Abstract
All vertebrate eggs are surrounded by an extracellular coat that supports growth of oocytes, protects oocytes, eggs, and early embryos, and participates in the process of fertilization. In mammals (platypus to human beings) the coat is called a zona pellucida (ZP) and in non-mammals (molluscs to birds), a vitelline envelope (VE). The ZP and VE are composed of just a few proteins that are related to one another and possess a common motif, called the zona pellucida domain (ZPD). The ZPD arose more than ~600 million years ago, consists of ~260 amino acids, and has 8 conserved Cys residues that participate in 4 intramolecular disulfides. It is likely that egg-coat proteins are derived from a common ancestral gene. This gene duplicated several times during evolution and gave rise to 3–4 genes in fish, 5 genes in amphibians, 6 genes in birds, and 3–4 genes in mammals. Some highly divergent sequences, N- and C-terminal to the ZPD, have been identified in egg-coat proteins and some of these sequences may be under positive Darwinian selection that drives evolution of the proteins. These and other aspects of egg-coat proteins, including their structure and synthesis, are addressed in this review.
Keywords: eggs, zona pellucida, vitelline envelope, ZP genes, ZP proteins, ZP domain
INTRODUCTION
“Evolutionary scenarios are an artform. They usefully exercise the brain, causing us to look at old data in new ways and stimulating us to collect new data. They do not have to be true”. W. Ford Doolittle, 2006 [1].
A relatively thick extracellular matrix or coat surrounds vertebrate eggs. In fish, amphibians, and birds the egg-coat is called a vitelline envelope (VE) and in mammals a zona pellucida (ZP) [2]. Vertebrate egg-coats differ considerably in size, but are quite uniform in composition since they consist of only a few proteins that possess a common motif, the zona pellucida domain (ZPD). ZP and VE proteins are a conserved group that share high sequence identity and have many structural features in common. The proteins assemble into fibrils and then into the matrix (egg-coat) that surrounds growing oocytes. Egg-coats protect oocytes, eggs, and early embryos, support oocyte growth, and play important roles during fertilization.
The mammalian ZP consists of 3–4 proteins, called ZP1–4 [3]. The amphibian and bird VE consists of 4–6 proteins, called ZP1-4, ZPd, and ZPax, and in many fish the VE consists of 2–4 proteins, called ZP1, ZP3, ZPax, and variants of ZP1 and ZP3 [4–6]. It has been proposed that ZP proteins are derived from a common ancestral gene, possibly ZP3, and that ancestral ZP3 protein functioned as an egg-coat protein. Perhaps a first duplication event of an ancestral ZP3 gave rise to ZP3 and a precursor to other ZP genes which subsequently duplicated several times and evolved into all other ZP genes.
In recent years, the repertoire of egg-coat proteins has been expanded by whole-genome sequencing and provided amino acid (aa) sequences for ZP proteins from non-traditional vertebrate models. These models include platypus (monotremata) [7], lizard [8], turtle [9], snake (reptilia) [10], shark (cartilaginous fish) [11], lamprey (jawless fish) [12], as well as amphioxus (fish-like chordate) [13]. The lamprey is thought to belong to the oldest living group of vertebrates (> 350 million years old) and probably is a good candidate with which to address questions concerning the evolutionary origin and duplication events of ancestral ZP genes.
Here we discuss some characteristic traits of egg-coats, ZP genes, and ZP proteins from vertebrate species that represent fish (> 40,000 species), amphibians (> 5,000 species), birds (> 10,000 species), and mammals (> 5,000 species), as well as some non-traditional vertebrate models (e.g., shark, turtle, snake, and platypus). Emphasis is placed on shared features and functions that indicate a common origin of ZP proteins dating back more than 500 million years [2, 14]. We also point out some of the characteristic structural features of the ZPD and discuss aspects of the synthesis and assembly of ZP proteins into fibrils and matrix (egg-coats).
Vertebrate eggs and egg-coats
Vertebrate oocytes, eggs, and embryos, from fish to human beings, are surrounded by an egg-coat. Fish and amphibian embryos hatch from the VE as free-swimming larvae and tadpoles, respectively, whereas among birds a newly hatched chick emerges that is fully developed. Mammalian embryos are surrounded by a ZP until the early blastocyst stage of development when they hatch from the ZP and implant in the uterus (with the exception of monotremes, such as the platypus, that lay eggs) [2, 15].
Mammalian eggs are ~100 μm in diameter with not much more than a 2-to-1 variation in size for eggs from different mammals (with the exception of monotremes, such as platypus, whose eggs are 20–30 times larger) [16–19]. Similarly, the ZP of various mammalian eggs averages ~15 μm in width, from ~6 μm in mice to ~20–25 μm in human beings [20–23]. Platypus eggs are an exception since they have a very thin ZP, ~1 μm in width, and the eggs are protected by a leathery shell.
Bird eggs are very large compared to mammalian eggs due to the accumulation of vast amounts of yolk. However, the region of the egg that gives rise to the bird embryo lies on the surface of the yolk and is microscopic. The VE of bird (e.g., chicken) eggs consists of 3 layers, of which the innermost layer, called the pervitelline layer (PVL), is a relatively thin matrix. The PVL is a functional homolog of the mammalian ZP and the amphibian and fish VE [24].
