SUMMARY
Studying 830 pre-B ALL cases from four clinical trials, we found that human ALL can be divided into two fundamentally distinct subtypes based on pre-BCR function. While absent in the majority of ALL cases, tonic pre-BCR signaling was found in 112 cases (13.5%). In these cases, tonic pre-BCR signaling induced activation of BCL6, which in turn increased pre-BCR signaling output at the transcriptional level. Interestingly, inhibition of pre-BCR-related tyrosine kinases reduced constitutive BCL6 expression and selectively killed patient-derived pre-BCR+ ALL cells. These findings identify a genetically and phenotypically distinct subset of human ALL that critically depends on tonic pre-BCR signaling. In vivo treatment studies suggested that pre-BCR tyrosine kinase inhibitors are useful for the treatment of patients with pre-BCR+ ALL.
INTRODUCTION
Bone marrow progenitor cells in mice produce approximately 10 million pre-B cells daily (Osmond, 1991), the vast majority of which is eliminated at the pre-B cell receptor (BCR) checkpoint (Sakaguchi and Melchers, 1986). Early pre-B cells are programmed to die unless they productively rearrange VHDJH gene segments and are rescued by ‘tonic’ pre-BCR signal activity into the long-lived pool of mature peripheral B cells (Rajewsky, 1996). Even in mature B cells, continuous tonic signaling from the BCR is required for B cell survival and maintenance and conditional ablation of tonic BCR signaling results in rapid B cell depletion (Kraus et al., 2004). Interestingly, however, loss of tonic BCR signaling can be rescued by activation of PI3K-AKT signaling (Srinivasan et al., 2009), identifying PI3K-AKT as a central survival pathway downstream of the (pre-) BCR. Tonic pre-BCR signaling involves constitutive activity of the proximal pre-BCR-associated SRC family kinases LYN, FYN and BLK (Saijo et al., 2003) as well as SYK and ZAP70 (Schweighoffer et al., 2003), which then activate PI3K (Guo et al., 2000; Okada et al., 2000). Recent work highlighted the particular importance of the PI3K p110δ (PIK3CD) isoform for pre-BCR survival signaling during early B cell development (Ramadani et al., 2010). The discovery that most subtypes of B cell lymphoma critically depend on BCR signaling (Davis et al., 2010; Schmitz et al., 2012) has led to the development of new targeting strategies that focus on BCR signaling at the level of SRC kinases (Lyn, Fyn and Blk), SYK/ZAP70 and PI3Kδ (Burger and Okkenhaug, 2014; Chen et al., 2006; Chen et al., 2013; Cheng et al., 2011; Ke et al., 2009; Yang et al., 2008). In addition, small molecule inhibition of BTK, which mediates ‘chronic active BCR signaling’ in activated B cell-like (ABC) diffuse large B cell lymphoma (DLBCL), chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) has achieved major clinical success in the treatment of these diseases (Byrd et al., 2013; Davis et al., 2010; Schmitz et al., 2012; Wang et al., 2013). While the role of BCR signaling in the biology and treatment has been elucidated in all major B cell lymphoma subtypes, the role of pre-BCR signaling has not been systematically studied in human pre-B acute lymphoblastic leukemia (ALL).
Goals of the present study were (i) to identify cases of human pre-B ALL with tonic or chronic active pre-BCR signaling, (ii) to estimate their frequency, (iii) to determine the role of pre-BCR signaling in specific pre-B ALL subtypes, (iv) to identify cooperating genetic lesions and (v) to develop a concept for therapeutic targeting of the pre-BCR pathway in human pre-B ALL.
RESULTS
Expression and Activity of the pre-BCR Defines a Distinct Subtype of Human ALL
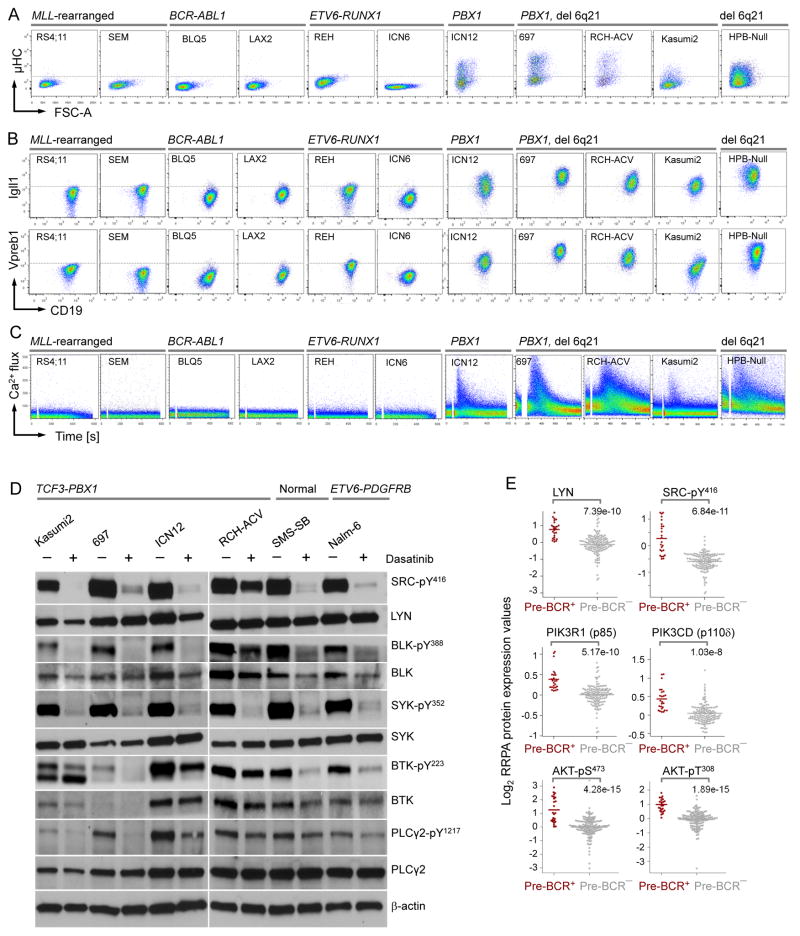
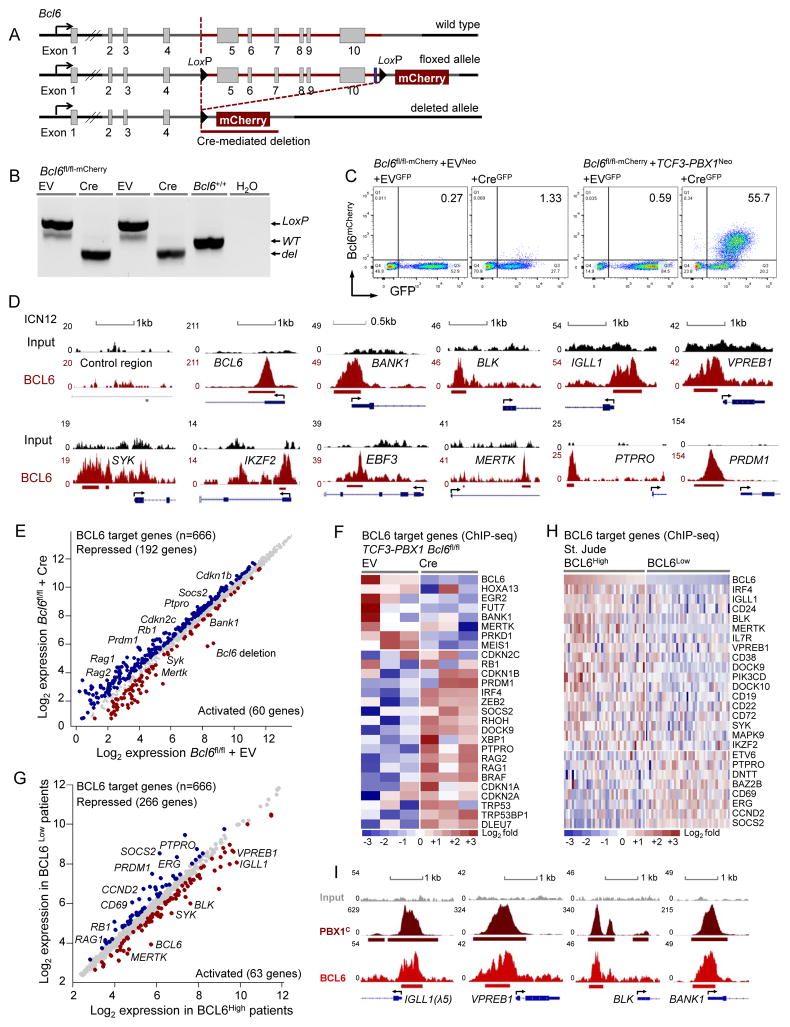
To elucidate pre-BCR expression and function in pre-B ALL cells, we measured expression of the immunoglobulin μ heavy chain (μHC), and the pre-BCR surrogate light chain components λ5 (IGLL1) and VpreB on a series of 31 patient-derived pre-B ALL xenograft samples and 15 ALL cell lines by flow cytometry (Table S1–S3). 28 of the 46 pre-B ALL samples and cell lines tested lacked surface pre-BCR expression including 5 MLL-rearranged (MLLr), 16 BCR-ABL1, 2 ETV6-RUNX1, and 5 ALL with other abnormalities. Of the 18 pre-BCR+ ALL cases, 14 harbored a PBX1 gene rearrangement (1q23), one carried a deletion at 6q21, one carried both PBX1 gene rearrangement and 6q21 deletion and two harbored PDGFRB gene rearrangements (Figure 1A–1B and S1A–S1I). Engagement of the pre-BCR using μHC-specific antibodies resulted in strong Ca2+ mobilization from cytoplasmic stores in all 7 pre-BCR+ ALL cases tested but not in any of the 19 other cases (Figure 1C and S1A–S1I). These findings suggest that most cases of human ALL lack pre-BCR signaling (pre-BCR−), whereas a distinct ALL subgroup (pre-BCR+) exists that is defined by pre-BCR expression and activity. Indeed, key components of the pre-BCR signaling, including SRC family kinases (LYN, BLK), SYK, BTK and PLCγ2 were constitutively active in 6 pre-BCR+ ALL samples (Figure 1D). Interestingly, phosphorylation of these molecules was sensitive to treatment of the dual ABL1/SRC-BTK inhibitor Dasatinib (Figure 1D).
