Summary
Maintaining genomic integrity during DNA replication is essential for stem cells. DNA replication origins are licensed by the MCM2–7 complexes, with most of them remaining dormant. Dormant origins (DOs) rescue replication fork stalling in S phase and ensure genome integrity. However, it is not known whether DOs exist and play important roles in any stem cell type. Here, we show that embryonic stem cells (ESCs) contain more DOs than tissue stem/progenitor cells such as neural stem/progenitor cells (NSPCs). Partial depletion of DOs does not affect ESC self-renewal but impairs their differentiation, including toward the neural lineage. However, reduction of DOs in NSPCs impairs their self-renewal due to accumulation of DNA damage and apoptosis. Furthermore, mice with reduced DOs show abnormal neurogenesis and semi-embryonic lethality. Our results reveal that ESCs are equipped with more DOs to better protect against replicative stress than tissue-specific stem/progenitor cells.
Highlights
-
•
ESCs possess more dormant origins than tissue stem/progenitor cells
-
•
The greater number of dormant origins in ESCs effectively protects genome integrity
-
•
Reduction of dormant origins impairs ESC differentiation, but not self-renewal
-
•
Reduction of dormant origins severely affects neurogenesis and embryonic viability
Genomic integrity is essential for stem cells. Dormant origins rescue replication fork stalling in S phase and ensure genome integrity. Lin and colleagues report that embryonic stem cells (ESCs) contain more dormant origins than tissue stem/progenitor cells, thereby more efficiently protect them against replicative stress. Reduction of dormant origins impairs ESC differentiation, neurogenesis, and embryonic viability.
Introduction
It is essential for stem cells, especially embryonic stem cells (ESCs), to maintain genome integrity. A key aspect of this is to ensure the fidelity of DNA replication. In eukaryotic genomes, DNA replication initiates at thousands of origins. Origins are licensed prior to S phase, a process that involves the recruitment of licensing factors MCM2, 3, 4, 5, 6, and 7 as double heterohexamers onto DNA (Evrin et al., 2009; Remus et al., 2009). During S phase, each MCM2–7 complex can initiate replication by acting as a helicase to unwind double-stranded DNA ahead of DNA polymerases (Bochman and Schwacha, 2009). MCM2–7 complexes are loaded onto the genome in 5- to 20-fold excess to the number utilized to initiate DNA replication. The excess MCM2–7 complexes usually remain dormant, but they initiate back-up replication forks to rescue replication when primary forks are slowed or stalled; therefore, they are called dormant origins (DOs) (Doksani et al., 2009; Ge and Blow, 2010; Ge et al., 2007; Ibarra et al., 2008). Replication forks frequently stall, for example, when encountering tightly bound protein-DNA complexes, transcription machinery, repetitive sequences, or DNA lesions (Makovets et al., 2004; Mirkin and Mirkin, 2007). Prolonged fork stalling increases the probability of fork collapse and double strand breaks, which could lead to chromosomal re-arrangements and genomic instability (Lambert et al., 2005). As a safeguard mechanism, DOs provide the first line of defense against fork stalling (Blow and Ge, 2009). Chromosomal fragile sites, which are prone to breakage upon replication stress, are shown to have lower capacity to activate DOs (Letessier et al., 2011). Mice with reduced DOs show genomic instability, age-related dysfunction, and develop tumors (Kunnev et al., 2010; Pruitt et al., 2007; Shima et al., 2007). Importantly, congenital hypomorphic MCM4 defects have been found in humans, associated with various abnormalities and elevated genomic instability (Gineau et al., 2012; Hughes et al., 2012).
Despite the importance of DOs, it is unknown whether they exist and function differently in stem cells. Here, we analyze DOs in ESCs and neural stem/progenitor cells (NSPCs) as an example of tissue stem/progenitor cells. We show that ESCs load more DOs onto the genome than NSPCs and that DOs play a significant role in defending against replication stress in both stem cell types.
