Abstract
Melanocytes, the pigment-producing cells, arise from multipotent neural crest (NC) cells during embryogenesis. Many genes required for melanocyte development were identified using mouse pigmentation mutants. The variable spotting mouse pigmentation mutant arose spontaneously at the Jackson Laboratory. We identified a G-to-A nucleotide transition in exon 3 of the Ets1 gene in variable spotting, which results in a missense G102E mutation. Homozygous variable spotting mice exhibit sporadic white spotting. Similarly, mice carrying a targeted deletion of Ets1 exhibit hypopigmentation; nevertheless, the function of Ets1 in melanocyte development is unknown. The transcription factor Ets1 is widely expressed in developing organs and tissues, including the NC. In the chick, Ets1 is required for the expression of Sox10, a transcription factor critical for the development of various NC derivatives, including melanocytes. We show that Ets1 is required early for murine NC cell and melanocyte precursor survival in vivo. Given the importance of Ets1 for Sox10 expression in the chick, we investigated a potential genetic interaction between these genes by comparing the hypopigmentation phenotypes of single and double heterozygous mice. The incidence of hypopigmentation in double heterozygotes was significantly greater than in single heterozygotes. The area of hypopigmentation in double heterozygotes was significantly larger than would be expected from the addition of the areas of hypopigmentation of single heterozygotes, suggesting that Ets1 and Sox10 interact synergistically in melanocyte development. Since Sox10 is also essential for enteric ganglia development, we examined the distal colons of Ets1 null mutants and found a significant decrease in enteric innervation, which was exacerbated by Sox10 heterozygosity. At the molecular level, Ets1 was found to activate an enhancer critical for Sox10 expression in NC-derived structures. Furthermore, enhancer activation was significantly inhibited by the variable spotting mutation. Together, these results suggest that Ets1 and Sox10 interact to promote proper melanocyte and enteric ganglia development from the NC.
Keywords: melanocyte, neural crest, Ets1, Sox10, enteric ganglia
Introduction
Melanocytes are derived from neural crest (NC) cells, a transient population of multipotent cells that arises from the dorsal aspect of the neural tube during vertebrate embryogenesis. In addition to melanocytes, other NC-derived lineages include endocrine cells, glial cells, neurons, craniofacial bone and cartilage, and cardiac cells (Le Douarin and Kalcheim, 1999). Defects in NC cell development result in a range of human disorders known as Neurocristopathies (Bolande, 1997). These include craniofacial malformations, cardio- and neurocutaneous syndromes, Waardenburg Syndrome, Hirschsprung's disease, and piebaldism, among others (reviewed in Hou and Pavan, 2008; Pingault et al., 2010). Neurocristopathies are often characterized by pigmentation defects, hence the study of mouse pigmentation mutants has been instrumental for the identification of genes and pathways involved in the development of various NC cell lineages, including melanocytes (Tassabehji et al., 1992; Hosoda et al., 1994; Puffenberger et al., 1994).
The deletion of the transcription factor Ets1 was recently shown to cause hypopigmentation in mice (Gao et al., 2010); however, the role of Ets1 in melanocyte development is unknown. Ets1 is a member of a large family of helix-turn-helix transcription factors that are characterized by the presence of the E26 transforming specific sequence (Ets) domain, which recognizes the core nucleotide sequence GGAA/T (reviewed in Dittmer, 2003). Ets1 is required for the survival of various cell lineages, including thymocytes, T cells (Muthusamy et al., 1995), and endothelial cells (Wei et al., 2009). During embryonic development, Ets1 is expressed in various organs and tissues, including the NC (Vandenbunder et al., 1989; Quéva et al., 1993; Kola et al., 1993; Maroulakou et al., 1994; Fafeur et al., 1997; Theveneau et al., 2007). Ets1 expression has also been correlated with pathological invasive processes. This transcription factor has been found to be upregulated in various tumor types, including melanoma (Keehn et al., 2003; Rothhammer et al., 2004; Rothhammer et al., 2005; reviewed in Garrett-Sinha, 2013), and its expression in certain tumors has been correlated with the upregulation of matrix metalloproteases (MMPs), particularly MMP1, MMP2 and MMP9 (reviewed in Dittmer, 2003; Rothhammer et al., 2004; Okuducu et al., 2006).
Several studies have implicated Ets1 in the proper development of NC cell derivatives. During chick embryonic development, Ets1 expression is correlated with epithelial-to-mesenchymal transition (EMT), (Fafeur et al., 1997), and although it is not required for EMT, Ets1 promotes cell mobilization, which is necessary for the initiation of cranial NC cell delamination (Theveneau et al., 2007). In the mouse, Ets1 deletion was found to produce cardiac malformations (Gao et al., 2010; Ye et al., 2010). Some of these cardiac malformations were found to arise as a result of the lack of NC-derived cells in the proximal aspects of the outflow tract endocardial cushions and the presence of an abnormal nodule of NC-derived cartilage within the heart. These defects suggest a role for Ets1 in proper NC cell migration and differentiation (Gao et al., 2010).
The exact mechanism by which Ets1 deficiency during embryonic development results in hypopigmentation has not been elucidated. In the chick cranial NC, Ets1 is required for the activation of the transcription factor sex determining region Y (SRY)-box 10 (Sox10) (Betancur et al., 2010), a gene required for the development of various NC cell derivatives, including melanocytes and enteric ganglia (Herbarth et al., 1998; Southard-Smith et al., 1998; Kapur, 1999; Potterf et al., 2001). In humans, mutations in Sox10 are responsible for Waardenburg syndrome type IV, which is characterized by pigmentation defects and aganglionosis of the distal colon (Hirschsprung's disease) (Pingault et al., 1998). The requirement of Ets1 for Sox10 expression in the chick embryo and the hypopigmentation phenotype of Ets1 null mutants, suggest that Ets1 may play an important role in the regulation of this and other melanocyte-specific genes in the mouse embryo.
In the present study, we report the identification of Ets1 as the mutated gene in the spontaneous variable spotting mouse pigmentation mutant. This finding, along with the observation that mice carrying a targeted deletion of Ets1 have belly spots (Gao et al., 2010), suggested a role for Ets1 in melanocyte development. We characterized the temporal requirement of Ets1 for melanocyte development and established a role for this transcription factor in NC and melanocyte precursor (melanoblast) survival. In light of the functional relationship that exists between Ets1 and Sox10 in the chick cranial NC, we hypothesized that, in the mouse, Ets1 may interact with Sox10 to promote melanocyte development. Analysis of the pigmentation phenotype of single and double heterozygous mutants for Ets1 and Sox10 revealed a synergistic genetic interaction between these transcription factors. Given the importance of Sox10 for enteric ganglia development, we also investigated the effect of Ets1 deficiency on enteric innervation. Analysis of the distal colons of Ets1 null mutant mice revealed defects in enteric ganglia patterning, which were further exacerbated by Sox10 heterozygosity, hinting to a potential interaction between Ets1 and Sox10 in the development of enteric ganglia. Using in vitro assays, we show that Ets1 is able to activate an enhancer critical for Sox10 expression in NC-derived lineages. Additionally, mutating Ets1, at the site we characterized in the variable spotting mutant, reduces the ability of Ets1 to activate this Sox10 enhancer. Together, our results indicate that Ets1 is required early for NC cell and melanoblast survival. Furthermore, the presence of a synergistic genetic interaction between Ets1 and Sox10 in melanocyte development, and the ability of Ets1 to activate a Sox10 regulatory region, suggests that at least some of the functions of Ets1 in melanocyte development may be mediated by Sox10.
Materials and Methods
Ethics statement
All animal work was approved by the Florida International University Institutional Animal Care and Use Committee (Protocol No. 13-064) and performed according to institutional guidelines established by the National Institutes of Health (NIH) (Guide for the Care and Use of Laboratory Animals, 2011).
Exome and direct sequencing of the variable spotting mutation
A single heterozygous variable spotting sample was processed for exome sequencing. Whole genome libraries with ~170 base inserts and paired-end index adapters were prepared from 3 μg DNA using a SPRI-TE robot and reagents (Beckman-Coulter). Libraries were enriched using the SureSelect Mouse All Exon Kit (Agilent) and pooled for sequencing in 2 lanes on a HiSeq2000 (Illumina) using version 3 chemistry. At least 50 million paired-end 100 base reads were obtained for each sample. Data was processed using RTA version 1.12.4.2 and CASAVA 1.7.0. Sequencing reads were aligned with ELAND, and then realigned with cross_match (http://www.phrap.org) to the UCSC mm9 mouse reference sequence. After removal of PCR duplicate reads, diploid genotypes were called using bam2mpg (Teer et al., 2010). Variants were annotated against the UCSC “known genes” gene set (Hsu et al., 2006) using ANNOVAR (http://www.openbioinformatics.org/annovar), and genotypes, gene annotations, and dbSNP (build 128) identifiers were reported in VarSifter format for viewing and filtering (Teer et al., 2012).
Filtering variants to those in coding exons or splice sites within the variable spotting interval (Chr 9 proximal to Apoa1; Spencer and Davisson, 1988) identified 2 variants with heterozygote genotype calls not present in dbSNP or 4 other unrelated mouse mutant exomes completed as part of the same project. The 2 variants were within Mtmr2 (c.1757G>A:p.R586Q; CDPred score 1) and Ets1 (c.305G>A:p.G102E; CDPred score -11). CDPred is an algorithm designed to predict the effect of amino acid substitutions (Johnston et al 2010) with scores above -3, as observed for the Mtmr2 variant, predicted to have a mild/neutral effect and scores below -7, as observed for Ets1, predicted to have a severe effect on protein function. The CDPred predictions are consistent with SIFT (Kumar et al. 2009), a second tool which predicts the Mtmr2 variant to be tolerated (SIFT score=0.07) compared to the deleterious Ets1 variant (SIFT score=0). To confirm Ets1 as the gene responsible for the variable spotting phenotype we used an independent targeted Ets1 allele to confirm that Ets1 deficiency alone is sufficient to cause hypopigmentation, providing strong evidence that the Ets1 variant is causative for the phenotype observed in variable spotting mice.