Figure 1. Expression and Activity of the pre-BCR Receptor in Subsets of pre-B ALL.
(A) Flow cytometry staining for cell cytoplasmic μHC, (B) cell surface expression of the surrogate light chain components λ5 (IGLL1) and VpreB, and (C) Ca2+ mobilization in response to pre–BCR engagement using μHC-specific antibodies in different subtypes of ALL cases. (D) Patient-derived Pre-BCR+ ALL cells were treated with or without Dasatinib (25 μmol/l, 24 hr). Phosphorylation of SRC, BLK, SYK, BTK and PLCγ2 were measured by Western blot. (E) Protein expression and phosphorylation levels of LYN, SRC, PIK3R1 (p85), PIK3CD (p110δ) and AKT were measure by reverse phase protein arrays (RPPA) in pre-BCR+ vs. pre-BCR− ALL patient samples (MDACC 1983–2007; n=208). Y axis shows log2 expression values of RPPA data. P values were calculated from two-sided Wilcoxon test. See also Figure S1 and Table S1–S3.
Tonic pre-BCR Signaling including Activation of SRC, SYK and PI3K in a Subset of Human ALL
To compare baseline signaling activity of pre-BCR− and pre-BCR+ ALL cells in a large cohort of patients (MDACC 1983–2007; n=208), we divided patient samples into two groups based on flow cytometry measurements of pre-BCR (μHC) expression and the Igα signaling chain CD79A (Figure S1J). None of the pre-BCR+ ALL cases in this cohort (n=26) expressed the stem/progenitor cell antigen CD34 and CD25, which are both expressed on the surface of most of the 182 pre-BCR− ALL cases (Figure S1K). Using a panel of 133 validated antibodies, we interrogated those 208 pre-B ALL samples at the time of diagnosis for expression and activity of 66 signaling molecules on reverse phase protein arrays (RPPA) (Tibes et al., 2006). RPPA analyses revealed that pre-BCR+ ALL cells exhibit significantly higher expression and activity of SRC family kinases including LYN, and strong constitutive activity of multiple components of the PI3K-AKT pathway, including PIK3R1 (p85α), PIK3CD (p110δ), AKT1-pS473/T308, MTOR-pS2448, RPS6KB-pS371 and RPS6-pS235/S236 (Figure 1E and S1L). Consistent with strong PI3K-AKT signaling, expression and activity of PTEN, a negative regulator of PI3K-AKT signaling, is low in pre-BCR+ ALL cells. These findings are consistent with the established central role of SRC/LYN kinase (Saijo et al., 2003) and PI3K-AKT signaling (Srinivasan et al., 2009), in particular PIK3d (Ramadani et al., 2010), in tonic pre-BCR signaling. Pre-BCR+ ALL cases are also distinct from pre-BCR− ALL cases by particularly low levels of STAT5 expression and activity (Figure S1L). We conclude that pre-BCR+ ALL cells are defined by a distinct signaling phenotype including constitutive SRC and PI3K-AKT signaling and lack of STAT5 activity.
Tonic pre-BCR Signaling Is Associated with a Distinct Gene Expression Profile in Human ALL
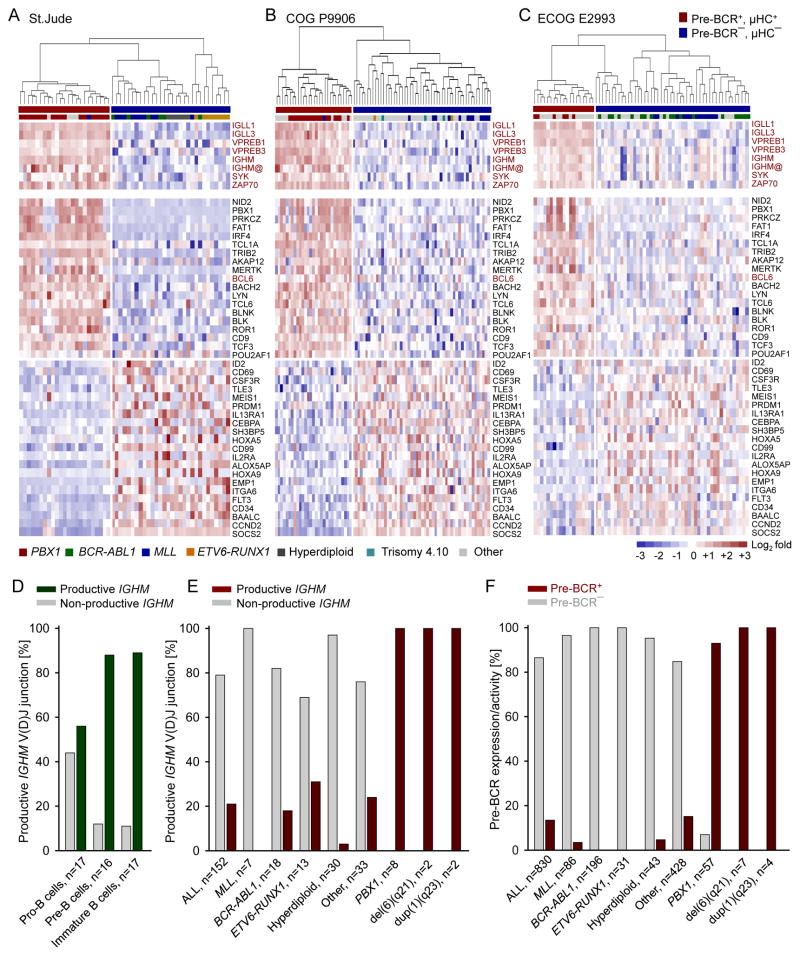
Upregulation of tonic pre-BCR signaling at the expense of STAT5 in pre-B ALL is reminiscent of negative regulation of IL7R/STAT5 upon pre-BCR activation during normal early B cell development (Ochiai et al., 2012). The shift from IL7R/STAT5 to pre-BCR signaling during early B cell differentiation triggers profound gene expression changes. Therefore, we analyzed the gene expression data from 3 pre-B ALL clinical trials for children (COG P9906, n =207 (Harvey et al., 2010) and St. Jude Research Hospital, n=132 (Ross et al., 2003)) and adults (ECOG E2993, n=191 (Geng et al., 2012)). Patients in these trials were classified as pre-BCR− or pre-BCR+ based on pre-BCR expression (IGLL1, IGLL3, VPREB1, VPREB3, IGHM) and its associated signaling molecules SYK and ZAP70 (Figure 2A–2C). To identify a common gene expression signature for pre-BCR+ ALL, we performed a supervised analysis of the microarray data and identified a set of 40 genes concordantly differentially expressed in pre-BCR+ vs. pre-BCR− ALL in all 3 clinical cohorts (Figure 2A–2C). The up-regulated genes include pre-BCR signaling molecules (PRKCZ, BLNK, BLK, LYN, MERTK, ROR1) and transcription factors that promote B-cell differentiation (BCL6, BACH2, IRF4, TCF3, POU2AF1). Down-regulated genes include IL2RA (CD25) and CD34, confirming flow cytometry data from the MDACC cohort (Figure S1K), the STAT5 target genes (ID2, CD69, CD99, ITGA6, CCND2, SOCS2) and PRDM1 (BLIMP1) (Figure 2A–2C).
Figure 2. Pre-BCR Signaling in ALL Is Associated with a Distinct Gene Expression and Signal Transduction Phenotype.
(A–C) Gene expression microarray data was analyzed from 3 cohorts of pre-B ALL patient samples for children (St. Jude Research Hospital; COG P9906) and adults (ECOG E2993). In each dataset, the patient samples were ranked based on their average mRNA expression levels of pre-BCR molecules (IGLL1, IGLL3, VPREB1, VPREB3, IGHM, SYK and ZAP70). The top 15% and the bottom 25% cases were considered as the pre-BCR+ and pre-BCR− and subject to the clustering analysis. Supervised analysis on pre-BCR+ vs. pre-BCR− ALL revealed a 40-gene expression signature that is significantly up- or down-regulated across all 3 cohorts. The color scale bar represents the relative log2 expression changes. (D) The configuration of the μ heavy chain locus (IGHM) was studied in human bone marrow pro-, pre- and immature B cells and (E) different subtypes of ALL patient samples (n=152). Y axis shows frequencies of normal B-cells or ALL clones with a functional or a non-functional IGHM gene rearrangement in these populations. (F) Frequency of different cytogenetic subtypes of pre-B ALL cases as pre-BCR+ or pre-BCR− based on 4clinical trials (n=830, MDACC, St. Jude, COG and ECOG). See also Table S4–S6.
Frequent Rearrangement of PBX1 in pre-BCR+ ALL
We studied the coding capacity of immunoglobulin heavy chain VHDJH gene rearrangements in 152 primary pre-B ALL samples and normal B cell precursors as reference (Trageser et al., 2009). Human bone marrow pro-B, pre-B and immature B cells from healthy donors (n=2) were studied by single-cell PCR. In marked contrast to other pre-B ALL subsets, most cases with TCF3-PBX1 rearrangement and/or 6q21 deletion carried productively rearranged IGHM alleles and showed evidence of selection for expression of in-frame VHDJH gene rearrangements, like normal pre-B cells (Figure 2D–2E; Table S4). In contrast, productive VHDJH gene rearrangements were negatively selected in all other ALL subsets. Combining data from 4 clinical trials (n=830, Table S5), we collectively found evidence of pre-BCR function in 112 cases (13.5%). Pre-BCR expression was frequent among ALL cases with PBX1-rearrangement or -duplication and 6q21 deletion, but rare or absent in cases with MLLr, BCR-ABL1, ETV6-RUNX1 and hyperdiploid ALL (Figure 2F; Table S5–S6).