Results
ESCs License More DOs Than NSPCs
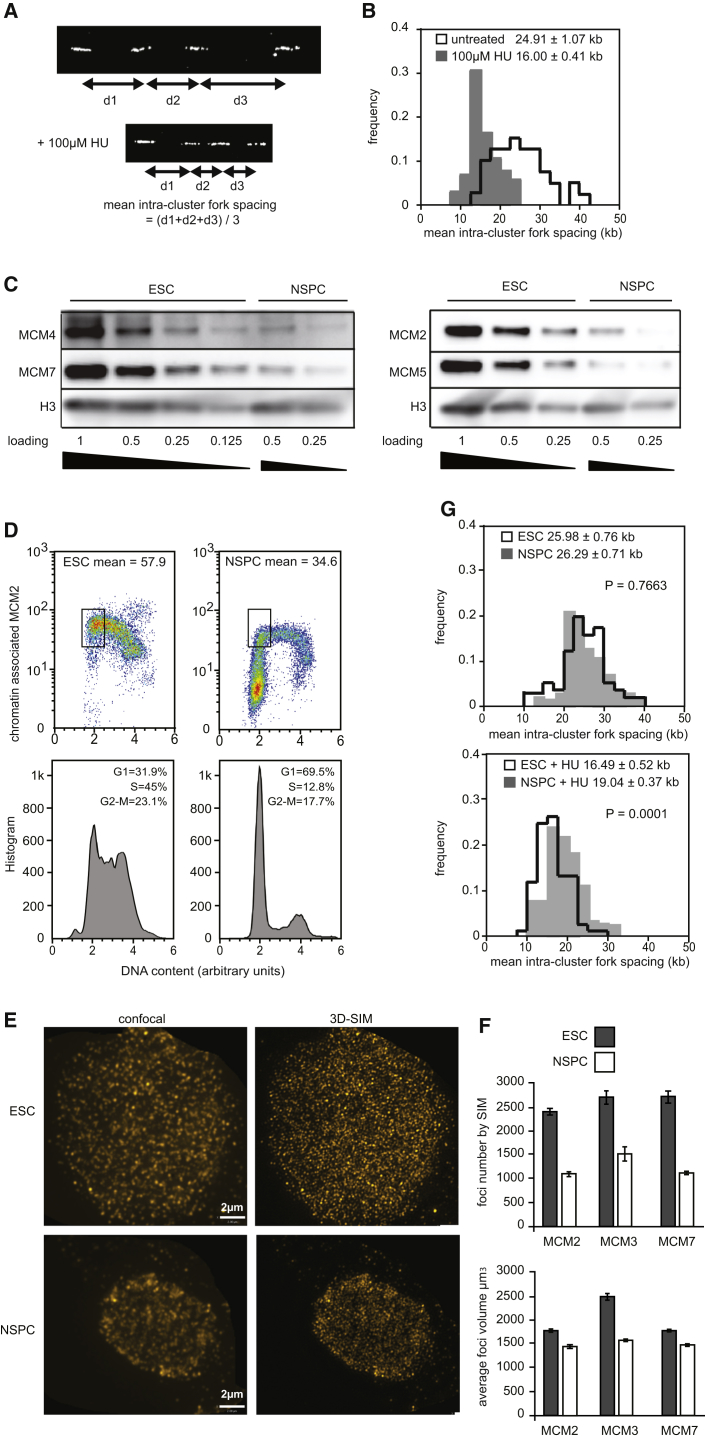
First, we investigated whether DOs exist in ESCs. DNA fiber assay was used to measure the density of replication forks, which involves labeling of the nascent strand DNA by BrdU pulse and visualization of labeled DNA after spreading on microscopic slides. DNA fibers containing at least a cluster of four consecutive BrdU-incorporated forks were chosen for analysis (e.g., Figure 1A). The average fork spacing within each cluster (i.e., mean intra-cluster fork spacing) was measured. The average fork spacing of the sample was calculated from the mean intra-cluster fork spacing of over 50 clusters (Figure 1B). ESCs have an average fork spacing of ∼25 kb, implying an average origin-to-origin distance of ∼50 kb within replicon clusters, consistent with replicon sizes in other mammalian cells (Berezney et al., 2000; Ge et al., 2007; Kawabata et al., 2011). After treatment with hydroxyurea (HU) that inhibits ribonucleotide reductase, replication forks in ESCs slowed down by ∼50% and the average fork spacing reduced to ∼16 kb (Figures 1A and 1B). These results show that DOs are activated in ESCs in response to replication stress.
Figure 1.
ESCs Possess More DOs Than NSPCs
(A and B) DNA fiber assay on mouse ESCs (CCE strain). For exclusion of artifacts arising from fork-to-fork fusion, cells were pulsed with BrdU for 10 min in the absence of HU and 20 min in the presence of HU to achieve similar replication fork length. (A) Examples of a DNA fiber containing a replicon cluster of four BrdU-labeled forks are shown. (B) Distribution of the mean intra-cluster fork spacing from >50 replicon clusters is shown. Overall fork spacing ± SEM is indicated in the chart.
(C–G) Comparisons between CCE cells derived from the 129/Sv mice and NSPCs from the E13.5 129/Sv embryo brains. (C) Immunoblotting of chromatin-bound MCM proteins with H3 as a loading control for quantification is shown. (D) Quantification of chromatin-bound MCM2 in G1-phase cells and cell-cycle distribution by FACS are shown. (E) 2D projection confocal and SIM images of chromatin-bound MCM2, MCM3, and MCM7 in G1 phase cells are shown. (F) Quantification of chromatin-bound MCM foci number and average focus volume imaged by SIM are shown. Error bars represent SEM of three independent experiments. (G) DNA fiber analysis of NSPCs and ESCs is shown. Cells were incubated with 100 μM HU for 4 hr before BrdU pulse. Overall fork spacing ± SEM from >50 replicon clusters is indicated. p values are from two-tailed t test.
Next, we compared the number of DOs in ESCs and tissue stem cells, using NSPCs as an example. Because 80%–95% of the chromatin-bound MCM2–7 complexes are DOs, we quantified the complexes on the chromatin by immunoblotting (Figure 1C). ESCs contain ∼2-fold more chromatin-bound MCM2–7 complexes than NSPCs. To exclude non-cycling cells from the analysis, we immunostained chromatin-bound MCM2 and analyzed the cells by flow cytometry. As licensing of replication origins starts at late mitosis and reaches the maximum at G1 phase, we quantified the chromatin-bound MCM2 in G1-phase ESCs and NSPCs. In line with the immunoblot results, ESCs contain ∼2-fold more chromatin-bound MCM2–7 complexes than NSPCs (Figure 1D). Furthermore, we used super-resolution 3D structured illumination microscopy (SIM) to quantify the chromatin-bound MCM2–7 complexes. SIM reaches 120 nm resolution in the x and y axis and 300 nm in the Z axis (Figure 1E), and a double hexameric MCM2–7 complex on DNA measures 25 × 16 nm (Evrin et al., 2009; Remus et al., 2009). Hence, each focus observed by SIM contains multiple MCM2–7 complexes. Quantification of chromatin-bound MCM2, MCM3, and MCM7 foci in G1 phase cells shows approximately twice more MCM2–7 complexes in ESCs than in NSPCs (Figures 1F, upper panel, and S5A). Because the average volume of MCM foci in ESCs is larger than in NSPCs, the difference of the chromatin-bound MCM2–7 complexes between ESCs and NSPCs is likely even greater (Figure 1F, lower panel). All the above data together demonstrate that ESCs possess ∼2-fold more chromatin-bound MCM2–7 complexes and therefore more DOs than NSPCs.
