Abstract
This study describes group A rotavirus (RVA) genotype prevalence in Belarus from 2008–2012. In 2008, data from 3 sites in Belarus (Brest, Mogilev, Minsk) indicated that G4P[8] was the predominant genotype. Data from Minsk (2008–2012) showed that G4P[8] was the predominant RVA genotype in all years except in 2011 when G3P[8] was most frequently detected. Other RVA genotypes common in Europe (G1P[8], G2P[4]) were detected each year of the study. This study reveals the dominance of genotype G4P[8] in Belarus and helps to establish the baseline genotype prevalence prior to RVA vaccine introduction in the country.
Keywords: rotavirus, genotype, VP4, VP7, Belarus
1. Introduction
Group A rotavirus (RVA) is a well-known etiological agent of acute gastroenteritis that mostly affects small children. The virus is widespread and plays a dominant role among causative agents of acute gastroenteritis both in developed and developing countries. It is believed that every child has an RVA infection at least once before 5 years of age. Moreover, RVA often causes a severe form of gastroenteritis (RVGE) with severe dehydration that often requires oral or intravenous rehydration (Sack et al., 1978). Inadequate therapy can lead to a lethal outcome which is a serious problem for developing countries and is estimated to result in 453,000 deaths annually prior to the introduction of vaccination (Tate et al., 2012).
RVA is a nonenveloped virus that belongs to the Reoviridae family. The virus genome contains 11 segments of double-stranded RNA that code 6 structural and 6 nonstructural proteins (Estes and Kapikian, 2007). RVA have two independently segregating serotype antigens, outer capsid proteins VP4 (P type) and VP7 (G type) coded by segments 4 and 9, respectively (Estes and Kapikian, 2007). There are at least 27 different G genotypes and 35 P genotypes known today (Matthijnssens et al., 2011). Among at least 12 G types and 15 P types identified in humans, more than 70 G-P antigen combinations have been detected (Matthijnssens et al., 2009).
Although RVA genotypes vary from year to year in particular regions, 5 common G/P combinations have been identified in Europe: G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] (Iturriza-Gomara et al., 2011; Iturriza-Gomara et al., 2009; Usonis et al., 2012). In the last 2 decades genotype G1P[8] has been predominant, being responsible for about 70% of RVA gastroenteritis in Europe (Santos and Hoshino, 2005). Recently, G9P[8] emerged in different parts of the world and now represents a globally common strain that is apparently becoming more widespread over time and it has been suggested that genotype G12 is also emerging to potentially become another globally important strain (Matthijnssens et al., 2009; Matthijnssens et al., 2010).
Development of new RVA vaccines and their recent licensing in many countries has resulted in the inclusion of RVGE as another vaccine preventable disease. Consequently, surveillance studies are needed in countries considering the use of RVA vaccines to monitor strain prevalence. In the Belarus laboratory, diagnosis of RVA infection by using enzyme immunoassays (commercial kits as well those produced in the Republican Research and Practical Center for Epidemiology and Microbiology) has been conducted for about 15 years but information concerning genotypic diversity of circulating viruses in Belarus is still very limited with only 4 reports published in Russian (Gudkov et al., 2011; Gudkov et al., 2008; Gudkov et al., 2010; Samoilovich et al., 2013). The purpose of this study was to determine circulating RVA genotypes in Belarus during 2008–12. This information will help establish the impact of future RVA vaccination programs in the country.
2. Materials and methods
All specimens were convenience samples from children admitted to infectious disease hospitals with a diagnosis of acute RVGE. The stool samples were tested first using a solid phase enzyme immunoassay (EIA for rotavirus antigen revealing, Republican Research and Practical Center for Epidemiology and Microbiology, Belarus). Specimens for genotyping were selected randomly from EIA-positive samples and total of 633 stool specimens were characterized. In 2008, specimens were collected in three regions of Belarus: 115 specimens from the capital Minsk-City (central part of the country), 16 specimens from Brest region (western part of the country), and 13 specimens from the Mogilev region (eastern part of the country). From 2009–2012, all specimens were from Minsk with 297 samples analyzed in 2009, 59 in 2010, 68 in 2011, and 65 in 2012.