Amphibian eggs typically are deposited in water in large groups or clusters and hatch within a relatively short time as free-swimming larvae. The eggs possess a VE that varies in thickness, from ~1 μm for Xenopus to ~8–15 μm for other amphibians. The eggs also have an additional gelatinous layer of protection, called the jelly coat [25].
Fish eggs are laid down in large numbers. They are surrounded by a VE that often consists of a thin outer layer and a thicker inner layer. The thin outer layer is possibly the result of crosslinking of VE proteins through their N-terminal proline-rich region (e.g., trout egg VE) [26–28]. Two kinds of fish eggs have been described; pelagic floating eggs that are highly hydrated and possess a “smooth” outer surface VE, and non-buoyant benthic eggs that have a “sticky” outer surface VE used to adhere to substrates [29]. Amphioxus (a fish-like marine chordate) eggs are ~150 μm in diameter and are surrounded by a VE that is ~6 μm thick [30].
Although the diameter of eggs and width of egg-coats can vary considerably among different organisms (Table 1), the relative elasticity and fibrillar nature of the egg-coat are characteristic features of all vertebrate VEs and ZPs.
Table 1.
Approximate egg-coat widths and egg diameters.
| Organism | Egg-Coat Width (μm) | Egg Diameter (μm) |
|---|---|---|
| Trouta | 25–50 | 3,000–6,000 |
| Frogb | 1 | 1,000 |
| Chicken | 1–3.5 | 30,000 |
| Platypusc | 1 | 4,000 |
| Possumd | 6 | 230 |
| Mouse | 6 | 80 |
| Rabbit, Sheep, Cat | 15 | 120–130 |
| Cow | 16 | 120 |
| Pig | 16 | 130 |
| Baboone | 13 | – |
| Human | 20–25 | 100–120 |
Oncorhynchus mykiss.
Xenopus laevis.
Ornithorhynchus anatinus.
Trichosurus vulpecula.
Papio hamadryas.
Evolution of ZP genes
A better understanding of the evolution of ZP genes and proteins has recently emerged. For example, comparisons of gene structure and organization made it possible to locate, in different species, comparable ZP genes (Table 2) and their adjacent regions within portions of chromosomes (synteny) [6]. ZP protein homologs are classified as either orthologs (e.g., mouse ZP3 vs chicken ZP3) or paralogs (e.g., mouse ZP3 vs mouse ZP2). It is likely that ZP proteins are derived from a common ancestral gene, possibly by an initial duplication event hundreds-of-millions of years ago. This event gave rise to ZP3 in one branch and the ZPd, ZPax, ZP2, and ZP1/ZP4 gene families, in this order, in other branches [4, 31]. ZP1/ZP4 has been cited as an example of duplication of a single gene because adjacent regions of these two genes in the mouse are located on the same chromosome (number 19) [6].
Table 2.
Location of ZP genes (Chromosome No.)*.
| Organism | ZP1 | ZP2 | ZP3 | ZP4 | ZPd | ZPax |
|---|---|---|---|---|---|---|
| Trout | x | x | (−)a | |||
| Frog | – | x | x | x | x | x |
| Chicken | 5 | 14 | 10 | 6 | 11 | 3 |
| Possum | – | x | x | x | ||
| Mouse | 19 | 7 | 5 | (13)b | ||
| Rat | 1 | 1 | 12 | 17 | ||
| Cow | (29)c | 25 | 25 | 28 | pseudod | |
| Pig | – | 3 | 3 | 14 | ||
| Dog | (18)c | 6 | 6 | 4 | ||
| Macacae | 14 | 20 | 3 | 1 | pseudod | |
| Chimpanzeef | 11 | 16 | 7 | 1 | pseudod | |
| Human | 11 | 16 | 7 | 1 | pseudod |
Taken from NCBI database.
x No information about location of ZP genes.
-Not found.
In some fish, e.g. gilthead seabream, a ZP2 homolog (ZPx) is present.
ZP4 is a pseudo-gene in mice.
ZP1 is a pseudo-gene in cows and dogs.
ZPax is a pseudo-gene in cows, macacas, chimpanzees, and humans.
Macaca mulatta.
Pan troglodytes.
ZP1-4 are found in mammals and other vertebrates, ZPd in amphibians and birds, and ZPax in fish, amphibians, and birds (Table 2) [6, 15]. The absence of ZPd and ZPax in mammals suggests that these genes were lost during evolution, as exemplified by non-functional ZPax (a pseudo-gene) found in some mammals (e.g., human beings, chimpanzees, macacas, and cows). In addition, ZP1 and ZP4 have been identified as pseudo-genes in several mammalian species (e.g., ZP1 in dogs and cows, and ZP4 in mice). This suggests that recent ZP gene evolution may have occurred by “gene death” [32].
Mammals, amphibians, and birds have 3–4, 5, and 6 ZP genes, respectively. Many fish (teleosts) have 2–4 ZP genes, whereas some fish (e.g., zebrafish) have multiple copies or tandem repeats of ZP1 and ZP3 genes.