Finally, DNA fiber assay shows similar overall fork spacing in both ESCs and NSPCs (∼26 kb; Figure 1G, left panel), suggesting a similar usage of primary origins. However, after HU treatment, average fork spacing reduces to ∼16 kb in ESCs and only to ∼19 kb in NSPCs (Figure 1G, right panel), confirming fewer DOs in NSPCs than ESCs.
Reducing DOs Impairs ESC Differentiation, but Not Self-Renewal
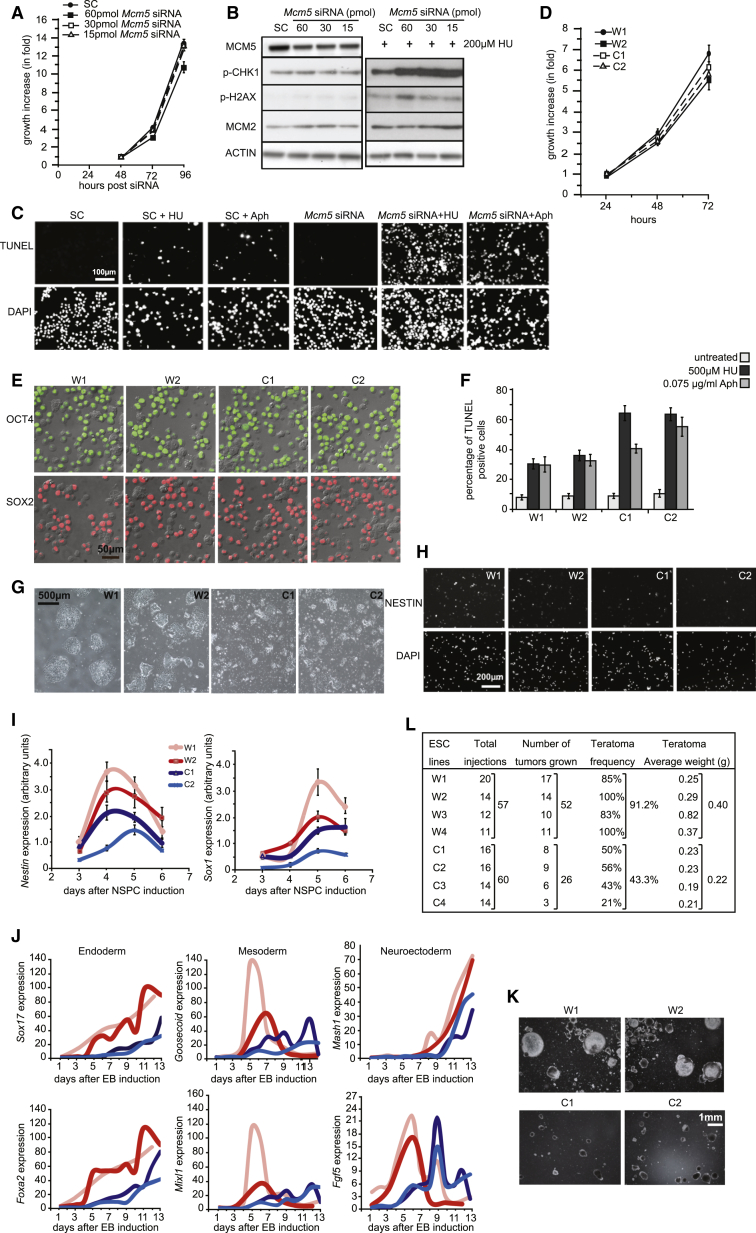
We next examined the function of DOs in ESCs by knocking down MCM5 using siRNAs. In order to reduce DOs while keeping primary origins intact, we titrated out the Mcm5 siRNAs to achieve ∼60% MCM5 knockdown in ESCs while maintaining a normal rate of cellular proliferation and DNA replication (Figures 2A, 2B, and S1A, upper panel). DNA fiber assay shows a similar average fork spacing between the siRNA-treated cells and the control, confirming that the usage of primary origins is unaffected. However, upon adding HU, activation of DOs is greatly reduced in the siRNA-treated cells (Figure S2A), and ESCs show hypersensitivity to replication inhibitors HU and aphidicolin, including a hyper-activation of DNA damage response proteins, a further reduction of the overall rate of DNA replication, and a significant increase of apoptosis (Figures 2B, 2C, and S1A, lower panel). These results demonstrate that DOs are required for ESCs to rescue replication fork stalling and to survive replication stress.
Figure 2.
Reducing DOs Impairs ESC Differentiation
(A) CCE cell proliferation at 48–96 hr after transfection with a serial dilution of Mcm5 siRNA. SC, scrambled siRNA. ∼90% transfection efficiency is achieved.
