Abstract
Adenocarcinoma, a type of non-small-cell lung cancer (NSCLC), is the most frequently diagnosed lung cancer and the leading cause of lung cancer mortality in the United States. It is well documented that biochemical changes occur early in the transition from normal to cancer cells, but the extent to which these alterations affect tumorigenesis in adenocarcinoma remains largely unknown. Herein we describe the application of mass spectrometry and multivariate statistical analysis in one of the largest biomarker research studies to date aimed at distinguishing metabolic differences between malignant and non-malignant lung tissue. Gas chromatography time-of-flight mass spectrometry was used to measure 462 metabolites in 39 malignant and non-malignant lung tissue pairs from current or former smokers with early stage (Stage IA–IB) adenocarcinoma. Statistical mixed effects models, orthogonal partial least squares discriminant analysis and network integration, were used to identify key cancer-associated metabolic perturbations in adenocarcinoma compared to non-malignant tissue. Cancer-associated biochemical alterations were characterized by: 1) decreased glucose levels, consistent with the Warburg effect, 2) changes in cellular redox status highlighted by elevations in cysteine and antioxidants, alpha- and gamma-tocopherol, 3) elevations in nucleotide metabolites 5,6-dihydrouracil and xanthine suggestive of increased dihydropyrimidine dehydrogenase and xanthine oxidoreductase activity, 4) increased 5'-deoxy-5'-methylthioadenosine levels indicative of reduced purine salvage and increased de novo purine synthesis and 5) coordinated elevations in glutamate and UDP-N-acetylglucosamine suggesting increased protein glycosylation. The present study revealed distinct metabolic perturbations associated with early stage lung adenocarcinoma which may provide candidate molecular targets for personalizing therapeutic interventions and treatment efficacy monitoring.
Keywords: lung cancer; metabolomics; biomarkers; 5'-deoxy-5'-methylthioadenosine; 5,6-dihydrouracil
INTRODUCTION
Lung cancer has been the leading cause of cancer death in the United States and worldwide for many decades. Low dose spiral computerized tomography (LDCT) is likely to become the first approved screening and early detection test in the upcoming year but it is plagued by a high false positive rate (1). There is a need to develop complimentary screening and early detection tools. A blood-based “lung cancer” signature is an attractive solution. Given that our knowledge of the molecular biology of smoking-induced lung cancer has dramatically increased over the past few years, this approach is plausible. To date this effort has been focused on the identification of genomic and proteomic signatures with limited success. A broader strategy that incorporates additional cancer traits is needed. It is well-recognized that wide coverage of cellular metabolism in cancer could help provide valuable diagnostic biomarkers and potentially identify molecular drivers of tumorigenesis. Recent advances in mass spectrometry have enabled comprehensive metabolomic analyses of lipids, carbohydrates, amino acids, and nucleotides within a variety of biological matrices. Early evidence from metabolomic investigation of cancer (2) have identified many altered biochemical profiles. However, to date, there have been few investigations of lung cancer, and most studies have looked at blood plasma or were limited by small sample sizes with mixed histologies (3–6).
In the current investigation, gas chromatography time-of-flight mass spectrometry (GC-TOF) was used to measure 462 lipid, carbohydrate, amino acid, organic acid and nucleotide metabolites in 39 malignant and non-malignant lung tissue pairs from current or former smokers with early stage adenocarcinoma. This study cohort represents patient characteristics and tumor histology most likely to be detected with LDCT screening. We hypothesize that identification of cancer induced cellular and tissue level biochemical changes can offer a robust method for identification of candidate circulating biomarkers and improve our understanding of biochemical changes involved in adenocarcinoma tumorigenesis.
MATERIALS AND METHODS
Sample Acquisition
De-identified malignant and adjacent non-malignant lung tissue was obtained from the New York University biorespository. Residual tumor and adjacent non-malignant tissue was harvested from the resected lung after routine pathological protocols were completed, following an approved IRB protocol with patient consent. Two to three tissue pieces were aliquoted into 1.5 ml Nunc vials, and then immediately placed in liquid nitrogen. After transport in liquid nitrogen, each vial was barcoded, and stored at −80°C until analyzed. All specimens were clinically annotated for age, gender, race, histology, smoking status, pack-years and stage of disease. For this clinical study, samples were selected that came from patients who met the following criteria: a) current or former smokers, b) adenocarcinoma histology, c) pathological stage IA or IB, and d) had understood and signed the IRB consent form.
Tissue Sample Preparation
Each tissue sample was weighed to approximately 5mg, and samples were kept frozen while weighing. After weighing, the samples were placed in a 2mL round bottom Eppendorf tube and stored at −20°C. Two small stainless steel grinding balls and 1mL of −20°C extraction solution (3:1 methanol:nano-pure water), degassed by sonication, were added to the samples. The tubes containing lung tissue, grinding balls, and extraction solution were placed in a −80°C freezer for one hour. The samples were then placed in a GenoGrinder 2010 for 5 minutes at 1000 RCF. Next the samples were placed into a −20°C freezer for 30 minutes to precipitate protein. Upon removal from −20°C samples were vortexed for 20 seconds and centrifuged at 16,100 RCF for 10 minutes. The supernatant was transferred to a clean 1.5mL Eppendorf tube, which was immediately centrifuged for 10 minutes at 16,100 RCF. All supernatant was transferred to a new 1.5mL Eppendorf tube and dried to completeness using a Labconco Centrivap.
Samples were derivatized by methoximation followed by silylation for GC-TOF-MS analysis. Once dried 10µL of methoxyamine hydrochloride (Aldrich: Cat. No. 226904) dissolved in pyridine (Acros Organics Cat. No. 270970) (40mg/mL) was added to the samples. Samples were shaken at maximum speed for 1.5 hours at 30°C. 50µL N-Methyl-N-(trimethylsilyl)trifluoroacetamide) (MSTFA) (Aldrich: Cat. No. 394866) spiked with internal standard mixture of fatty-acid methyl esters (FAMEs) was added and samples were shaken at maximum speed for 30 minutes at 37°C. Samples were then placed in the autosampler and 0.5µL of derivatized sample was injected on the GC-TOF.
GC-TOF Data Collection and Analysis
Samples were analyzed using GC-TOF mass spectrometry. The study design was entered into the MiniX database (7). A Gerstel MPS2 automatic liner exchange system (ALEX) was used to eliminate cross-contamination from sample matrix occurring between sample runs. 0.5 microliter of sample was injected at 50°C (ramped to 250°C) in splitless mode with a 25 sec splitless time. An Agilent 6890 gas chromatograph (Santa Clara, CA) was used with a 30 m long, 0.25 mm i.d. Rtx5Sil-MS column with 0.25 µm 5% diphenyl film; an additional 10 m integrated guard column was used (Restek, Bellefonte PA) (8–10). Chromatography was performed at a constant flow of 1 ml/min, ramping the oven temperature from 50°C for to 330°C over 22 min. Mass spectrometry used a Leco Pegasus IV time of flight mass (TOF) spectrometer with 280°C transfer line temperature, electron ionization at −70 V and an ion source temperature of 250°C. Mass spectra were acquired from m/z 85–500 at 17 spectra/sec and 1850 V detector voltage.