DNA samples from 2 heterozygous and 2 homozygous variable spotting mutant mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). A 496bp DNA fragment surrounding the suspected mutation was amplified via PCR using the primers 5’-AAGGTGTAGAGTAACTAGCATCGTCAG and 5’-AACTCCTAAGGCAGAGAAGAAAATAAG. PCR products were purified using ExoSAP-IT (Affimetryx), following the manufacturer's instructions, and directly sequenced. Sequences were analyzed using the Applied Biosystems Sequence Scanner v1.0 software (Applied Biosystems) and sequence alignments against the Mus musculus genomic database were carried out via the Nucleotide BLAST server (NCBI, NIH). Multiple alignments to the Ets1 protein sequences for human, rat, zebrafish, dog, and mouse were carried out using ClustalW (via ExPASy.org).
Animals and Genotyping
Ets1 mutant mice were a kind gift from Dr. Eric Svensson (Universtiy of Chicago, Chicago, IL). They were generated as previously described (Barton et al., 1998), and re-derived by outcrossing to C57BL/6J mice for eight generations (Gao et al., 2010). They have been maintained on C57BL/6J genetic background by intercrossing and by outcrossing to C57BL/6J mice (Jackson Laboratories). Mice carrying the Dct-LacZ transgene (Tg(Dct-lacZ)#Ove) were obtained from Dr. Paul Overbeek (Baylor College of Medicine, Houston, TX) and generated on an FVB/N genetic background, as previously described (Zhao and Overbeek, 1999). Dct-LacZ mice were maintained by intercrossing and bred with Ets1 heterozygotes to generate Ets1+/−::Dct-LacZ+ mice, which were intercrossed for the generation of embryos. Sox10tmlWeg/Sox10+ mice (hereafter referred to as Sox10LacZ/+) were obtained from Dr. Michael Wegner (Institut für Biochemie, Universität Erlangen-Nürnberg, Germany) and generated on a mixed genetic background (129S1/Sv; C3HeB/FeJ; C57BL/6J), as previously described via insertion of the LacZ gene into the Sox10 coding sequence (Britsch et al., 2001). Sox10LacZ/+ mice were maintained by intercrossing and bred with Ets1 heterozygotes to generate double heterozygotes.
Genomic DNA was isolated from tail biopsies or yolk sacs, and used for genotyping by PCR. Genotyping for Ets1 was performed as previously described (Gao et al., 2010). For Sox10LacZ, genotyping of was performed using the primers 5’-CAGGTGGGCGTTGGGCTCTT, 5’-CAGAGCTTGCCTAGTGTCTT, and 5’-TAAAAATGCGCTCAGGTCAA (Britsch et al., 2001). Dct-LacZ mice were genotyped via β-galactosidase staining of ear tips, using standard protocols (Nagy, 2003).
β-galactosidase and Lysotracker Red staining of whole mouse embryos
Embryos from intercrosses between Ets1+/−::Dct-LacZ+ mice were harvested between embryonic days (E) 10.75-E15.5. Embryos from intercrosses between Ets1+/−::Sox10LacZ/+ mice were harvested at E9.5, E11.5, and E12.5. Whole embryos were subjected to β-galactosidase (LacZ) staining following standard protocols (Nagy, 2003). For each stage, at least 3 embryos of each genotype were stained and photographed. The staining was visualized under a Leica MZ6 dissecting scope and photographed using a Leica DC500 camera. In the case of E10.75 embryos, Lysotracker Red staining was carried out prior to β-galactosidase staining by incubating embryos in 5μM Lysotracker Red DND-99 (Life Technologies) in PBS, protected from light, at 37°C for 30 minutes. Subsequently, embryos were washed with PBS (pH7.2) and LacZ staining was performed.
Immunofluorescence
Freshly dissected E10.25 and E12.5 embryos were fixed in 4% paraformaldehyde for 3 and 16 hours, respectively, at 4°C, while E10.75, LacZ and Lysotracker Red-stained, embryos were post-fixed in 4% paraformaldehyde for 20 minutes at room temperature. Embryos were then washed with PBS and incubated at 4°C in 10% sucrose for 4 hours and in 20% sucrose, overnight. Embryos were embedded in Tissue-Tek Tissue Freezing Medium (Sakura), frozen at -80°C, and sectioned at 10μm. Sections were blocked with 1% Bovine Serum Albumin (BSA), 0.1% Triton X-100 in PBS for 45 minutes at room temperature and incubated in primary antibodies (diluted in blocking solution) for 1 hour at room temperature. Primary antibodies include rabbit anti-phospho-Histone H3 (Ser10) (1:100; Millipore, 06-570), goat polyclonal anti-Sox10 (1:100; Santa Cruz Biotechnology, sc-17342), and rabbit anti-cleaved Caspase-3 (1:200; Cell Signaling, 9664S). After washing, sections were incubated with a suitable fluorescently-tagged secondary antibody (goat anti-rabbit Alexa Fluor 488 (1:200; Life Technologies, A11008), or donkey anti-goat Alexa Fluor 594 (1:200; Life Technologies, A11058)) for 1 hour at room temperature. Sequential incubation in secondary antibodies (donkey anti-goat Alexa Fluor 594 followed by goat anti-rabbit Alexa Fluor 488) was carried out for Sox10 and cleaved Caspase-3 double-labeling in sections from E10.25 embryos. Cell nuclei were counterstained with 1μg/ml Hoechst Dye (BioRad). The staining was visualized using a Leica DMRB compound fluorescent microscope. Sections were photographed using a Leica DC500 camera.
For E10.75 Ets1−/−::Dct-LacZ+ (n=3) and Ets1+/−::Dct-LacZ+ (n=3) embryos, melanoblasts (LacZ positive cells), dying melanoblasts (LacZ and Lysotracker Red positive cells), and proliferating melanoblasts (LacZ and phospho-Histone H3 positive cells) were counted in 80 sections spanning the trunk region of each embryo. For E10.25 Ets1−/− (n=3) and Ets1+/− (n=3) embryos, NC cells (Sox10 positive cells) and dying NC cells (Sox10 and cleaved Caspase-3 positive cells) were counted in 35 sections spanning the anterior half of the trunk region of each embryo. For E12.5 Ets1−/− (n=3) and Ets1+/+ (n=3) embryos, Sox10 positive cells in the dorsal root ganglia were counted in 15 sections spanning the trunk region of each embryo. Data are shown as mean ± standard deviation. The Student's t-test was used to determine statistical significance. Data were considered statistically significant at p<0.05
Phenotypic and statistical analyses
Ets1+/− mice were crossed to Sox10LacZ/+ to generate progeny that were either Ets1+/−, Sox10LacZ/+, or Ets1+/−::Sox10LacZ/+. The single heterozygous progeny of the first generation (F1) were intercrossed to generate the second generation (F2) animals. Third generation animals (F3) were generated by intercrossing single heterozygous as well as double heterozygous F2 mice. For each generation, the ventral aspects of each mouse were photographed at 6 weeks of age. The areas of hypopigmentation were measured using the Image J 1.44p program (NIH).
Statistical differences for the frequency of belly spot appearance were determined using the Chi-square test. In order to compare the mean areas of hypopigmentation among the different genotypes, a Box-Cox transformation was applied to the belly spot area data (measured in mm2). For each generation, the transformed areas of hypopigmentation of Ets1+/−::Sox10LacZ/+ mice were compared to the sum of the transformed areas of hypopigmentation of Ets1+/− and Sox10LacZ/+ mice using a One-Way ANOVA with a contrast. Scatter plots of the hypopigmentation areas showing the mean and standard deviations for each group were generated using GraphPad Prism 6 (GraphPad Software, Inc). Data were considered statistically significant at p<0.05
Colon dissection and Acetylcholinesterase staining
Ets1+/−, Sox10LacZ/+, and Ets1+/−::Sox10LacZ/+ mice were crossed to generate progeny of the following genotypes: Ets1+/−::Sox10+/+, Ets1+/−::Sox10LacZ/+, Ets1−/−::Sox10+/+, and Ets1−/−::Sox10LacZ/+. Littermates of the genotypes listed above, ranging from 4 to 7 months of age, were euthanized and their colons dissected. Approximately 30mm of the distal aspect of the colons of each animal were removed, cleaned, and fixed in 4% paraformaldehyde, containing 10mM CaCl2, overnight, at 4°C. Subsequently, the colons were briefly washed with PBS (pH7.4) and incubated in 5% sucrose for 24 hours. The colons were then washed with PBS and subjected to Acetylcholinesterase staining as previously described (Gariepy et al., 1998). The staining was visualized under a Leica MZ6 dissecting scope and photographed using a Leica DC500 camera. Colons from at least 5 animals of each genotype were stained and photographed. The extent of enteric innervation was quantified using the images captured, by measuring the white (nerve-free) area in 8 squares of 0.25mm2 each (for a total of 2mm2 per colon), within a region located 15mm anterior to the anus. The ImageJ 1.44p program (NIH) was used to measure the white areas in each segment of colon. The Student's t-test was used to compare the nerve-free areas of colons from mice of the various genotypes listed above. Scatter plots of the nerve-free areas in 2mm2 of colon for each genotype, showing the mean and standard deviations were generated using GraphPad Prism 6 (GraphPad Software, Inc).
Vectors for Luciferase assays
The Ets1 overexpression vector (pCMV-HA-Ets1) and the pCMV-HA empty vector (Clontech) were kindly provided by Dr. Satrajit Sinha (State University of New York (SUNY) at Buffalo, Buffalo, NY). The pCMV-HA-Ets1 vector contains a Hyaluronic acid (HA)-tagged version of the full length mouse Ets1 coding sequence (Nagarajan et al., 2009). The Ets1 overexpression vector carrying the variable spotting mutation (pCMV-HA-Ets1Mut) was prepared from the PCMV-HA-Ets1 vector, using the Phusion Site-Directed Mutagenesis Kit (Thermo Scientific), following the manufacturer's instructions. The purified plasmid was sequenced to confirm the presence of the mutation. The Sox10-MCS4 luciferase vector (Sox10-MCS4 fragment in pLGF-E1b vector) was generated as previously described (Antonellis et al., 2008). The pRL-TK Renilla luciferase control vector (Promega) was used as an internal control for transfection efficiency.