(B) Immunoblotting of total cell lysate at 72 hr after Mcm5 siRNA transfection. Fifteen picomoles Mcm5 siRNA knocked down MCM5 and had no effect on cell growth or DNA replication; hence, it was used for further analysis.
(C) TUNEL assay of ESCs after treatment with 500 μM HU or 0.075 μg/ml aphidicolin (Aph) for 48 hr. Fifteen picomoles Mcm5 or scrambled siRNA was transfected into the cells 72 hr prior to the HU and Aph treatment.
(D–K) Assays on Mcm4C/C ESCs (C1 and C2) and wild-type Mcm4+/+ ESCs (W1 and W2). (D) Cell proliferation rate analyzed over 72 hr is shown. (E) Overlay of OCT4 or SOX2 immunofluorescence images with DIC images is shown. OCT4- or SOX2-negative cells, larger than ESCs, are mostly MEF contamination in the ESC culture. (F) TUNEL assay of ESCs after treatment with HU or Aph for 48 hr is shown. (G and H) DIC images and immunofluorescence of NESTIN, respectively, of NSPCs at 96 hr after induced differentiation from ESCs are shown. (I) qRT-PCR analysis of Nestin and Sox1 expression during induced NSPC differentiation from ESCs is shown. (J) qRT-PCR data of the expression of three germ layer markers from days 1 to 13 during embryoid body (EB) differentiation from ESCs are shown. (K) DIC images of EBs at day 13 after induced differentiation from ESCs are shown.
(L) Frequency and average weight of teratomas generated from the wild-type (W1–W4) and Mcm4C/C (C1–C4) ESCs.
Error bars in (A), (D), (F), (I), and (J) all represent SEM of three independent experiments. See also Figures S1 and S2.
To avoid the transient effect of siRNAs, we derived ESCs from the Mcm4Chaos3 mice that contain a point mutation within the Mcm4 gene, resulting in the unstable MCM2–7 complexes and thus reduced DOs on chromatin (Kawabata et al., 2011). We assayed four Mcm4Chaos3/Chaos3 (Mcm4C/C) ESC lines and four wild-type controls (Mcm4+/+). Immunoblotting shows a partial reduction of the chromatin-bound MCM2–7 complexes in the Mcm4C/C ESCs (Figure S1C). Consistent with the Mcm5-siRNA-treated ESCs, the overall rates of proliferation and DNA replication of the Mcm4C/C ESCs are normal compared with the wild-type ESCs (Figures 2D, S1D, and S1E). The Mcm4C/C ESCs also maintain pluripotency: there are 80%–95% of Oct4, Sox2, and SSEA-1-positive cells in the Mcm4C/C ESC culture, similar to the control (Figures 2E, S1F, and S1G). Expectedly, the Mcm4C/C ESCs are hypersensitive to replication fork inhibitors HU and aphidicolin (Figures 2F, S1H, and S1I).
Because Mcm4C/C ESCs maintain normal self-renewal, we examined their differentiation. As they differentiate into NSPCs, they show increased cell death and decreased expression of NSPC markers NESTIN and SOX1 (Figures 2G–2I, S2A, and S2B). In addition, they show defective differentiation toward embryoid bodies, displaying abnormal morphology and compromised expression of neuroectoderm, endoderm, and mesoderm markers (Figures 2J, 2K, and S2C). To further assess their differentiation capability in vivo, we injected them into immune-compromised mice and allowed them to form teratomas. Although the cellular composition of the Mcm4C/C and the wild-type ESC-derived teratomas is similar (Figure S2D), the Mcm4C/C ESCs generate 50% fewer teratomas and these teratomas weigh 50% less than those derived from the wild-type controls (Figures 2L and S2E). Together, these data suggest that, upon reduction of DOs, ESCs maintain normal self-renewal but are impaired in differentiation. This is consistent with our observation that ESCs load more DOs than NSPCs. As a result, the self-renewal of ESCs is more robust against DO reduction than differentiation.