Result files were exported to our servers and further processed by our metabolomics BinBase database. All database entries in BinBase were matched against the Fiehn mass spectral library of 1,200 authentic metabolite spectra using retention index and mass spectrum information or the NIST11 commercial library. Identified metabolites were reported if present in at least 50% of the samples per study design group (as defined in the MiniX database); output results were exported to the BinBase database and filtered by multiple parameters to exclude noisy or inconsistent peaks (10). Quantification was reported as peak height using the unique ion as default (11). Missing values were replaced using the raw data netCDF files from the quantification ion traces at the target retention times, subtracting local background noise (7). The unit norm normalization (12) was carried out on a sample specific basis to correct for analytical variance in total tissue mass analyzed. Briefly, sample-wise metabolite intensities were expressed as a ratio to the total ion intensity for all annotated analytes. This is a simple and powerful normalization approach, which in the absence of appropriate analytical surrogates, can account for a variety of analytical sources of variance (e.g. extraction or derivatization), but can also affect biological interpretation (13) and should be evaluated on a study specific basis. Daily quality controls, standard plasma obtained from NIST and evaluation of signal intensities for FAME internal standards were used to monitor instrument performance over the length of the data acquisition.
Data Analysis
Statistical analysis was implemented on log2 transformed metabolite values using mixed effects models to identify differentially-regulated metabolites between adenocarcinoma and normal tissues. Mixed effects models were generated for observed metabolite values given patient age, gender, pack-years of smoking history and cancer status with patient identifiers included as a random factor to account for the correlation of measurements from the same patient. A chi-squared test was used to assess the significance of metabolic differences through comparison of the full model to a reduced model not including a cancer term. The significance levels (i.e. p-values) were adjusted for multiple hypothesis testing according to Benjamini and Hochberg (14) at a false discovery rate (FDR) of 5% (abbreviated pFDR <0.05).
Multivariate Modeling was carried out using orthogonal signal correction partial least squares discriminant analysis (O-PLS-DA) (15) to identify robust predictors of metabolic changes in adenocarcinoma tumor compared to non-malignant lung tissue. O-PLS-DA modeling was conducted on covariate adjusted (gender, age and packs of cigarettes smoked per year), log2 transformed and autoscaled data. The 39 patients’, tumor and control tissue pairs, were split between 2/3 training and 1/3 test data sets. The training set was used to carry out feature selection and model optimization, and the final model performance was determined by predicting the class labels (tumor or control) for the held out test set. Model latent variable (LV) number and orthogonal LV (OLV) number was selected using leave-one-out cross-validation. A preliminary 2 OLV (2 total LV) model was developed and used to carry out feature selection. Feature selection was implemented to identify the top ~10% (42 out of 462) of all metabolic predictors for cancer. The full variable set was filtered to retain metabolites which displayed significant correlation with model scores (Spearman’s pFDR ≤ 0.05) (16) and model loadings on OLV1 in the top 90th quantile in magnitude (17).
The top 10% feature model was evaluated using Monte Carlo training and testing cross-validation and permutation testing (18). Internal training and testing was done by further splitting the training set into 2/3 pseudo-training and 1/3 pseudo-test sets, while preserving individual patients’ tumor and control tissue pairs. This was randomly repeated 100 times and used to estimate the distributions for the O-PLS-DA model performance statistics: the model fit to the training data (Q2) and root mean squared error of prediction for the test data (RMSEP). The probability of achieving the model's predictive performance was estimated through comparison of Q2 and RMSEP distributions to 100 randomly permuted models (random class labels), calculated by replicating the internal training and testing procedures described above. The described approach was also used to determine model performance for the excluded (bottom 90%) feature set (n=420).
Optimized model classification performance was validated through prediction of class labels for the originally held out test set, and are reported as sensitivity, specificity and the area under the receiver operator characteristic curve.
Network analysis was used to investigate statistical and multivariate modeling results within a biochemical context and to help estimate functional roles for structurally unknown metabolites. A biochemical and chemical similarity network (19) was developed for all measured metabolites with KEGG (20) and PubChem CIDs (21) identifiers (n = 178). Enzymatic interactions were determined based on product-precursor relationships defined in the KEGG RPAIR database. Molecules not directly participating in biochemical transformations, but sharing many structural properties, based on PubChem Substructure Fingerprints (22), were connected at a threshold of Tanimoto similarity ≥ 0.7.
A partial correlation network was calculated to analyze empirical dependencies among metabolic discriminants between adenocarcinoma tumor and non-malignant lung tissues. Partial correlations were calculated between covariate adjusted data (gender, age and packs of cigarettes smoked per year) for all structurally identified O-PLS-DA selected features (top 10% feature set). Metabolite relationships were determined based on significant FDR adjusted (14) partial correlations (pFDR ≤ 0.05).
A mass spectral similarity and partial correlation network was used to estimate relationships between covariate adjusted annotated and structurally unknown O-PLS-DA selected features. Mass spectral similarities were calculated based on cosine correlations ≥0.75 of annotated and unknown metabolites electron ionization mass spectral profiles (23). Partial correlations were calculated between all known and unknown species and limited two connections per unknown (pFDR≤0.05).
Network mapping was used in Cytoscape (24) to encode statistical and multivariate modeling results through network edge and node attributes.
RESULTS
Paired tissue samples were obtained from 39 patients with adenocarcinoma histology (Table 1). The majority of patients were elderly white female former smokers. The average age was 72 with a mean of 36 pack years; all patients were diagnosed with Stage IA or IB disease.
Table 1.