Viral RNA was extracted from 10% stool suspensions prepared in PBS using a KingFisher Extraction system (Thermo Electron Corp, Finland). G (VP7) and P (VP4) typing were carried out by reverse transcription-polymerase chain reaction (RT-PCR) genotyping as described previously (Hull et al., 2011). Genotype determination of non-typeable samples was performed by nucleotide sequencing as described previously (Hull et al., 2011).
3. Results
Of the 633 specimens, G types could be assigned to 628 samples and 5 were G non-typeable (NT). P types were determined for 629 specimens. The predominant G types detected were G1, G2, G3, and G4. Among them G4 type was predominant (336 strains, 53.5%) followed by G2 (105 strains, 16.7%), G3 (101 strains, 16.1%) and G1 (72 strains, 11.5%). Genotypes G9 (5 strains, 0.8%), and G8 (2 strains, 0.3%) were less frequently detected. Among the VP4 specificities, genotype P[8] was most frequent and represented by 80.9% (509 strains) and P[4] was detected in 106 samples (16.9%). Genotypes P[6] and P[9] were present at rates of 1.3 and 0.8%, respectively. The most common G-P combination among the 624 G and P-typed samples was G4P[8] accounting for 52.4% (327) of the pediatric gastroenteritis cases followed by G2P[4] (103 cases, 16.5%) and G3P[8] (96, 15.4%). Genotype G1P[8] was detected in 11.7% of samples (72 cases). Other combinations (G2P[6], G4P[6], G3P[9], G8P[4], G9P[8]) were rare and found only in 1–5 cases each during the study period.
3.1. RVA genotype distribution in different regions of Belarus
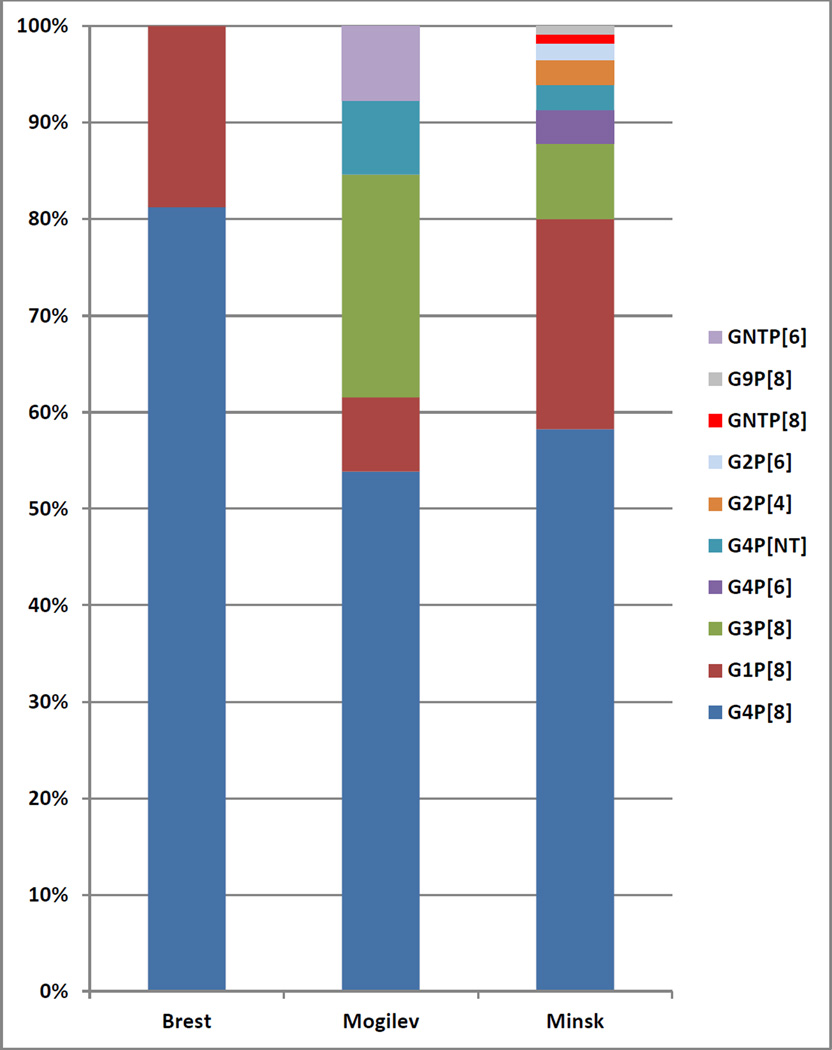
