and genomic DNA was purified from the lysates by isopropanol precipitation. Genomic DNA in proximity to the guide sequence of sgRNA was amplified by PCR using the primers listed in Supplemental Table 2 and then gel purified. The purified PCR products were digested with MboI, and sequenced using the same forward and/or reverse primer.
For blastocyst genotyping, Scr7-coinjected blastocysts were transferred to PCR tubes containing 5 μL of QuickExtract buffer (Epibio; #QE09050). The samples were incubated at 65 °C for 6 min, and then at 98°C for 2 min. PCR were performed as described in the section of Cell-based Genotyping using an LPETG-specific primer, Kell-forward and -reverse primers (Supplementary Table 3).
Online Methods
Designing CRISPR target sequence and prediction of off-target effects
Guide sequences of sgRNA for CRISPR interference were designed as detailed in 9. The target sequence preceding the PAM motif was obtained from the exon region of the indicated genes. Potential off-target effects of guide sequence were predicted using NCBI Nucleotide BLAST. The guide sequences of sgRNAs used are shown in Supplementary Table 4. The targeting templates for human TSG101 and Tap1 contain the 100 bp and 213 bp homology sequences on each side, respectively.
Cell-based Genotyping
Cells were lysed at 55°C for 2 hours using tail lysis buffer (1% SDS, 100 mM NaCl, 50mM Tris, and 1mM EDTA) containing proteinase K (Roche Diagnostics; #03115879001). Genomic DNA was purified from the lysates by isopropanol precipitation. Genomic DNA in the vicinity of the sgRNA target was amplified using the primers listed in supplementary table 2 and the following PCR cycles: [KOD Xtreme (EMD millipore; #71975), Condition: 95°C for 2 min; 10x(98°C for 10 s, 50°C for 30 s, 68°C for 30 s); 15x(98°C for 10 s, 60°C for 30 s, 68°C for 30 s);68°C for 2 min; hold at 4°C].
Construction for sgRNA vectors and plasmid donors
The sgRNA template sequence for site-directed mutagenesis was designed as in Supplementary Table 5, and was incorporated into the NheI/BamHI site of CMV promoter-deleted pCDH-EF1-Hygro (SBI; CD515B-1). The vectors for expressing sgRNA targeting human TSG101 and the mouse Tap1 genes (pCDH-EF1-Hygro-hTSG101-sgRNA and pCDH-EF1-Hygro-mTap1-sgRNA) were constructed using the primers listed in Supplementary Table 6 for site-directed mutagenesis (GM Biosciences, Inc.; #7001).
The stop cassette for the hTSG101 gene and the Venus reporter gene for the mTap1 gene were designed to be in the proximity of each guide sequence as shown in Fig. 1b and Supplementary Table 4. The targeting template for the hTSG101 locus and the mTap1 locus (Supplementary Table 7) were incorporated into pCDH-CMV-MCS-EF1-RFP (SBI; CD512B-1) and pCDH-EF1-Hygro (SBI; CD515B-1), respectively, using NheI and BamHI sites on the targeting templates and SpeI and BamHI sites on the vectors (Supplementary Table 7).
Cell culture
MelJuSo and DC2.4 cells were grown in RPMI supplemented with 10% heat-inactivated fetal bovine serum (IFS) at 37°C and 5% CO2. A549 cells were cultured in DMEM supplemented with 10% IFS at 37°C and 5% CO2.
Lentivirus generation
pCW-Cas9 (provided by Dr. David Sabatini)4 or pCDH-EF1-Hygro-sgRNA were co-transfected into HEK293T cells with the virus packaging vector, pMD2.G and psPAX2. After 48 hours, supernatants were collected and the virus was concentrated using PEG (SBI; #LV810A-1) and dissolved in PBS.
Lentivirus transduction and selection for sgRNA stable Cas9 inducible cells
A549, MelJuSo and DC2.4 cells were infected with lentivirus expressing Cas9 cDNA, and were cultured in puromycin-containing medium (Sigma Aldrich; #P9620) (A549: 1.5 μg/mL, MelJuSo: 2 μg/mL, DC2.4: 7 μg/mL). These Cas9-inducible cells were re-infected with lentivirus carrying sgRNA-coding DNA, and were cultured in medium containing hygromycin B (Life tech.; #10687010) (A549: 100 μg/mL, MelJuSo: 100 μg/mL, DC2.4: 250 μg/mL)
sgRNA and Cas9 mRNA synthesis
Cas9 mRNA was prepared as described in 6. The T7 promoter-fused mKell-CT- or Igκ-CT-targeting guide sequence was added to 5’ of the sgRNA generic tail sequence by PCR amplification, using the pX330 vector (http://www.addgene.org/42230/) as a template. The primers used are listed in Supplemental Table 6. The T7-sgRNA template PCR product for in vitro transcription (IVT) was gel-purified (Promega; PAA9282), and the purified PCR product was used as a template for IVT using MEGAshortscript T7 kit (Life Technologies; AM1354). Both Cas9 mRNA and sgRNAs were purified using MEGAclear kit (Life Technologies; AM1908) and eluted in RNase-free water.