Cell culture, transfections, and Luciferase activity assays
The B16-F10 mouse melanoma cell line (ATCC, CRL-6322) was cultured under standard conditions. Cells were seeded in 12 well plates at a density of 1.5 × 105 cells per well, roughly 24 hours prior to transfection. Once the cells reached 70-90% confluency they were co-transfected with 0.5μg/well Sox10-MCS4 luciferase reporter vector, 0.5μg/well pCMV-HA-Ets1, pCMVHA-Ets1Mut or pCMV-HA empty vector, and 0.05μg/well pRL-TK Renilla luciferase control vector, using the Lipofectamine 2000 lipid transfection reagent (Life Technologies), following the manufacturer's instructions. The cells were then incubated at 37°C, 5% CO2 for 24 hours. After 24 hours, the cells were gently washed with PBS (Thermo Scientific) and lysed with 1x Passive Lysis Buffer (Promega), at room temperature. For each well, 20μl of cell lysate were used to measure firefly and Renilla luciferase activities with the Dual Luciferase Reporter Assay System (Promega) following the manufacturer's instructions. Firefly and Renilla luciferase activities were measured using a TD-20/20 Luminometer (Turner Designs). For each experiment, the ratio of firefly to Renilla luciferase activity and the fold increase in this ratio over the ratio for the cells transfected with the PCMV-HA empty vector were calculated. Data represent mean fold activation over the PCMV-HA empty vector (bar height) and standard deviation (error bars) of three independent experiments carried out in quadruplicates. The Student's t-test was used to compare the mean fold activation in cells transfected with the Ets1 overexpression vector to that of cells transfected with the Ets1 overexpression vector carrying the variable spotting mutation.
Results
Variable spotting is caused by a novel missense mutation in the Ets1 gene
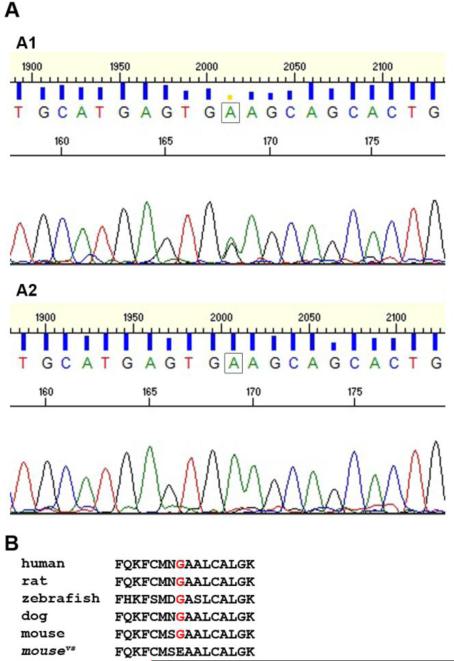
The spontaneous variable spotting mouse mutant exhibits a similar pigmentation phenotype to that of mice carrying a targeted deletion of the Ets1 gene (Barton et al., 1998), which were used in this study. Variable spotting mutants present with white feet and tail, and occasionally head blaze and ventral spotting, which may be large (Jackson Laboratories). The Ets1 null mutants always have large belly spots, hypopigmented tail and feet, and occasionally head blaze, while only roughly 40% of Ets1 heterozygotes have belly spots, which tend to be small in size. As part of an exome sequencing project at the National Institutes of Health (NIH), a possible mutation within the open reading frame (ORF) of Ets1 was identified in variable spotting. Through direct sequencing of heterozygous and homozygous mutants, we confirmed a nucleotide G-to-A transition in exon 3 of the Ets1 gene (Fig. 1A), which results in a Glycine to Glutamate substitution at residue 102 (G102E) of the Ets1 protein. Multiple sequence alignment showed that this Glycine residue is highly conserved among different species (Fig. 1B), which suggests that it may be important for Ets1 protein function.
Figure 1. Variable spotting is caused by a novel missense mutation in the Ets1 gene.
(A) Mutation analysis of variable spotting mice. Sequencing of heterozygous (A1) and homozygous (A2) variable spotting mutant mice revealed a G-to-A transition (boxes) in exon 3 of the Ets1 gene at nucleotide position 647 (RefSeq NM_011808.2). This mutation results in a Glycine to Glutamate change at amino acid 102 (G102E) of the Ets1 protein. (B) Multiple sequence alignment of the Ets1 protein shows that the substituted Glycine residue (red font) is highly conserved among different species.
Ets1 is required for melanoblast and neural crest cell survival
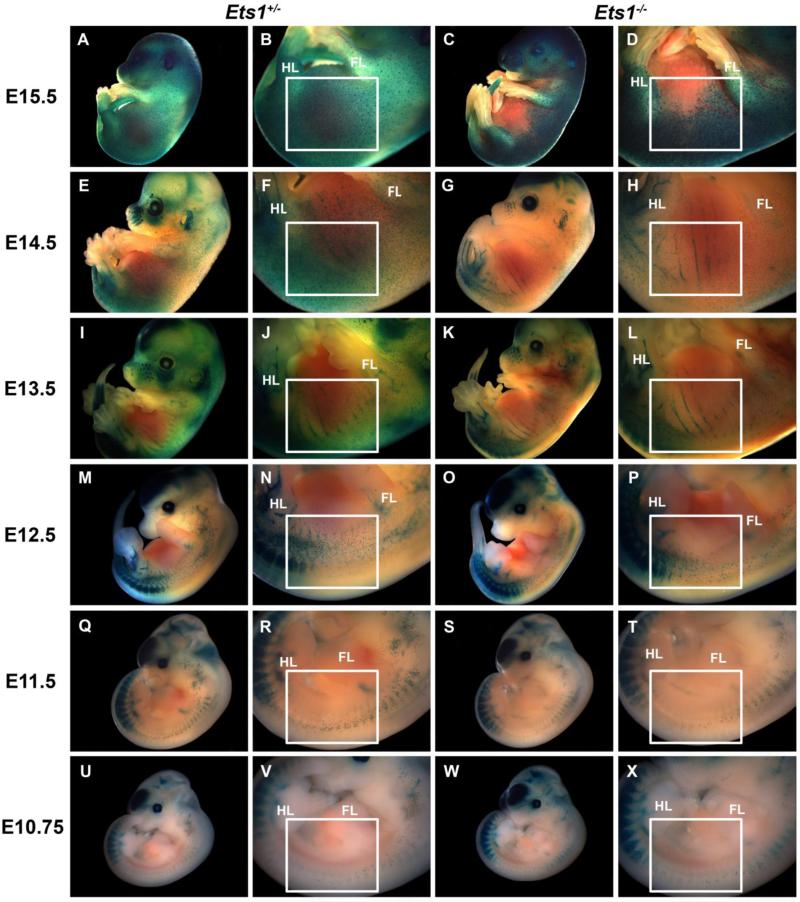
The hypopigmentation phenotype observed in Ets1 heterozygous and null mutant mice hinted to a potential role for Ets1 in melanocyte development. In order to establish the earliest time point at which Ets1 deficiency results in impaired melanocyte development, Dct-LacZ transgenic mice, in which LacZ expression is driven to melanoblasts under the control of the Dopachrome tautomerase (Dct) promoter (Zhao and Overbeek, 1999), were used to label melanoblasts via β-galactosidase activity in Ets1 null, heterozygous, and wild type embryos. An apparent decrease in the number of trunk melanoblasts was observed in E11.5-E15.5 Ets1−/− embryos compared to Ets1+/− (Fig. 2, Fig. S1) and wild type littermates (data not shown).
Figure 2. Ets1 null mutant embryos have decreased numbers of melanoblasts.
Dct-LacZ transgenic mice, in which LacZ expression is driven by the Dopachrome tautomerase (Dct) promoter that drives expression to melanoblasts, were crossed to Ets1+/− mice to produce Ets1+/−::Dct-LacZ+ mice. Embryos from intercrosses between Ets1+/−::Dct-LacZ+ mice were harvested between embryonic days (E)10.75-15.5, subjected to LacZ staining and analyzed for the numbers and position of melanoblasts (blue dots) in the trunk region. At all these ages, Ets1−/− embryos have reduced numbers (predominantly in the areas contained within the white frames) of melanoblasts (C, D, G, H, K, L, O, P, S, T, W, X) compared to Ets1+/− (A, B, E, F, I, J, M, N, Q, R, U, V) and wild type (not shown) littermates (HL: Hindlimb, FL: Forelimb).