In 2008, specimens were collected in 3 regions of Belarus, permitting a comparison of genotype prevalence in different parts of the country (Table 1, Figure 1). Genotype G4P[8] was predominant in each of three locations studied: Brest region – 81.3% (13 cases), Mogilev region – 53.8% (7 cases), Minsk - 58.3% 9 (67 cases). In the Brest region, the other genotype detected was G1P[8] (3 of 18 cases; Figure 1). In the Mogilev region, aside from the aforementioned G4P[8], 2 other genotypes were detected, G3P[8] and G1P[8], along with 2 partially-typed strains (Figure 1). In Minsk, 7 genotypes were detected (Figure 1). Along with G4P[8] strains, G1P[8] and G3P[8] were present at notable levels (21.7% and 7.8%, respectively; Figure 1). Genotypes G2P[4], G2P[6], G4P[6], and G9P[8] were also detected at low levels along with some G or P non-typeable strains (Figure 1). Although the data obtained demonstrate some differences in genotype distribution in 3 regions, the small number of samples tested makes it difficult to ascribe significance to the noted variation in genotypes.
Table. 1.
Distribution of RVA G and P types in Belarus, 2008
| P[4] | P[6] | P[8] | NT | Total No. | |
|---|---|---|---|---|---|
| G1 | 29 | 29 | |||
| G2 | 3 | 2 | 5 | ||
| G3 | 12 | 12 | |||
| G4 | 4 | 87 | 4 | 95 | |
| G9 | 1 | 1 | |||
| NT | 1 | 1 | 2 | ||
| Total No. | 3 | 7 | 130 | 4 | 144 |
Figure 1.
Percentage prevalence of RVA genotypes at 3 sites in Belarus in 2008. The number of cases per site is: 13 (Brest), 7 (Mogilev), and 67 (Minsk).
3.2. Temporal variation in RVA genotype distribution-Minsk
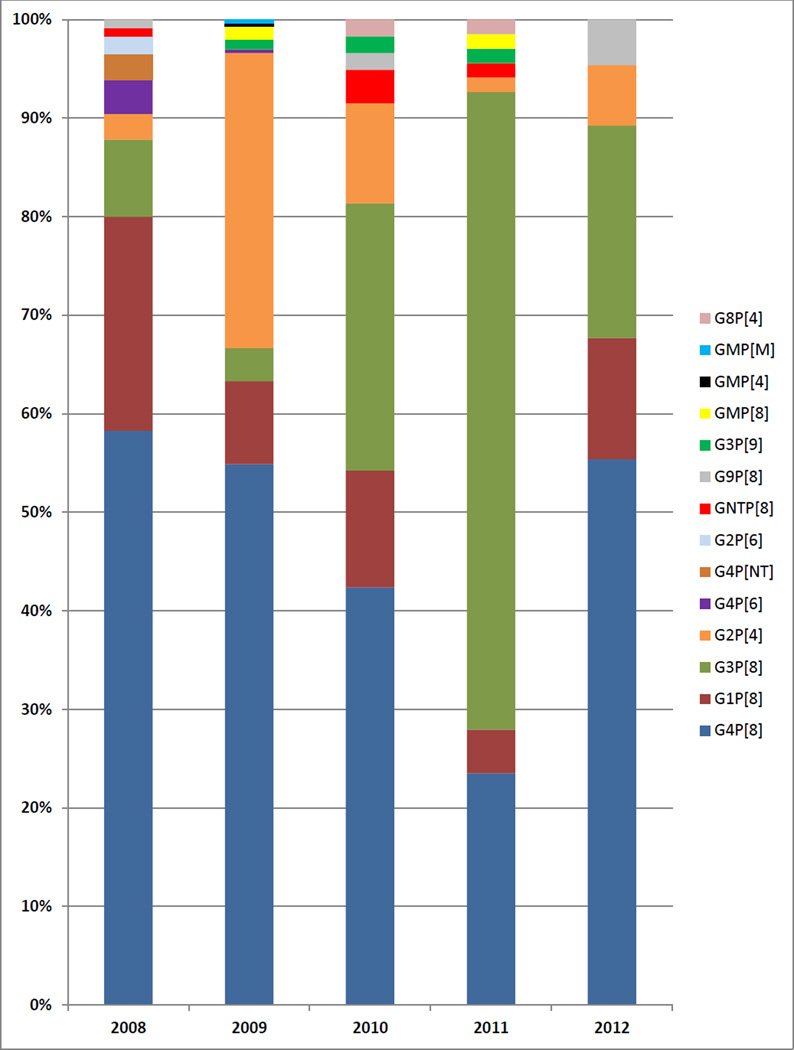