ZP1-4 genes are also present in non-traditional vertebrate models, such as shark [11], turtle [9], snake [10], and platypus [7] (Table 3). The aa sequences of these ZP proteins have been derived by conceptual translation from whole-genome sequences and have not been characterized further. For amphioxus, 5 ZP proteins have been identified as components of its egg-coat (called BbZP1-5) and all have a type-II ZPD [30] (Fig. 1b). Lamprey ZP genes have been predicted based on their similarity to fish ZP genes and identified as ZP protein homologs [30]. However, the predicted proteins are not full-length due to incomplete coverage of the genome.
Table 3.
ZP genes of non-traditional vertebrate models.
| Organism | ZP Genes |
|---|---|
| Amphioxusc | (5 ZP genes)a |
| Lampreyd | (ZP2, ZP4, ZPax)b |
| Sharke | ZP1-4, ZPd |
| Turtlef | ZP1-4 |
| Snakeg | ZP1-3 |
| Platypus | ZP1-4 |
Named 5 ZP proteins based on mass spectrometric analyses.
ZP proteins not full-length and tentatively named.
Branchiostoma belcheri.
Petromyzon marinus.
Callorhynchus milii.
Chelonia mydas.
Ophiophagus hannah.
Figure 1b.
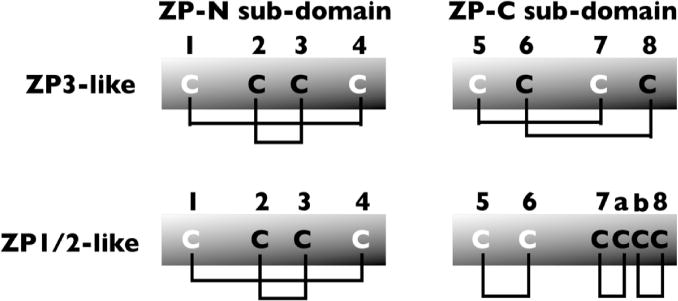
Schematic representation of intramolecular disulfides of ZP3- like (type I) and ZP1/2-like (type II) ZPD proteins.
Sequence identity of ZP proteins
The ZP/VE of vertebrate eggs, from fish to human beings, consists of only a few ZP proteins, called ZP1-4, ZPd, and ZPax. ZPd is only found in Xenopus and chicken, and ZPax is found in some fish, Xenopus, and chicken. ZP3 is always the smallest of the four ZP proteins and ZP4 is always smaller than ZP1 (Table 4).
Table 4.
Sizes of polypeptide precursors (No. aa Residues)*.
| Organism | ZP1 | ZP2 | ZP3 | ZP4 | ZPd | ZPax |
|---|---|---|---|---|---|---|
| Trout | 563/524a | 441 | (698)b | |||
| Frog | 699 | 460 | 544 | 376 | 905 | |
| Chicken | 934 | 695 | 446 | 543 | 418 | 837 |
| Possum | 712 | 422 | 527 | |||
| Mouse | 623 | 713 | 424 | |||
| Rat | 617 | 695 | 424 | 545 | ||
| Cow | 713 | 421 | 534 | |||
| Pig | 716 | 421 | 536 | |||
| Dog | 715 | 426 | 531 | |||
| Macaca | 640 | 745 | 424 | 539 | ||
| Chimpanzee | 638 | 745 | 424 | 540 | ||
| Human | 638 | 745 | 424 | 540 |
Taken from NCBI database.
ZP1α/ZP1β.
Gilthead seabream ZPx.
Sequences of vertebrate ZP proteins (ZP1-4) have been compared with orthologs from human beings and, although aa sequences between species vary in length, there is a high degree of sequence identity in overlapping sequences (Table 5). The average % identity suggests that the twelve organisms examined can be divided into four groups, I–IV: I - trout, 33% ave. identity; II - frog, chicken, and possum, 43–51% ave. identity; III - mouse, rat, cow, pig, and dog, 64–69% ave. identity; IV - macaca, chimpanzee, and human beings, 93–99% ave. identity. It has been suggested that proteins with sequence identities of 40% or more perform the same function (ZP proteins in groups II, III, and IV) and those with identities of 25–40% perform similar functions (ZP proteins in group I) [33]. In groups II–IV, with sequence identities of 40% or more, ZP proteins are structural and sperm-binding proteins, whereas in group I, with sequence identities of 25–40%, ZP proteins play only a structural role (e.g., fish sperm do not bind to the VE, but enter the egg via an opening in the VE, called the micropyle).
Table 5.
Comparison of vertebrate and human ZP1-4.
| Average % Identitya | ||||
|---|---|---|---|---|
| Organism | hZP1b | hZP2 | hZP3 | hZP4 |
| Trout | 33/31 | 36 (40)c | ||
| Frog | 42 | 44 (49) | 42 | |
| Chicken | 45 | 44 | 48 (58) | 52 |
| Possum | 54 | 46 (54) | 54 | |
| Mouse | 67 | 58 | 68 (75) | |
| Rat | 68 | 58 | 67 (73) | 64 |
| Cow | 67 | 72 (82) | 69 | |
| Pig | 63 | 73 (84) | 67 | |
| Dog | 67 | 71 (80) | 69 | |
| Macaca | 94 | 93 | 93 (97) | 93 |
| Chimpanzee | 99 | 99 | 99 (100) | 99 |
Whole sequences of ZP proteins (ZP1, 2, 3 and 4) are compared with sequences of human ZP1-4 (see Table 4). The numbers represent % identity of each of the ZP proteins with the corresponding human ortholog. Note that aa sequences vary in length.