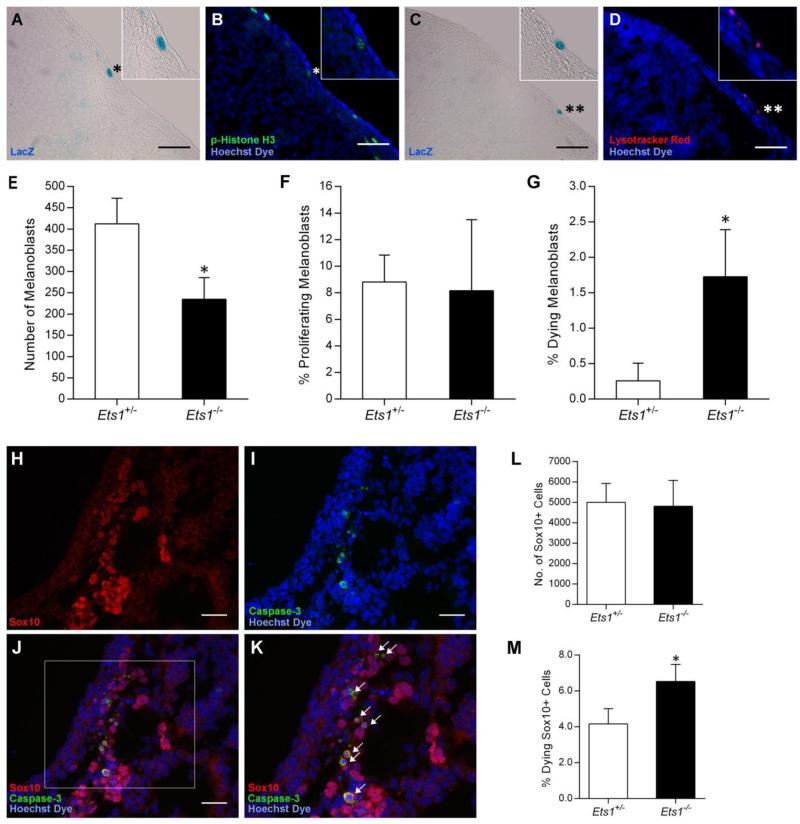
The marked decrease in melanoblast numbers observed in Ets1−/− embryos implied that melanoblast survival and/or proliferation were likely impaired as a result of Ets1 deletion. To establish the mechanism of action of Ets1 in murine melanocyte development, the total numbers of trunk melanoblasts, as well as the numbers of dying and proliferating melanoblasts, were quantified at E10.75 (Fig. 3A-G). This embryonic stage was chosen because it was the earliest time point during which an apparent difference in melanoblast numbers could still be detected via LacZ staining. A significant decrease in the number of trunk melanoblasts was observed in Ets1−/− embryos compared to Ets1+/− littermates (p=0.018, Fig. 3E). At this stage, the percent of proliferating melanoblasts, which were identified by LacZ and phospho-Histone H3 double-labeling, did not differ significantly between Ets1+/− and Ets1−/− littermates (p=0.852, Fig. 3F). These results indicate that Ets1 deficiency does not result in impaired melanoblast proliferation at E10.75. A significant increase in the percent of dying melanoblasts, which were labeled by LacZ and Lysotracker Red staining, was observed in Ets1−/− embryos compared to Ets1+/−littermates (p=0.023, Fig. 3G), suggesting that Ets1 is required for melanoblast survival at this stage.
Figure 3. Ets1 is required for melanoblast and neural crest cell survival.
(A, C) Melanoblasts traveling along the surface of the embryo are marked via LacZ staining (black arrows). (B) Proliferating cells are positive for the phospho-Histone H3 (p-Histone H3) antibody (green fluorescence). (D) Lysotracker Red marks lysosomes in dying cells (red fluorescence). (A, B) Proliferating melanoblasts (*) are LacZ and p-Histone H3 positive. (C, D) Dying melanoblasts (**) are LacZ and Lysotracker Red positive. (E) Melanoblasts, (F) proliferating melanoblasts and (G) dying melanoblasts were counted in 80 sections for E10.75 Ets1+/− (n=3) and Ets1−/− (n=3) embryos. (E) Ets1−/− embryos have significantly reduced numbers of melanoblasts compared to Ets1+/− littermates (*p<0.05). (F) The percent of proliferating melanoblasts did not differ significantly between Ets1−/− and Ets1+/− littermates (p=0.852), (G) while the percent of dying melanoblasts was significantly higher (*p<0.05) in Ets1−/− embryos compared to Ets1+/− littermates. (H-K) Sox10 and cleaved Caspase-3 double-label immunofluorescence of E10.25 mouse embryos. Neural crest (NC) cells were labeled via Sox10 antibody staining (red fluorescence). Dying cells were labeled using a cleaved Caspase-3 antibody (green fluorescence). (J, K) Dying NC cells are positive for Sox10 and cleaved Caspase-3 (white arrows). (L) NC cells and (M) dying NC cells were counted in 35 sections spanning the anterior half of the trunk region for E10.25 Ets1+/− (n=3) and Ets1−/− (n=3) embryos. (J) The number of NC cells in Ets1−/− embryos is not significantly different from that of Ets1+/− littermates (p=0.848). (K) The percent of dying NC cells is significantly higher (*p<0.05) in Ets1−/− embryos compared to Ets1+/− littermates (scale bars = 50 μm).
Given the increase in melanoblast cell death observed in Ets1−/− mutant embryos at E10.75, we investigated the possibility that Ets1 may be required for NC cell survival prior to the specification of melanoblasts. To this end, we labeled NC cells and dying cells via Sox10 and cleaved Caspase-3 immunostaining at E10.25 in Ets1−/− and Ets1+/− littermates (Fig. 3H and I). The transcription factor Sox10 was chosen as a NC cell marker because it is initially expressed in NC cells as they begin to delaminate from the neural tube and persists in cells that will give rise to peripheral nervous system components (Kuhlbrodt et al., 1997) and melanocytes (Southard-Smith et al., 1998). The total number of NC cells (Sox10 positive cells) present in the anterior half of the trunk region, did not differ significantly between Ets1−/− and Ets1+/− embryos (p=0.848, Fig. 3J). Nonetheless, the percent of dying NC cells in this region (Sox10 and cleaved Caspase-3 positive cells) was significantly higher in Ets1−/− embryos compared to Ets1+/− littermates (p=0.033, Fig. 3K), which suggests that Ets1 is required for NC cell survival at this stage.
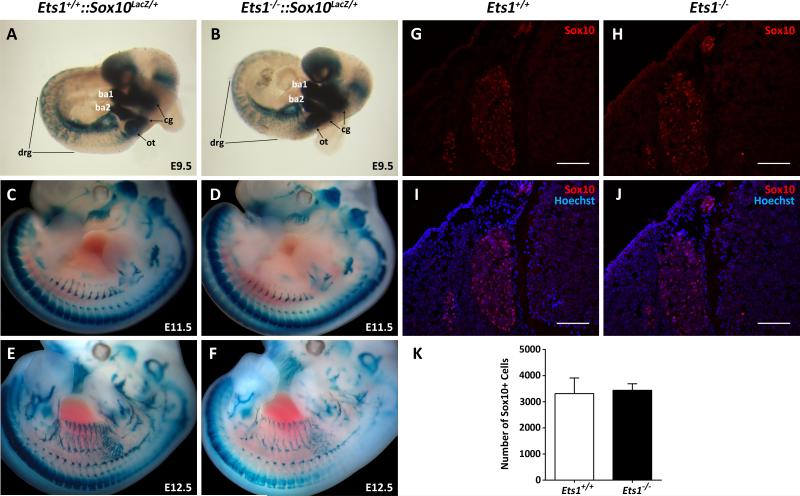
Sox10 expression was also examined at E9.5 via in situ hybridization (Fig. S2) and LacZ staining (Fig. 6A and B) of whole embryos from intercrosses between Ets1+/− mice or Ets1+/−::Sox10LacZ/+ mice, respectively. At this stage, the pattern and levels of Sox10 expression do not appear to differ between Ets1−/− embryos and Ets1+/+ or Ets1+/− littermates. Nevertheless, Sox10 in situ hybridization revealed ectopic Sox10 expression in the frontal aspect of the head in Ets1−/− embryos (Fig. S2B, red arrowhead). The lack of a significant difference in the numbers of NC cells between Ets1−/− and Ets1+/− embryos at E10.25, in spite of the increased number of dying NC cells in Ets1−/− embryos, implies that Ets1 appears to act precisely around this time point so that the effects of Ets1 deficiency become apparent soon after, when a sharp decrease in melanoblast numbers is observed. The fact that Sox10 expression did not appear to be affected in Ets1 null mutant mice at E9.5, suggests that the critical time point at which Ets1 regulates NC cell and melanoblast survival is somewhere between E10 and E11.
Figure 6. Sox10 expression is not altered at E9.5 but is reduced at E12.5 in some Ets1 null mutant embryos.
(A-F) Sox10 expression was visualized via LacZ staining in embryos from intercrosses between Ets1+/−::Sox10LacZ/+ mice. (A, B) At E9.5, no gross differences were observed in LacZ expression between Ets1−/− and Ets1+/+ or Ets1+/− (not shown) littermates. (C, D) At E11.5, LacZ expression was reduced in 75% of Ets1−/− embryos (n=4) (D), compared to Ets1+/− (n=3) (not shown) and Ets1+/+ (n=3) (C) littermates. (E, F) At E12.5, a decrease in LacZ expression was observed in 50% of Ets1−/− (n=6) (F) embryos compared to Ets1+/− (n=9) (not shown), and Ets1+/+ (E) (n=3) littermates. (G-K) Sox10 positive cells in the dorsal root ganglia were quantified in 15 sections spanning the trunk region for Ets1+/+ (n=3) (G, I) and Ets1−/− (n=3) (H, J) embryos at E12.5. (K) At this stage, no significant differences in the numbers of Sox10 positive cells present within the dorsal root ganglia were observed between Ets1+/+ and Ets1−/−littermates (p=0.753). (Magnifications: A, B= 40X, C, D= 25x, E, F= 21x) (cg: cranial ganglia, ot: otocyst, drg: dorsal root ganglia, ba1: first branchial arch, ba2: second branchial arch) (scale bars = 100μm).
