Abstract
Cardiovascular diseases (CVD) are commonly thought to be complex, non-Mendelian diseases that are influenced by genetic and environmental factors. A growing body of evidence suggests that epigenetic pathways play a key role in vascular biology and might be involved in defining and transducing CVD inheritability. In this review, we argue the importance of epigenetics in vascular biology, especially from the perspective of endothelial cell (EC) phenotype. We highlight and discuss the role of epigenetic modifications across the transcriptional unit of protein coding genes, especially the role of intragenic chromatin modifications, which are underappreciated and not well characterized in the current era of genome wide studies. Importantly, we describe the practical application of epigenetics in CVD therapeutics.
Keywords: Epigenetics, long non-coding RNA, endothelial cells, intragenic epigenetic modifications, DNA methylation, histone post-translational modifications
Introduction
Prevalent cardiovascular diseases (CVD), such as coronary artery disease (CAD), are thought to be complex, non-Mendelian diseases in which genetic and environmental factors figure prominently1. Traditionally, the inherited aspect of CVD is attributed to genetic variations in multiple protein-coding genes. Hence, studies have tested this hypothesis by conducting genome wide association studies (GWAS), often with subsequent meta-analysis, to identify SNPs that are present or absent in patients with CAD. Although the earliest collected studies indicate that 46 genome-wide significant loci are associated with CAD, they only account for ~10% of CAD inheritability, suggesting that other factors are involved 1. One other major factor is epigenetics, which can be defined as alterations in chromatin that are not due to changes in the DNA sequence per se2. These chromatin-based pathways may or may not be faithfully inherited with mitotic or meiotic cell division.
What is Epigenetics?
DNA methylation, histone density, variants, and post-translational modifications, and RNA-based mechanisms represent the 3 interrelated pathways that figure prominently in the molecular basis of epigenetics. We will briefly discuss their commonly known regulatory mechanisms and role in endothelial cell (EC) biology.
DNA Methylation
DNA methylation is an epigenetic modification that occurs at the 5-position of cytosine to generate 5-methylcytosine and is commonly observed to occur in the context of a cytosine followed by a guanine (CpG)3. Although CpG sites are relatively sparse in the genome, due to deamination of 5-methylcytosine to thymine during the evolution of the mammalian genome, certain regions are not CpG depleted and these regions are referred to as CpG Islands (CGIs)3. The addition of a methyl group to cytosine is catalyzed by DNA methyltransferases (DNMTs), namely DNMT1, DNMT3a and DNMT3B4. In contrast, DNA demethylation can occur passively or actively4. The latter mechanism is currently under investigation and thought to be mediated, in part, by the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine and subsequent oxidative products by the ten-eleven translocation enzyme family (TET1, TET2, and TET3)5. These oxidative products are thought to be substrates for base excision repair glycosylases that subsequently restores the initial cytosine6.
At gene promoters, DNA methylation mediates transcriptional repression. It is mechanistically thought to mediate repression by preventing the binding of transcription factors (TF), including the CCCTC-binding factor (CTCF), and recruiting methyl binding proteins, such as MeCP2, that can bind 5-methylcytosine to prevent the recruitment of transcriptional machinery4. Methyl binding proteins can also recruit other chromatin modifying enzymes, including histone deacetylases (HDACs) and histone methyltransferases, to further reduce chromatin accessibility4.
Although studies of DNA methylation in CVD and EC disease phenotypes are still early in their infancy, tantalizing new studies suggest that these are key next questions. For example, recent studies on the role of DNA methylation in ECs have shown that aberrant DNA methylation at the promoters of flow-inducible genes contributes to atherosclerosis. Prominently, an in vivo model of disturbed blood flow was performed by partial carotid ligation in mice. This model mimics the hemodynamic state of regions in the aorta that are prone to atherosclerotic plaque formation. DNMT1 was induced by disturbed flow and was associated with inflammation. Importantly, pharmacological inhibition of DNMTs in this in vivo model showed reduced atherosclerotic plaque growth. DNMT1 is argued to promote inflammation by hypermethylating a set of flow-sensitive genes, including HoxA5, under conditions of disturbed flow7. Consistent with these findings, a comparison of swine ECs from the athero-susceptible inner curvature of the aortic arch and the athero-protected descending thoracic aorta found that genes with differentially DNA methylated regions (DMRs) have functions associated with atherosclerosis, including oxidative stress and inflammation8. Interestingly, the same group found that DNMT3a was induced by disturbed flow9. This was associated with increased DNA methylation of the promoter and inhibition of the transcription of KLF4, a flow-inducible factor required for maintenance of the anti-inflammatory and anti-thrombotic properties of healthy vascular ECs9. Collectively, these studies argue that DNMTs and aberrant DNA methylation promote a pro-inflammatory state for atherosclerotic plaque formation. Additional studies will need to clarify the key regulators of aberrant DNA methylation at disturbed flow-inhibited genes. Studies will also need to ask which genes are activated or repressed by flow-mediated epigenetic changes.