Reducing DOs Impairs ESC Differentiation to NSPCs
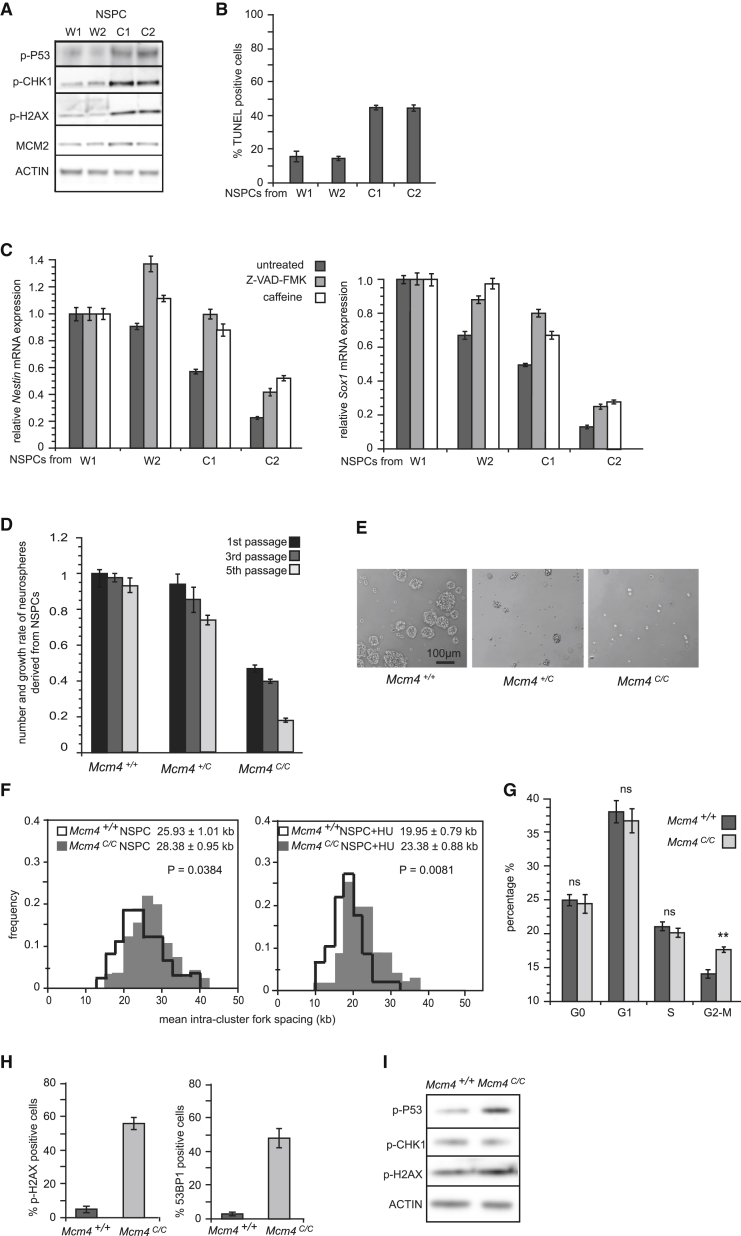
We further investigated the differentiation of the Mcm4C/C ESCs into NSPCs. Mcm4C/C ESC-derived NSPCs show hyper-activation of phosphorylated CHK1, P53, and H2AX and increased apoptosis (CASPASE 3 cleavage and 3-fold increase in TUNEL staining; Figures 3A, 3B, and S3A–S3C). Addition of caffeine, an ATM/ATR inhibitor, or the CASPASE inhibitor Z-VAD-FMK during NSPC differentiation largely rescued the differentiation efficiency, as shown by the increased expression of NESTIN and SOX1 (Figures 3C and S3C). The partial nature of the rescue could be due to the key role of ATR kinase during DNA replication and cell-cycle progression (Jirmanova et al., 2005; Ruzankina et al., 2007). Despite this, the above data clearly illustrate a functional relationship between reduced DOs and impaired neural differentiation of the Mcm4C/C ESCs due to elevated DNA damage response and cell death. The defect in the neural differentiation of the Mcm4C/C ESCs is likely due to compromised survival of differentiating cells.
Figure 3.
Reducing DOs Impairs the Differentiation of NSPCs
(A–C) Analysis of the NSPC differentiation from the Mcm4+/+ (W1 and W2) and Mcm4C/C (C1 and C2) ESCs. (A) Immunoblot of the NSPC total lysate is shown. (B) TUNEL assay on NSPCs at 96 hr after induced differentiation is shown. (C) qRT-PCR of Nestin and Sox1 expression in NSPCs is shown. Treatment with caffeine (4 mM) or Z-VAD-FMK (40 μM) started at 48 hr after induction, and NSPCs were harvested at 96 hr for analysis.
(D–I) Analysis of neurospheres clonally derived from NSPCs isolated from the E13.5 mouse forebrain. (D) Neurospheres were passaged every 6 days to give a new round of clonogenic assay. Number and growth rate of neurospheres were measured by counting the neurospheres and the total number of cells at each passage. Error bars represent SEM from four independent experiments and each experiment containing five embryos of each genotype. (E) Representative images of neurospheres at fifth passage are shown. (F) DNA fiber analysis is shown. Cells were treated with 100 μM HU for 4 hr before analysis. Overall average fork spacing ± SEM from >50 replicon clusters is shown. p values are from two-tailed t test. (G) Cell-cycle analysis of neurospheres at fifth passage by FACS after pyronin Y and DAPI staining is shown. Note G2-M blockage of the cells in the Mcm4C/C neurospheres. Two-tailed t test: non-significant (ns); p < 0.005 (∗∗). (H) Immunofluorescence quantifying the percentage of γH2AX or 53BP1 positive cells in neurospheres is shown. (I) Immunoblot of total cell lysate of neurospheres is shown.
Error bars in (B), (C), (G), and (H) all represent SEM of three independent experiments. See also Figure S3.
To confirm our in vitro findings on neural differentiation, we isolated NSPCs from the Mcm4C/C mice during embryogenesis. NSPCs from the forebrain of the E13.5 Mcm4C/C embryos generated 50% fewer neurospheres than the wild-type NSPCs, even though both expressed similar level of NESTIN and SOX2 (Figures 3D, S3D, and S3E). In addition, NSPCs from the Mcm4C/C embryos lost clonogenic ability after four to six passages, whereas the wild-type NSPCs continue to form neurospheres (Figure 3E). DNA fiber assay revealed a significant lack of DOs in the Mcm4C/C NSPCs as compared with wild-type NSPCs (Figure 3F). Furthermore, there is increased cell death in the Mcm4C/C neurosphere culture and a blockage of these NSPCs at the G2-M phase (Figures 3G and S3F). Elevated DNA damage markers γH2AX, 53BP1, and phospho-P53 were also observed in the Mcm4C/C NSPCs (Figures 3H and 3I). Together, these data show abnormal proliferation and differentiation of the NSPCs in the Mcm4C/C embryonic mice brains.