Ets1 genetically interacts with Sox10 in melanocyte and enteric ganglia development
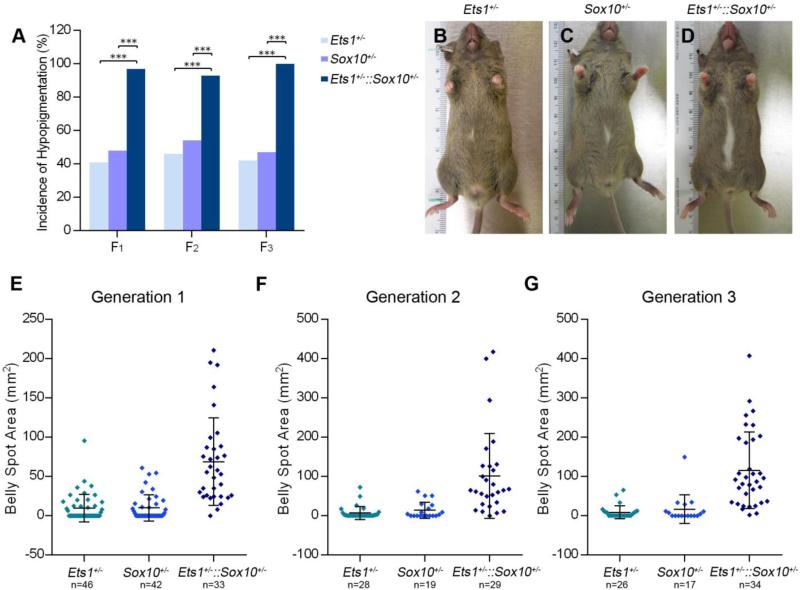
The increase in melanoblast and NC cell apoptosis observed in Ets1 null mutants, indicates that Ets1 is required early for NC cell and melanoblast survival. Other genes required for melanoblast survival, include those that code for the transcription factors Mitf (microphthalmia-associated transcription factor), Sox10, and Pax3 (Paired-box 3), and for the signaling molecules Endothelin-3 (Edn3) and Kit-ligand (Kitl), and their receptors Endothelin receptor b (Ednrb) and Kit (reviewed in Silver et al., 2006). Evidence from studies in the chick embryo pointed to a potential genetic interaction between Ets1 and Sox10 in melanocyte development. In the chick embryo, Ets1, along with Sox9 and c-Myb, directly binds to and activates a Sox10 enhancer element that drives Sox10 expression to cranial NC cells (Betancur et al., 2010). In order to determine whether a genetic interaction exists between Ets1 and Sox10 in murine melanocyte development, Ets1+/− mice were crossed to Sox10LacZ/+ mice and the hypopigmentation phenotypes of the progeny were examined for three generations. For all generations, the frequency of belly spot appearance in double heterozygotes (Ets1+/−::Sox10LacZ/+) was significantly higher than in single heterozygotes (Ets1+/− and Sox10LacZ/+) (p<0.001 for all generations, Fig. 4A). In the three generations examined, almost all Ets1+/−::Sox10LacZ/+ mice had belly spots (F1: 97%, F2: 96%, F3: 100%), compared to approximately 40% for Ets1+/− mice and 48% of Sox10LacZ/+ mice. In addition to showing increased hypopigmentation frequency, double heterozygous mice had belly spots that were significantly larger than would be expected from the addition of the areas of the belly spots of the single heterozygotes (p<0.001 for all generations, Fig. 4B-G). These results suggest that a synergistic genetic interaction exists between Ets1 and Sox10 in melanocyte development.
Figure 4. Ets1 and Sox10 interact synergistically in melanocyte development.
(A) The incidence of hypopigmentation in Ets1+/−::Sox10LacZ/+ mice was significantly greater than the incidence of hypopigmentation in Ets1+/− and Sox10LacZ/+ mice (p<0.001) for all three generations analyzed. (B, C, D) Representative pictures of ventral hypopigmentation for each genotype show that double heterozygous mice had larger areas of hypopigmentation compared to Ets1 and Sox10 single heterozygotes. (E, F, G) The areas of hypopigmentation in Ets1+/−, Sox10+/−, and Ets1+/−::Sox10LacZ/+ were measured for progeny from 3 generations. The transformed areas of the belly spots of Ets1+/−::Sox10LacZ/+ mice were significantly greater than the sum of the transformed areas of the belly spots of Ets1+/− and Sox10LacZ/+ mice (p<0.001) in all three generations examined.
Mutations in the transcription factor Sox10 are responsible for Waardenburg syndrome type IV (WS4), which is characterized by pigmentation defects and aganglionosis of the distal colon (Hirschsprung's disease) (Pingault et al., 1998). Mutations in Sox10 have also been associated with non-syndromic Hirschsprung's disease (Sanchez-Mejias et al., 2010). Given the importance of Sox10 for proper enteric innervation, and the synergistic genetic interaction between Ets1 and Sox10 in melanocyte development, we hypothesized that Ets1 deficiency might also negatively affect enteric innervation. Megacolon was observed in 2 Ets1 null mice, both of which were heterozygous for Sox10 (Ets1−/−::Sox10LacZ/+) (Fig. S3). In both instances, the megacolon manifested early (3.5 and 12 weeks of age) and was found to be the result of severe hypoganglionosis of the distal colon (Fig. S3B and D). Nevertheless, megacolon was only observed in 2 of the 7 animals that were obtained for this genotype.
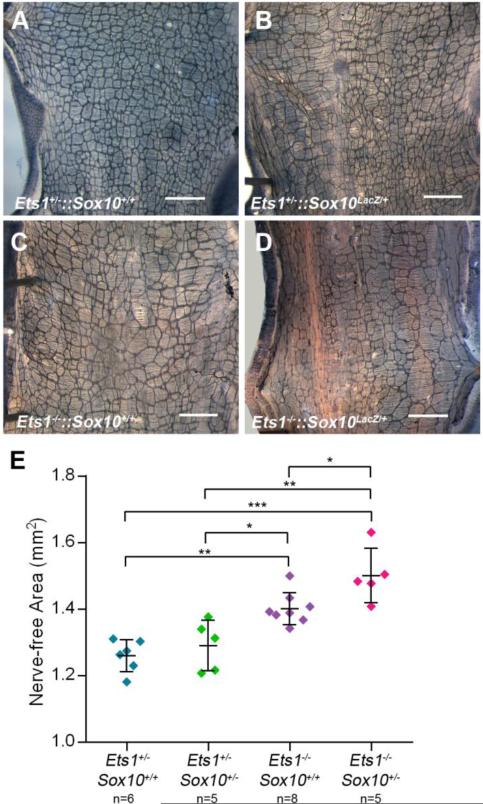
To further characterize the effect of Ets1 deficiency on enteric ganglia patterning, the distal colons of animals from crosses between Ets1+/−, Sox10LacZ/+ and Ets1+/−::Sox10LacZ/+ mice were dissected and subjected to Acetylcholinesterase staining to label enteric ganglia (Fig 5AD). For each colon, the nerve-free (white) area in 2mm2 within a region located 15mm anterior to the anus, was measured (Fig. 5E). The mean nerve-free area of colons from Ets1−/−::Sox10+/+ mice (n=8, mean=1.402±0.048mm2) was significantly larger than the mean nerve-free areas of colons from Ets1+/−::Sox10+/+ (n=6, mean=1.260±0.048mm2, p=0.001) and Ets1+/−:: Sox10LacZ/+ (n=5, mean=1.291±0.075mm2, p=0.010) mice (Fig. 5E), suggesting that Ets1 deficiency results in impaired enteric ganglia patterning in the distal colon.
Figure 5. Ets1 null mutants show deffects in enteric ganglia patterning.
(A-D) Representative images of enteric ganglia labeled via Acetylcholinesterase staining in the distal colons from (A) Ets1+/−::Sox10+/+, (B) Ets1+/−::Sox10LacZ/+, (C) Ets1−/−::Sox10+/+, and (D) Ets1−/−::Sox10LacZ/+ mice (scale bars = 1mm). Scatterplot showing the nerve-free areas (including means and standard deviations) in 2mm2 regions of the distal colons from mice of each genotype. The mean nerve-free area in colons from Ets1+/−::Sox10+/+ mice did not differ significantly from that of Ets1+/−::Sox10LacZ/+ (p=1.000) mice. The mean nerve-free area of colons from Ets1−/−::Sox10+/+ mice was significantly larger than the mean nerve-free areas of colons from Ets1+/−::Sox10+/+ (**p<0.005) and Ets1+/−::Sox10LacZ/+ (*p<0.05) mice. The mean nerve-free area of the colons from Ets1−/−::Sox10LacZ/+mice was significantly greater compared to that of colons from Ets1+/−::Sox10+/+ (***p<0.001), Ets1+/−::Sox10LacZ/+ (**p<0.005), and Ets1−/−::Sox10+/+ (*p<0.05) mice.
In order to explore the effect of Sox10 heterozygosity, we compared the nerve-free areas of colons from Ets1+/− and Ets1−/− mice that were either wild type or heterozygous for Sox10. The mean nerve-free area in colons from Ets1+/−::Sox10+/+ mice did not differ significantly (p=1.000) from that of colons from Ets1+/−::Sox10LacZ/+ mice (Fig. 5E). Although Sox10 heterozygosity did not significantly impact enteric ganglia patterning in Ets1 heterozygotes, the colons of Ets1 null mutants that were also heterozygous for Sox10 had significantly larger nerve-free areas (n=5, mean=1.501±0.081mm2) compared to those of Ets1 null mutants that were wild type for Sox10 (p=0.017, Fig. 5E). Thus, Ets1 null mutants had nerve free areas that were significantly larger than Ets1 heterozygotes when they were wild type or heterozygous for Sox10.
These results provide evidence to support the presence of a genetic interaction between Ets1 and Sox10 in the development of enteric ganglia. Although the colons from the two Ets1−/−::Sox10LacZ/+ animals in which megacolon was detected (Fig. S2) were not included in this analysis, as these were considered extreme cases, the occurrence of megacolon in these mice further supports the possibility that Ets1 and Sox10 interact in enteric ganglia development.
The synergistic genetic interaction observed between Ets1 and Sox10 pointed to a potential functional interaction between these transcription factors in melanocyte and enteric ganglia development. Given that Ets1 is required for Sox10 expression in the chick cranial NC (Betancur et al., 2010), it is possible that it could also be an important regulator of Sox10 expression in the mouse embryo. In order to visualize gross changes in Sox10 expression in Ets1 null mutants, embryos from intercrosses between Ets1+/−::Sox10LacZ/+ mice were harvested at E9.5, E11.5, and E12.5 and Sox10 expression was analyzed via LacZ staining. At E9.5, no apparent changes in LacZ staining were detected in Ets1−/− embryos compared to Ets1+/+ (Fig. 6A and B) and Ets1+/− littermates (not shown). At E11.5, LacZ staining was reduced in 75% of the Ets1−/− (n=4) embryos (Fig. 6D) examined compared to Ets1+/+ (n=3) (Fig. 6C) and Ets1+/− (n=3) (not shown) littermates. At E12.5, a decrease in LacZ staining was observed in 50% of Ets1−/− (n=6) embryos (Fig. 6F) compared to Ets1+/+ (n=3) (Fig. 6E) and Ets1+/− (n=9) (not shown) littermates. The changes in LacZ staining observed in Ets1 null mutant embryos suggest that Ets1 may regulate Sox10 expression during embryonic development. However, these changes could also be the result of a decrease in the number of NC cell derivatives, thus the numbers of Sox10 positive cells in the dorsal root ganglia, where LacZ staining was reduced, were quantified for E12.5 Ets1−/− (n=3) and Ets1+/+ (n=3) embryos. The average number of Sox10 positive cells in the dorsal root ganglia was not significantly different (p=0.753) between Ets1−/− and wild type embryos, indicating that the decreased intensity in LacZ staining observed in Ets1−/− embryos is not caused by a decrease in the number of Sox10-positive NC derivatives.