Histone Biology
Histones are proteins that package DNA into chromatin by forming octamers composed of 2 H3-H4 and 2 H2A-H2B heterodimers. These octamers are wrapped by 146 nucleotides of DNA to form a nucleosome. Nucleosomes are connected to each other by linker DNA to form a “beads-on-the-string” chromatin structure that can be further condensed into higher order structures2, 10. Histone proteins can be modified by a myriad of post-translational modifications (PTM) to regulate transcription2, 10. The most well-characterized classes of histone PTM are lysine methylation and acetylation2.
Histone acetylation is dynamically regulated by histone acetyltransferases (HATs) and HDACs. Histone acetylation is thought to activate transcriptional initiation by neutralizing the basic charges on the lysine tails and recruiting bromodomain-containing proteins, including other HATs and chromatin remodeling complexes, to increase chromatin accessibility at the promoter and enhancer (Figure 1). Subsequently, TFs and the transcriptional machinery can be recruited to activate transcription initiation11.
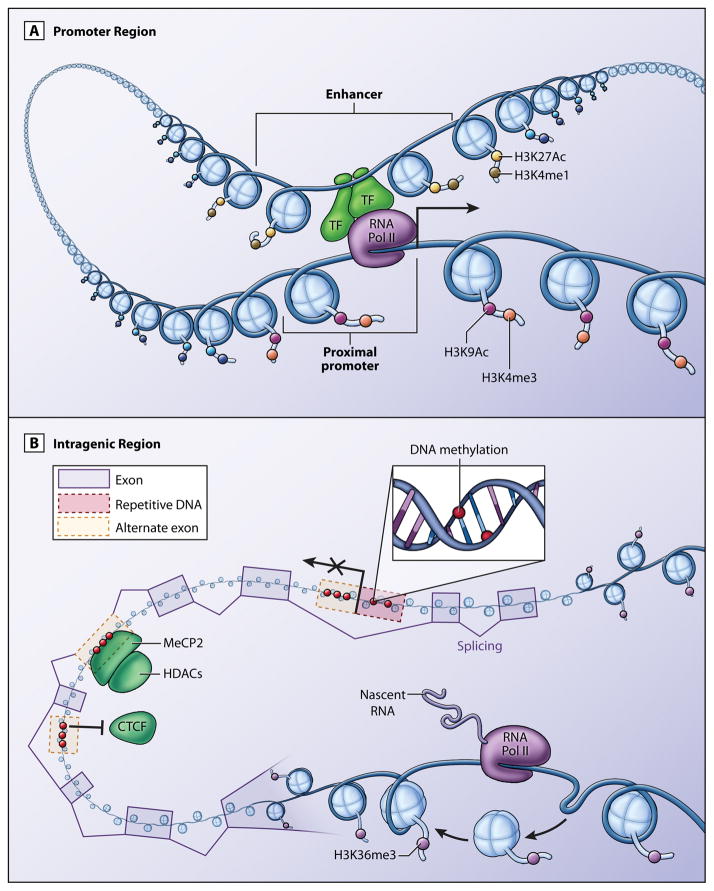
Figure 1.
Epigenetic-based mechanisms of gene regulation. (A) The epigenetic signatures at the promoter and enhancer of a gene contribute to the recruitment of trans factors (TF) and RNA Polymerase II (Pol II) for transcription initiation regulation. Epigenetic modifications that are observed frequently at proximal promoter regions include H3K9Ac (pink) and H3K4me3 (orange). Common epigenetic modifications that are associated with an active enhancer are H3K4me1 (olive) and H3K27Ac (yellow). (B) Intragenic epigenetic modifications are critical for transcription and mRNA variant expression. H3K36me3 (purple) is commonly associated with transcription elongation by RNA polymerase II through intragenic regions. HK36me3 is believed to be necessary for the inhibition of spurious transcription initiation. Intragenic DNA methylation (red) is positively correlated with gene expression and may prevent spurious transcript initiation from DNA repetitive elements or alternative promoters. Intragenic DNA methylation also affects alternative exon inclusion. DNA methylation may interfere with CTCF recruitment to inhibit exon inclusion. Alternatively, DNA methylation may recruit MeCP2 and HDACs to promote exon inclusion.