Reduction of DOs Impairs Embryonic Neurogenesis and Compromises Embryonic Viability
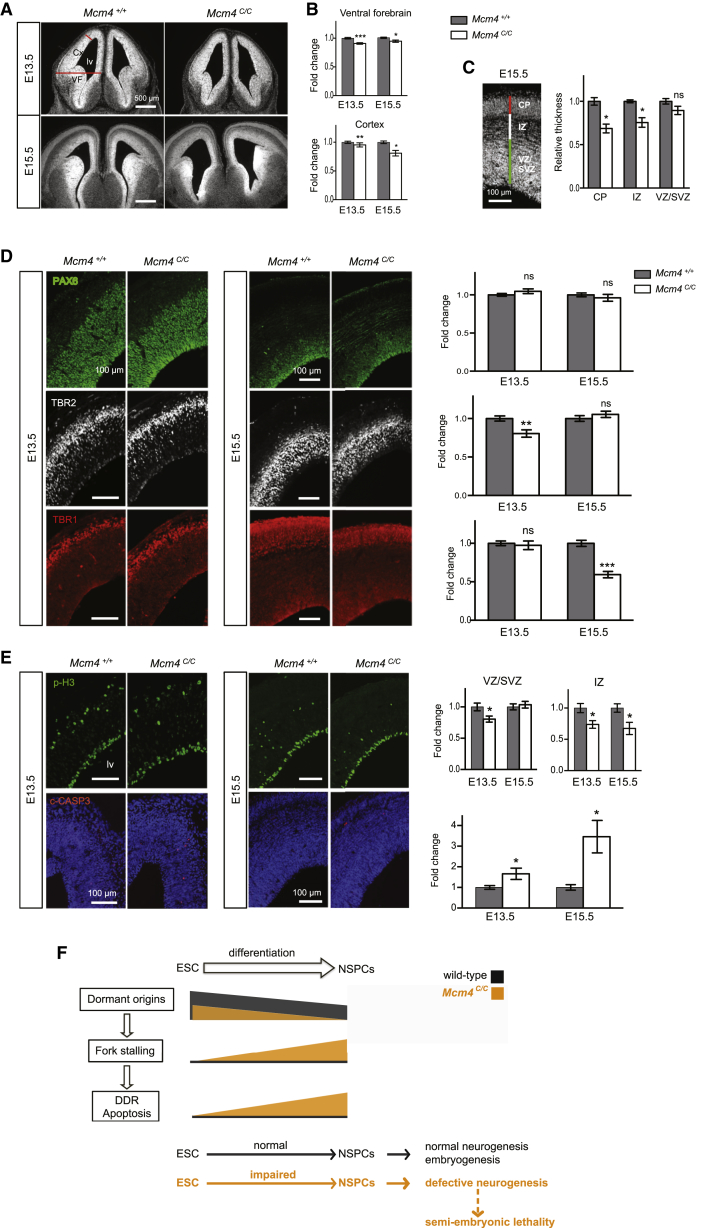
To investigate the in vivo properties of Mcm4C/C NSPCs, we examined different stages of neurogenesis in the Mcm4C/C embryos. At E13.5 and E15.5, Mcm4C/C embryos show a reduction in the size of the ventral forebrain and the cortex compared to the wild-type (Figures 4A–4C). The discrete atrophy of ganglionic eminences and the thinning of cortex indicate that early neurogenesis is globally impaired in the Mcm4C/C embryos. The ventricular layer of the PAX6+ radial glia cells, i.e., neural stem cells, is however of similar size in the Mcm4C/C and wild-type embryos (Figure 4D, upper panel), suggesting that the formation and self-renewal of neural stem cells are not significantly altered by the partial loss of MCM4 function. In contrast, the number of intermediate progenitor cells (TBR2+) in the cortical wall of the E13.5 Mcm4C/C embryos is significantly reduced compared to the wild-type (Figure 4D, middle panel), indicating a defect in neural stem cell differentiation and/or migration during early neurogenesis. Intermediate progenitor cells proliferate rapidly and migrate to give rise to post-mitotic neurons of the cerebral cortex. Consistently, the number of early-born post-mitotic neurons (TBR1+) was reduced in the cortical plate of the E15.5 Mcm4C/C brains (Figure 4D, lower panel). Furthermore, the Mcm4C/C mutants displayed anomalies in cell proliferation and survival within the cortex (Figure 4E): a reduction of mitotic cells (phospho-HISTONE H3+) and an increase of apoptotic cells (cleaved-CASPASE3+) were detected in the sub-ventricular and intermediate zones, suggesting that cell death contributes initially to the attrition of intermediate progenitor cell pool and then to the reduction of cortical neurons. Consequently, a thinning of the cerebral cortex was observed in the E19.5 Mcm4C/C brains (Figure S4A). However, at this late stage of development, intermediate progenitor cell formation has recovered and the Mcm4C/C-caused defects in neurogenesis other than cortex were no longer detectable, likely due to tissue homeostasis during development (Figure S4B).
Figure 4.