The 2008 data for Minsk are described in the previous section. In 2009, 297 samples from Minsk were genotyped and 59–68 samples were genotyped in years 2010, 2011, and 2012 (Tables 2–5, Figure 2). G4P[8] was the predominant genotype in 2008, 2009, 2010, and 2012 with 42.4–58.3% prevalence during these years and it was the second most common genotype in 2011 (23.5%; Figure 2). In 2011, G3P[8] was the predominant genotype (64.7%) and it was the second most frequently detected genotype in 2010 (27.1%) and 2012 (21.5%; Figure 2). G2P[4] reached 30.0% prevalence in 2009 and 10.2% in 2010 but was detected less frequently in other years (Figure 2). The highest detection frequency for G1P[8] was 21.7% in 2008 and its prevalence ranged from 4.4–12.3% in 2009–12 (Figure 2). Other genotypes (G8P[4], G9P[8], G3P[9]), as well as G or P non-typeable strains, were detected at low frequencies in multiple years (Tables 2–5, Figure 2). Rare P[6] RVA strains were detected in 2008 (G2P[6], 2 cases; G4P[6], 4 cases) and 2009 (G4P[6], 1 case) but not in other years (Table 1, Table 2, Figure 2). Six mixed genotype infections were detected in 2009 and 1was detected in 2011 (Table 2, Table 4, Figure 2).
Table. 2.
Distribution of rotavirus G and P types in Minsk, 2009
| P[4] | P[6] | P[8] | P[9] | Mixed | Total No. | |
|---|---|---|---|---|---|---|
| G1 | 25 | 25 | ||||
| G2 | 89 | 89 | ||||
| G3 | 10 | 3 | 13 | |||
| G4 | 1 | 163 | 164 | |||
| Mixed* | 1 | 4 | 1 | 6 | ||
| Total No. | 90 | 1 | 202 | 3 | 1 |
includes 1 G2,4P[4,8], 1 G1,4P[8], 1 G2,4P[4], and 3 G1,3P[8]
Table. 5.
Distribution of rotavirus G and P types in Minsk, 2012
| P[4] | P[8] | Total No. | |
|---|---|---|---|
| G1 | 8 | 8 | |
| G2 | 4 | 4 | |
| G3 | 14 | 14 | |
| G4 | 36 | 36 | |
| G9 | 3 | 3 | |
| Total No. | 4 | 61 | 65 |
Figure 2.
Percentage prevalence of RVA genotypes in Minsk, 2008–2012. The number of cases per year is: 67 (2008), 297 (2009), 59 (2010), 68 (2011), and 65 (2012).
Table. 4.