Patient characteristics.
| Variable | Lung Cancer patients |
|---|---|
| Total sample size, N | 39 |
| Age, mean (± SD) | 72.33 (± 8.78) |
| Pack/Year, mean (± SD) | 35.91 (± 26.05) |
| Female, No. (%) | 24 (61.54%) |
| Current Smoker, No. (%) | 5 (12.82%) |
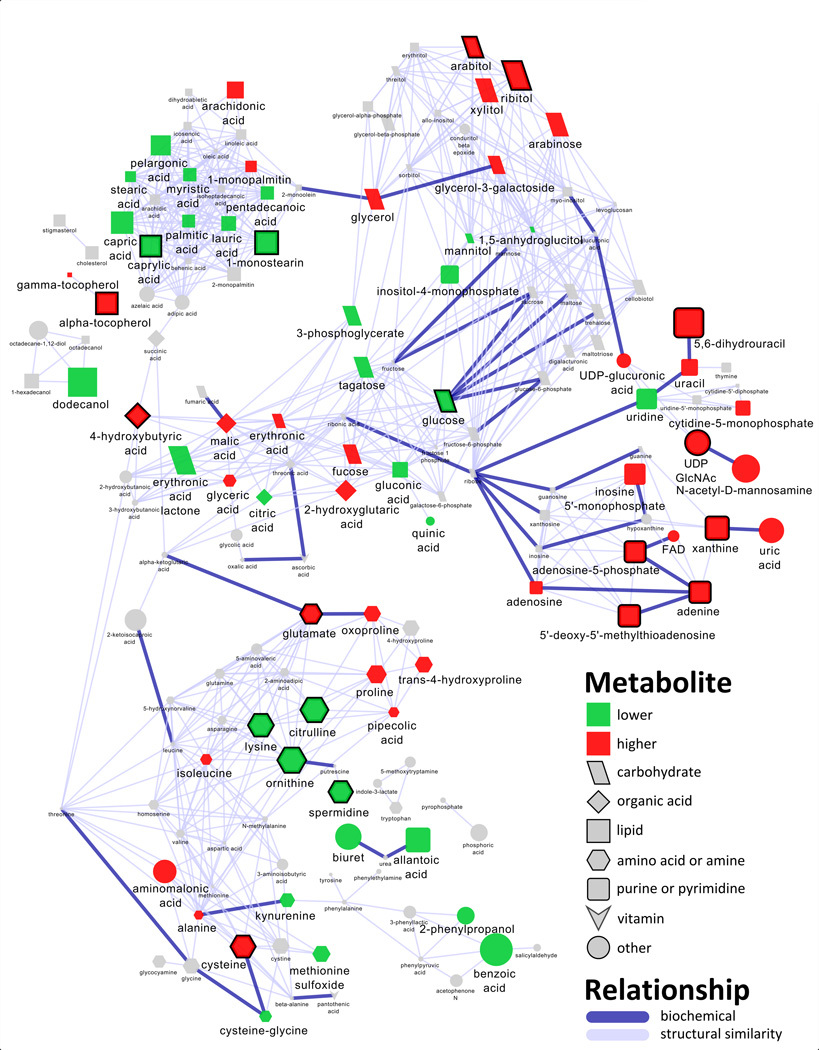
Metabolomic profiling was performed after extraction and derivatization using gas chromatography time-of-flight (GC-TOF) mass spectrometry. A total of 462 compounds were measured, and 183 of these were annotated with known molecular structures. Additional metabolite information: retention time, mass spectra, etc. and the proportion and percentage of patient-matched comparisons in which metabolite levels were increased in tumors compared to normal tissue are reported in Table S4. A large number of differences were found between normal and malignant tissue. Mixed effects models were used to identify 70 significantly different metabolites between non-malignant and adenocarcinoma tissues after adjusting for the false discovery rate (pFDR < 0.05) (Table 2). The compounds were equally divided, with 35 increasing and 35 decreasing in tumor, compared with control tissue. A metabolomic network was calculated to display enzymatic transformations and structural similarities among the 183 structurally identified compounds in the context of their relative changes between adenocarcinoma tumor and non-malignant tissue (Figure 1). In addition to the classical statistical approach, O-PLS-DA multivariate classification modeling was used to select the top 10% multivariate discriminants between cancer and control tissues. Monte Carlo cross-validation and permutation testing were used to validate the models predictive performance for classification of cancer vs. control tissues (Table S3). The top metabolic changes between tumor and control tissues were comprised of 20 annotated (Table 2) and 22 structurally unknown metabolites (Table S2). Prediction of cancer or control class labels for an originally held out test set (1/3 of the data) was used to confirm that an O-PLS-DA model calculated from the selected features (top 10%) displayed improved predictive performance (area under the curve (AUC) 88.5%; sensitivity, 92.3%; specificity, 84.6%) compared to a model constructed from all excluded features (bottom 90%) (AUC, 80.77%; sensitivity, 76.92%; specificity, 84.62%) (Supplemental Table S3) (see methods for details of calculations).
Table 2.
Significantly altered metabolites and key discriminants of biochemical changes in adenocarcinoma compared to non-malignant tissue.
| Name | Fold changea |
Directionb | pFDRc | Rankd |
|---|---|---|---|---|
| Amino Acids and Derivatives | ||||
| ornithine | 0.5 | DOWN | 9.61E-06 | 2 |
| citrulline | 0.7 | DOWN | 0.003944 | 4 |
| lysine | 0.4 | DOWN | 6.49E-05 | 5 |
| cysteine | 1.6 | UP | 0.002516 | 6 |
| spermidine | 0.7 | DOWN | 0.001625 | 8 |
| glutamate | 1.4 | UP | 0.001694 | 12 |
| trans-4-hydroxyproline | 1.3 | UP | 0.007531 | |
| proline | 1.4 | UP | 0.008061 | |
| methionine sulfoxide | 0.6 | DOWN | 0.045096 | |
| isothreonic acid | 1.3 | UP | 0.007637 | |
| glyceric acid | 1.4 | UP | 0.017117 | |
| alanine | 1.2 | UP | 0.013497 | |
| histidine | 0.6 | DOWN | 0.012104 | |
| Organic Acids | ||||
| 4-hydroxybutyric acid | 1.3 | UP | 0.004333 | 9 |
| malic acid | 1.3 | UP | 0.045285 | |
| citric acid | 0.6 | DOWN | 0.007308 | |
| 2-hydroxyglutaric acid | 1.7 | UP | 0.003292 | |
| Carbohydrates and Related Compounds | ||||
| ribitol | 1.8 | UP | 6.64E-07 | 3 |
| glucose | 0.5 | DOWN | 0.000366 | 16 |
| Fucose + rhamnose | 2.1 | UP | 0.000177 | 18 |