Similar to histone acetylation, histone lysine methylation is regulated dynamically by histone lysine methyltransferases (KMTs) and histone lysine demethylases (KDMs)3. Unlike histone acetylation, mono-, di- and tri-methylation of histone lysine residues can occur. Importantly, at any specific lysine residue, histone acetylation and methylation are mutually exclusive. The specific lysine residue and even the number of methyl groups on that specific lysine residue can have different downstream effects on transcription initiation3. For instance, H3K4 monomethylation is localized to both promoters and enhancers (Figure 1). This modification is commonly enriched at repressed, inducible genes and it is involved in spatially restricting the recruitment of factors that recognize trimethylated H3K4, including ING1, to active promoters12. In contrast, H3K4 trimethylation is enriched at the promoter of active genes and is known to recruit factors important for transcription, including TFIID13 (Figure 1).
Although most nucleosomes consist of the 4 “canonical” core histones, histone variants can be incorporated into nucleosomes, typically in a replication independent manner, to mediate transcription regulation 14. One prominent histone variant is H2A.Z. H2A.Z is enriched at the promoters and enhancers of transcriptionally active genes and the promoters of inactive genes that are poised for expression14–16. The transcriptional activation of H2A.Z enriched promoters may, in part, be regulated by H2A.Z histone acetylation16.
Much effort has focused on characterizing the role of histone modifications in EC biology through studies on histone modifying enzymes, especially HDACs. Pharmacological inhibition and siRNA depletion of HDACs have shown both pro- and anti-angiogenic activity in models of postnatal angiogenesis suggesting that unique HDACs promote and inhibit angiogenesis17–19. Furthermore, HDAC activity has been shown to be important for endothelial differentiation of embryonic stem cells (ESCs) and adult endothelial progenitor cells20, 21.
Some HDACs have also been reported to contribute to atherosclerosis. HDAC3 expression was observed to increase in the aorta of ApoE−/− mice, especially at branching areas, and the inner curvature of the aortic arch in rats. These regions experience disturbed blood flow and are prone to atherosclerotic plaque formation. Consistent with this observation, ECs that are subjected to in vitro models of disturbed flow show increased HDAC3 expression 22, 23. Importantly, HDAC3 depletion in aortic isograft models of ApoE−/− mice results in the formation of severe atherosclerotic lesions and even vessel rupture. This is interesting in that, in contrast to DNMTs, increased HDAC3 is protective against atheroprone flow. HDAC3 contributes to atherosclerosis in part, through its interaction with Nrf2 and MEF2, which prevents these TFs from binding and activating the expression of target downstream genes with anti-inflammatory and -oxidative activities22. It is currently unknown if HDAC3 can also act directly on chromatin to promote atherosclerotic progression. Similarly, HDAC2 can contribute to atherosclerotic progression as suggested by its increased expression in the inner curvature of the aortic arch in rat and ECs subjected to disturbed flow in vitro. Furthermore, HDAC2 can also interact and deacetylate Nrf222. Moreover, oxidized low-density lipoprotein (OxLDL), a proatherosclerotic molecule, is found to downregulate HDAC2 expression resulting in the transcriptional activation of Arg2, which results in eNOS uncoupling and increased eNOS-dependent reactive oxidative species production24. Genetic deletion of HDAC2 in EC will need to be conducted to assess whether HDAC2 is also a protective response to atheroprone flow.
SIRT1, which belongs to an unique family of TSA-insensitive HDACs with a requirement for nicotinamide adenine dinucleotide (NAD+), may have an atheroprotective role 25. Importantly, SIRT1 expression is reduced in atherosclerotic plaques. Furthermore, studies in ApoE−/− SIRT1+/− mice show that SIRT1 prevents endothelial activation26 SIRT1 is thought to mediate its atheroprotective role by various mechanisms. This suggests that decrease in SIRT1 in atheroprone flow is mechanistically important. SIRT1 can deacetylate eNOS protein to increase nitric oxide production and promote an anti-inflammatory and -oxidative environment under atheroprotective flow conditions27. At the transcriptional level, SIRT1 can activate the expression of anti-oxidant genes in ECs by deacetylating and increasing the activity of the TFs FoxO3a and PGC-1α at these genes28. Surprisingly, reduced H4K16 acetylation and increased elongating Pol II recruitment occur at these genes under oxidative stress conditions28. This observation suggests that the H4K16 deacetylase activity of SIRT1 may possibly activate these genes by reducing spurious transcript production. Additionally, the loss of SIRT1 in atherosclerotic plaque may de-repress pro-atherosclerotic genes in a H4K16 acetylation dependent manner as suggested for plasminogen activator inhibitor-129. These latter concepts are important in that they highlight a key point. Later in this review, we expand on the concept of intragenic epigenetic modification and contrast their functional importance with epigenetic modifications in the proximal promoter regions. Increases or decreases in HDACs may have contrasting effects on transcription if they are recruited to the proximal promoter versus the intragenic regions.