TCF3-PBX1 Binds to and Upregulates pre-BCR Genes in Human ALL Cells
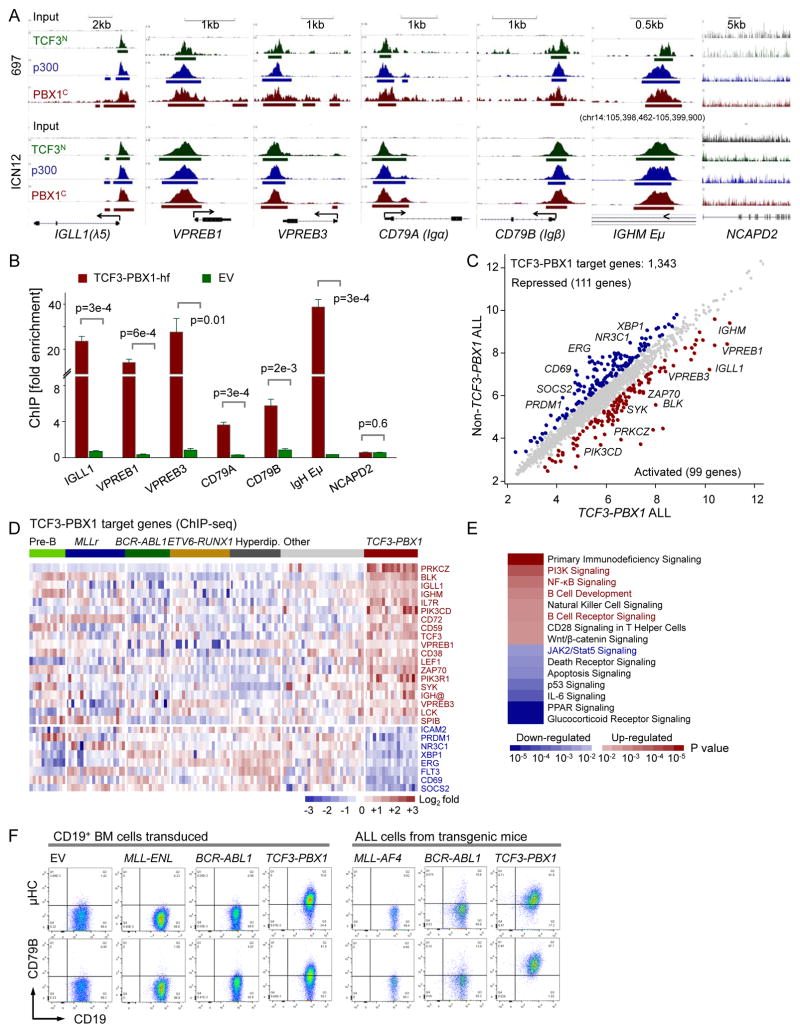
PBX1 is a proto-oncogene with a critical role in hematopoiesis and lymphopoiesis (Sanyal et al., 2007). In pre-B ALL, PBX1 (1q23) is frequently rearranged to the B cell-specific TCF3 (19p13) locus, encoding E2A (E12 and E47) transcription factors. In these cases, the N-terminal transcriptional activation domains of TCF3 (exons 1–16, TCF3N) are fused to the C-terminal Hox cooperative motif and homeodomain of PBX1 (exons 4-9, PBX1C) (Nourse et al., 1990). To identify targets of oncogenic TCF3-PBX1 activity, we performed chromatin immunoprecipitation (ChIP), followed by massively parallel DNA sequencing (ChIPseq), using antibodies against the PBX1C, the TCF3N and the TCF3-PBX1 co-activator p300 (Bayly et al., 2004) in a TCF3-PBX1 ALL cell line (697) and a primary TCF3-PBX1 ALL sample (ICN12) (Figure S2A). ChIPseq data revealed that TCF3-PBX1, in concert with its coactivator p300, strongly bound to promoter regions of genes that encode key components of the pre-BCR, including IGLL1, VPREB1, VPREB3, CD79A and CD79B, as well as μHC enhancer regions (Figure 3A). Specific binding of TCF3-PBX1 to these 6 loci was confirmed by single-locus quantitative ChIP (qChIP), using the fusion-specific TCF3-PBX1-hf (Figure 3B and S2B) and the TCF3N, PBX1C and p300 antibodies (Figure S2C–S2F). To identify genome-wide target genes that are directly regulated by TCF3-PBX1, we matched TCF3-PBX1 binding (ChIPseq, Figure S2D–S2F) with specific gene expression changes that distinguish TCF3-PBX1 cases from other pre-B ALL subsets (Ross et al., 2003). Among the 1,343 TCF3-PBX1 (PBX1C, TCF3N and p300) target genes in promoter regions (within 2kb distance from transcription start site), 99 were specifically up-regulated, including multiple pre-BCR components (IGLL1, VREB1, VREB3, IGHM) and pre-BCR downstream signaling molecules (BLK, LCK, SYK, ZAP70, PIK3CD, PIK3R1). In addition, 111 targets were significantly downregulated in TCF3-PBX1 compared to other pre-B ALL subtypes, including the STAT5 target genes CD69 and SOCS2 (Figure 3C–3D). A pathway analysis of TCF3-PBX1 ChIPseq/gene expression targets suggested that TCF3-PBX1 is a positive regulator of B cell development and BCR signaling and negatively regulated STAT5 (Figure 3E).
Figure 3. TCF3-PBX1 fusion protein binds to and upregulates genes encoding pre-BCR components.
(A) ChIPseq tracks for TCF3N, PBX1C and p300 antibodies vs. input in a TCF3-PBX1 ALL line 697 and a primary sample ICN12 on IGLL1, VPREB1, VPREB3, CD79A and CD79B promoter regions and the IGHM enhancer regions (Eμ). Gene models were shown in UCSC genome browser hg18. B () QChIP validation using an HA antibody that is specific for the HA-Flag-tagged TCF3-PBX1 fusion or an EV as control. NCAPD2 serves as a negative control gene. Data represent means ± SEM (n=3). P values from t-test. (C) The scatter plot shows the average gene expression values of the TCF3-PBX1-rearranged (x-axis) vs. the non-TCF3-PBX1-rearranged ALL patient samples (y-axis) from the St. Jude dataset on the TCF3-PBX1 target genes (n=1,343). Blue and red highlighted are the down- (n=111) and up- (n=99) regulated genes (>1.5 fold change). (D) The heatmap representation for some up- or down-regulated genes. The color scale bar represents the log2 expression changes. (E) Ingenuity pathway analysis for the up- and down-regulated genes. The color scale indicates p values calculated from Ingenuity. (F). Flow cytometry staining for cytoplasmic μHC, surface CD79B and CD19 in full-blown ALL populations that had developed after latencies of 90–180 days in the bone marrow from the MLL-AF4-, BCR-ABL1- or TCF3-PBX1-transgenic mice. See also Figure S2.
To further test whether gene rearrangement and oncogenic activation of TCF3-PBX1 induces pre-BCR activity, we studied full-blown ALL populations that had developed after latencies of 90–180 days in the bone marrow from the TCF3-PBX1-, BCR-ABL1- and MLL-AF4-transgenic mice. Flow cytometry of surface μHC and CD79B (Igβ) expression confirmed that ALL cells developing in TCF3-PBX1-transgenic mice are characterized by pre-BCR expression in contrast to pre-BCR− ALL in BCR-ABL1-and MLL-AF4-transgenic mice (Figure 3F).
Pre-BCR Signaling Regulates the Balance between BCL6 and STAT5 Activity in Human ALL
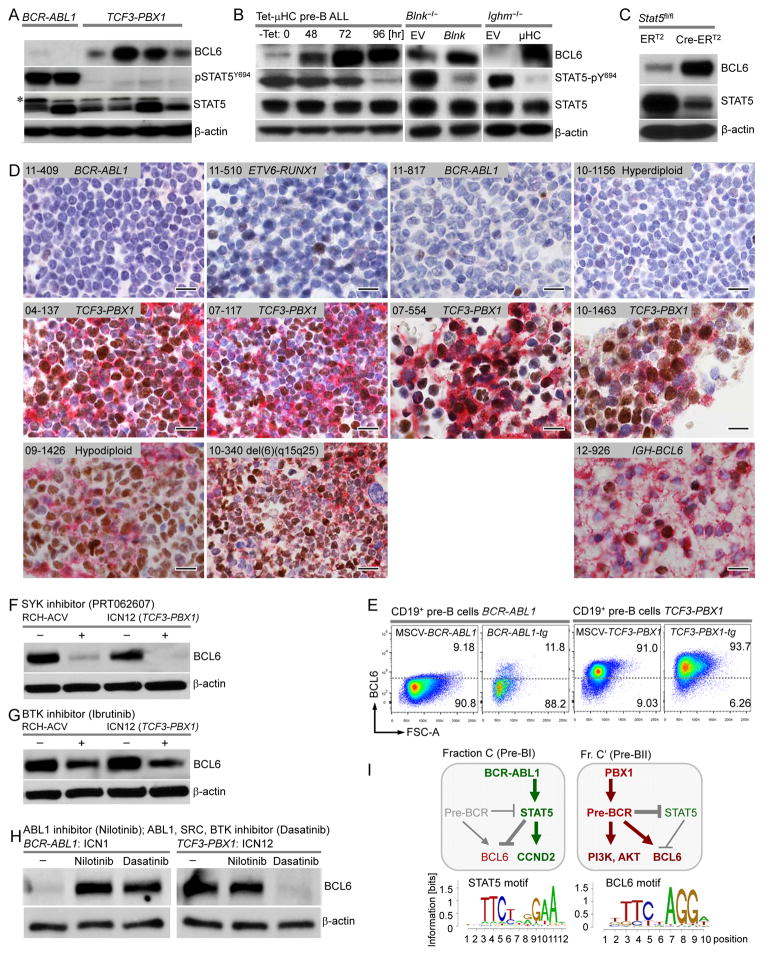
Studying gene expression changes in 530 ALL cases from 3 clinical trials (St Jude, COG P9906, ECOG E2993; Table S5), we found that pre-BCR+ ALL cells (n=71, 13.4%) exhibit consistently higher expression levels of BCL6 than pre-BCR− cases (n=459, 86.4%; Figure 2A–2C). Quantitative RT-PCR analyses confirmed 6- to 30-fold higher mRNA and protein levels of BCL6 in TCF3-PBX1 ALL compared to other ALL subsets (Figure 4A and S3A). These findings suggested that BCL6 may be a direct target of TCF3-PBX1 and other oncogenic transcription factors that drive pre-BCR+ ALL. However, ChIPseq and qChIP analysis showed no evidence of binding of TCF3-PBX1 to the BCL6 locus (Figure S3B–S3C), indicating that TCF3-PBX1 induces BCL6 expression through an indirect mechanism. Recent studies by us and others (Duy et al., 2010; Swaminathan et al., 2013) revealed a critical role of BCL6 as a survival factor at the pre-BCR checkpoint during normal B cell development. In BCR-ABL1 ALL (pre-BCR−), inhibition of STAT5 activity by treatment with tyrosine kinase inhibitors induced strong expression of BCL6 (Duy et al., 2011). Likewise, in our Western blot analysis, while pre-BCR+ (TCF3-PBX1) ALL cases expressed high levels of BCL6 in the absence of STAT5 activity, we observed the converse in pre-BCR− (BCR-ABL1) ALL cells (Figure 4A). To test whether pre-BCR signaling directly affects the balance between BCL6 and STAT5, we used an inducible system for pre-BCR activation in mouse ALL cells. To this end, Rag2−/− mouse ALL cells, lacking the ability of endogenous pre-BCR expression, were engineered to express a pre-rearranged tetracyclin-inducible μHC, which results in subsequent pre-BCR expression (Trageser et al., 2009). Inducible pre-BCR activation resulted in a massive increase of BCL6 protein levels (40-fold) and concomitant silencing of STAT5 activity (p-Y694; Figure 4B). Similarly, complementation of a defective pre-BCR signaling chain in Blnk−/− or Ighm−/− mouse ALL cells by reconstitution of Blnk or μHC-expression caused dramatic upregulation of BCL6 at the expense of STAT5 activity (p-Y694; Figure 4B). Likewise, Cre-mediated deletion of Stat5 in Stat5a/bfl/fl ALL cells was sufficient to increase BCL6 protein levels (Figure 4C). These experiments provide genetic evidence that the balance between BCL6 and STAT5-activity is directly influenced by pre-BCR signaling and, hence, suggests that high expression levels of BCL6 represent as a surrogate marker for pre-BCR activity in human ALL cells.