Distribution of rotavirus G and P types in Minsk, 2011
| P[4] | P[8] | P[9] | Total No. | |
|---|---|---|---|---|
| G1 | 3 | 3 | ||
| G2 | 1 | 1 | ||
| G3 | 44 | 1 | 45 | |
| G4 | 16 | 16 | ||
| G8 | 1 | 1 | ||
| Mixed* | 1 | 1 | ||
| NT | 1 | 1 | ||
| Total No. | 2 | 65 | 1 | 68 |
1 G3,4P[8]
4. Discussion
Worldwide RVA surveillance has permitted establishment of the most common genotypes for different geographic areas (Banyai et al., 2012). At the same time, current studies that are being carried out in many countries that have not previously been surveyed, provide new data and constantly update our knowledge of the prevalence and epidemiological significance of individual RVA genotypes.
Investigation of clinical samples from patients with acute RVGE in Belarus from 2008–2012 revealed high genetic diversity in circulating RVA strains. It was confirmed that RVA of all the most predominant genotypes from Europe circulate in Belarus, namely G1P[8], G2P[4], G3P[8], G4P[8], G9P[8]. These genotypes were responsible for 94.8% (600/633) of RVA cases countrywide. In addition, some rare genotypes (i.e., G4P[6], G8P[4], G3P[9]) were detected in multiple study years. Genotype G4P[8] was the predominant etiological agent of RVGE in Belarus during the time of investigation, which was responsible for about 52% of cases in the country and confirms finding of previous studies in Belarus (Gudkov et al., 2011; Gudkov et al., 2008; Gudkov et al., 2010; Samoilovich et al., 2013). This is in contrast to results of contemporary studies conducted in other European countries which demonstrated that genotype G1P[8] was the predominant strain in the region in most years (Laszlo et al., 2012; Ruggeri et al., 2011; Vesikari et al., 2013). However, molecular epidemiological data indicate that in the last three decades genotype G4P[8] played an important role in the European region and other parts of the world as a causative agent of RVGE, being responsible for up to 60% of annual cases in various countries (Arista et al., 1997; Bon et al., 2000; Buesa et al., 2000; Iturriza-Gomara et al., 2011; Iturriza-Gomara et al., 2009; Koopmans and Brown, 1999; Mirzayeva et al., 2009; Ogilvie et al., 2011; Usonis et al., 2012). In certain periods, G4P[8] strains were identified as the most prevalent genotype in Spain in 1996 (Buesa et al., 2000), in Italy at the beginning of 1990s (Arista et al., 1997), in Albania, Bulgaria, and Russia in 2005–06 and Hungary in 2007–11 (Laszlo et al., 2012; Ogilvie et al., 2011). In this study, G4P[8] was identified as a predominant genotype in all years except 2011 and in all 3 regions investigated in 2008, thus confirming the high epidemiological significance of this genotype for Belarus.
Genotype G2P[4] was the second most frequently identified genotype in Belarus; it was detected in 16.5% of specimens in the study and comprised 30% of genotyped samples in 2009. It is interesting to note that the peak prevalence of this genotype in 2009 was preceded by very low level activity in 2008 (2.0% prevalence, with all cases from Minsk) and followed by a decrease in prevalence to 10.2% in 2010. Large annual fluctuations in G2P[4] prevalence have been noted in other countries (Dulgheroff et al., 2012; Esteban et al., 2010; Giammanco et al., 2014; Mladenova et al., 2010). A similar trend albeit with a broader temporal peak was observed for the 3rd most frequently identified genotype, G3P[8], which exhibited a peak level of detection in 2011 (65% prevalence) and detection frequencies of 27% and 22% in 2010 and 2012, respectively. Before 2010, the genotype G3P[8] prevalence was less than 10%. The peak of genotype G1P[8] detection was 20% in 2008 and the prevalence of this genotype did not exceed 12% in the remaining years of the study. It is worth mentioning that genotype G9P[8], which has been reported as a common strain in other European countries (Iturriza-Gomara et al., 2011; Iturriza-Gomara et al., 2009) was found rarely with only 5 detections in Belarus during the 5-year course of this study.