Reducing DOs Impairs Embryonic Neurogenesis and Affects Embryonic Viability
(A and B) Coronal sections of the Mcm4+/+ and Mcm4C/C embryonic forebrain. (A) Phase contrast views are shown. Red lines show width of ventral forebrain (VF) and thickness of cortex (Cx). (B) Quantification of forebrain size is shown.
(C) DAPI staining and measurement of the E15.5 cortex: ventricular/sub-ventricular zone (VZ/SVZ); intermediate zone (IZ); and cortical plate (CP).
(D) Cortex coronal sections with immunolabeling of VZ/SVZ stem/progenitor cells (PAX6+), intermediate progenitor cells (TBR2+), and early born, deep-layer cortical neurons (TBR1+).
(E) Immunolabeling of phospho-H3 and cleaved-CASPASE 3 (c-CASP3) cells on cortex coronal sections. lv, lateral ventricles. Error bars represent SEM of three independent experiments comprising in total five Mcm4+/+ and four Mcm4C/C embryos at E13.5 and seven Mcm4+/+ and seven Mcm4C/C embryos at E15.5. Two-tailed t test: ns; p < 0.05 (∗); p < 0.005 (∗∗); p < 0.0005 (∗∗∗).
(F) Model. Black and orange colors indicate the conditions of the wild-type and the partial depletion of DOs (as in the Mcm4C/C mice), respectively. In the wild-type ESCs, DOs initiate back-up replication forks to rescue fork stalling and maintain genome integrity. ESCs possess a greater number of DOs than NSPCs. Upon reduction of DO, there is a further reduction of DOs in the NSPCs, likely reaching the threshold required to rescue the endogenous fork stalling during DNA replication. As a result, DNA damage is accumulated and cell death incurs, eventually impairing NSPC proliferation and differentiation. This explains the neurogenic defect in the Mcm4C/C embryos, which could contribute to the semi-embryonic lethality of the Mcm4C/C mice.
See also Figure S4.
Beyond neurogenic defects, only 40% of Mcm4C/C mice are viable (Figure S4C). Because the homozygotes are present at the correct ratio at E13.5, E15.5, and E19.9, the Mcm4C/C fetus likely dies shortly after birth. The semi-lethality of the Mcm4C/C mice is consistent with the in vitro differentiation defect of the Mcm4C/C ESCs.
Discussion
We have demonstrated that ESCs recruit ∼2-fold more DOs onto the genome than NSPCs. Upon reduction of DOs, the self-renewal of ESCs is unaffected, whereas their differentiation including toward NSPCs is impaired. This is due to a further reduction of DOs in NSPCs, presumably below the threshold required to rescue the endogenous fork stalling during DNA replication (Figure 4F). As a result, DNA damage is accumulated and cell death incurs, eventually leading to impaired neurogenesis in the Mcm4C/C mice. ESCs have been shown to employ unique mechanisms to maintain a more-stable genome than somatic cells, including efficient DNA repair, elimination of damaged cells, antioxidant defense, and suppression of mutagenesis (Giachino et al., 2013). Our study adds a new dimension to these unique properties by showing that ESCs use more DOs to effectively protect their genomes from replication stress and ensure their genome integrity.
It remains elusive how ESCs recruit a larger number of DOs than tissue stem/progenitor cells during DNA licensing. It is possible that ESCs express a higher level of proteins that mediate DNA licensing. Alternatively, it could be due to their open and hyper-dynamic chromatin structure (Mattout and Meshorer, 2010), which facilitates MCM2–7 loading (Miotto and Struhl, 2010; Sugimoto et al., 2011; Swarnalatha et al., 2012; Wong et al., 2010).
Because NSPCs possess fewer DOs than ESCs, when DOs are reduced, neurogenesis is more severely affected. Our findings may be related to the severe neurogenic defect in the Meier-Gorlin syndrome patients, who are characterized by mutations in replication licensing components and reduction in origin licensing (Bicknell et al., 2011; Kerzendorfer et al., 2013). In addition to NSPCs, other tissue stem/progenitor cells may possess fewer DOs than ESCs, because Mcm4C/C ESCs display broad in vitro differentiation defects. The neurogenic defect together with other organ abnormalities could collectively contribute to the semi-embryonic lethality of the Mcm4C/C mice. Future studies in other tissue stem cells in the Mcm4C/C mice will allow further understanding of the growth retardation and other deficiencies associated with the hypomorphic MCM4 conditions in human patients.
Experimental Procedures
The Mcm4chaos3/chaos3 and wild-type mouse ESC lines were derived from the blastocysts by crossing the Mcm4chaos3/+ mice. They were maintained on MEF feeder in standard ESC culture medium. CCE mouse ESCs were grown without feeder. siRNA was transfected into ESCs by Lipofectamine 2000 according to manufacturer’s instructions. NSPCs were isolated from the forebrain of E13.5 mice and cultured as described in Supplemental Information. Neurosphere culture on day 6 was dissociated into single cells and plate at 1,000 cells/ml to initiate clonal neurosphere assays. DNA fiber, chromatin-bound MCMs analysis by immunoblotting, and FACS were carried out as previously described (Ge et al., 2007). Embryonic brains from the E13.5, 15.5, and 19.5 mice were dissected, cryosectioned, and immunostained with various antibodies, including TBR2, P-H3, and cleaved-CASPASE 3 where positive cells were counted and PAX6 and TBR1 where the thickness of cell layer was measured. The rate of DNA synthesis was measured by pulse labeling cells with Click-iT EdU Alexa Fluor 647 Flow Cytometry kit (Invitrogen) according to manufacturer’s instruction. Cell growth rate was assayed with the alamarBlue CellViability Reagent (Invitrogen), and apoptosis was assayed by ApopTag Fluorescein In Situ Apoptosis Detection Kit (Millipore) according to the manufacturer’s instructions. NSPC differentiation was performed as previously described using the N2B27 medium (Ying et al., 2003). Embryoid body differentiation was achieved by hanging drop method (Jackson et al., 2010). The differentiation efficiency into the ectoderm, mesoderm, and endoderm was assessed by qRT-PCR using lineage-specific primers (Nestin, Sox1, Sox17, Goosecoid, Mash1, Fgf5, Mixl1, and Foxa2). For imaging of the chromatin-bound MCM foci, cells were synchronized by thymidine followed by nocodazole. Delta Vision OMX imaging system (Applied Precision) was used to collect 3D images, followed by analysis with Volocity (PerkinElmer), where a threshold-based segmentation was applied.