Figure 4. Pre-BCR Signaling in ALL Cells Drives Expression of BCL6.
(A) BCL6, STAT5, and pSTAT5Y694 Western blotting with β-actin as a loading control in human BCR-ABL1 or TCF3-PBX1 ALL samples. Asterisk denotes non-specific band. (B) Protein expression levels of BCL6, STAT5 and pSTAT5Y694 by Western blot with β-actin as the loading control at different time points of tetracycline culture of Rag2−/− tTA/μ-chain ALL cells (left panel), before and after pre-BCR signaling reconstitution by retroviral expression of Blnk-GFP or GFP EV in the Blnk−/− ALL cells (middle panel), or CD8/μ-chain or CD8 EV in Ighm−/− ALL cells (right panel). (C) BCL6 and STAT5 Western blot in the presence or absence of Cre-mediated deletion of Stat5 in Stat5fl/fl pre-B ALL cells. (D) Immunohistochemistry double-stainings for BCL6 (nuclear, brown) and μHC (cytoplasmic/membrane, red) on the same slides of paraffin-embedded bone marrow samples from Pre-BCR− and Pre-BCR+ ALL patients. A Burkitt’s lymphoma sample (12-926) carrying an IGH-BCL6 gene rearrangement was used as a positive control. Scale bars, 20 um. (E) Bone marrow pre-B cells transduced with BCR-ABL1 or TCF3-PBX1 vectors, or from full-blown ALL populations that had developed after latencies of 90–180 days from the BCR-ABL1- or TCF3-PBX1-transgenic mice were stained for intracellular Bcl6 protein. Cells were gated as live CD19+ cells. The Bcl6 expression threshold was set according to an isotype staining control. (F–G) BCL6 protein expression by Western blotting in presence or absence of SYK inhibitor (PRT062607, 10 μmol/l), BTK inhibitor (Ibrutinib, 10 μmol/l) for 24 hr in a TCF3-PBX1 ALL cell line (RCH-ACV) and a primary sample (ICN12), or (H) in presence or absence of Nilotinib (200 nmol/l) or Dasatinib (25 nmol/l) for 24 hr in primary BCR-ABL1 (ICN1) or TCF3-PBX1 (ICN12) ALL cells. β-actin was used as a loading control. (I) A schematic of pre-BCR, STAT5 and BCL6 regulation in BCR-ABL1 and TCF3-PBX1 ALL. See also Figure S3 and Table S7-S8.
BCL6 Expression Represents a Surrogate Marker for pre-BCR Activity in Human ALL
Immunohistochemical analyses for expression of the pre-BCR (μHC) and BCL6 were performed on bone marrow biopsies from 72 ALL patients. These analyses confirmed that all 12 pre-BCR+ (μHC+) ALL cases express BCL6 at high levels (Figure S3D; Table S7). These cases carried lesions affecting PBX1, through TCF3-PBX1 and/or gain of 1q23 encompassing PBX1. By contrast, 60 other cases lacked both BCL6 and μHC surface expression (Figure S3E). We next performed double-stainings for BCL6 and μHC on the same slides, confirming that pre-BCR− ALL cells express neither μHC nor BCL6. In pre-BCR+ ALL cases, virtually all ALL cells expressed both surface μHC and BCL6 at levels comparable to mature B cell lymphoma carrying IGH-BCL6 rearrangement (Figure 4D). Bone marrow biopsies from pre-BCR+ ALL cases included various degrees of admixtures of normal bone marrow cells that lacked both μHC and BCL6 expression.
SYK Tyrosine Kinase Signaling Is Required for BCL6 Activation downstream of Tonic pre-BCR Signaling
Based on the finding that BCL6 is induced by pre-BCR signaling and is a potential target for drug-treatment in pre-BCR+ ALL cells, we measured the effects of small molecule inhibitors of proximal pre-BCR tyrosine kinases on BCL6 expression. While 57 of 61 ALL cases with PBX1-rearrangement or duplication showed evidence of tonic pre-BCR signaling (Figure 2F; Table S5), we did not find pre-BCR activity in any of 196 cases with Ph+ ALL (BCR-ABL1). Consistent with divergent pre-BCR activity in these subsets, we found that TCF3-PBX1 but not BCR-ABL1 fusion oncogenes induced expression of BCL6 in pre-B ALL cells (Figure 4E). Treatment with small molecule tyrosine kinase inhibitors of SYK (PRT062607) and BTK (Ibrutinib) reduced BCL6 expression in pre-BCR+ TCF3-PBX1 ALL cells (Figure 4F and 4G), demonstrating that SYK tyrosine kinase activity is required for BCL6 expression in pre-BCR+ ALL cells. While TCF3-PBX1 induces constitutive BCL6 expression via tonic pre-BCR signaling, BCR-ABL1 represses BCL6 expression. Ph+ ALL lack BCL6 expression in the presence of active BCR-ABL1, however, ABL1 tyrosine kinase inhibitors (Nilotinib, Dasatinib) relieve BCR-ABL1/STAT5-mediated repression of BCL6 (Figure 4H). Interestingly, the specific ABL1 kinase inhibitor Nilotinib, which does not affect pre-BCR signaling, has no effect on constitutive BCL6 expression in TCF3-PBX1 ALL cells. However, the dual ABL1/SRC kinase inhibitor Dasatinib, which inhibits the SRC kinases BLK, LYN and FYN upstream of SYK, almost entirely abolished BCL6 expression in pre-BCR+ TCF3-PBX1 ALL cells (Figure 4H). These findings reveal that based on divergent functions of pre-BCR/BCL6 signaling, human ALL can be divided into two genetically and phenotypically distinct subsets.
We propose that in the majority of ALL cases, the transforming oncogene (e.g. BCR-ABL1) mimics constitutive active cytokine signaling via activation of STAT5 and repression of BCL6. In 10–15% of the cases, oncogenic lesions (e.g. TCF3-PBX1; Table S8) result in tonic pre-BCR signaling and activation of BCL6. These differences may reflect different cells of origins of the two subsets. BCR-ABL1 engages IL7R-dependent survival and proliferation signals via STAT5 that are active in pro-B and early pre-B cells (Fraction C); by contrast, activation of pre-BCR signaling (e.g. by TCF3-PBX1) in late pre-B cells (Fraction C′) suppresses IL7R and cytokine signaling (Ochiai et al., 2012). We propose that the mutually exclusive nature of cytokine receptor/STAT5 and pre-BCR/BCL6 pathways in human ALL reflects distinct developmental origins from early (Fraction C) and late (Fraction C′) pre-B cells (Figure 4I). BCL6 ChIPseq analysis revealed that BCL6 and STAT5 binding sites were the most highly enriched DNA elements (p=10−60 for both) associated with BCL6 binding peaks in TCF3-PBX1 ALL cells (Figure 4I), suggesting that BCL6 and STAT5 may compete for binding to those promoters, which would be consistent with mutual exclusive activity of STAT5 and BCL6 in pre-BCR− and pre-BCR+ ALL subtypes, respectively.