There were two uncommon genotypes in Belarus and both of them were characterized by presence of P[6] type VP4 protein in combination with G4 and G2 types. In general, P[6] type is not frequently detected in Europe (Iturriza-Gomara et al., 2011; Iturriza-Gomara et al., 2009; Papp et al., 2013; Usonis et al., 2012). The P[6] genotype, paired with a variety of G types (G1, G2, G4, G8, G9), has been detected in RVGE specimens primarily from Africa and India (Gentsch et al., 2005; Santos and Hoshino, 2005). The very low level frequency of detection of G2/4P[6] strains in this study is consistent with other RVA prevalence reports from Europe.
A limitation of the study was that the samples were collected and tested during all the years of the study for only one site (Minsk City) and that data for multiple regions were available for 2008 only. This is due to fact that the stool samples collected were convenience samples collected through passive surveillance. Thus we could not collect equivalent numbers of samples from all regions of Belarus to better define the predominant RVA each year. If future funding permits, we would perform active RVA surveillance with recruitment of cases, paid staff for sample collection and shipment to increase the number of samples and annual study sites. Ideally this type of surveillance can be introduced before RVA vaccine introduction and maintained during the vaccine use era.
5. Conclusion
This study represents the first detailed multi-year report of RVA genotype prevalence in Belarus. The results of this study will help assess future vaccine effectiveness studies in Belarus by potentially identifying changes in genotype distribution post-vaccine introduction and the emergence of strains that have escaped immunity elicited by RVA vaccines.
Table. 3.
Distribution of rotavirus G and P types in Minsk, 2010
| P[4] | P[8] | P[9] | Total No. | |
|---|---|---|---|---|
| G1 | 7 | 7 | ||
| G2 | 6 | 6 | ||
| G3 | 16 | 1 | 17 | |
| G4 | 25 | 25 | ||
| G8 | 1 | 1 | ||
| G9 | 1 | 1 | ||
| NT | 2 | 2 | ||
| Total No. | 7 | 51 | 1 | 59 |
Acknowledgment
We wish to thank Rashi Gautam and Sunando Roy for their critical review of this manuscript. Funding for this study was provided by the World Health Organization.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
Footnotes
Conflict of interest statement
The authors of this study declare that they have no conflict of interest, financial or otherwise, related to this article.
References
- Arista S, Vizzi E, Ferraro D, Cascio A, Di Stefano R. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Archives of virology. 1997;142:2065–2071. doi: 10.1007/s007050050224. [DOI] [PubMed] [Google Scholar]
- Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(Suppl 1):A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- Bon F, Fromantin C, Aho S, Pothier P, Kohli E. G and P genotyping of rotavirus strains circulating in france over a three-year period: detection of G9 and P[6] strains at low frequencies. The AZAY Group. Journal of clinical microbiology. 2000;38:1681–1683. doi: 10.1128/jcm.38.4.1681-1683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesa J, de Souza CO, Asensi M, Martinez C, Prat J, Gil MT. VP7 and VP4 genotypes among rotavirus strains recovered from children with gastroenteritis over a 3-year period in Valencia, Spain. European journal of epidemiology. 2000;16:501–506. doi: 10.1023/a:1007618215377. [DOI] [PubMed] [Google Scholar]
- Dulgheroff AC, Figueiredo EF, Moreira LP, Moreira KC, Moura LM, Gouvea VS, Domingues AL. Distribution of rotavirus genotypes after vaccine introduction in the Triangulo Mineiro region of Brazil: 4-Year follow-up study. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2012;55:67–71. doi: 10.1016/j.jcv.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Esteban LE, Rota RP, Gentsch JR, Jiang B, Esona M, Glass RI, Glikmann G, Castello AA. Molecular epidemiology of group A rotavirus in Buenos Aires, Argentina 2004–2007: reemergence of G2P[4] and emergence of G9P[8] strains. Journal of medical virology. 2010;82:1083–1093. doi: 10.1002/jmv.21745. [DOI] [PubMed] [Google Scholar]