| arabitol | 1.4 | UP | 0.003463 | 20 |
| xylitol | 1.4 | UP | 0.000939 | |
| tagatose | 0.8 | DOWN | 0.022467 | |
| mannitol | 0.7 | DOWN | 0.016961 | |
| glycerol-3-galactoside | 2.2 | UP | 0.003224 | |
| glycerol | 1.2 | UP | 0.018636 | |
| fucose | 1.5 | UP | 0.007527 | |
| erythronic acid lactone | 0.6 | DOWN | 0.006205 | |
| erythronic acid | 1.2 | UP | 0.048138 | |
| arabinose | 1.3 | UP | 0.000454 | |
| 1,5-anhydroglucitol | 0.8 | DOWN | 0.037117 | |
| Monoglycerols | ||||
| 1-monopalmitin | 1.1 | UP | 0.047396 | |
| 1-monostearin | 0.8 | DOWN | 0.047268 | 11 |
| Tocopherols | ||||
| alpha-tocopherol | 2.2 | UP | 4.12E-07 | 13 |
| gamma-tocopherol | 1.2 | UP | 0.010335 | |
| Fatty Acids | ||||
| arachidonic acid | 1.5 | UP | 0.006889 | |
| capric acid | 0.8 | DOWN | 0.004333 | |
| caprylic acid | 0.8 | DOWN | 0.001644 | 19 |
| lauric acid | 0.9 | DOWN | 0.034948 | |
| palmitic acid | 0.9 | DOWN | 0.027453 | |
| pelargonic acid | 0.8 | DOWN | 0.033083 | |
| pentadecanoic acid | 0.8 | DOWN | 0.014977 | |
| stearic acid | 0.8 | DOWN | 0.047396 | |
| Miscellaneous Lipids | ||||
| dihydrosphingosine | 0.6 | DOWN | 0.000544 | |
| dodecanol | 0.8 | DOWN | 0.00205 | |
| gluconic acid | 0.5 | DOWN | 0.000381 | |
| triethanolamine | 0.7 | DOWN | 0.002243 | |
| 3-phosphoglycerate | 0.4 | DOWN | 0.001049 | |
| Purines and Pyrimidines | ||||
| 5,6-dihydrouracil | 2.4 | UP | 2.42E-07 | 1 |
| xanthine | 2.7 | UP | 0.000381 | 10 |
| 5'-deoxy-5'-methylthioadenosine | 1.8 | UP | 6.49E-05 | 14 |
| adenine | 1.5 | UP | 0.000366 | 15 |
| adenosine-5-phosphate | 2.5 | UP | 0.00132 | 17 |
| uridine | 0.7 | DOWN | 0.002044 | |
| uracil | 1.4 | UP | 0.036508 | |
| inositol-4-monophosphate | 0.7 | DOWN | 0.020452 | |
| inosine 5'-monophosphate | 3.5 | UP | 0.00085 | |
| cytidine-5-monophosphate | 1.8 | UP | 0.001397 | |
| allantoic acid | 0.6 | DOWN | 0.005247 | |
| Miscellaneous Metabolites | ||||
| UDP GlcNAc | 2.3 | UP | 2.24E-05 | 7 |
| uric acid | 1.4 | UP | 0.019398 | |
| UDP-glucuronic acid | 1.2 | UP | 0.00667 | |
| quinic acid | 0.8 | DOWN | 0.020487 | |
| nicotinamide | 1.4 | UP | 0.006078 | |
| N-acetyl-D-mannosamine | 1.5 | UP | 0.000333 | |
| hydroxylamine | 0.8 | DOWN | 0.013145 | |
| creatinine | 1.4 | UP | 0.01111 | |
| biuret | 0.6 | DOWN | 0.002136 | |
| benzoic acid | 0.9 | DOWN | 0.038501 | |
| aminomalonic acid | 1.3 | UP | 0.030249 | |
ratio of means relative to control
direction of change in means relative to control
false discovery rate adjusted mixed effects model p-value
importance of metabolic change based on O-PLS-DA model loading
Figure 1.
Metabolomic network of biochemical differences between adenocarcinoma and non-malignant lung tissue. Edge color and width denote the type (enzymatic, purple; structural similarity, gray) and strength of relationships between metabolites. Node color displays significance (mixed effects model, pFDR ≤ 0.05) and direction of the change in tumor relative to non-malignant tissue (green, decrease; red, increase; gray, insignificant change) (Table 2 and Table S1). Node size displays O-PLS-DA loadings (empirical importance), and thick borders indicate O-PLS-DA selected discriminants for adenocarcinoma (Table 2). See metabolite O-PLS-DA model loading in Table S4 for quantitative differences in metabolites and corresponding node sizes. Node shape denotes the biochemical super class of each molecule.
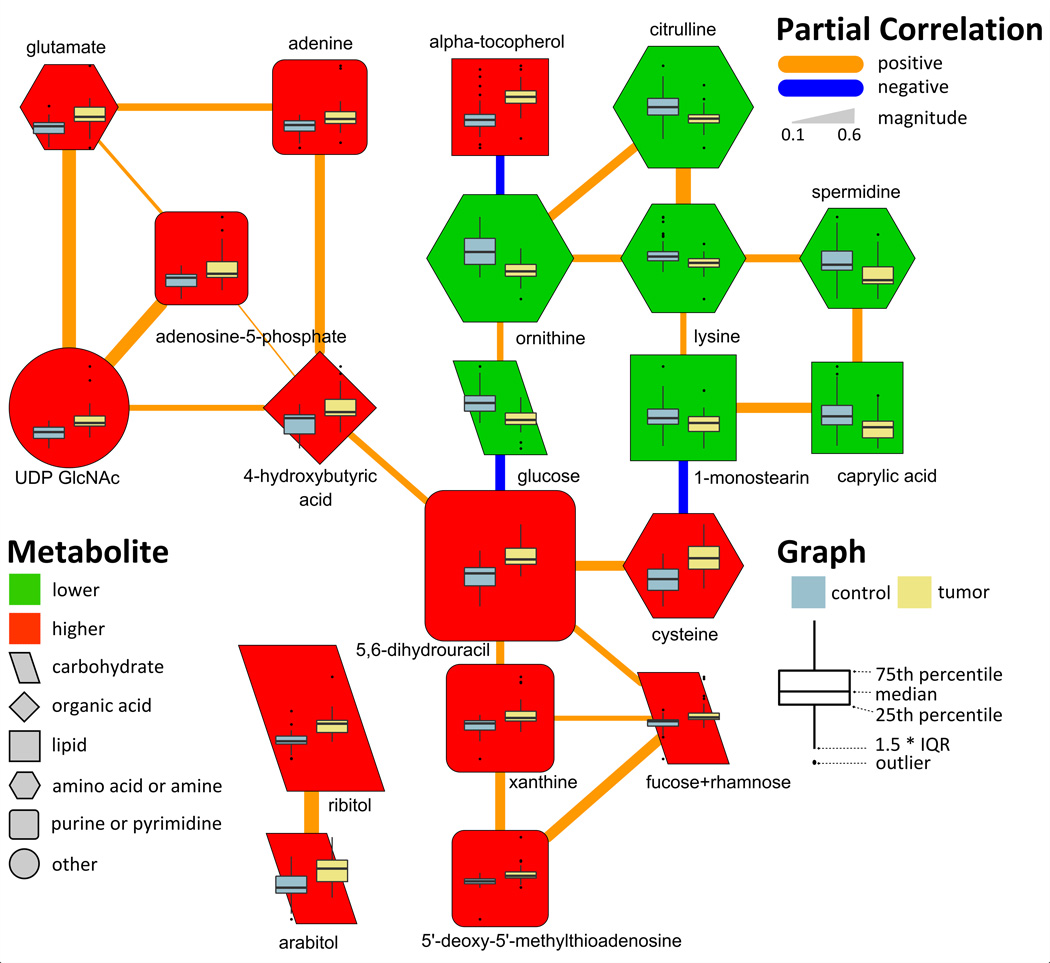
Partial correlations and combined with mass spectral similarity networks were used to analyze empirical relationships among all annotated and structurally unknown O-PLS-DA selected features (Figure S1 and Table S2).