Genetic Mouse Model of Conditional BCL6 Ablation in pre-BCR+ TCF3-PBX1 ALL
BCL6 functions as a classical proto-oncogene in germinal center-derived B cell lymphoma (Ye et al., 1993) and represents a powerful survival factor at the pre-BCR checkpoint during early B cell development (Duy et al., 2010). For these reasons, we developed a genetic mouse model to determine the role of BCL6 in pre-BCR+ ALL cells. In Bcl6fl/fl-mCherry mice, exons 5–10 were flanked by LoxP sites and Cre-mediated deletion of these exons results in expression of a truncated Bcl6 protein fused to mCherry (Figure 5A). Thereby mCherry expression reflects transcriptional activity of the Bcl6 promoter and Cre-mediated deletion can be used for inducible ablation of Bcl6 function and as mCherry-based reporter of Bcl6 expression. In a genetic experiment, we transduced Bcl6fl/fl-mCherry pre-B cells with TCF3-PBX1 or an empty vector (EV) control. Cre-mediated deletion (Figure 5B) activated the Bcl6-mCherry reporter (Figure 5C). While only ~1% of pre-B cells carrying an EV control showed transcriptional activation of the Bcl6 promoter, more than 50% of TCF3-PBX1-transduced cells actively transcribed Bcl6 (Figure 5C). These findings demonstrate that TCF3-PBX1 indirectly induces Bcl6 expression through transcriptional activation of pre-BCR signaling.
Figure 5. BCL6 Is a Key Regulator of the Transcriptional Program in pre-BCR+ ALL Cells.
(A) A genetic model for inducible ablation of BCL6 and a mCherry-based Bcl6 reporter (Bcl6fl/fl-mCherry). Exons 5–10 of the Bcl6 locus were targeted for inducible Cre-mediated deletion. LoxP sites are indicated as black triangles. (B) PCR validation of the floxed, deleted and wild type Bcl6 allele from Bcl6fl/fl-mCherry mouse bone marrow pre-B cells. (C) FACS analysis of EVNeo or TCF3-PBX1Neo retrovirally transduced Bcl6fl/fl-mCherry pre-B cells that were transduced with either EVGFP or CreGFP vector for Bcl6 deletion. Y-axis indicates mCherry expression and x-axis indicates GFP expression. (D) BCL6 ChIPseq binding tracks of target genes in patient-derived TCF3-PBX1 ALL cells (ICN12). Y axis represents the number of reads for peak summit normalized by the total number of reads per track. Gene models are shown in UCSC genome browser hg18. A control intragenic region and BCL6 were used as negative and positive controls. (E–F) A meta-analysis of BCL6 ChIPseq target genes (n=666) with gene expression microarray data for the EV vs. Cre transduced Bcl6fl/fl TCF3-PBX1 ALL cells or (G–H) for the BCL6High vs. BCL6Low ALL patient samples from St. Jude. The scatter or heatmap plots showed up- and down-regulated BCL6 target genes in each data set. The color scale bar represents relative log2 expression changes. (I) PBX1 and BCL6 ChIPseq binding tracks vs. input in ICN12 cells on pre-BCR-related genes (IGLL1, VPREB1, BLK, BANK1). See also Figure S4.
BCL6 Transcriptionally Activates pre-BCR Components in TCF3-PBX1 ALL
To identify BCL6 transcriptional targets in pre-BCR+ ALL cells, we performed ChIPseq analysis in patient-derived TCF3-PBX1 ALL cells (Figure S4A–S4D). As in DLBCL and Ph+ ALL (Duy et al., 2011), BCL6 directly binds to a number of checkpoint molecules including RB1, CDKN2C and CDKN1B in pre-BCR+ ALL (Figure S4A). A previous study demonstrated that BCL6 increases tonic BCR signaling in DLBCL cells by transcriptional repression of the inhibitory phosphatase PTPRO (Juszczynski et al., 2009). Likewise, PTPRO was also identified as a transcriptional target of BCL6 in pre-BCR+ ALL cells. In addition, BCL6 also binds to pre-BCR components (IGLL1, VPREB1) and downstream signaling molecules (BLK, BANK1, SYK) (Figure 5D). QChIP validated BCL6 binding to these loci (Figure S4B). To determine how BCL6 binding affects gene expression in PBX1-rearranged ALL cells, we measured gene expression changes in response to acute ablation of Bcl6 in a Bcl6fl/fl-mCherry mouse model for TCF3-PBX1 ALL. Upon Cre-mediated deletion of Bcl6, multiple BCL6 target genes were de-repressed, including checkpoint molecules RB1, CDKN1A, CDKN1B, CDKN2A and CDKN2C (Figure 5E and 5F). Also PTPRO were among the de-repressed BCL6 target genes, suggesting that BCL6 increases tonic pre-BCR signaling by transcriptional repression of the (pre-)BCR signaling inhibitor PTPRO as described in DLBCL (Juszczynski et al., 2009). Interestingly, BCL6 targets with transcriptional activation by Bcl6 include multiple pre-BCR related molecules (Bank1, Syk; Figure 5E and 5F). To test BCL6-dependent gene regulation in ALL patient samples, we integrated BCL6 ChIPseq target genes with a BCL6-dependent signature of gene expression in 118 cases of childhood pre-B ALL (St. Jude). This analysis showed transcriptional activation of 63 BCL6 target genes including multiple pre-BCR related molecules (IGLL1, BLK, VPREB1, PIK3CD, SYK; Figure 5G and 5H). Collectively, these findings suggest a self-enforcing activation loop between tonic pre-BCR signaling and BCL6 activity in a subset of human ALL: Oncogenic activation of tonic pre-BCR signaling (e.g. as a result of PBX1-rearrangement and –duplication) induces strong activation of BCL6, which further activates pre-BCR signaling (e.g. through transcriptional repression of PTPRO and PRDM1 and activation of IGLL1, VPREB1, BLK, SYK). Interestingly, PBX1 binding to promoter regions of pre-BCR-related genes frequently co-localizes with binding of BCL6, suggesting that BCL6 and PBX1 cooperate in the regulation of these genes (Figure 5I).
Pre-BCR+ ALL Cells Are Dependent on BCL6-Activity
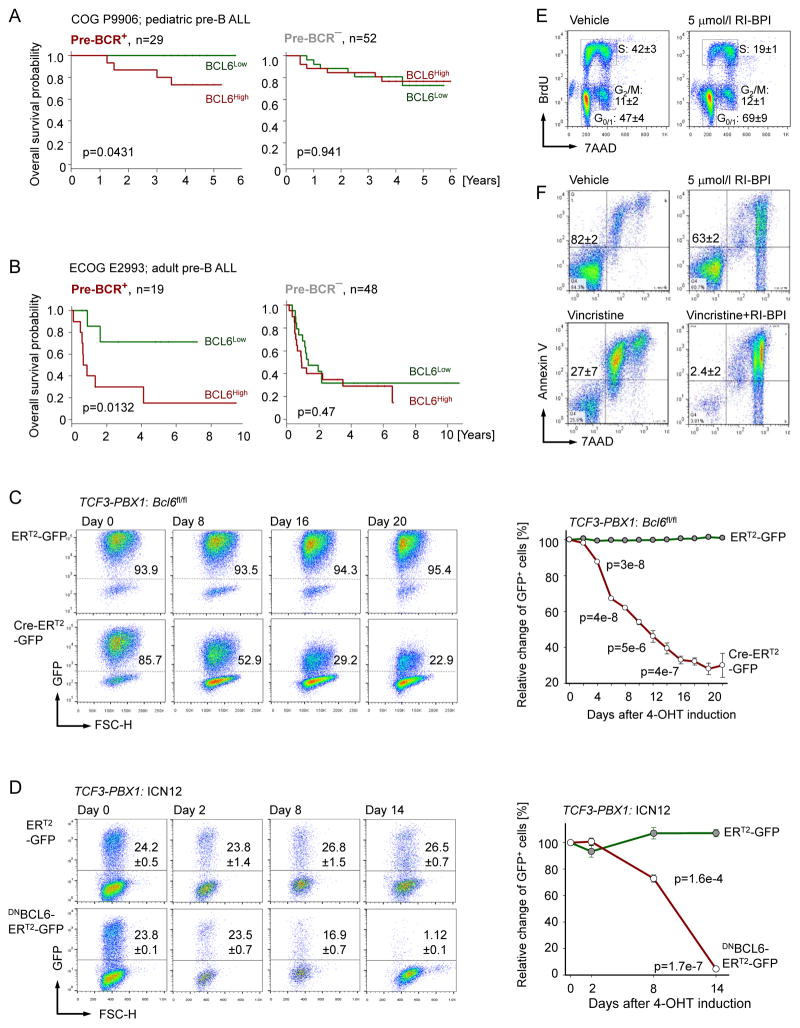
Pre-BCR+ ALL and BCR-dependent B cell lymphoma share constitutively high BCL6 expression levels. To determine whether high expression levels of BCL6 correlate with clinical outcome of ALL patients, we analyzed mRNA levels of BCL6 at the time of diagnosis in children with high-risk ALL (COG P9906) and adults with ALL (ECOG E2993). Only patients of known pre-BCR expression status were included in the analysis. Since BCL6 was specifically upregulated by tonic pre-BCR signaling, we performed clinical outcome analyses separately for pre-BCR+ and pre-BCR− ALL cases. In both the pediatric (COG P9906; p=0.043) and adult (ECOG E2993; p=0.013) clinical trials, higher than median expression levels of BCL6 were predictive of poor outcome among patients with pre-BCR+ but not pre-BCR− ALL (Figure 6A and 6B). These findings suggest that high expression levels of BCL6 affect the course of disease in pre-BCR+ but not pre-BCR− ALL. To determine the potential contribution of BCL6 signaling to survival and proliferation of pre-BCR+ ALL cells, we studied inducible ablation of Bcl6 in a mouse model (Figure 5A–5C). Bcl6fl/fl TCF3-PBX1 ALL cells were transduced with 4-hydroxy-tamoxifen (4-OHT)-inducible Cre (Cre-ERT2) or an EV control (ERT2). While 4-OHT mediated deletion of Bcl6 resulted in progressive depletion of Cre-ERT2-transduced cells, it had no measurable effects in ERT2-transduced Bcl6fl/fl TCF3-PBX1 ALL cells (Figure 6C). To verify BCL6-dependency of pre-BCR+ ALL in a patient-derived setting, we transduced xenografted TCF3-PBX1 ALL cells with an inducible dominant-negative BCL6 mutant lacking the BCL6-BTB domain (DNBCL6-ERT2) (Shaffer et al., 2000). While the fraction of ICN12 cells carrying ERT2 empty vectors remained stable after 4-OHT treatment, cells transduced with DNBCL6-ERT2 were rapidly depleted (Figure 6D). These findings collectively identify BCL6 as a potential target for the treatment of pre-BCR+ ALL.