Human ZP proteins are designated with an h.
Comparison of ZPD sequences of ZP3. Note that the % identity of the ZPDs is 1–11% higher than the % identity of whole sequences.
Structure of the ZPD
A structural element present in ZP proteins and in a variety of other proteins was defined by pattern-based sequence analysis and called the “zona pellucida domain” (ZPD) [34]. It was suggested that the ZPD has a common tertiary structure and plays a common biological role. Proteins with the same domain combination tend to share an ancestor and have functional features in common [35]. The ZPD arose more than ~600 million years ago and is found in a variety of animal species, in vertebrates and invertebrates, and in a variety of tissues and organs. It is a component of all ZP proteins and is present in hundreds of other proteins [36]. The latter include secreted proteins that function as receptors or mechanical transducers between adjacent cells, in intermediators during differentiation, morphogenesis and signaling, or in proteins of extracellular matrices other than egg-coats.
A ZPD consists of ~260 aa and has 8 conserved Cys residues that participate in 4 intramolecular disulfides. It is a bi-partite structure consisting of 2 sub-domains, ZP-N (~120 aa) and ZP-C (~130 aa), linked by a protease-sensitive region (Fig. 1a). Each sub-domain has 4 conserved Cys residues, however, the ZP-C sub-domain can have 2 additional Cys residues. ZP-N and ZP-C share a common immunoglobulin (Ig)-like topology with β-sheet arrangements symmetrical to each other, although the sub-domains have significantly different primary structures and intramolecular disulfide bonds [37]. There are two types of ZPDs: Type-I (ZP3-like) with 8 Cys residues and type-II (ZP1/2-like) with 10 Cys residues. The type-I ZPD has a ZP-N sub-domain with 4 Cys residues, linked 1,4 and 2,3, and a ZP-C sub-domain with 4 Cys residues, linked 5,7 and 6,8 (Fig. 1b). The type-II ZPD has a ZP-N sub-domain with 4 Cys residues, linked 1,4 and 2,3, and a ZP-C sub-domain with 6 Cys residues, linked 5,6, 7,a, and b,8 (Fig. 1b).
Figure 1a.
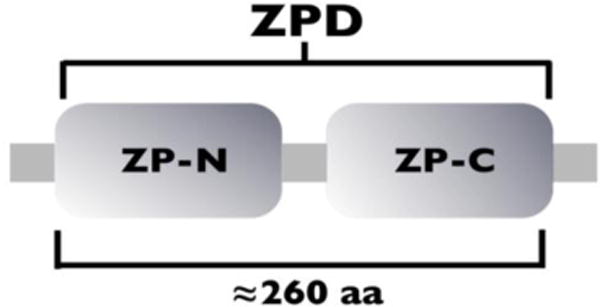
Schematic representation of the ZPD composed of a ZP-N and a ZP-C sub-domain.
There are exceptions to the two types of ZPD described above, including ZPDs with 12 Cys residues. The two additional Cys residues, referred to as Cx and Cy and linked x,y, are present in fish (e.g., trout) ZP1-like proteins (ZP1α and ZP1β) and are located between the ZP-N and ZP-C sub-domains [27]. In chicken and pig ZP3, Cys residue clustering in ZP-C sub-domains is variable [37]; there is a disulfide linkage to a Cys residue C-terminal to the ZPD, linked 6,11 and 8,9, whereas in mouse ZP3 it is 6,8 and 9,11 (Cys residues 9 and 11 are located downstream of the ZPD). It is likely that the additional disulfide linkage in trout and the different disulfide bonding patterns in chicken and pig cause the polypeptides to adopt different conformations, as compared to ZP1/2-like and ZP3-like proteins, which in turn could alter the specificity of egg coat assembly in these species.
Function of the ZPD and adjoining regions
Protein domains like the ZPD are evolutionary units that can be duplicated and recombined, and typically, pairs of domains are found in one sequential order (A→B or B→A), almost never in both [33]. Considering the two ZP sub-domains, ZP-N and ZP-C, as separate units, the sequential order is always ZP-N→ZP-C and never ZP-C→ZP-N, although the ZP-N sub-domain can be present in the absence of ZP-C.
The ZPD is common to all vertebrate egg-coat proteins and the domain conservation is reflected in the high percentage of its sequence identity within vertebrate ZP proteins (Table 5). It has been proposed that the ZPD is a conserved module for polymerization of extracellular proteins (e.g., to form ZP fibrils) [38, 39] and it is likely that the bi-partite structure of the ZPD (ZP-N and ZP-C sub-domains) endows it with dual functions. For example, the ZP-N sub-domain can serve as an independent structural domain in the absence of ZP-C (e.g., in proteins Oosp1 [40], Plac1 [41], and papillote [42]), and the ZP-N sub-domain alone has been shown to be an active folding unit that can polymerize into fibrils [43]. In addition, divergent copies of ZP-N sub-domains are found in single or multiple copies in the N-terminal regions of ZP1, ZP2, ZP4, and ZPax [44]. On the other hand, no protein has been found to consist only of the ZP-C sub-domain which suggests that its role is dependent on its partner ZP-N. Perhaps ZP-C plays a regulatory role during assembly of ZPD protein complexes.