Polyamine pathway-related compounds were altered in tumor compared to non-malignant tissue. 5'-deoxy-5'-methylthioadenosine (MTA) was elevated 1.8-fold in cancer compared with normal (pFDR = 6.27 × 10−5) tissue, and this change was significantly correlated with 2.1- and 2.7-fold increases in the carbohydrates fucose/rhamnose and nucleotide xanthine, respectively (Figure 2 and Table 2). The polyamine spermidine, and related urea cycle intermediates, ornithine and citrulline, showed concerted decreases in cancer compared to control tissue (Figure 2), while another polyamine, putrescine was unchanged (Table S1). Additionally decreases in four other structurally unknown metabolites were also significantly correlated with changes in spermidine and ornithine (Figure S1).
Figure 2.
Partial correlation network displaying conditionally independent relationships between O-PLS-DA selected discriminants for adenocarcinoma (Table 2). Edge color and width denote the direction and magnitude of partial correlations (pFDR≤0.05). Node color displays the direction of the change in tumor relative to non-malignant tissue (green, decrease; red, increase; pFDR ≤ 0.05). Node size displays the metabolite loading (empirical importance) in the O-PLS-DA model and shape denotes the biochemical super class of each molecule. See metabolite O-PLS-DA model loading in Table S4 for quantitative differences in metabolites and corresponding node sizes. Node inset boxplots summarize differences in z-scaled measurements between tumor and non-malignant tissue.
Many compounds associated with purine and pyrimidine biosynthesis were significantly increased in tumor compared to control tissue (Table 2). Of these metabolites, 5,6-dihydrouracil was significantly elevated by 2.4-fold in cancer compared to control tissue (Table 2), and constituted the single best multivariate predictor for cancer. The cancer-dependent increase in, 5,6-dihydrouracil, an oxidation product of uracil was also positively correlated with similar changes in MTA, xanthine and 4-hydroxybutyric acid (Figure 2).
Carbohydrates showed variable changes in cancer compared to non-malignant tissue (Table 2). Glucose was significantly reduced by 0.5-fold in cancer compared to control tissue (Table 2), and this change was positively correlated with a decrease in ornithine and the increase in 5,6-dihydrouracil (Figure 2). Conversely, ribitol and arabitol showed correlated elevations in cancer compared to control tissue which were unrelated other O-PLS-DA selected discriminants for cancer.
There were significant differences in lipid profiles between cancer and non-malignant tissues (Table 2). Particularly striking was that the majority of fatty acids were all significantly decreased in cancer relative to non-malignant tissue (Figure 1 and Table 2) with the exception of arachidonic acid which was elevated by 1.5-fold. The correlated decreases in caprylic acid and 1-monostearin (Table 2) were also positively correlated with reductions in lysine and spermidine, and (through cysteine) negatively correlated with the elevation in 5,6-dihydrouracil (Figure 2). Both of the vitamin E-related compounds, α- and γ-tocopherol, were significantly elevated in adenocarcinoma (Table 2). The 2.2-fold increase in α-tocopherol was also correlated with the decrease in ornithine (Figure 2).
Similar to carbohydrates, amino acids showed variable changes in cancer (Table 2). In contrast to the noted decreases ornithine, citrulline and lysine, glutamate was significantly elevated by 1.4-fold in cancer compared to control tissue (Table 2). The increase in glutamate was positively correlated with increases in adenine, adenosine-5-phosphate (AMP) and uridine diphosphate N-acetylglucosamine (UDP GlcNAc) (Figure 2).
DISCUSSION
We report one of the largest clinical studies of lung cancer tissue to date to demonstrate a differential metabolomic signature between patient-matched lung adenocarcinoma and non-malignant tissues. One limitation of this study, and others like it, is the potential for tissue microheterogeneity at the sub-biopsy level. Previously, Hori and coauthors evaluated seven lung cancer and non-cancer tissues, of which 4 were adenocarcinoma and GC-MS analysis was used to identify significant changes in 48 metabolites (5). More recently, Kami and colleagues used capillary electrophoresis time-of-flight MS to evaluate 9 tumor and normal lung tissue pairs; three adenocarcinoma samples were evaluated with general increases in amino acids (n=19) in lung tumors compared to normal tissues (25). However these studies were not designed to specifically investigate biochemical perturbations in early stage adenocarcinoma and due to the pathological heterogeneity of the samples in the earlier studies, a detailed comparison to the current investigation is not possible. Here we present the most comprehensive analysis to date of metabolic differences between early stage adenocarcinoma and non-malignant tissue.
Compared to non-malignant tissue, adenocarcinoma displayed significant elevations in ribitol, arabitol, and fucose/rhamnose and a reduction in glucose (Table 2 and Figures 1&2). The observed 2-fold reduction in glucose in adenocarcinoma relative to normal tissue is consistent with the Warburg effect, wherein a high rate of aerobic glycolysis is linked to cytosolic lactic acid fermentation, rather than mitochondrial pyruvate oxidation (26). While glucose was reduced, other members of the glucuronate and pentose interconversion pathway (KEGG (20)), arabitol, ribitol, UDP-GlcNAc and xylitol, all showed significant elevations in cancer compared to non-malignant tissue (Table 2). These observations suggest that compared to control, adenocarcinoma displays increased pentose phosphate metabolism and an elevated glucuronidation status. The pentose phosphate pathway is involved in nucleotide synthesis for DNA replication and is used to provide reducing equivalents for a variety cellular reactions (27).
We observed a direct correlation between the noted reduction in glucose and a 2.4-fold elevation in 5,6-dihydrouracil, which constituted the single best discriminant of adenocarcinoma compared to non-malignant tissue (Table 2). 5,6-dihydrouracil is an oxidation product of the nucleotide uracil, and may provide a stable marker of altered nucleotide metabolism in adenocarcinoma compared to non-malignant tissue. In humans, catabolism of uracil to 5,6-dihydrouracil is mediated by dihydropyrimidine dehydrogenase (DPD). Both DPD activity and expression have been shown to be increased in lung adenocarcinoma compared to control tissue (28). The degree of patients’ DPD activity has also been related to improved efficacy of cytotoxic effects from common postoperative adjuvant therapy for non-small cell lung cancer (NSCLC) anti-cancer drug, 5-fluorouracil and its derivatives (28, 29). Given our data and the supporting literature, we hypothesize that monitoring the ratio between 5,6-dihydrouracil and uracil in patients with NSCLC may provide a personalized diagnostic to identify cohorts of patients with high DPD activity who may particularly benefit from anti-cancer therapy with DPD inhibitors.