Figure 6. Pre-BCR+ ALL Cells Are Dependent on BCL6-Activity.
(A–B) Patients were segregated into two groups based on higher or lower than the median expression of BCL6 in pre-BCR+ and pre-BCR− ALL in 2 clinical trials: (A) COG P9906 and (B) ECOG E2993. Kaplan-Myer estimates were used to plot the survival probabilities. P values were calculated from the log-rank test. (C) Bcl6fl/fl TCF3-PBX1 pre-B ALL cells were transduced with 4-OHT-inducible Cre (Cre-ERT2-GFP) or EV control (ERT2-GFP). Percentage of GFP-positive cells were measure by flow cytometry at different time points following 4-OHT treatment and time course data are depicted. (D) Patient-derive Pre-BCR+ ALL cells (ICN12) were transduced with 4-OHT-inducible dominant-negative BCL6 (DNBCL6-ERT2-GFP) or EV control (ERT2-GFP). Percentages of GFP-positive cells were measured by the flow cytometry at different time points following 4-OHT treatment. (E) ICN12 cells were treated with vehicle or 5 μmol/l RI-BPI for 24 hr and then subjected to cell-cycle analysis (BrdU and 7-AAD staining). (F) ICN12 cells were exposed to vehicle, Vincristine (1 nmol/l), RI-BPI (5 μmol/l), or combination of Vincristine (1 nmol/l) and RI-BPI (5 μmol/l) for 3 days, followed by flow cytometry for Annexin V and 7-AAD staining. Data represent means ± SEM (n=3). P values from t-test. See also Figure S5.
Pharmacological Inhibition of BCL6 in pre-BCR+ ALL Cells
In a complementary approach, we tested the consequences of shRNA-mediated knockdown of BCL6 in pre-BCR+ ALL cells. Upon doxycycline-inducible expression of BCL6-specific shRNAs, we observed moderate reduction of BCL6 protein expression and impaired colony formation capacity in pre-BCR+ ALL cells (Figure S5A–S5C). For proof-of-principle experiments, a specific retro-inverso BCL6 peptide-inhibitor (RI-BPI) was used to inhibit BCL6 function (Cerchietti et al., 2009). Treatment of patient-derived pre-BCR+ ALL cells with 5 μmol/l RI-BPI induced cell cycle arrest within 1 day of treatment (Figure 6E and S5D) and caused impaired colony formation capacity (Figure S5E). Whereas RI-BPI treatment alone did not induce significant acute toxicity, it strongly sensitized pre-BCR+ ALL cells to Vincristine (1 nmol/l; p=0.003; Figure 6F and S5F), a mitotic spindle inhibitor that is part of the chemotherapy backbone in most current clinical trials for ALL. In conclusion, BCL6 represents a predictor of poor clinical outcome and a potential target for therapy of patients with pre-BCR+ ALL.
Pre-BCR+ and pre-BCR− ALL Cells Exhibit Distinct Kinase-Inhibitor Sensitivity Profiles
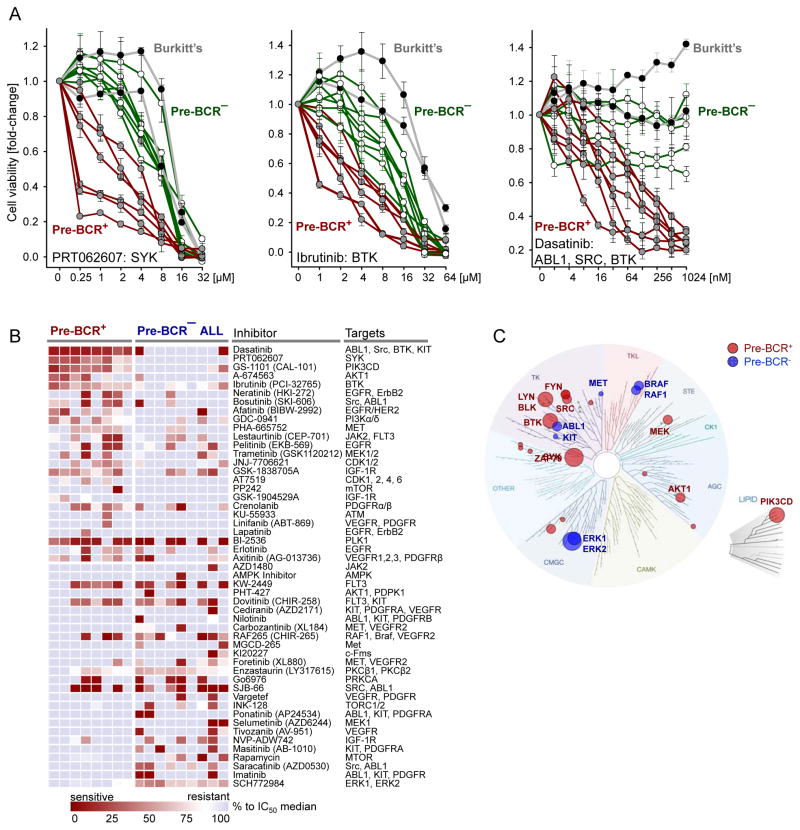
Besides inhibition of BCL6 expression (Figure 4F–4H and S6A), PRT062607 (SYK), Ibrutinib (BTK) and Dasatinib (SRC) induced selective toxicity in pre-BCR+ ALL cells (Figure 7A). Pre-BCR+ ALL (n=7), pre-BCR− ALL (n=8) and mature B cell lymphoma cells (n=2) were treated with inhibitors of pre-BCR tyrosine kinases PRT062607 (SYK), Ibrutinib (BTK), Dasatinib (SRC) as well as inhibitors of pre-BCR downstream signaling, including GS-1101 (PI3Kδ), AZD05363 (AKT1), Rapamycin (mTOR), AZD6244 (MEK1), SCH772984 (ERK1, ERK2), Enzastaurin (PKCβ), MI2 (MALT1) and BMS345541 (IKK, NF-κB) (Figure S6B). For treatment with the dual SRC/ABL1 kinase inhibitor Dasatinib, only Ph+ ALL cells expressing mutant BCR-ABL1T315I were studied to rule out effects of BCR-ABL1 tyrosine kinase inhibition. Compared to pre-BCR− ALL and mature B cell lymphoma cells, pre-BCR+ ALL cells were selectively sensitive to inhibition of SYK and SRC (BLK, LYN) and, to some lesser degree, to inhibition of BTK (Figure 7A) and PI3Kδ (Figure S6B). On the other hand, pre-BCR− ALL cells were selectively sensitive to inhibitors of MEK1 (AZD6244) and ERK1/2 (SCH772984), suggesting that pre-BCR+ and pre-BCR− ALL cells can be distinguished based on a specific profile of kinase-inhibitor sensitivity.
Figure 7. Validation of Pharmacological Inhibition of BCL6-pre-BCR Signaling as Therapeutic Target in pre-BCR+ ALL Cells.
(A) Cell viability was measured using CCK-8 in presence or absence of PRT062607 (SYK), Ibrutinib (BTK) or Dasatinib (ABL1/SRC/BTK) for 72 hr with gradients of concentrations as indicated in the X axis in pre-BCR+ ALL (n=7), pre-BCR− ALL (n=8) and 2 Burkitt lymphoma (MN60, MHH-preB). Y axis shows the percentage of viable cells with the untreated cells as control (set to 100%). Data represent means ± SD (n=3). (B) Pre-BCR+ (n=8) and pre-BCR− (n=9) patient-derived ALL samples were treated with a diverse panel of 51 kinase inhibitors as described previously (Tyner et al., 2013). The heatmap represents the IC50 values for each sample relative to the observed median IC50 value for over 400 primary leukemia samples interrogated by this assay at OHSU. Red or blue colors denote higher or lower than the median sensitivity of the pre-B ALL cells tested. Pre-BCR+ ALL: 07-112, 11-064, ICN12, 697, RCH-ACV, Kasumi-2, HPB-null, Nalm6. Pre-BCR− ALL: BV173, SUPB15, BLQ5, LAX2, SEM, RS4;11, REH, LAX7R, SFO3. (C) Kinase dendrogram of pre-BCR+ and pre-BCR− ALL based on experimentally measured sensitivities to individual inhibitors and their known inhibitory profile based on biochemical IC50 values for individual kinase targets. TREESpots software was used (KINOMEscan, http://www.discoverx.com/). See also Figure S6.
To address this possibility in a formal experiment, we treated pre-BCR+ (n=8) and pre-BCR− (n=9) ALL cells with a diverse panel of 51 kinase inhibitors that are currently being studied for the treatment of hematological malignancies and solid tumors (Figure 7B). This analysis confirmed that pre-BCR+ ALL cells are particularly sensitive to inhibitors of SYK, SRC and PIK3δ, whereas pre-BCR− ALL cells are more responsive to inhibition of MEK1 and ERK1/2 (Figure 7B). Integrating experimentally measured sensitivities to individual inhibitors with their known biochemical IC50 values for individual kinase targets, we established a kinase dendrogram of specific vulnerabilities of pre-BCR+ and pre-BCR− ALL (Figure 7C). While pre-BCR+ ALL cells were most dependent on SYK, ZAP70, BTK, and LYN activity, pre-BCR− ALL cells were most vulnerable to inhibition of ABL1, PDGFR, ERK1, ERK2, MET and KIT kinase activity (Figure 7C).