- Estes MK, Kapikian A. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5th ed. Philadelphia, PA: Kluwer/Lippincott, Williams and Wilkins; 2007. pp. 1917–1974. [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. The Journal of infectious diseases. 2005;192(Suppl 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Giammanco GM, Bonura F, Zeller M, Heylen E, Van Ranst M, Martella V, Banyai K, Matthijnssens J, De Grazia S. Evolution of DS-1-like human G2P[4] rotaviruses assessed by complete genome analyses. The Journal of general virology. 2014;95:91–109. doi: 10.1099/vir.0.056788-0. [DOI] [PubMed] [Google Scholar]
- Gudkov VG, Tchistenko GN, Fisenko EG, Klyuchareva AA, Virinskaya AS, Biskina NM, Plotnikova KY, Malyavko DV, Zapolskaya VV, N NU. Epidemiological survey of rotavirus infection in the Republic of Belarus. Zdravoohranenie. 2011;11:20–26. [Google Scholar]
- Gudkov VG, Virinskaya AS, Malyavko DV, Klyuchareva AA, Sebut NV, Biskina NM, Plotnikova KU, Novatskaya UN, Pashkovich VV, Golotik DM, Grinvich OV, Klyuiko NL, Budnik YA, Zueva VL. Rotavirus infection in Belarus. Zdravoohranenie. 2008;11:8–13. [Google Scholar]
- Gudkov VG, Virinskaya AS, Plotnikova KU, Biskina NM, Novatskaya UN, Malyavko DV, Klyuchareva AA, Karpuk GV, NL K. Rotavirus infection in the republic of Belarus: epidemic process characteristics, disease burden evaluation and pathogenic agent population G-P structure. Zdravoohranenie. 2010;11:28–33. [Google Scholar]
- Hull JJ, Teel EN, Kerin TK, Freeman MM, Esona MD, Gentsch JR, Cortese MM, Parashar UD, Glass RI, Bowen MD National Rotavirus Strain Surveillance, S. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. The Pediatric infectious disease journal. 2011;30:S42–S47. doi: 10.1097/INF.0b013e3181fefd78. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Dallman T, Banyai K, Bottiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Koopmans M, Korsun N, Koukou D, Kroneman A, Laszlo B, Lappalainen M, Maunula L, Marques AM, Matthijnssens J, Midgley S, Mladenova Z, Nawaz S, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Steyer A, Sidaraviciute-Ivaskeviciene I, Syriopoulou V, Tran AN, Usonis V, Van Ranst M, De Rougemont A, Gray J. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiology and infection. 2011;139:895–909. doi: 10.1017/S0950268810001810. [DOI] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Dallman T, Banyai K, Bottiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Korsun N, Kroneman A, Lappalainen M, Laszlo B, Maunula L, Matthinjnssens J, Midgley S, Mladenova Z, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Schreier E, Steyer A, Sidaraviciute I, Tran AN, Usonis V, Van Ranst M, de Rougemont A, Gray J. Rotavirus surveillance in europe, 2005–2008: web-enabled reporting and real-time analysis of genotyping and epidemiological data. The Journal of infectious diseases. 2009;200(Suppl 1):S215–S221. doi: 10.1086/605049. [DOI] [PubMed] [Google Scholar]
- Koopmans M, Brown D. Acta paediatrica. Supplement 88. Oslo, Norway: 1999. 1992. Seasonality and diversity of Group A rotaviruses in Europe; pp. 14–19. [DOI] [PubMed] [Google Scholar]