The cancer-dependent elevation in 5,6-dihydrouracil was also positively correlated with an increase in xanthine (Figure 2). The degradation of hypoxanthine to xanthine followed by the conversion of xanthine to uric acid, also elevated in adenocarcinoma (Table 2), is mediated by xanthine oxidoreductase (XOR) (30). XOR, a key enzyme in the metabolism of purine nucleotides, has also been linked to the production of reactive oxygen species (ROS) (30, 31). Assessment of XOR activity has been suggested as a diagnostic for NSCLC (32). In particular, reduced XOR expression was associated with shortened survival times (30, 32), while XOR-induced ROS products have been linked with an increased risk for developing various forms of cancer, including NSCLC (30, 33–35). Given this evidence, assessment of xanthine and uric acid levels may provide novel indicators of XOR activity and serve as diagnostic marker of tumorigenesis.
Cancer dependent elevations in cysteine (Table 2) were positively correlated with changes in 5,6-dihydrouracil (Figure 2). This observation may reflect an increase in glutathione synthesis in response to elevated ROS production in cancer compared to control tissue. Previously, Krepela and coauthors identified significant elevations of cysteine in squamous cell lung tumor compared to non-involved tumor tissue (36). Cysteine, an important precursor in glutathione production (37), has been shown to be elevated in various types of cancer including: breast, ovarian, head and neck, brain and lung cancer (37). Consistent with the evidence of increased oxidative stress in adenocarcinoma, we also observed cancer-associated elevations in the vitamin E isoforms α-tocopherol and γ-tocopherol (Table 2). These well-known anti-oxidants have been extensively studied as chemopreventive agents; however, the relationship between serum levels of tocopherols and lung cancer have yielded conflicting results (38). To date there appears to be little, if any, data on tocopherol levels in solid tumors or cancer cells. However their observed elevation in adenocarcinoma compared to non-malignant tissue is in line with other observations supporting an increase in compensatory mechanisms to deal with oxidative stress in tumors. Furthermore, the CARET study suggested that antioxidant supplementation does not help in preventing lung cancer (39).
Related to the previously noted increases in nucleotide metabolites (Figure 2), and directly correlated with changes in xanthine (Figure 2), 5'-deoxy-5'-methylthioadenosine (MTA) was 1.8-fold elevated in cancer compared with non-tumorous tissue (Table 2). MTA, through the action of 5'-deoxy-5'-methylthioadenosine phosphorylase (MTAP), is involved in S-adenosylmethionine (AdoMet) salvage, purine salvage and spermidine synthesis (40). MTAP activity has been shown to be reduced in a wide variety of tumor types including NSCLC (41). Tumors with reduced in MTAP expression or activity, instead of purine salvage (42), are highly dependent on de novo purine synthesis for production of DNA, RNA and purine containing energy molecules (e.g. ATP). Consequently, the use of de novo purine synthesis inhibitors as anti-cancer therapy for MTAP-deficient tumors (42) has gained considerable attention. The observed increase in MTA in lung adenocarcinoma suggests a decrease in MTAP-dependent purine salvage and an increased reliance on de novo purine synthesis. This is further supported by the observed 3.5-fold increase in inosine-5-monophosphate (IMP), an important component in de novo purine synthesis (43), in malignant compared to control tissue (Table 2). In addition to de novo purine synthesis, MTA is also involved in polyamine synthesis.
Two polyamine-related metabolites, ornithine and spermidine, were reduced in cancer compared to control tissue (Table 2). This is somewhat surprising given previous reports of increased polyamines in various types of cancers including breast (44) and colorectal cancer (45). However, it has also been previously shown that increased levels of MTA can inhibit MTAP activity, and lead to decreased levels of polyamines in non-small cell lung carcinoma (40). In addition to the previously-noted evidence for decreased MTAP activity, cancer-associated reduction in ornithine may also contribute to the observed decrease in spermidine in adenocarcinoma compared to non-malignant tissue (Table 2). Ornithine, an important intermediate in nitrogen disposal through conversion to citrulline, is involved in proline synthesis (46). Compared to normal tissue, adenocarcinoma displayed a significant reduction in citrulline and an elevation in proline. This evidence supports the hypothesis that in adenocarcinoma, ornithine may be diverted away from citrulline and spermidine synthesis toward proline production.
Adenine, which can be produced from the cleavage of MTA by MTAP, and its metabolite adenosine-5-monophosphate (AMP) were increased in adenocarcinoma compared to control tissue (Figure 2 and Table 2). Given previous evidence of reduced MTAP activity, we expect that adenine originates from plasma or surrounding tissues (47). Furthermore, we did not see a direct correlation between MTA and adenosine levels (Figure 2), but instead both adenine and AMP were significantly correlated with a 1.4-fold cancer-dependent elevation in glutamate (Table 2 and Figure 2). During de novo purine synthesis, glutamine (unchanged, Table S1) acts as an amido donor through phosphoribosyl pyrophosphate (PRPP) to produce ribosylamine-5-phosphate and glutamate (48). PRPP can participate in both de novo purine synthesis or salvage pathways and lead to production of IMP and AMP (48). While we observed a significant positive correlation between glutamate and AMP (Figure 2), both glutamate and AMP showed a far stronger relationship with UDP-N-acetylglucosamine (UDP-GlcNAc) (Figure 2).
Known cancer related perturbations in hexosamine biosynthesis (49) may explain the observed correlation between glutamate and a 2.3-fold increase in UDP-GlcNAc in adenocarcinoma compared with non-malignant tissue (Table 2 and Figure 2). Glutamate is a byproduct UDP-GlcNAc synthesis, which can be converted to N-acetyl mannosamine, which was also increased by 1.5-fold in adenocarcinoma (Table 2). Previous investigations have suggested that increases in protein glycosylation with GlcNAc are common aspects of cancer cells and tumors (50). O-GlcNAc protein glycosylation is suggested to be a protective response, and increase the tolerance of cells to a variety of sources of stress (49). If the increase in UDP-GlcNAc is a marker of increased protein glycosylation in cancer, then monitoring the levels of this molecule may provide a marker of changes in cellular protein function in adenocarcinoma compared with non-malignant tissue. For example, the activity of protein kinase C (PKC), implicated in tumorigenesis, has been shown to be up-regulated by increased flux through the hexosamine biosynthetic pathway (51). The PKC-alpha isoform is highly expressed in NSCLC and preferentially increased in adenocarcinoma compared with squamous cell carcinoma (52). Together, these observations suggest the need to combine proteomic, glycomic and metabolomic analyses in order to fully understand molecular mechanisms of protein glycosylation in tumorigenesis.