Long Noncoding RNA (LncRNA)
LncRNAs are distinct from short noncoding RNAs, such as miRNAs, which are also known to play a role in EC function and atherosclerosis and are thought to predominantly regulate gene expression through post-transcriptional mechanisms in the cytoplasm30. LncRNAs are arbitrarily defined by their length of >200 nucleotides. This newly described heterogeneous class of transcribed regions of the non-protein coding genome may or may not be 5′-capped, spliced, or polyadenylated31. They are in the process of being characterized and are currently observed to act in cis or trans. They can be localized and functional in either the nucleus or cytoplasm. LncRNAs functions can be mediated by RNA-RNA, RNA-DNA, and RNA-protein interactions31. An important number of them regulate chromatin function in the nucleus31. Hence, they can be considered to function in a chromatin-based fashion.
Although current efforts show that epigenetic modifiers play a critical role in ECs under disease and physiological conditions, little is known about the repertoire of unique EC-enriched lncRNAs, their function and mechanisms of action, and the extent of their contribution to CVDs. However, studies have begun to identify lncRNAs that play a prominent role in ECs. A recent genome-wide analysis was conducted to identify the most highly expressed lncRNAs in ECs32. One of these lncRNAs is metastasis-associated lung adenocarcinoma transcript 1 (MALAT1). Although highly expressed, MALAT1 is widely expressed in tissues representing all germ layers and is found to be upregulated in various tumours33. Therefore, it is not EC-enriched. In ECs, MALAT1 is thought to regulate cell proliferation in vitro and in vivo and modulate cell migration32. Importantly, MALAT1 is upregulated in ECs under hyperglycemic conditions and contributes to diabetic retinopathy in rodent models of diabetes mellitus, in part, by increasing the expression of inflammatory cytokines in ECs 34. The mechanistic role of MALAT1 is currently not clear. However, MALAT1 binds intragenic regions of target expressed genes and associate with chromatin modifiers involved in transcription activation, such as SET2, and pre-mRNA splicing factors35, 36. Future studies will have to identify EC-enriched lncRNAs and assess if they are functionally important epigenetic modifiers of EC gene expression.
Epigenetic regulation of a prototypic endothelial-enriched gene: eNOS
Early studies have focused on characterizing the role of epigenetic pathways in the vascular endothelium by focusing on the promoters of EC-expressed genes. One of the most well-characterized examples of this approach is eNOS (NOS3)3. eNOS plays a prominent role in vascular biology as shown by eNOS null mice that are characterized by systemic and pulmonary hypertension, abnormal vascular remodeling, defective angiogenesis, and pathological healing to vascular injury37. Importantly, eNOS is thought to be atheroprotective and its expression is reduced in the ECs of atheroprone regions of mouse aorta38. EC-enriched expression of eNOS is, in part, regulated by an unique epigenetic signature. Specifically, the eNOS proximal promoter is enriched with H3K9Ac, H4K12Ac, H3K4me2, H3K4me3, and the H2A.Z histone variant in ECs, relative to non-ECs3. These histone modifications and histone variant are functionally important for eNOS transcription activation. In contrast, the eNOS proximal promoter is DNA hypermethylated in non-EC to mediate transcription repression. Importantly, dynamic changes in the epigenetic signature of the eNOS promoter are involved in the activation and repression of eNOS expression in ECs subjected to environmental stimuli, especially hypoxia3. These early studies suggest that epigenetic pathways play a functional role in healthy and diseased vascular endothelium. As discussed below, future studies should not be limited in their bias towards cis regulatory elements at the proximal promoter, which are thought to be important for gene expression. They should also address intragenic epigenetic modifications.