Ets1 transactivates a neural crest-lineage specific Sox10 regulatory region
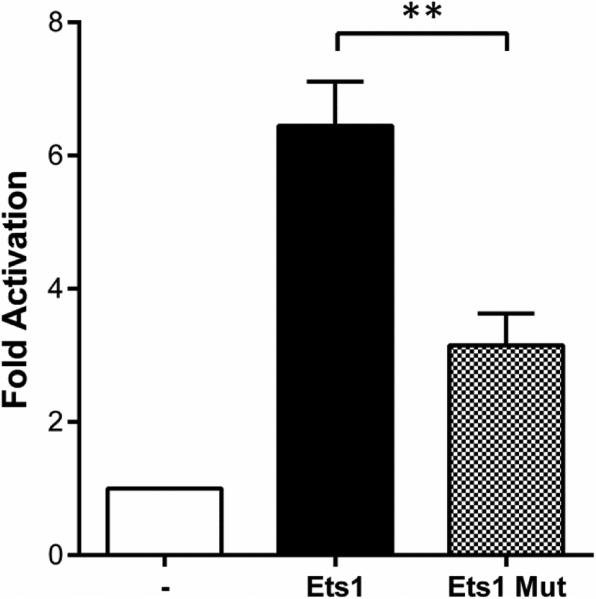
Although many transcriptional targets of Sox10 have been identified, the various aspects of Sox10 regulation are only now beginning to be understood. The Sox10 promoter, which has not been well characterized, does not direct the expression of Sox10 to most of its typical expression sites (Deal et al., 2006). However, 14 multiple conserved sequences (MCS) that can drive Sox10 expression to various cell lineages have been described (Antonellis et al., 2006, Deal et al., 2006, Antonellis et al., 2008). A subset of these sequences was first identified by mutation analysis of the transgene-insertion Sox10Hry mutant, a mouse model of WS4. In this mutant, a 16kb area located 47kb upstream of Sox10 is deleted (Antonellis et al., 2008). This deletion severely impairs but does not completely abrogate Sox10 expression, suggesting that this 16kb region contains cis-regulatory elements necessary for Sox10 expression. Of these elements, Sox10-MCS1c, Sox10-MCS4, Sox10-MCS5, Sox10-MCS7, and Sox10-MCS9 can drive reporter gene expression in cultured melanocytes. Furthermore, both Sox10-MCS4 and Sox10-MCS7 are able to recapitulate Sox10 expression in vivo, in almost all NC-derived structures, including melanocytes and enteric ganglia, where Sox10 is normally expressed (Antonellis et al., 2008). Analysis of the 820bp sequence of the Sox10-MCS4 enhancer using the MatInspector software (Genomatix, Inc.) revealed the presence of 4 potential Ets1 binding sites (Table S1). The ability of Ets1 to activate the Sox10-MCS4 enhancer was tested via luciferase reporter assay in B16-F10 mouse melanoma cells. Compared to cells transfected with the empty PCMV-HA vector, cells transfected with an Ets1 overexpression vector (PCMV-HA-Ets1) showed approximately a 6.5-fold increase in luciferase activity (Fig. 7).
Figure 7. Activation of the Sox10-MCS4 enhancer by wild type and mutant Ets1.
B16 mouse melanoma cells were co-transfected with the Sox10-MCS4 luciferase reporter vector, the pRL-TK Renilla luciferase control vector, and either the PCMV-HA empty vector (-), or an expression vector encoding an HA-tagged version of the full length wild type (Ets1) or mutant (Ets1 Mut) mouse Ets1 coding sequence. Ets1 was found to activate the Sox10-MCS4 enhancer. The presence of the variable spotting mutation resulted in a significant decrease in Sox10-MCS4 enhancer activation (**p<0.005). The firefly luciferase activity was normalized by Renilla luciferase activity. Data represent mean fold activation over the PCMV-HA empty vector (bar height) and standard deviation (error bars) of 3 independent experiments carried out in quadruplicates.
In light of the fact that Ets1 is able to transactivate the Sox10-MCS4 enhancer in mouse melanoma cells, Sox10 expression, as well as the expression of other melanoblast-specific genes, was evaluated in mouse melanocytes (melan-a) and melanoma cells (B16-F10) in which Ets1 was either overexpressed or knocked-down. In mouse melanocytes, we failed to detect substantial changes in the expression levels of Kit, Ednrb, Pax3, Mitf, and Sox10 in cells transfected with an Ets1 overexpression vector, compared to cells transfected with an empty vector (Fig. S4B). Similarly, no considerable changes in the expression levels of Kit, Pax3, Mitf, and Sox10 were detected in mouse melanoma cells overexpressing Ets1 (Fig. S4D).
Given the lack of substantial changes in the expression of melanoblast-specific genes as a result of Ets1 overexpression, we assessed the effect of Ets1 knockdown on the expression of these genes. For both cell lines, no substantial changes on the expression levels of Pax3, Mitf, Sox10, and Kit were detected (Fig. S5). We observed a slight upregulation in Ednrb expression (fold change = 2.006±0.087), in mouse melanocytes transfected with 2 individual Ets1 siRNAs (Fig. S5C); nonetheless, this upregulation was not observed in cells transfected with an Ets1 siRNA pool (Fig. S5B). These results suggest that at least within the context of melanoma cells and differentiated melanocytes, Ets1 does not appear to regulate the expression levels of Sox10, Pax3, Mitf, Kit, and Ednrb.
The variable spotting mutation inhibits Sox10-MCS4 enhancer activation
The variable spotting mutation results in the substitution of a highly conserved Glycine (G102) residue. The G102 residue is present within the Pointed (PNT) domain of the Ets1 protein. This domain enhances the transactivation activity of Ets1 and contains a docking site for the mitogen-activated protein kinase (MAPK) ERK2 (Seidel and Graves, 2002). Docking of ERK2 at this site is critical for the phosphorylation of a threonine residue (T38) that is required for enhanced transcriptional activation by Ets1 via recruitment of the transcription factor CREB-binding protein (CBP) (reviewed in Garrett-Sinha, 2013). In order to test whether the variable spotting mutation affects the ability of Ets1 to activate the Sox10-MCS4 enhancer, the mutation was inserted into the Ets1 ORF via site-directed mutagenesis. A significant decrease in relative luciferase activity was observed in cells transfected with the Ets1 overexpression vector carrying the variable spotting mutation compared to those transfected with the wild type Ets1 overexpression vector (p=0.0022) (Fig. 7). This decrease corresponds to approximately a 49% reduction in Sox10-MCS4 enhancer activation.
Discussion
In the present study, we provide evidence to support a role for the transcription factor Ets1 in NC cell and melanoblast survival, and show that it interacts with Sox10 to promote proper melanocyte and enteric ganglia development. In vivo analysis revealed a significant increase in melanoblast cell death and a concomitant decrease in melanoblast numbers as early as E10.75 in Ets1 null mutants compared to heterozygous littermates. The early survival defect observed in the melanoblast population in Ets1 null mutant mice suggests that Ets1 may be involved in the regulation of one or more early melanoblast-specific genes. Given the increase in melanoblast cell death observed in Ets1 null mutants at E10.75, it was plausible that Ets1 may regulate the survival of NC cells prior to their specification into the melanocyte lineage. Although no significant differences in the numbers of trunk NC cells were observed between E10.25 Ets1 null and heterozygous littermates, a significant increase in the number of dying NC cells was observed in Ets1 null mutants. These results indicate that Ets1 acts as a survival factor for both NC cells, prior to their differentiation into the melanocytic lineage, and melanocyte precursors. Although the presence of viable melanocytes in adult Ets1 null mutants indicates that Ets1 is not essential for proper melanocyte differentiation, it is possible that the large decrease in melanoblast cell numbers observed in these mice could in part arise as a consequence of altered NC cell fate decisions. The presence of ectopic NC cell-derived cartilage nodules in the hearts of Ets1 null mutant mice suggests that Ets1 may act as a negative regulator of cartilage fate in NC cells (Gao et al., 2010). Consequently, in spite of the melanoblast survival defect observed in Ets1 null mutants, we cannot discard the possibility that in addition to being required for melanoblast survival, Ets1 may also be involved in NC migration and or in cell lineage specification.
Other pigmentation mutants in which melanoblast survival defects are observed as early as E10.5-E11.5 include mutants of the transcription factors Sox10, Pax3, and Mitf as well as mutants of the G-coupled Endothelin receptor b (Ednrb) and the tyrosine kinase receptor Kit. Mutations in these genes not only affect melanoblast survival, but also proliferation, migration and final differentiation (reviewed in Baxter et al., 2004; Silver et al., 2006). The transcription factors Pax3 and Sox10 are initially expressed in NC cells at E8.5 prior to their commitment into the melanocytic fate. Mitf, Kit, and Ednrb, on the other hand, are first expressed between E10-E10.5, around the time of melanoblast specification from the NC (reviewed in Silver et al., 2006). All of these genes are required for melanoblast survival and it is possible that the Ets1-induced effect on melanoblast survival is mediated via one or more of these critical melanocytic genes. Of these genes, we considered Sox10 as the most likely downstream effector of Ets1 based on evidence from studies in the avian system. In the chick embryo, Ets1 drives Sox10 expression in cranial NC cells by directly binding to and activating the Sox10E2 enhancer, which is critical for the initiation of Sox10 expression in cranial NC cells (Betancur et al., 2010). The hypopigmentation phenotype observed in Ets1 null mutant mice results from defects in the development of melanocytes derived from trunk NC cells. As such, it is conceivable that in the mouse Ets1 regulates Sox10 expression in the trunk NC. In order to determine whether a genetic interaction exists between Ets1 and Sox10 in melanocyte development, we compared the pigmentation phenotypes of single and double heterozygous progeny from crosses between Ets1+/− and Sox10LacZ/+ mice. Ets1+/− and Sox10LacZ/+ mice were generated in different genetic backgrounds; for this reason, three generations of mice were examined in order to take into account the potential effect of background modifiers. A significant increase in the frequency of belly spot appearance was observed in double heterozygous mice compared to single heterozygotes. Analysis of the belly spot areas of each mouse revealed that the mean areas of hypopigmentation in double heterozygous mice were significantly larger than the sum of the areas of hypopigmentation in single heterozygotes. These results were consistent and significant across all generations and indicate that a synergistic genetic interaction exists between Ets1 and Sox10 in melanocyte development. The fact that the results were consistent across all generations suggests that the presence of strain-specific alleles did not seem to significantly affect the penetrance of the hypopigmentation phenotype in these mice.