Lastly, we also observed adenocarcinoma-dependent elevations in 2-hydroxyglutarate. 2-hydroxyglutarate has recently been found to be an “oncometabolite”, resulting from mutation of the enzyme isocitrate dehydrogenase 1 and 2 (IDH1, IDH2) in cancer cells, modifying its catalytic function to produce 2-hydroxyglutarate from the substrate isocitrate instead of the normal alpha-ketoglutarate product (53, 54). Increased levels of 2-hydroxyglutarate have been found in leukemia (55), glioma (56), thyroid carcinoma (57). The finding of elevated 2-hydroxyglutarate suggests that lung adenocarcinoma tissue should be examined for IDH1 and IDH2 mutations, a question that we plan to investigate in future studies.
The observed perturbations of molecular mechanisms and biochemical pathways in adenocarcinoma compared to non-malignant tissue were consistent with known cancer-dependent increases in energy utilization and proliferation. The current investigation identified new biochemical pathways altered in adenocarcinoma, which may aid the development of diagnostic markers for cancer screening, early detection and treatment efficacy monitoring as shown by the validated performance of the multivariate classification model (AUC, 88.5%; sensitivity, 92.3%; specificity, 84.6%). This study suggests that measuring the ratios of 5,6-dihydrouracil/uracil and xanthine/uric acid may serve as valuable new biomarkers for tumorigenesis in lung cancer.
Supplementary Material
Acknowledgments
Financial Support: We acknowledge the following grants, W81XWH-10-1-0635 (Gandara), LUNGevity Foundation 201118739 (Miyamoto), NIH 1 U24 DK097154 NIH West Coast Metabolomics Center award (Fiehn), Tobacco Related Disease Research Program 20PT0034 (Kelly).
Footnotes
Conflict-of-Interest: The authors declare no conflict-of-interest.
REFERENCES CITED
- 1.National Lung Screening Trial Research T. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboud OA, Weiss RH. New opportunities from the cancer metabolome. Clinical chemistry. 2013;59:138–146. doi: 10.1373/clinchem.2012.184598. [DOI] [PubMed] [Google Scholar]
- 3.Lokhov PG, Trifonova OP, Maslov DL, Archakov AI. Blood plasma metabolites and the risk of developing lung cancer in Russia. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation. 2013;22:335–341. doi: 10.1097/CEJ.0b013e32835b3898. [DOI] [PubMed] [Google Scholar]
- 4.Rocha CM, Carrola J, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. Journal of proteome research. 2011;10:4314–4324. doi: 10.1021/pr200550p. [DOI] [PubMed] [Google Scholar]
- 5.Hori S, Nishiumi S, Kobayashi K, Shinohara M, Hatakeyama Y, Kotani Y, et al. A metabolomic approach to lung cancer. Lung cancer. 2011;74:284–292. doi: 10.1016/j.lungcan.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ, et al. Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Molecular bioSystems. 2013;9:2370–2378. doi: 10.1039/c3mb70138g. [DOI] [PubMed] [Google Scholar]
- 7.Scholz M, Fiehn O. SetupX--a public study design database for metabolomic projects. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2007:169–180. [PubMed] [Google Scholar]
- 8.Weckwerth W, Wenzel K, Fiehn O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics. 2004;4:78–83. doi: 10.1002/pmic.200200500. [DOI] [PubMed] [Google Scholar]
- 9.Fiehn O. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends in analytical chemistry : TRAC. 2008;27:261–269. doi: 10.1016/j.trac.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Analytical biochemistry. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Lee do Y, Fiehn O. High quality metabolomic data for Chlamydomonas reinhardtii. Plant methods. 2008;4:7. doi: 10.1186/1746-4811-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholz M, Gatzek S, Sterling A, Fiehn O, Selbig J. Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics. 2004;20:2447–2454. doi: 10.1093/bioinformatics/bth270. [DOI] [PubMed] [Google Scholar]
- 13.Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards . BMC bioinformatics. 2007;8:93. doi: 10.1186/1471-2105-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 15.Svensson OKT, MacGregor JF. An investigation of orthogonal signal correction algorithms and their characteristics. J Chemom. 2002;16:176–188. [Google Scholar]
- 16.Wiklund S, Johansson E, Sjostrom L, Mellerowicz EJ, Edlund U, Shockcor JP, et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical chemistry. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 17.Palermo G, Piraino P, Zucht HD. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. Advances and applications in bioinformatics and chemistry : AABC. 2009;2:57–70. doi: 10.2147/aabc.s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grapov D, Fahrmann J, Hwang J, Poudel A, Jo J, Periwal V, et al. Diabetes associated metabolomic perturbations in NOD mice. Metabolomics : Official journal of the Metabolomic Society. 2014:1–13. doi: 10.1007/s11306-014-0706-2. 2014/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barupal DK, Haldiya PK, Wohlgemuth G, Kind T, Kothari SL, Pinkerton KE, et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolton E, Wang Y, Thiessen PA, SH B. Annual Reports in Computational Chemistry. Washington, DC: American Chemical Society; 2008. PubChem: Integrated Platform of Small Molecules and Biological Activities. [Google Scholar]
- 22.Cao Y, Charisi A, Cheng LC, Jiang T, Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kami K, Fujimori T, Sato H, Sato M, Yamamoto H, Ohashi Y, et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics : Official journal of the Metabolomic Society. 2013;9:444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature reviews Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 27.Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein & cell. 2014;5:592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi T, Kondo K, Fujino H, Takahashi Y, Sawada N, Sakiyama S, et al. Thymidylate synthase and dihydropyrimidine dehydrogenase in non-small cell lung cancer: relationship between mRNA expression and activity. Anticancer research. 2005;25:923–930. [PubMed] [Google Scholar]
- 29.Shintani Y, Inoue M, Funakoshi Y, Matsumura A, Ohta M, Maeda H, et al. Low dihydropyrimidine dehydrogenase correlates with prolonged survival in patients with lung adenocarcinoma treated with 5-fluorouracil. Anticancer research. 2011;31:4665–4671. [PubMed] [Google Scholar]