N- and C-terminal to the ZPD are highly divergent aa sequences that have been identified in mouse and chicken ZP3 [45, 46]. For example, in mouse ZP3 there are 2 clusters of sites N-terminal (aa 25–50) and C-terminal (aa 331–373) (Table 6) to the ZPD and, similarly, in chicken ZP3 (aa 2–30, aa 381–436), that apparently are under positive selection (including a few mutations within the ZPD). Regions that are N- and C-terminal to the polymerization module are more likely to be involved in species-specific functions (e.g., sperm-egg interactions) and it is possible that selective pressure drives the mutations in these regions and thereby promotes divergence and evolution of ZP3 proteins. Mutations within the ZPD module and adjoining regions can also affect post-translational modifications, such as N- (Asn-X-Ser/Thr) or O- (Ser, Thr) linked glycosylation. For example, 2 potential O-linked glycosylation sites (Ser331 and Ser333) are within the highly divergent C-terminal cluster of mouse ZP3 and might promote divergence between mouse ZP3 and other closely related mammalian species due to differences in glycosylation pattern.
Table 6.
Divergent ZP3 sequences in mammals.

|
Highly divergent aa’s (↓) in mouse ZP3 as compared to homologous C-terminal sequences of rat, cow, pig, dog, macaca, chimpanzee, and human. Note that mouse S334 (underlined) is conserved in all eight species.
In general, the ZPD module is a conserved evolutionary unit essential for polymerization of proteins [38], whereas adjoining regions contribute functional diversification that may be caused by selective pressure related to species-specific functions.
ZP Protein synthesis, secretion, and assembly
ZP proteins are secreted proteins and as such are synthesized as precursor polypeptides with a signal sequence (SS) at the N-terminus and a C-terminal propeptide (CTP) at the C-terminus. They have a ZPD with 8 or more conserved Cys residues present as intramolecular disulfides, a consensus furin-cleavage site (CFCS) located close to the C-terminus, and a transmembrane domain (TMD) downstream of the CFCS (some fish lack a TMD or a hydrophobic C-terminus). ZP1 and ZP4 also have a trefoil domain adjacent to the ZPD.
For mammals and amphibians the ovary is the only site of synthesis of ZP proteins [47, 48] (Table 7). However, in fish that release several tens-of-thousands of relatively small eggs at a time, and in birds that have very large, yolk-laden eggs, there are two sites of ZP protein synthesis, the ovary and the liver [49–52] (Table 7). Perhaps the additional sites of ZP protein synthesis in fish and birds reflect the necessity of synthesizing large amounts of ZP proteins in a relatively short time. For some fish, it might therefore be advantageous to synthesize ZP proteins in the liver. On the other hand, small fish like zebrafish release relatively small clutches of eggs (~hundreds) and do not express ZP genes in the liver. In birds, ZP proteins are synthesized in the liver and in granulosa cells that surround growing oocytes. A fully-grown chicken oocyte is ~200–300 times larger in diameter than a mammalian egg and thus the ZP (although thinner than a mammalian ZP) covers an area that is tens-of-thousands times larger than the mammalian counterpart.
Table 7.
Site(s) of ZP protein synthesis.
Oocytes and/or granulosa cells.
Rainbow trout - liver; zebrafish – ovary; medaka - liver and oocytes.
Xenopus, mice, humans - oocytes.
Chicken - liver and ovary.
All vertebrate ZP proteins are synthesized as precursor polypeptides that have a SS and a CTP downstream of the ZPD, and it has been suggested that sequences within the CTP are required for secretion and assembly of ZP proteins [48, 53]. The CTP includes a CFCS, a hydrophobic peptide, called an external hydrophobic patch (EHP), a TMD, followed by a short hydrophilic C-terminus. Another hydrophobic peptide, called an internal hydrophobic patch (IHP), is present within the ZPD (ZP-N or ZP-C sub-domain). The IHP and EHP are well conserved units within vertebrate ZP proteins as revealed in sequence alignments of ZP1-4 orthologs from trout, frog, chicken, possum, mouse, and human beings (with the possible exception of trout ZP1α) (Table 8). Based on experiments with mouse ZP3 it has been suggested that IHP-EHP elements regulate assembly of ZP proteins [54]. In ZP protein precursors the IHP interacts with the EHP thereby preventing premature polymerization and assembly of ZP fibrils and matrix within oocytes. When the CTP is removed by proteolysis at the CFCS during secretion of ZP precursor proteins, the EHP dissociates from the IHP, “activated” ZP protein is released from the oocyte, and ZP assembly (i.e., formation of fibrils and matrix) can take place [36].
Table 8.