- Laszlo B, Konya J, Dandar E, Deak J, Farkas A, Gray J, Grosz G, Iturriza-Gomara M, Jakab F, Juhasz A, Kisfali P, Kovacs J, Lengyel G, Martella V, Melegh B, Meszaros J, Molnar P, Nyul Z, Papp H, Patri L, Puskas E, Santha I, Schneider F, Szomor K, Toth A, Toth E, Szucs G, Banyai K. Surveillance of human rotaviruses in 2007–2011, Hungary: exploring the genetic relatedness between vaccine and field strains. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2012;55:140–146. doi: 10.1016/j.jcv.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Banyai K, Rahman M, Zeller M, Beutels P, Van Damme P, Van Ranst M. Rotavirus disease and vaccination: impact on genotype diversity. Future microbiology. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Archives of virology. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Molecular biology and evolution. 2010;27:2431–2436. doi: 10.1093/molbev/msq137. [DOI] [PubMed] [Google Scholar]
- Mirzayeva R, Cortese MM, Mosina L, Biellik R, Lobanov A, Chernyshova L, Lashkarashvili M, Turkov S, Iturriza-Gomara M, Gray J, Parashar UD, Steele D, Emiroglu N. Rotavirus burden among children in the newly independent states of the former union of soviet socialist republics: literature review and first-year results from the rotavirus surveillance network. The Journal of infectious diseases. 2009;200(Suppl 1):S203–S214. doi: 10.1086/605041. [DOI] [PubMed] [Google Scholar]
- Mladenova Z, Korsun N, Geonova T, Iturriza-Gomara M. Molecular epidemiology of rotaviruses in Bulgaria: annual shift of the predominant genotype. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2010;29:555–562. doi: 10.1007/s10096-010-0895-1. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Khoury H, El Khoury AC, Goetghebeur MM. Burden of rotavirus gastroenteritis in the pediatric population in Central and Eastern Europe: serotype distribution and burden of illness. Human vaccines. 2011;7:523–533. doi: 10.4161/hv.7.5.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp H, Borzak R, Farkas S, Kisfali P, Lengyel G, Molnar P, Melegh B, Matthijnssens J, Jakab F, Martella V, Banyai K. Zoonotic transmission of reassortant porcine G4P[6] rotaviruses in Hungarian pediatric patients identified sporadically over a 15 year period. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013;19:71–80. doi: 10.1016/j.meegid.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Ruggeri FM, Delogu R, Petouchoff T, Tcheremenskaia O, De Petris S, Fiore L. Molecular characterization of rotavirus strains from children with diarrhea in Italy, 2007–2009. Journal of medical virology. 2011;83:1657–1668. doi: 10.1002/jmv.22163. [DOI] [PubMed] [Google Scholar]
- Sack DA, Chowdhury AM, Eusof A, Ali MA, Merson MH, Islam S, Black RE, Brown KH. Oral hydration rotavirus diarrhoea: a double blind comparison of sucrose with glucose electrolyte solution. Lancet. 1978;2:280–283. doi: 10.1016/s0140-6736(78)91687-2. [DOI] [PubMed] [Google Scholar]
- Samoilovich EO, Logunova NV, Semeiko GV, Yermalovich MA, Kluiko NL, Astapov AA, Luzhinskii VS, Lisitskaya TI, Kanashkova TA, Biskina NM. Rotavirus Gastroenteritis among children in Minsk City in 2012: incidence, seasonality, age distribution, genotype landscape of causitive agent. Medical Journal. 2013;4:95–98. [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Reviews in medical virology. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, Network WHcGRS. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. The Lancet infectious diseases. 2012;12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- Usonis V, Ivaskeviciene I, Desselberger U, Rodrigo C. The unpredictable diversity of co-circulating rotavirus types in Europe and the possible impact of universal mass vaccination programmes on rotavirus genotype incidence. Vaccine. 2012;30:4596–4605. doi: 10.1016/j.vaccine.2012.04.097. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Uhari M, Renko M, Hemming M, Salminen M, Torcel-Pagnon L, Bricout H, Simondon F. Impact and Effectiveness of RotaTeq(R) Vaccine Based on 3 Years of Surveillance Following Introduction of a Rotavirus Immunization Program in Finland. The Pediatric infectious disease journal. 2013;32:1365–1373. doi: 10.1097/INF.0000000000000086. [DOI] [PubMed] [Google Scholar]