Validation of pre-BCR-BCL6 Signaling as Therapeutic Target in pre-BCR+ ALL Cells
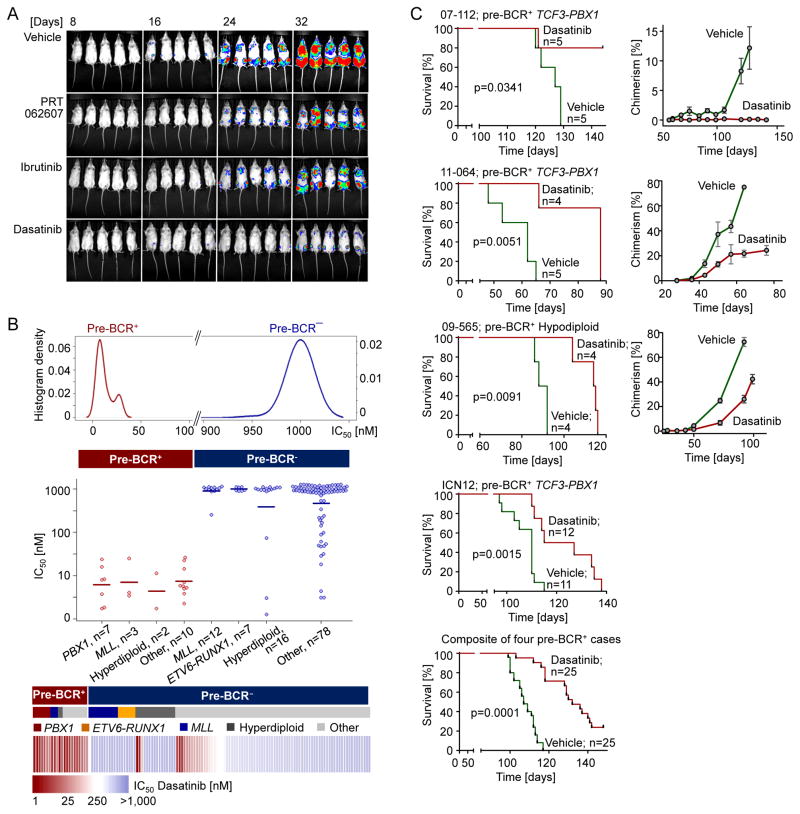
The usefulness of pre-BCR signaling inhibitors PRT062607 (SYK), Ibrutinib (BTK) and Dasatinib (SRC, BTK) for the treatment of pre-BCR+ ALL was tested in a proof-of-principle experiment. To this end, one million patient-derived TCF3-PBX1 pre-BCR+ ALL cells (ICN12) labelled with luciferase were injected intravenously into sublethally irradiated NOD/SCID mice. Recipient mice were then treated with vehicle (n=11), PRT062607 (100 mg/kg; n=7), Ibrutinib (75 mg/kg; n=7) and Dasatinib (40 mg/kg; n=12). All the inhibitors were well tolerated and achieved significant reduction of leukemia burden in vivo, as determined by luciferase bioimaging (Figure 8A). These experiments, together with in vitro testing (Figure 7A and 7B) suggested that Dasatinib has the strongest anti-leukemic effect among the 3 pre-BCR signaling inhibitors. For this reason, we prioritized Dasatinib for detailed in vitro validation experiments for a group of 135 patient-derived ALL samples, including various ALL subtypes but not Ph+ ALL. Patient-derived ALL cells were cultured over 72 hr in the absence or presence of Dasatinib in concentrations ranging from 1 nmol/l to 1,000 nmol/l to calculate Dasatinib IC50 values for these cases. Our previous kinase inhibitor screening assays (Tyner et al., 2013) showed that the median IC50 for Dasatinib of all hematologic malignancy samples received at OHSU (n>400) was 900 nmol/l. Importantly, all 22 pre-BCR+ ALL cases were sensitive to Dasatinib (IC50<50 nmol/l). By contrast, only 6 of 113 pre-BCR− ALL cases had IC50<50 nmol/l and the vast majority of these cases did not respond to Dasatinib at concentrations up to 1,000 nmol/l (Figure 8B). Based on a large data set from 135 patient-derived ALL samples, we conclude that Dasatinib has strong selective anti-leukemia effects on pre-BCR+ ALL cases. To confirm efficacy of Dasatinib in vivo, we transplanted leukemia cells from 4 patients with pre-BCR+ ALL into sublethally irradiated NOD/SCID mice. Three of these cases carried a TCF3-PBX1 gene rearrangement, and 1 case is hypodiploid. In 1 case (07-112), treatment with Dasatinib alone was sufficient to eradicate ALL and to cure mouse transplant recipients, whereas vehicle-treated mice died within 130 days after injection (Figure 8C). In the 3 other cases, Dasatinib treatment significantly delayed leukemic expansion and prolonged overall survival of transplant recipient mice. A composite Kaplan-Meier analysis of the 4 cases, injected into a total of 25 mice per group showed a substantial benefit of Dasatinib treatment (p=0.0001, Figure 8C). To monitor expansion of human leukemia cells (expressing human CD19 and CD45) in the recipient mice, peripheral blood from these mice was drawn weekly and analyzed for chimerism. The Dasatinib-treated cohorts showed minimal signs of circulating human chimerism comparing to the untreated cohorts (Figure 8C). These results demonstrate feasibility and efficacy of leukemia clearance in vivo of pre-BCR+ ALL by Dasatinib based on 4 patient-derived samples. We expect that combinations with other pre-BCR signaling inhibitors (e.g. PRT062607, Ibrutinib) may have synergistic effects, which will be the focus of future studies by our group.
Figure 8. Validation of Pharmacological Inhibition of BCL6-pre-BCR Signaling in Patient-Derived pre-BCR+ ALL Cells.
(A) Patient-derived pre-BCR+ ALL cells (ICN12) were labelled with luciferase and 1,000,000 cells were injected intravenously to sublethally irradiated NOD/SCID mice. The mice were randomly separated into 4 groups and treated with vehicle, the SYK inhibitor PRT062607 (100 mg/kg), the BTK inhibitor Ibrutinib (75 mg/kg), or Dasatinib (40 mg/kg) respectively. Bioimages were taken at different time points for each of the groups. (B) Plot of Dasatinib IC50’s from primary ALL patient samples (n=135, OHSU). ALL samples were cultured over 72 hr in the presence or absence of Dasatinib in concentrations ranging from 1 nmol/l to 1,000 nmol/l to calculate the IC50 values. (C) Patient-derived pre-BCR+ ALL cells (n=4) were injected to sublethally irradiated NOD/SCID mice. Mice were treated with vehicle or Dasatinib (40–50 mg/kg). Kaplan-Meier estimates were used to plot the survival probabilities for each treatment group vs. vehicle control. P values were calculated from log-rank test. Peripheral blood was drawn weekly and analyzed for chimerism with either human CD19 or human CD45 vs. murine CD45 and plotted over time. Data represent means ± SD (n=4 or 5).
DISCUSSION
Recent analyses revealed that BCR-dependent B cell lymphomas can be divided based on tonic or chronic active BCR signaling (Davis et al., 2010; Rickert, 2013; Schmitz et al., 2012). Tonic BCR signaling involves activation of one single pathway, namely PI3Kδ signaling downstream of SRC (LYN, FYN, BLK) and SYK (Burger and Okkenhaug, 2014; Chen et al., 2013), and is associated with germinal center B cell-like (GCB-) DLBCL and Burkitt’s lymphoma (Young and Staudt, 2014). On the other hand, chronic active BCR signaling in ABC-DLBCL, CLL and MCL engages multiple downstream pathways including BTK (and its downstream targets PKCβ, MALT1 and NF-κB), calcineurin/NFAT and MEK/Erk (Young and Staudt, 2014). While the majority of B cell lymphoma cases are BCR-dependent and sensitive to BCR signaling antagonists, classical Hodgkin’s lymphoma (Kanzler et al., 1996) and PMBL (Leithauser et al., 2001) together account for 15–20% of human B cell lymphoma and lack BCR function. The classification of B cell lymphoma based on (i) chronic active, (ii) tonic and (iii) lack of BCR signaling informed the development of new treatment strategies and has led to the successful introduction of BTK (chronic active) (Byrd et al., 2013; Byrd et al., 2014; Wang et al., 2011) and PI3Kδ (tonic BCR signaling) inhibitors (Gopal et al., 2013) into patient care.
Like mature B cell lymphoma, pre-B ALL originates from B cell precursors that critically depend on survival signals emanating from a functional (pre-)BCR. In contrast to B cell lymphoma, however, a classification of human ALL based on pre-BCR function and activity is not available. Studying 830 ALL cases from four clinical trials, we found no functional equivalent of chronic active BCR signaling in B cell lymphoma. In the majority of ALL cases, ALL cells lacked pre-BCR signaling (pre-BCR−), a functional equivalent to classical Hodgkin’s lymphoma and PBML among mature B cell malignancies. However, in about 13.5% of human ALL cases, ALL cells expressed a functional pre-BCR (pre-BCR+), and were highly sensitive to inhibition of SYK and SRC kinases and relatively resistant to inhibition of PKCβ, MALT1 and IKK/NF-κB.
Unlike ABC-DLBCL (chronic active BCR signaling), GCB-DLBCL express high levels of BCL6 in the context of tonic BCR signaling (Juszczynski et al., 2009). Here we found constitutive and pre-BCR-dependent expression of BCL6 in all pre-BCR+ ALL cases studied. Conversely, as a marker in immunohistochemistry staining, BCL6 reliably identified pre-BCR+ ALL cases. BCL6 expression is indeed dependent on tonic pre-BCR signaling, as small molecule inhibition of SYK and SRC family kinases abolished BCL6 expression in pre-BCR+ ALL cells. Since functional assays to measure tonic pre-BCR signaling in ALL patient samples may not be practical for diagnostic purposes, we propose that BCL6 and μHC immunostaining may be a feasible alternative to rapidly identify patients that might benefit from treatment with inhibitors of tonic pre-BCR signaling (e.g. SYK, SRC, PIK3δ).
EXPERIMENTAL PROCEDURES
Primary Human Samples and Cell Lines
Primary ALL cases were obtained with the approval of the Institutional Review Boards of the University of California San Francisco (UCSF) and Oregon Health and Science University (OHSU). Primary human ALL samples were cultured on OP9 stroma in Minimum Essential Medium (MEMα, Life Technologies) with GlutaMAX containing 20% FBS, 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin and 1mM sodium pyruvate at 37°C in a humidified incubator with 5% CO2. The human cell lines were purchased from DSMZ (Braunschweig, Germany). See Table S1–S3.