Conservation of IHP and EHPa.
| IHP | EHP | ||
|---|---|---|---|
| ZP1/4 | |||
| Trout | zp1α | 370DAVLHVE376 | 551SQKVIMI557 |
| zp1β | 330PGPLIVE336 | 511SGELILT517 | |
| Frog | zp1 | – | – |
| zp4 | 308PGPLMLE314 | 480DGPVDFI486 | |
| Chicken | zp1 | 746PGPLQLQ752 | 920RGRIVLP926 |
| zp4 | 321PGPLSLE327 | 491KGPVIFL497 | |
| Possum | zp1 | – | – |
| zp4 | 309PGPLALE315 | 479QGPIFFL485 | |
| Mouse | zp1 | 394SGPLRLE400 | 566PGAVGFE572 |
| zp4 | – | – | |
| Human | zp1 | 402PGPLRLE408 | 573PGPVGFE579 |
| zp4 | 312PGPLTLE318 | 482KGPMILL488 | |
| ZP2 | |||
| Gilthead seabreamb | 521RGELQIT527 | 691SGPILVN698 | |
| Frog | 468DGPLTLV474 | 634SGPILIV640 | |
| Chicken | 467QGPLSLI473 | 633QGPVLLV639 | |
| Possum | 486PGPLSLV492 | 651PGPVFLV657 | |
| Mouse | 483PGPLVLV489 | 648PGPILLL654 | |
| Human | 490LGPFTLI496 | 657PGPILLL663 | |
| ZP3 | |||
| Trout | 231YFSMRLMT238 | 424WEGDVQLGPIFIS436 | |
| Frog | 198AFSLRLMT205 | 390EHSLATIGPILVV402 | |
| Chicken | 192VFSLRLMS199 | 377VAADVVIGPVLLS389 | |
| Possum | 175KFSLRLMA182 | 354FEADLMLGPLVLS366 | |
| Mouse | 170AFSLRLME177 | 357DEADVTVGPLIFL369 | |
| Human | 171TFSLRLME178 | 356EEADVTVGPLIFL368 |
In fish and birds some ZP precursor proteins are synthesized in the liver and are transported via circulating blood to the oocyte for uptake (similar to the synthesis of egg-yolk precursor, vitellogenin, in the liver of amphibians and birds [55]) [56]. It is not known how these proteins are transported to their site of assembly or how they are assembled into an egg-coat. Synthesis of individual ZP proteins must be coordinated in a timely manner to ensure that they are present simultaneously so that they can assemble first into fibrils and then into matrix. It is possible that ZP precursors are transported in the bloodstream in small vesicles, perhaps bound to specific receptors, that dock on to the oocyte’s plasma membrane for uptake into the oocytes. The precursor proteins would then follow the biosynthetic pathway described above.
Final comments
During the past 30 years or so, identification and characterization of vertebrate egg-coat genes and proteins, from organisms as different as fish and human beings, has come into its own. ZP proteins apparently are derived from a common ancestral gene that gave rise to ZP3 and 4 other ZP gene families (ZPd, ZPax, ZP2, and ZP1/ZP4). One of the gene families (ZPax) is found in fish, amphibians, and birds, and another (ZPd) in amphibians and birds. ZPax is found in a few mammals, but is always present as a pseudo-gene. ZP1-4 are found in mammals and other vertebrates. All of the corresponding ZP proteins possess a ZPD that participates in polymerization of processed ZP/VE precursor proteins into higher-order structures, such as fibrils and matrix (egg-coats). The presence of a ZPD in proteins from jellyfish (cnidarians), a member of the oldest of the true metazoan phyla, suggests that it arose more than ~600 million years ago. Furthermore, a ZPD is present in a host of organisms, including echinoderms, worms, and flies, in hundreds of different proteins [2, 36], including TGF-β receptor type III (betaglycan), hensin, vomeroglandin, tectorin-α and -β, uromodulin, mesoglein, DMBT-1, cuticlins, oikosins, DYF-7 and RAM-5, and two-to-three dozen proteins in Drosophila [57]. It is very likely that as more and more organisms are subjected to whole-genome sequencing the number and variety of ZPD proteins will continue to increase and evolutionary relationships between the relevant genes/proteins will be clarified further.
Acknowledgments
We are grateful to the Bodleian Library, Oxford University, for providing a stimulating and comfortable environment in which to write this review. Our research was supported in part by the NIH (NICHD).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no disclosures.