Whether synergistic genetic interactions indicate the presence of an underlying functional relationship remains controversial (Perez-Perez et al., 2009). Synergistic genetic interactions identified through the study of double heterozygous pigmentation mutants have often been found to be the result of molecular interactions between the genes in question. The presence of a synergistic genetic interaction between the transcription factor Mitf and the tyrosine kinase receptor Kit was the first indication that these genes work together in the modulation of pigment production. Mice heterozygous for a semi-dominant Mitf mutation (Mitfmi-wh/+) have a uniform ventral spot and a light coat color. Crossing Mitfmi-wh/+ mice to mice heterozygous for a semi-dominant mutation in Kit (KitW-36H), which only have ventral spots and occasionally white feet and tail, resulted in an exacerbated pigmentation phenotype, which was more severe than expected if the hypopigmented areas of the single heterozygotes were added (Beechey et al., 1994). Similar results were obtained when Mitfmi-wh/+ mice were crossed to heterozygous mice in which the LacZ gene was inserted into the Kit locus (KitW-LacZ/+). Additional experiments demonstrated that signaling via the Kit receptor modulates Mitf and is required for Mitf-dependent induction of Tyrosinase (Tyr), a rate limiting enzyme in melanin production (Hou et al., 2000). The study of double heterozygous pigmentation mutants has also revealed the presence of synergistic interactions resulting from underlying molecular interactions between Mitf and the anti-apoptotic factor Bcl2 (McGill et al., 2002) and between the transcription factors Pax3 and Kit (Guo et al., 2010)
To determine whether the synergistic genetic interaction between Ets1 and Sox10 in melanocyte development could be the result of an underlying functional interaction, we examined Sox10 expression in Ets1 mutant mice using the Sox10LacZ transgenic system. A decrease in LacZ expression was observed in at least half of all E11.5 and E12.5 Ets1 null embryos compared to Ets1 heterozygous and wild type littermates suggesting that Ets1 may play a role in the regulation of Sox10 expression. In order to gain a better understanding of the underlying functional interaction that may exist between Ets1 and Sox10, the ability of wild type and mutated Ets1 protein to activate the Sox10-MCS4 enhancer, which is critical for Sox10 expression in mouse melanocytes (Antonellis et al., 2008) was investigated. Analysis of the Sox10-MCS4 sequence revealed the presence of four potential binding sites for Ets1 suggesting the possibility that Ets1 could regulate Sox10 expression via this enhancer. Through luciferase reporter assays we showed that Ets1 is able to drive reporter gene expression through Sox10-MCS4. We also showed that the variable spotting mutation, which we determined to be a mutation in the Ets1 ORF, results in a significant reduction in Sox10-MCS4 enhancer activation. These data suggest that the pigmentation defect observed in Ets1 mutant mice may, at least in part, be the result of decreased Sox10 expression. In spite of this, we did not observe substantial changes in Sox10 expression in mouse melanocytes or melanoma cells in which Ets1 was knocked-down or overexpressed. It is possible that in the mouse melanocyte and melanoma cell lines used for these experiments other factors might be able to compensate for Ets1 deficiency. In the case of Ets1 overexpression, the absence of a significant change in the expression of Sox10 or other potential downstream targets could result if endogenous Ets1 protein is sufficient to drive the expression of said targets, and thus the regulatory regions of these genes are already saturated by the endogenous protein. Additional experiments in NC cells and melanoblasts, aimed at understanding the effects of Ets1 overexpression and knockdown on Sox10 expression, are needed to further characterize the functional relationship between Ets1 and Sox10 in melanocyte development.
The presence of a spontaneous mutation in Ets1 which results in a pigmentation phenotype provided the opportunity to validate the finding that Ets1 can drive expression through Sox10-MCS4. Although the G102E Ets1 mutant protein is able to activate the Sox10-MCS4 enhancer, reporter gene expression is reduced by approximately 49% compared to the wild type Ets1 protein. These results suggest that the variable spotting mutation is hypomorphic, because it reduces but does not completely abrogate Ets1 protein function. The Glycine residue mutated in the spontaneous variable spotting mutant is present within the PNT domain, an important functional domain of the Ets1 protein that enhances the transactivation activity of Ets1. The PNT domain contains a docking site for ERK2 (Seidel and Graves, 2002). Docking of ERK2 at this site is critical for the phosphorylation of a threonine residue that is required for enhanced transcriptional activation by Ets1 via recruitment of the transcription factor CBP (reviewed in Garrett-Sinha, 2013). Mutating this ERK2 docking site was previously found to abrogate Ras/MAPK-mediated enhancement of Ets1 transactivation activity to the same degree as mutating the phosphoacceptor T38 residue (Seidel and Graves, 2002). Three residues within the PNT domain of Ets1, Leucine 114 (L114), L116 and Phenylalanine 120 (F120) have been shown to play a role in ERK2 binding (Seidel and Graves, 2002). Given the reduced Ets1 transactivation activity of the variable spotting mutant protein, it is possible that the G102 residue may be important for docking of ERK2 and T38 phosphorylation; nonetheless, the hypomorphic nature of the G102E mutation suggests that this residue although important, is likely not essential. It is also possible that the effect of the variable spotting mutation on Sox10-MCS4 activation is not related to ERK2 docking and subsequent T38 phosphorylation. Phosphorylation of T38 is important for Ets1 activation of Ras-responsive genes (Seidel and Graves, 2002). To date, no evidence exists to support a direct role for the MAPK pathway in Sox10 activation in the melanocyte lineage. Additionally the G102E mutation is located outside of the (114) LXLXXXF(120) domain, which is crucial for ERK docking (Dittmer, 2003). Additional studies are needed to dissect the specific mechanism via which the variable spotting mutation results in decreased Sox10-MCS4 activation.
In addition to being required for proper melanocyte development, Sox10 is also required for the development of enteric ganglia (Southard-Smith et al., 1998). In the spontaneous embryonic lethal Sox10Dom/Dom mutant mouse, NC cells fail to colonize the intestine as a result of increased apoptosis (Kapur, 1999). This may be the result, at least in part, of decreased activation of the tyrosine kinase receptor c-Ret, which is also required for the development of enteric ganglia, and has been shown to be activated by Pax3 and Sox10 (Lang et al., 2000). The requirement of Sox10 for the survival of enteric ganglia precursors prompted us to examine the colons of Ets1 null mutant mice to determine whether Ets1 deficiency would negatively affect enteric innervation. Ets1 null mice had significantly decreased enteric innervation in the distal colon, compared to heterozygotes, indicating that Ets1 may also be important for the development of enteric ganglia. In view of the role of Ets1 in NC cell survival, it is possible that the defect in enteric ganglia patterning observed in Ets1 null mutant mice may be the result of a reduction in the number of NC cells available to colonize the distal colon.
Throughout the course of the present study, two instances of severe hypoganglionic megacolon were observed in Ets1 null mutant mice, which were also heterozygous for Sox10 (Ets1−/−::Sox10LacZ/+) (Fig. S2). Although no significant difference in the extent of enteric innervation was observed between Ets1+/−::Sox10+/+ and Ets1+/−::Sox10LacZ/+ mice, Ets1−/−::Sox10LacZ/+ mice were found to have colons with greater nerve-free areas compared to Ets1−/− mice that were wild type for Sox10. The significant decrease in enteric innervation observed in Ets1−/−::Sox10LacZ/+ mice compared to Ets1−/−::Sox10+/+ may be the result of a synergistic interaction between Ets1 and Sox10. In light of the absence of a significant difference in enteric innervation between Ets1+/−::Sox10+/+ and Ets1+/−::Sox10LacZ/+ mice and the embryonic lethality of Sox10LacZ/LacZ mutants, it was not possible to directly test the presence of a synergistic genetic interaction using double null mutant mice. Nevertheless, the decreased enteric innervation observed in Ets1−/−::Sox10LacZ/+ mice compared to Ets1−/−::Sox10+/+ mice and the presence of hypoganglionic megacolon in 2 of the 7 Ets1−/−::Sox10LacZ/+ mice examined suggests that Ets1 and Sox10 may interact in the development of enteric ganglia. Our finding that Ets1 promotes the survival NC cells is also consistent with this hypothesis. A decrease in enteric progenitor cell survival has been described in Sox10 heterozygous mice that are also homozygous for the mutant Ednrbsl/sl (piebald) or Edn3ls/ls (lethal spotting) alleles. Compared to single mutants, these animals show a dramatic decrease in the number of enteric neuronal progenitor cells in the digestive tract , which results from increased apoptosis in vagal NC cells prior to the time when they begin to invade the digestive tract at E9.5-10 (Stanchina et al., 2006). In Sox10 homozygous mutants, increased apoptosis of enteric ganglia precursor cells also results in the failure of these cells to colonize the intestines (Kapur, 1999).