Targeting single-stranded DNA template
The template single stranded oligonucleotides for the insertion of LPETG into the Kell and Igκ constant region loci are listed in Supplementary Table 7.
Quantification of band intensities
The area corresponding to the band of interest in Fig. 1c, a primer set #1 of upper panel, was selected in the Scr7-treated lanes (DMSO, 0.01-1 μM in Fig. 1c) of A549 and MelJuSo cells, and a similar area with no obvious material was selected as background, and subtracted. Intensities measured for each lane were normalized by the intensities of the bands in the panel of a primer set #2, (background subtracted for primer set #1). For DC2.4, total intensity of both inserted and unmodified bands are measured. Ratios of insertions over total (inserted and unmodified) were calculated, and then the -fold change was calculated by comparing the ratio of insertions with Scr7 over that without Scr7.
Antibody
The anti-Tsg101 antibody and anti-Tap1 antibody were purchased from Abcam (ab83) and Santa Cruz Biotechnology (sc-11465), respectively. For GFP protein detection, GFP (B-2) HRP (Santa Cruz Biotechnology, SC-9996 HRP) was used.
MTT assay
Cell viability of A549, MelJuSo and DC2.4 were determined by MTT assay kit (Roche, #11465007001). The cells transfected with the plasmid donor were grown in the presence of Scr7 (0.01, 0.1, 1, and 10 uM) for 24, 48 or 72 hrs, and subjected to MTT assays. Each experiment was repeated a minimum of three independent times.
Preparation of samples generic for chip-based capillary electrophoresis and deep sequencing
The Cas9-inducible sgRNA stable MelJuSo cells were transfected with the 200 bp homology arm-containing donor plasmid used in Figs. 1b and 1d. About 100 - 200 bp of amplicons are sufficient to distinguish the size difference between the endogenous and the stop cassette-inserted TSG101 locus in analyses of chip-based capillary electrophoresis. Since the cells were transfected with the plasmid donor, which contains 200 bp of the TSG101 homologous region (Fig. 1b), 650 bp of the TSG101 genomic region was amplified using primer set (#2) as in Figs. 1c and 1d. The PCR products containing only the TSG101 genomic region were gel-purified. Using the purified 650 bp amplicon, we re-amplifed ~200 bp of the TSG101 genomic region (a primer set: see Supplementary Table 3, TSG101 partial P5FW/P7RV). Adapter sequences and barcodes were then added using the primers listed in Supplementary Table 3 (MiSeq P5 universal FW, and MiSeq P7 RV barcode #1/#2).
Quantification of indel frequency by chip-based capillary electrophoresis
The PCR products, containing the wild-type amplicon (318 bp), inserted amplicon (336 bp) and deleted amplicon (<318 bp), were loaded onto an Agilent 2100 BioAnalyzer. In the chromatography data obtained, the region of 313 bp to 323 bp, 288 bp to 313 bp, or 326 bp to 346 bp was gated to observe the peak of wt (= 318 bp), deletion (<318 bp) or insertion (= 336 bp), respectively. Insertion [%] was calculated by determining the ratio of chromatographic intensity in the insertion region over the total intensity. Deletion [%] was calculated in a similar fashion. Indel [%] is the sum of deletion [%] and insertion [%].
Quantification of indel frequency by deep sequencing
For deep sequencing, the prepared PCR products, containing the adaptors and barcodes, were run on Illumina MiSeq (150X150-paired end read). All reads from MiSeq were analyzed with Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)27, then read counts of deletion and insertion at the TSG101 locus were extracted using wild-type and stop cassette-inserted TSG101 sequence, respectively. Statistically analyzed data were visualized with visualizing software, IGV (http://www.broadinstitute.org/igv/home)28.
Detection of Venus fluorescence by microscope and imaging analysis by CellProfiler
Images of the cells treated with or without Scr7 in Fig. 1g were obtained by non-confocal mode of Andor Revolution Spinning Disk Confocal, FRAPPA and TIRF system microscope. The images obtained were analyzed by CellProfiler (http://www.cellprofiler.org/)29. The pipeline to recognize cells and measure the fluorescence intensities of the cells recognized is described in the supplementary file, ‘Takeshi_DIC_GFP.cppipe’.
Off-target prediction
Potential off-target site were determined by the algorithm from the Zhang lab (http://www.genome-engineering.org). The 23 bps including the PAM motif of Kell sgRNA guide sequence was utilized since the algorithm allows more than 23 bp. The top 5 off-target sites predicted are shown in Supplementary Fig. 7 (a).
Southern blot
Genomic DNA was separated on a 0.5% agarose gel after digesting with EcoRI restriction enzyme, transferred to a nylon membrane (Millipore; INYC00010) and hybridized with PCRDIG (Roche Diagnostics; 11636090910)-labeled probes. The Kell-LPETG-targeted alleles were detected using anti-Dioxigenin-AP Fab fragment (Roche Diagnostics; 11093274910).