References
- 1.Koonin EV, Senkevich TG, Dolja VV. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litscher ES, Wassarman PM. A Guide to Zona Pellucida Domain Proteins. Wiley and Sons; Hoboken: 2015. [Google Scholar]
- 3.Wassarman PM. J Biol Chem. 2008;283:24285. doi: 10.1074/jbc.R800027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spargo SC, Hope RM. Biol Reprod. 2003;68:358. doi: 10.1095/biolreprod.102.008086. [DOI] [PubMed] [Google Scholar]
- 5.Okumura H, Kohno Y, Iwata Y, Mori H, Aoki N, Sato C, Kitajima K, Nadano D, Matsuda T. Biochem J. 2004;384:191. doi: 10.1042/BJ20040299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith J, Paton IR, Hughes DC, Burt DW. Mol Reprod Dev. 2005;70:133. doi: 10.1002/mrd.20197. [DOI] [PubMed] [Google Scholar]
- 7.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grützner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang SP, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, López-Otín C, Ordóñez GR, Eichler EE, Chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AF, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ES, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefèvre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux JL, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Nefedov M, de Jong PJ, Renfree MB, Mardis ER, Wilson RK. Nature. 2008;453:175. [Google Scholar]
- 8.Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, Ray DA, Boissinot S, Shedlock AM, Botka C, Castoe TA, Colbourne JK, Fujita MK, Moreno RG, Ten Hallers BF, Haussler D, Heger A, Heiman D, Janes DE, Johnson J, de Jong PJ, Koriabine MY, Lara M, Novick PA, Organ CL, Peach SE, Poe S, Pollock DD, de Queiroz K, Sanger T, Searle S, Smith JD, Smith Z, Swofford R, Turner-Maier J, Wade J, Young S, Zadissa A, Edwards SV, Glenn TC, Schneider CJ, Losos JB, Lander ES, Breen M, Ponting CP, Lindblad-Toh K. Nature. 2011;477:587. [Google Scholar]
- 9.Wang Z, Pascual-Anaya J, Zadissa A, Li W, Niimura Y, Huang Z, Li C, White S, Xiong Z, Fang D, Wang B, Ming Y, Chen Y, Zheng Y, Kuraku S, Pignatelli M, Herrero J, Beal K, Nozawa M, Li Q, Wang J, Zhang H, Yu L, Shigenobu S, Wang J, Liu J, Flicek P, Searle S, Wang J, Kuratani S, Yin Y, Aken B, Zhang G, Irie N. Nature Genet. 2013;45:701. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, Wüster W, Woods AE, Logan JM, Harrison RA, Castoe TA, de Koning AP, Pollock DD, Yandell M, Calderon D, Renjifo C, Currier RB, Salgado D, Pla D, Sanz L, Hyder AS, Ribeiro JM, Arntzen JW, van den Thillart GE, Boetzer M, Pirovano W, Dirks RP, Spaink HP, Duboule D, McGlinn E, Kini RM, Richardson MK. Proc Natl Acad Sci, USA. 2013;110:20651. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, Hoon S, Gangu V, Roy SW, Irimia M, Korzh V, Kondrychyn I, Lim ZW, Tay BH, Tohari S, Kong KW, Ho S, Lorente-Galdos B, Quilez J, Marques-Bonet T, Raney BJ, Ingham PW, Tay A, Hillier LW, Minx P, Boehm T, Wilson RK, Brenner S, Warren WC. Nature. 2014;505:174. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SM, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson YW, Nanlohy KG, Libants SV, Yeh CY, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, de Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W. Nature Genet. 2013;45:415. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutiérrez EL, Dubchak I, Garcia-Fernàndez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. Nature. 2008;453:1064. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 14.Benton MJ, Ayala FJ. Science. 2003;300:1698. doi: 10.1126/science.1077795. [DOI] [PubMed] [Google Scholar]
- 15.Litscher ES, Wassarman PM. Histol Histopathol. 2007;22:337. doi: 10.14670/HH-22.337. [DOI] [PubMed] [Google Scholar]
- 16.Hartman CG. Quart Rev Biol. 1929;4:373. [Google Scholar]
- 17.Hamilton WJ, Laing JA. J Anat. 1946;80:194. [PubMed] [Google Scholar]
- 18.Selwood L. Cell Tissue Organs. 2000;166:208. doi: 10.1159/000016733. [DOI] [PubMed] [Google Scholar]
- 19.Gruetzner F, Nixon B, Jones RC. Sex Dev. 2008;2:115. doi: 10.1159/000143429. [DOI] [PubMed] [Google Scholar]
- 20.Dunbar BS, Prasad SV, Timmons TM. In: A ComparativeOverview of Mammalian Fertilization. Dunbar BS, O’Rand MG, editors. Plenum; NY: 1991. p. 97. [Google Scholar]
- 21.Selwood L, Robinson ES, Pedersen RA, Vandeberg JI. Intl J Dev Biol. 1997;41:397. [PubMed] [Google Scholar]
- 22.Wassarman PM, Qi H, Litscher ES. Proc Roy Soc London, Biol Sci. 1997;264:323. doi: 10.1098/rspb.1997.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mate KE, McCartney CA. Mol Reprod Dev. 1998;51:322. doi: 10.1002/(SICI)1098-2795(199811)51:3<322::AID-MRD12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Bellairs R, Harkness M, Harkness RD. J Ultrastr Res. 1963;8:339. [Google Scholar]