Taken together, the results of this study suggest that Ets1 is required for proper NC cell and melanoblast survival, possibly acting upstream of Sox10. Our data indicate that Ets1 and Sox10 act synergistically during melanocyte development and may also interact in enteric ganglia development. Furthermore, the ability of Ets1 to activate a Sox10 regulatory region that is required for Sox10 expression in melanocytes and enteric ganglia, suggests that the synergistic genetic interaction that exists between Ets1 and Sox10 may be the result of an interaction at the molecular level. Additional studies are needed to determine if this interplay also acts in other neural crest derivatives. In the present study, we also identified a spontaneous Ets1 mouse pigmentation mutant, and characterized the negative effect of this mutation on the ability of Ets1 to activate a Sox10 regulatory region. Additional studies are warranted to characterize the molecular aspects of the interaction between Ets1 and Sox10 and the mechanism via which the variable spotting mutation may impair this interaction. The results of this study in combination with the future work proposed will provide a better understanding of the gene regulatory network directing melanocyte development. A detailed characterization of this network is critical because alterations in genes required for the development of melanocytes and other NC cell derivatives have been recognized in several human pathological conditions, including cancer.
Supplementary Material
Highlights.
- Ets1 is the mutated gene in the variable spotting mouse pigmentation mutant.
- Ets1 is required for murine neural crest cell and melanocyte precursor survival.
- Ets1 and Sox10 interact synergistically in melanocyte development
- Ets1 deficiency negatively affects enteric innervation
- Ets1 can activate a Sox10 enhancer and the variable spotting mutation inhibits enhancer activation
Acknowledgments
We thank Dr. Satrajit Sinha (State University of New York (SUNY) at Buffalo, Buffalo, NY) for providing the PCMV-HA-Ets1 and PCMV-HA vectors, and Dr. Eric Svensson (University of Chicago, Chicago, IL) for providing the Ets1 mutant mice. We also thank Dr. Paul Sharp from the Florida International University (FIU, Miami, FL) DNA Core for his help with direct sequencing, and Drs Paulette Johnson and Jianbin Zhu from the FIU Statistical Consulting Department for their help with the statistical analyses of the hypopigmentation area data. We are also grateful to Dr. Fernando Noriega (FIU) for kindly allowing us to use his laboratory's real-time PCR system. This work was supported by the National Institutes of Health (NIH (NIAMS) R15AR062331 to L.K). Additional support for this project was provided by NIH/NIGMS R25GM061347.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonellis A, Huynh JL, Lee-Lin S-QQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, Pavan WJ. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 2008;4:e1000174. doi: 10.1371/journal.pgen.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin S-QQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 2006;15:259–71. doi: 10.1093/hmg/ddi442. [DOI] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Hou L, Loftus SK, Pavan WJ. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 2004;17:215–24. doi: 10.1111/j.1600-0749.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- Beechey CV, Harrison MA. A new spontaneous W allele, W36H. Mouse Genome. 1994;92:502. [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3570–5. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolande RP. Neurocristopathy: its growth and development in 20 years. Pediatr Pathol Lab Med. 1997;17:1–25. [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal KK, Cantrell VA, Chandler RL, Saunders TL, Mortlock DP, Southard-Smith EM. Distant regulatory elements in a Sox10-beta GEO BAC transgene are required for expression of Sox10 in the enteric nervous system and other neural crest-derived tissues. Dev. Dyn. 2006;235:1413–32. doi: 10.1002/dvdy.20769. [DOI] [PubMed] [Google Scholar]
- Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafeur V, Tulasne D, Quéva C, Vercamer C, Dimster V, Mattot V, Stéhelin D, Desbiens X, Vandenbunder B. The ETS1 transcription factor is expressed during epithelial-mesenchymal transitions in the chick embryo and is activated in scatter factor-stimulated MDCK epithelial cells. Cell Growth Differ. 1997;8:655–65. [PubMed] [Google Scholar]
- Gao Z, Kim GH, Mackinnon AC, Flagg AE, Bassett B, Earley JU, Svensson EC. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137:1543–51. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariepy CE, Williams SC, Richardson JA, Hammer RE, Yanagisawa M. Transgenic expression of the endothelin-B receptor prevents congenital intestinal aganglionosis in a rat model of Hirschsprung disease. J. Clin. Invest. 1998;102:1092–1101. doi: 10.1172/JCI3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha L. Review of Ets1 structure, function, and roles in immunity. Cellular and Molecular Life Sciences. 2013;70 doi: 10.1007/s00018-012-1243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X-LL, Ruan H-BB, Li Y, Gao X, Li W. Identification of a novel nonsense mutation on the Pax3 gene in ENU-derived white belly spotting mice and its genetic interaction with c-Kit. Pigment Cell Melanoma Res. 2010;23:252–62. doi: 10.1111/j.1755-148X.2010.00677.x. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Bénard C, Branicky R, Burgess J, Hihi AK, Rea S. Why only time will tell. Mech. Ageing Dev. 2001;122:571–94. doi: 10.1016/s0047-6374(01)00218-4. [DOI] [PubMed] [Google Scholar]
- Herbarth B, Pingault V, Bondurand N, Kuhlbrodt K, Hermans-Borgmeyer I, Puliti A, Lemort N, Goossens M, Wegner M. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5161–5. doi: 10.1073/pnas.95.9.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–76. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–76. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Hou L, Panthier JJ, Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–89. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–46. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, NIH Intramural Sequencing Center (NISC) Chong K, Mullikin JC, Biesecker LG. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86:743–8. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur RP. Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatr. Dev. Pathol. 1999;2:559–69. doi: 10.1007/s100249900162. [DOI] [PubMed] [Google Scholar]
- Keehn CA, Smoller BR, Morgan MB. Expression of the ets-1 proto-oncogene in melanocytic lesions. Mod. Pathol. 2003;16:772–7. doi: 10.1097/01.MP.0000082395.59356.4F. [DOI] [PubMed] [Google Scholar]
- Kola I, Brookes S, Green AR, Garber R, Tymms M, Papas TS, Seth A. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7588–92. doi: 10.1073/pnas.90.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 1998;18:237–50. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Invest. 2000;106:963–71. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. 2nd ed. Cambridge University Press; Cambridge: 1999. p. 472. [Google Scholar]
- Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–65. [PubMed] [Google Scholar]
- McGill G, Horstmann M, Widlund H, Du J, Motyckova G, Nishimura E, Lin Y-L, Ramaswamy S, Avery W, Ding H-F. Bcl2 Regulation by the Melanocyte Master Regulator Mitf Modulates Lineage Survival and Melanoma Cell Viability. Cell. 2002;109:707–18. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- Nagarajan P, Parikh N, Garrett-Sinha LA, Sinha S. Ets1 induces dysplastic changes when expressed in terminally-differentiating squamous epidermal cells. PLoS ONE. 2009;4:e4179. doi: 10.1371/journal.pone.0004179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R, editors. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2003. [Google Scholar]
- Okuducu AF, Zils U, Michaelis SA, Mawrin C, Deimling A. von. Increased expression of avian erythroblastosis virus E26 oncogene homolog 1 in World Health Organization grade 1 meningiomas is associated with an elevated risk of recurrence and is correlated with the expression of its target genes matrix metalloproteinase-2 and MMP-9. Cancer. 2006;107:1365–72. doi: 10.1002/cncr.22130. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Candela H, Micol JL. Understanding synergy in genetic interactions. Trends Genet. 2009;25:368–76. doi: 10.1016/j.tig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Préhu MO, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith JC, Read AP, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–3. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Mollaaghababa R, Hou L, Southard-Smith EM, Hornyak TJ, Arnheiter H, Pavan WJ. Analysis of SOX10 function in neural crest-derived melanocyte development: SOX10-dependent transcriptional control of dopachrome tautomerase. Dev. Biol. 2001;237:245–57. doi: 10.1006/dbio.2001.0372. [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, Chakravart A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–66. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Quéva C, Leprince D, Stéhelin D, Vandenbunder B. p54c-ets-1 and p68c-ets-1, the two transcription factors encoded by the c-ets-1 locus, are differentially expressed during the development of the chick embryo. Oncogene. 1993;8:2511–20. [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff A-KK. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 2005;65:448–56. [PubMed] [Google Scholar]
- Rothhammer T, Hahne JC, Florin A, Poser I, Soncin F, Wernert N, Bosserhoff A-KK. The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell. Mol. Life Sci. 2004;61:118–28. doi: 10.1007/s00018-003-3337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Mejías A, Watanabe Y, M Fernández R, López-Alonso M, Antiñolo G, Bondurand N, Borrego S. Involvement of SOX10 in the pathogenesis of Hirschsprung disease: report of a truncating mutation in an isolated patient. J. Mol. Med. 2010;88:507–14. doi: 10.1007/s00109-010-0592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel JJ, Graves BJ. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 2002;16:127–37. doi: 10.1101/gad.950902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Hou L, Pavan WJ. The genetic regulation of pigment cell development. In: Saint-Jeannet J-P, editor. Neural Crest Induction and Differentiation: Advances in Experimental Medicine and Biology. Vol. 589. Landes Bioscience and Springer Science+Business Media, LLC; New York: 2006. pp. 155–169. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998;18:60–4. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Davisson MT. Variable spotting (vs). Mouse News Lett. 1988;81:71. [Google Scholar]
- Stanchina L, Baral V, Robert F, Pingault V, Lemort N, Pachnis V, Goossens M, Bondurand N. Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev. Biol. 2006;295:232–49. doi: 10.1016/j.ydbio.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Read AP, Newton VE, Harris R, Balling R, Gruss P, Strachan T. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992;355:635–6. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, Collins FS, Mullikin JC, Biesecker LG. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to mesenchyme transition to collective cell migration. Dev. Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- Vandenbunder B, Pardanaud L, Jaffredo T, Mirabel MA, Stehelin D. Complementary patterns of expression of c-ets 1, c-myb and c-myc in the blood-forming system of the chick embryo. Development. 1989;107:265–74. doi: 10.1242/dev.107.2.265. [DOI] [PubMed] [Google Scholar]
- Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, Muthusamy N, Man AK, Oshima RG, Leone G, Ostrowski MC. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–30. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, Ivy D, Perryman MB, Garrett-Sinha LA, Grossfeld P. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum. Mol. Genet. 2010;19:648–56. doi: 10.1093/hmg/ddp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Overbeek PA. Tyrosinase-related protein 2 promoter targets transgene expression to ocular and neural crest-derived tissues. Dev. Biol. 1999;216:154–63. doi: 10.1006/dbio.1999.9480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.