Author Contributions
X.Q.G. designed and performed the experiments and wrote the manuscript. J.H. and J.-L.T. did embryonic brain analysis. E.-C.C. conducted teratoma assay. S.Y. and N.S. provided Mcm4chaos3 and wild-type ESCs. H.L. provided guidance on the project and revised the manuscript.
Acknowledgments
We thank Felix Rivera-Molina for assistance with SIM. This work was funded by a grant from the Connecticut Stem Cell Research Fund (10SCA05) to X.Q.G. and the G. Harold and Leica Y. Mathers Award to H.L.
Published: July 16, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.06.002.
Supplemental Information
References
- Berezney R., Dubey D.D., Huberman J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Bicknell L.S., Bongers E.M., Leitch A., Brown S., Schoots J., Harley M.E., Aftimos S., Al-Aama J.Y., Bober M., Brown P.A. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Ge X.Q. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep. 2009;10:406–412. doi: 10.1038/embor.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M.L., Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y., Bermejo R., Fiorani S., Haber J.E., Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Q., Blow J.J. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 2010;191:1285–1297. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Q., Jackson D.A., Blow J.J. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C., Orlando L., Turinetto V. Maintenance of genomic stability in mouse embryonic stem cells: relevance in aging and disease. Int. J. Mol. Sci. 2013;14:2617–2636. doi: 10.3390/ijms14022617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineau L., Cognet C., Kara N., Lach F.P., Dunne J., Veturi U., Picard C., Trouillet C., Eidenschenk C., Aoufouchi S. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J. Clin. Invest. 2012;122:821–832. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.R., Guasti L., Meimaridou E., Chuang C.H., Schimenti J.C., King P.J., Costigan C., Clark A.J., Metherell L.A. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J. Clin. Invest. 2012;122:814–820. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Schwob E., Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M., Taylor A.H., Jones E.A., Forrester L.M. The culture of mouse embryonic stem cells and formation of embryoid bodies. Methods Mol. Biol. 2010;633:1–18. doi: 10.1007/978-1-59745-019-5_1. [DOI] [PubMed] [Google Scholar]
- Jirmanova L., Bulavin D.V., Fornace A.J., Jr. Inhibition of the ATR/Chk1 pathway induces a p38-dependent S-phase delay in mouse embryonic stem cells. Cell Cycle. 2005;4:1428–1434. doi: 10.4161/cc.4.10.2055. [DOI] [PubMed] [Google Scholar]
- Kawabata T., Luebben S.W., Yamaguchi S., Ilves I., Matise I., Buske T., Botchan M.R., Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzendorfer C., Colnaghi R., Abramowicz I., Carpenter G., O’Driscoll M. Meier-Gorlin syndrome and Wolf-Hirschhorn syndrome: two developmental disorders highlighting the importance of efficient DNA replication for normal development and neurogenesis. DNA Repair (Amst.) 2013;12:637–644. doi: 10.1016/j.dnarep.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Kunnev D., Rusiniak M.E., Kudla A., Freeland A., Cady G.K., Pruitt S.C. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010;29:3630–3638. doi: 10.1038/onc.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S., Watson A., Sheedy D.M., Martin B., Carr A.M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Letessier A., Millot G.A., Koundrioukoff S., Lachagès A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Makovets S., Herskowitz I., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A., Meshorer E. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010;22:334–341. doi: 10.1016/j.ceb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Miotto B., Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010;37:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin E.V., Mirkin S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt S.C., Bailey K.J., Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- Remus D., Beuron F., Tolun G., Griffith J.D., Morris E.P., Diffley J.F. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., Brown E.J. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., Schimenti J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Yugawa T., Iizuka M., Kiyono T., Fujita M. Chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with Cdc10 protein-dependent transcript 1 (Cdt1) and promotes pre-replication complex formation. J. Biol. Chem. 2011;286:39200–39210. doi: 10.1074/jbc.M111.256123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarnalatha M., Singh A.K., Kumar V. The epigenetic control of E-box and Myc-dependent chromatin modifications regulate the licensing of lamin B2 origin during cell cycle. Nucleic Acids Res. 2012;40:9021–9035. doi: 10.1093/nar/gks617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.G., Glozak M.A., Cao T.V., Vaziri C., Seto E., Alexandrow M. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11-geminin. Cell Cycle. 2010;9:4351–4363. doi: 10.4161/cc.9.21.13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.