- 25.Hedrick JL. Intl J Dev Biol. 2008;52:683. doi: 10.1387/ijdb.082580jh. [DOI] [PubMed] [Google Scholar]
- 26.Brivio MF, Bassi R, Cotelli F. Mol Reprod Dev. 1991;28:85. doi: 10.1002/mrd.1080280114. [DOI] [PubMed] [Google Scholar]
- 27.Darie CC, Biniossek ML, Jovine L, Litscher ES, Wassarman PM. Biochemistry. 2004;43:7459. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- 28.Darie CC, Janssen WG, Litscher ES, Wassarman PM. Biochim Biophys Acta. 2008;1784:385. doi: 10.1016/j.bbapap.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Berois N, Arezo MJ, Papa NG. Biol Res. 2011;44:119. [PubMed] [Google Scholar]
- 30.Xu Q, Li G, Cao L, Wang Z, Ye H, Chen X, Yang X, Wang Y, Chen L. BMC Evol Biol. 2012;12:239. doi: 10.1186/1471-2148-12-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claw KG, Swanson WJ. Annu Rev Genomics Hum Genet. 2012;13:109. doi: 10.1146/annurev-genom-090711-163745. [DOI] [PubMed] [Google Scholar]
- 32.Goudet G, Mugnier S, Callebaut I, Monget P. Biol Reprod. 2008;78:796. doi: 10.1095/biolreprod.107.064568. [DOI] [PubMed] [Google Scholar]
- 33.Chothia C, Gough J, Vogel C, Teichmann SA. Science. 2003;300:1701. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 34.Bork P, Sander C. FEBS Letts. 1992;300:237. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 35.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Curr Opin Struct Biol. 2004;14:208. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Jovine L, Darie CC, Litscher ES, Wassarman PM. Annu Rev Biochem. 2005;74:83. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- 37.Han L, Monné M, Okumura H, Schwend T, Cherry AL, Flot D, Matsuda T, Jovine L. Cell. 2010;143:404. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 38.Jovine L, Qi H, Williams Z, Litscher E, Wassarman PM. Nature Cell Biol. 2002;4:457. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- 39.Litscher ES, Janssen WG, Darie CC, Wassarman PM. J Cell Physiol. 2008;214:153. doi: 10.1002/jcp.21174. [DOI] [PubMed] [Google Scholar]
- 40.Yan C, Pendola FL, Jacob R, Lau AL, Eppig JJ, Matzuk MM. Genesis. 2001;31:105. doi: 10.1002/gene.10010. [DOI] [PubMed] [Google Scholar]
- 41.Cocchia M, Huber R, Pantano S, Chen EY, Ma P, Forabosco A, Ko MS, Schlessiger D. Genomics. 2000;68:305. doi: 10.1006/geno.2000.6302. [DOI] [PubMed] [Google Scholar]
- 42.Bökel C, Prokop A, Brown NH. J Cell Sci. 2005;118:633. doi: 10.1242/jcs.01619. [DOI] [PubMed] [Google Scholar]
- 43.Jovine L, Janssen WG, Litscher ES, Wassarman PM. BMC Biochem. 2006;7:11. doi: 10.1186/1471-2091-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callebaut I, Mornon JP, Monget P. Bioinformatics. 2007;23:1871. doi: 10.1093/bioinformatics/btm265. [DOI] [PubMed] [Google Scholar]
- 45.Swanson WJ, Yang Z, Wolfner MF, Aquadro CF. Proc Natl Acad Sci, USA. 2001;98:2509. doi: 10.1073/pnas.051605998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calkins JD, El-Hinn D, Swanson WJ. J Mol Evol. 2007;65:555. doi: 10.1007/s00239-007-9034-8. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi S, Hedrick JL, Katagiri C. Dev Growth Differ. 1989;31:85. doi: 10.1111/j.1440-169X.1989.00085.x. [DOI] [PubMed] [Google Scholar]
- 48.Wassarman PM, Litscher ES. Curr Top Dev Biol. 2013;102:243. doi: 10.1016/B978-0-12-416024-8.00009-X. [DOI] [PubMed] [Google Scholar]
- 49.Tesoriero JV. J Ultrastr Res. 1978;64:315. doi: 10.1016/s0022-5320(78)90040-0. [DOI] [PubMed] [Google Scholar]
- 50.Bausek N, Waclawek M, Schneider WJ, Wohlrab F. J Biol Chem. 2000;275:28866. doi: 10.1074/jbc.275.37.28866. [DOI] [PubMed] [Google Scholar]
- 51.Arukwe A, Goksøyr A. Comp Hepatol. 2003;2:4. doi: 10.1186/1476-5926-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sano K, Kawaguchi M, Watanabe S, Nagakura Y, Hiraki T, Yasumasu S. J Exp Zool (Mol Dev Evol) 2013;320B:332. doi: 10.1002/jez.b.22507. [DOI] [PubMed] [Google Scholar]
- 53.Qi H, Williams Z, Wassarman PM. Mol Biol Cell. 2002;13:530. doi: 10.1091/mbc.01-09-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jovine L, Qi H, Williams Z, Litscher ES, Wassarman PM. Proc Natl Acad Sci, USA. 2004;101:5922. doi: 10.1073/pnas.0401600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace RA. Developmental Biology, A Comprehensive Synthesis. In: Browder LW, editor. Oogenesis. Plenum; NY: 1985. p. 127. [Google Scholar]
- 56.Darie CC, Biniossek ML, Gawinowicz MA, Milgrom Y, Thumfart JO, Jovine L, Litscher ES, Wassarman PM. J Biol Chem. 2005;280:37585. doi: 10.1074/jbc.M506709200. [DOI] [PubMed] [Google Scholar]
- 57.Plaza S, Chanut-Delalande H, Fernandes I, Wassarman PM, Payre F. Trends Cell Biol. 2010;20:524. doi: 10.1016/j.tcb.2010.06.002. [DOI] [PubMed] [Google Scholar]