- 30.Konno H, Minamiya Y, Saito H, Imai K, Kawaharada Y, Motoyama S, et al. Acquired xanthine dehydrogenase expression shortens survival in patients with resected adenocarcinoma of lung. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:1727–1732. doi: 10.1007/s13277-012-0431-2. [DOI] [PubMed] [Google Scholar]
- 31.Boueiz A, Damarla M, Hassoun PM. Xanthine oxidoreductase in respiratory and cardiovascular disorders. American journal of physiology Lung cellular and molecular physiology. 2008;294:L830–L840. doi: 10.1152/ajplung.00007.2008. [DOI] [PubMed] [Google Scholar]
- 32.Kim AW, Batus M, Myint R, Fidler MJ, Basu S, Bonomi P, et al. Prognostic value of xanthine oxidoreductase expression in patients with non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands) 2011;71:186–190. doi: 10.1016/j.lungcan.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Kaynar H, Meral M, Turhan H, Keles M, Celik G, Akcay F. Glutathione peroxidase, glutathione-S-transferase, catalase, xanthine oxidase, Cu-Zn superoxide dismutase activities, total glutathione, nitric oxide, and malondialdehyde levels in erythrocytes of patients with small cell and non-small cell lung cancer. Cancer letters. 2005;227:133–139. doi: 10.1016/j.canlet.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Tsao SM, Yin MC, Liu WH. Oxidant stress and B vitamins status in patients with non-small cell lung cancer. Nutrition and cancer. 2007;59:8–13. doi: 10.1080/01635580701365043. [DOI] [PubMed] [Google Scholar]
- 35.Wright RM, McManaman JL, Repine JE. Alcohol-induced breast cancer: a proposed mechanism. Free radical biology & medicine. 1999;26:348–354. doi: 10.1016/s0891-5849(98)00204-4. [DOI] [PubMed] [Google Scholar]
- 36.Krepela E, Prochazka J, Karova B, Cermak J, Roubkova H. Cathepsin B, thiols and cysteine protease inhibitors in squamous-cell lung cancer. Neoplasma. 1997;44:219–239. [PubMed] [Google Scholar]
- 37.Gamcsik MP, Kasibhatla MS, Teeter SD, Colvin OM. Glutathione levels in human tumors. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2012;17:671–691. doi: 10.3109/1354750X.2012.715672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodson K, Tangrea JA, Barrett MJ, Virtamo J, Taylor PR, Albanes D. Serum alpha-tocopherol and subsequent risk of lung cancer among male smokers. Journal of the National Cancer Institute. 1999;91:1738–1743. doi: 10.1093/jnci/91.20.1738. [DOI] [PubMed] [Google Scholar]
- 39.Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL, Jr, Omenn GS, et al. The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. Journal of the National Cancer Institute. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 40.Basu I, Locker J, Cassera MB, Belbin TJ, Merino EF, Dong X, et al. Growth and metastases of human lung cancer are inhibited in mouse xenografts by a transition state analogue of 5'-methylthioadenosine phosphorylase. The Journal of biological chemistry. 2011;286:4902–4911. doi: 10.1074/jbc.M110.198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe F, Takao M, Inoue K, Nishioka J, Nobori T, Shiraishi T, et al. Immunohistochemical diagnosis of methylthioadenosine phosphorylase (MTAP) deficiency in non-small cell lung carcinoma. Lung cancer (Amsterdam, Netherlands) 2009;63:39–44. doi: 10.1016/j.lungcan.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Kindler HL, Burris HA, 3rd, Sandler AB, Oliff IA. A phase II multicenter study of L-alanosine, a potent inhibitor of adenine biosynthesis, in patients with MTAP-deficient cancer. Investigational new drugs. 2009;27:75–81. doi: 10.1007/s10637-008-9160-1. [DOI] [PubMed] [Google Scholar]
- 43.Kelley WN, Holmes EW, Van der Weyden MB. Current concepts on the regulation of purine biosynthesis de novo in man. Arthritis and rheumatism. 1975;18:673–680. doi: 10.1002/art.1780180706. [DOI] [PubMed] [Google Scholar]
- 44.Kingsnorth AN, Wallace HM, Bundred NJ, Dixon JM. Polyamines in breast cancer. The British journal of surgery. 1984;71:352–356. doi: 10.1002/bjs.1800710513. [DOI] [PubMed] [Google Scholar]
- 45.Kingsnorth AN, Lumsden AB, Wallace HM. Polyamines in colorectal cancer. The British journal of surgery. 1984;71:791–794. doi: 10.1002/bjs.1800711019. [DOI] [PubMed] [Google Scholar]
- 46.Morris SM., Jr Enzymes of arginine metabolism. The Journal of nutrition. 2004;134:2743S–2747S. doi: 10.1093/jn/134.10.2743S. discussion 65S–67S. [DOI] [PubMed] [Google Scholar]
- 47.Astrid Ruefli-Brasse DS, Jessica Orf, Minqing Rong, Jianxia Shi, Tim Carlson, Kim Quon, Alexander Kamb, Dineli Wickramasinghe. Methylthioadenosine (MTA) Rescues Methylthioadenosine Phosphorylase (MTAP)-Deficient Tumors from Purine Synthesis Inhibition in Vivo via Non-Autonomous Adenine Supply Journal of Cancer Therapy. 2011;2:523–534. [Google Scholar]
- 48.Moffatt BA, Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. The Arabidopsis book/American Society of Plant Biologists. 2002;1:e0018. doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual review of biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Z, Vosseller K. O-GlcNAc in cancer biology. Amino acids. 2013;45:719–733. doi: 10.1007/s00726-013-1543-8. [DOI] [PubMed] [Google Scholar]
- 51.Matthews JA, Acevedo-Duncan M, Potter RL. Selective decrease of membrane-associated PKC-alpha and PKC-epsilon in response to elevated intracellular O-GlcNAc levels in transformed human glial cells. Biochimica et biophysica acta. 2005;1743:305–315. doi: 10.1016/j.bbamcr.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Lahn M, Su C, Li S, Chedid M, Hanna KR, Graff JR, et al. Expression levels of protein kinase C-alpha in non-small-cell lung cancer. Clinical lung cancer. 2004;6:184–189. doi: 10.3816/clc.2004.n.032. [DOI] [PubMed] [Google Scholar]
- 53.Garber K. Oncometabolite? IDH1 discoveries raise possibility of new metabolism targets in brain cancers and leukemia. Journal of the National Cancer Institute. 2010;102:926–928. doi: 10.1093/jnci/djq262. [DOI] [PubMed] [Google Scholar]
- 54.McCarthy N. Metabolism: unmasking an oncometabolite. Nature reviews Cancer. 2012;12:229. doi: 10.1038/nrc3248. [DOI] [PubMed] [Google Scholar]
- 55.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakheja D, Boriack RL, Mitui M, Khokhar S, Holt SA, Kapur P. Papillary thyroid carcinoma shows elevated levels of 2-hydroxyglutarate. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2011;32:325–333. doi: 10.1007/s13277-010-0125-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.