Sortase-mediated labeling of immunoglobulin light chains with biotin
Pentamutant Sortase A was expressed and purified as described (David Liu et al., 2011 30. Approximately 150 uL of serum from a wild-type mouse and from a mouse carrying Igκ-LPETG mutation was used for labeling. The reaction mixture was assembled as follows: 500 μM GGG-biotin, 5 μM sortase A, and 4 uL serum in 50 mM Tris-HCl, 150 mM NaCl, and 10 mM CaCl2. The reaction was incubated at 4°C for 1 hr. The reaction mixture was then adsorbed onto protein-G beads (Roche Diagnostics; #11719416001) to remove non-Ig serum components as well as the enzyme. Bound materials were released by incubation with SDS sample buffer at 98°C and analyzed by SDS-PAGE. To detect biotin-modified Ig light chains, Avidin-HRP (Fisher Scientific; 45000765) was used following transfer to a PVDF membrane.
Statistical analysis
Fisher's exact test in Prism software was used to calculate p-values when comparing the number of insertions in blastocysts co-injected with or without NHEJ inhibitor treatment. Fisher's exact test in R software was utilized to evaluate the significance in comparing the frequency of deletions, insertions, and wildtype alleles in the mouse cohort generated with or without NHEJ inhibitor treatment. An input example for Fisher's test in R software is below:
| Kell | wildtype | deletion | insertion |
|---|---|---|---|
| Scr7 (−) | 11 | 9 | 8 |
| Scr7 (+) | 10 | 0 | 14 |
Supplementary Material
Acknowledgements
We thank members of the Ploegh lab, especially Novalia Pishesha and Fikadu Tafesse, for critical reading of the manuscript; Paul A. Koenig, Lee Kim Swee, Chikdu S. Shivalila, Haoyi Wang, Hui Yang and Rudolf Jaenisch for discussions; Tim Wang and David M. Sabatini for providing materials; Prathapan Thiru and George Bell of BARC (WIBR) for assistance with statistical analysis; Feng Zhang (Addgene) for pX330.
This work was supported by the National Institutes of Health (RO1 grant AI087879-01 to H.L.P.), Japan Society for the Promotion of Science (to T.M.), Japan Heart Foundation (to T.M.), AACRPancreatic Cancer Action Network (to S.K.D. and H.L.P), SNSF Early Postdoc Mobility fellowship (to M.T.).
Footnotes
Contribution: T.M. and H.L.P. conceived of and designed experiments; T.M., S.K.D., and A.M.B. performed experiments; T.M. and M.T. analyzed data; T.M., J.R.I. and H.L.P. wrote the manuscript.
The authors have no conflicting financial interests.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell stem cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature biotechnology. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nature reviews. Molecular cell biology. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 11.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & development. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boboila C, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. The Journal of experimental medicine. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA repair. 2014;17:81–97. doi: 10.1016/j.dnarep.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger. Nucleases. 2013;G3 doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12109–12113. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank KM, et al. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 18.Sanyal S, et al. Type I interferon imposes a TSG101/ISG15 checkpoint at the Golgi for glycoprotein trafficking during influenza virus infection. Cell host & microbe. 2013;14:510–521. doi: 10.1016/j.chom.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 20.Christianson JC, Shaler TA, Tyler RE, Kopito RR. S-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nature cell biology. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CC, Freinkman E, Sabatini DM, Ploegh HL. The Protein Synthesis Inhibitor Blasticidin S Enters Mammalian Cells via Leucine-rich Repeat12 containing Protein 8D. The Journal of biological chemistry. 2014;289:17124–17131. doi: 10.1074/jbc.M114.571257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinkevich Y, et al. Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nature cell biology. 2012;14:1251–1260. doi: 10.1038/ncb2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovenden M, et al. IgG subclass and heavy chain domains contribute to binding and protection by mAbs to the poly gamma-D-glutamic acid capsular antigen of Bacillus anthracis. PLoS pathogens. 2013;9:e1003306. doi: 10.1371/journal.ppat.1003306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J, et al. Engineered red blood cells as carriers for systemic delivery of a wide array of functional probes. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1409861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.So S, Adachi N, Lieber MR, Koyama H. Genetic interactions between BLM and DNA ligase IV in human cells. The Journal of biological chemistry. 2004;279:55433–55442. doi: 10.1074/jbc.M409827200. [DOI] [PubMed] [Google Scholar]
- 26.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L, Florea L, Langmead B. Lighter: fast and memory-efficient sequencing error correction without counting. Genome biology. 2014;15:509. doi: 10.1186/s13059-014-0509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popp MW, Antos JM, Ploegh HL. Site-specific protein labeling via sortase-mediated transpeptidation. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1503s56. Chapter 15, Unit 15 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.