In Vivo Leukemia Cell Transplantation and Treatment
All mouse experiments were subject to institutional approval by the University of California San Francisco Institutional Animal Care and Use Committee. 106 cells from primary TCF3-PBX1 pre-BCR+ ALL were inoculated via intravenous injection into sublethally irradiated (250 cGy) adult female NOD/SCID mice (n=28). After injection of leukemia cells, the mice were randomly separated into 4 groups and treated with 1) Dasatinib (40 mg/kg); 2) Ibrutinib (75 mg/kg); 3) PRT062607 (100 mg/kg); 4) vehicle. Leukemic infiltration was confirmed by flow cytometry and bioimaging.
Statistical Analysis
The Kaplan-Meier method was used to estimate overall survival. Log-rank test was used to compare survival differences between patient groups. The R package ‘survival’ version 2.35–8 was used for the survival analysis.
Supplementary Material
HIGHLIGHTS.
ALL can be divided into two distinct subtypes based on pre-BCR function
Pre-BCR-induced activation of BCL6 further increased pre-BCR signaling output
Pre-BCR inhibitors reduced BCL6 levels and selectively killed pre-BCR+ ALL cells
BCL6 represents a biomarker to identify patients with pre-BCR+ ALL
SIGNIFICANCE.
Recent work successfully introduced BCR signaling inhibitors into patient care for various subtypes of mature B cell lymphoma, e.g. Ibrutinib (BTK) and Idelalisib (PI3Kδ) for germinal center-derived B cell lymphoma. However, it is not known whether pre-BCR signaling represents a therapeutic target in pre-B ALL. Here we report the identification of a subset of human ALL cases that critically depend on tonic pre-BCR signaling and are selectively sensitive to small molecule inhibitors of SYK and SRC tyrosine kinases downstream of the pre-BCR.
Acknowledgments
We thank Michael L Cleary (Stanford) for critical discussions, Arthur L Shaffer and Louis M Staudt for inducible BCL6 constructs, Mark Kamps and David B. Sykes for inducible TCF3-PBX1 vectors, Lothar Hennighausen for Stat5abfl/fl mice, and Julia Gastier-Foster and I-Ming Chen for fresh samples from the COG ALL Biology Bank (proposal #2008-08). This work is supported by grants from the NIH/NCI through R01CA137060, R01CA139032, R01CA157644, R01CA169458, R01CA172558 (to M.M.), CA178765 (to R.G.R.), R00CA151457 and R01CA183947 (to J.W.T.), U10 CA98543 (COG Chair’s grant), U10 CA98413 (COG Statistical Center), and U24 CA114766 (COG Specimen Banking), the Hyundai Hope on Wheels, the St. Baldrick’s Foundation, the Leukemia & Lymphoma Society Specialized Center of Research (LLS SCOR) and Tucker’s Toy Box Foundation (to B.H.C.), the William Lawrence and Blanche Hughes Foundation, the California Institute for Regenerative Medicine (CIRM; TR2-01816), Leukaemia and Lymphoma Research (to M.M.), the Medical Research Council (UK) and the National Institute for Health Research Oxford Biomedical Research Centre Program (to T.M. and E.B). B.J.D. is supported by the Howard Hughes Medical Institute. M.M. is a Scholar of the Leukemia and Lymphoma Society and a Senior Investigator of the Wellcome Trust.
Footnotes
ACCESSION NUMBERS
The gene expression microarray and ChIPseq data reported in this paper have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database with the GEO accession numbers GSE59332 and GSE59538.
Supplemental Information including Supplemental Experimental Procedures, six Figures and eight Tables can be found with this article online.
AUTHOR CONTRIBUTIONS
H.G., C.H. and K.B.L. designed and performed the majority of the analysis and experiments. Z.C., D.B., S.T., N.G., W.Y.C., J.H., D.L, E.B., G.X., J.L., A.D., Z.Q., E.P., C.H., R.N., S.M.K., S.S., L.N.C., J.Y., J.W.T., B.H.C., S.M.K. and T.A.M. performed experiments and analyzed data. H.G., A.D., S.M.K., E.P., A.M., S.P.H., M.L.L. and B.H.C. provided and characterized patient samples and clinical outcome data. J.J.B., B.H.Y., M.A., A.M., J.A.B., R.G.R. and M.M. provided important reagents and mouse samples. M.M. and B.H. designed experiments and conceived the study. M.M. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, LeBrun DP. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279:55362–55371. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- Burger JA, Okkenhaug K. Haematological cancer: idelalisib-targeting PI3Kdelta in patients with B-cell malignancies. Nat Rev Clin Oncol. 2014;11:184–186. doi: 10.1038/nrclinonc.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, O’Brien S, James DF. Ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2014;369:1278–1279. doi: 10.1056/NEJMc1309710. [DOI] [PubMed] [Google Scholar]
- Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, Dowdy SF, Melnick A. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D, Doench JG, Bogusz AM, Habermann TM, Dogan A, et al. SYK inhibition modulates distinct PI3K - dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer Cell. 2013;23:826–838. doi: 10.1016/j.ccr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Juszczynski P, Takeyama K, Aguiar RC, Shipp MA. Protein tyrosine phosphatase receptor-type O truncated (PTPROt) regulates SYK phosphorylation, proximal B-cell-receptor signaling, and cellular proliferation. Blood. 2006;108:3428–3433. doi: 10.1182/blood-2006-03-013821. [DOI] [PubMed] [Google Scholar]
- Cheng S, Coffey G, Zhang XH, Shaknovich R, Song Z, Lu P, Pandey A, Melnick AM, Sinha U, Wang YL. SYK inhibition and response prediction in diffuse large B-cell lymphoma. Blood. 2011;118:6342–6352. doi: 10.1182/blood-2011-02-333773. [DOI] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, Klemm L, Kweon SM, Nahar R, Braig M, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, Kweon SM, Zickl L, Shojaee S, Neuberg D, et al. Integrative Epigenomic Analysis Identifies Biomarkers and Therapeutic Targets in Adult B-Acute Lymphoblastic Leukemia. Cancer Discov. 2012;2:1004–1023. doi: 10.1158/2159-8290.CD-12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal AK, Kahl BS, de Vos S, Wagner-Johnston ND, Schuster SJ, Jurczak WJ, Flinn IW, Flowers CR, Martin P, Viardot A, et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N Engl J Med. 2013;370:1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Kato RM, Garcia-Lloret M, Wahl MI, Rawlings DJ. Engagement of the human pre-B cell receptor generates a lipid raft-dependent signaling complex. Immunity. 2000;13:243–253. doi: 10.1016/s1074-7613(00)00024-8. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczynski P, Chen L, O’Donnell E, Polo JM, Ranuncolo SM, Dalla-Favera R, Melnick A, Shipp MA. BCL6 modulates tonic BCR signaling in diffuse large B-cell lymphomas by repressing the SYK phosphatase, PTPROt. Blood. 2009;114:5315–5321. doi: 10.1182/blood-2009-02-204362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Chelvarajan RL, Sindhava V, Robertson DA, Lekakis L, Jennings CD, Bondada S. Anomalous constitutive Src kinase activity promotes B lymphoma survival and growth. Mol Cancer. 2009;8:132. doi: 10.1186/1476-4598-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Leithauser F, Bauerle M, Huynh MQ, Moller P. Isotype-switched immunoglobulin genes with a high load of somatic hypermutation and lack of ongoing mutational activity are prevalent in mediastinal B-cell lymphoma. Blood. 2001;98:2762–2770. doi: 10.1182/blood.v98.9.2762. [DOI] [PubMed] [Google Scholar]
- Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13:817–827. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- Osmond DG. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr Opin Immunol. 1991;3:179–185. doi: 10.1016/0952-7915(91)90047-5. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Ramadani F, Bolland DJ, Garcon F, Emery JL, Vanhaesebroeck B, Corcoran AE, Okkenhaug K. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci Signal. 2010;3:ra60. doi: 10.1126/scisignal.2001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol. 2013;13:578–591. doi: 10.1038/nri3487. [DOI] [PubMed] [Google Scholar]
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- Saijo K, Schmedt C, Su IH, Karasuyama H, Lowell CA, Reth M, Adachi T, Patke A, Santana A, Tarakhovsky A. Essential role of Src-family protein tyrosine kinases in NF-kappaB activation during B cell development. Nat Immunol. 2003;4:274–279. doi: 10.1038/ni893. [DOI] [PubMed] [Google Scholar]
- Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–582. doi: 10.1038/324579a0. [DOI] [PubMed] [Google Scholar]
- Sanyal M, Tung JW, Karsunky H, Zeng H, Selleri L, Weissman IL, Herzenberg LA, Cleary ML. B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood. 2007;109:4191–4199. doi: 10.1182/blood-2006-10-054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighoffer E, Vanes L, Mathiot A, Nakamura T, Tybulewicz VL. Unexpected requirement for ZAP-70 in pre-B cell development and allelic exclusion. Immunity. 2003;18:523–533. doi: 10.1016/s1074-7613(03)00082-7. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med. 2013;19:1014–1022. doi: 10.1038/nm.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, Park E, Schuh W, Gruber T, Herzog S, et al. Pre-B cell receptor-mediated cell cycle arrest in Ph+ acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206:1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner JW, Yang WF, Bankhead A, 3rd, Fan G, Fletcher LB, Bryant J, Glover JM, Chang BH, Spurgeon SE, Fleming WH, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li L, Zhang H, Luo X, Dai J, Zhou S, Gu J, Zhu J, Atadja P, Lu C, et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J Biol Chem. 2011;286:38725–38737. doi: 10.1074/jbc.M111.262410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, Jurczak W, Advani RH, Romaguera JE, Williams ME, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369:507–516. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Lu P, Lee FY, Chadburn A, Barrientos JC, Leonard JP, Ye F, Zhang D, Knowles DM, Wang YL. Tyrosine kinase inhibition in diffuse large B-cell lymphoma: molecular basis for antitumor activity and drug resistance of dasatinib. Leukemia. 2008;22:1755–1766. doi: 10.1038/leu.2008.163. [DOI] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2014;12:229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.