ENCODE and Intragenic Modifications
Defining the cell-specific epigenetic modifications that encompass EC-enriched genes, including their proximal promoters, enhancers, and intragenic regions, is a current goal of many groups. Although studies that focus on the promoter of protein coding genes, such as eNOS, are valuable, we need a comprehensive road map for how endothelial cells are distinct from other cell types. This cannot be inferred indirectly from studying peripheral blood genomic DNA or other solid cell types. Is the heterogeneity of EC phenotypes mediated by cis/trans paradigms, epigenetic marks or both? Recent whole genome profiles of various epigenetic modifications have been generated for cells of the cardiovascular system, including the vascular endothelium. These studies begin the process of addressing this question and circumvent the bias of assessing the role of epigenetic modifications at the promoter7, 8, 39, 40. One of the most prominent efforts in dissecting the epigenetic landscape of the vascular endothelium came from the Encyclopedia of DNA elements (ENCODE) project41.
It is estimated that the ~20,000 – 25,000 protein-coding genes in the human genome is represented by 1.5% of the genome. The primary goal of the ENCODE project was to determine the role of the remaining non-protein coding components. In a series of landmark papers, the ENCODE consortium argued that 80% of the genome had biochemical function, much of which was involved in controlling the expression of the protein-coding genome41. This finding is based on the chromatin-based datasets that were generated in the ENCODE project. These include, but are not limited to, RNA sequencing, TF binding, long-range interaction, DNA methylation and histone modification profiles. By integrating the ENCODE epigenetic datasets, cell-specific promoters and putative enhancers have been identified in ECs that play a role in the EC gene expression profile39 (Figure 1). In particular, the EC specific promoters and putative enhancers that are identified by their epigenetic profiles are associated with genes that are enriched for EC functions, including angiogenesis and blood vessel morphogenesis39. Although a large amount of epigenomic data was generated for ECs, the role of epigenetic pathways in EC received relatively modest attention compared to other cell types. For instance, ChIP-seq datasets for ECs from ENCODE are generated at relatively low resolution and some genes are not accurately annotated, such as eNOS. Furthermore, various datasets, including DNA methylation profiling and long-range interaction data, were not generated for ECs by ENCODE41.
Aside from characterizing promoters and putative enhancers, the vast amount of epigenomic data that have been, and will be, generated can be used to characterize under-appreciated aspects of gene regulation that may have a functional role in EC biology. One such aspect is intragenic epigenetic modifications. In the following sections, we will describe their currently known regulatory roles and potential contribution to vascular biology.
DNA methylation
Although the role of intragenic DNA methylation is not well understood, it is likely to play a critical role in the vasculature based on the observation that the majority of human atherosclerosis-specific differentially methylated CpGs are mapped to intragenic regions40. Similarly, athero-susceptible DMRs in swine are also enriched in exons8. Surprisingly, intragenic DNA methylation is positively correlated with gene expression (Figure 1) 42. However, genomic integration of a transgene with patch intragenic DNA methylation into a murine cell line show reduced expression and Pol II occupancy relative to its unmethylated counterpart43. Thus, the positive correlation of intragenic DNA methylation with gene expression might be related to its function in preventing spurious expression of repetitive elements44 while a gene is actively transcribed (Figure 1). Although little is known about the function of intragenic DNA methylation, recent literature has suggested that regional intragenic DNA methylation can regulate the expression of mRNA variants42, 44–47 (Figure 1).
DNA methylation has been observed in mammals to globally repress transcription from intragenic CpG islands (CGIs). The absence of this intragenic methylation allows spurious transcriptional initiation at non-canonical sites. These regions now become alternative promoters of genes, which may or may not affect the open reading frame42, 44, 46. Interestingly, differential DNA methylation at these intragenic CGIs can regulate the tissue-specific expression of mRNA variants and evidence of transcription initiation has been observed at these regions 46.
Alternative splicing might also be regulated by DNA methylation. Studies on CD45, whose splice variants are tightly correlated with lymphocyte development, have shown that DNA methylation can affect exon retention through its effect on the recruitment of CTCF44, 47. This observation is interesting as CTCF is more known for its role in promoter activation and repression, blocking of enhancer-promoter interaction, and regulation of three-dimensional chromatn organization48. Exon 5 retention in CD45 is associated with the lack of methylation of its respective intragenic genomic region. The lack of methylation allows the binding of CTCF to the exon, promotes Pol II pausing, and possibly allows co-transcriptional spliceosome assembly for exon 5 inclusion (Figure 1). Although recent observations suggest that DNA methylation has little effect on CTCF binding on a global scale, methylation sensitive CTCF-binding sites have been noted and are more frequently observed in exons49. This observation suggests that DNA methylation-sensitive, CTCF-dependent regulation of splicing may occur at an unique set of genes47. Contrary to these findings, intragenic DNA methylation has been observed by others to be enriched at alternative exons that are included in many mRNA variants45. In these instances, inclusion of these alternative exon is mediated in a Methyl CpG binding protein (MeCP2)- and HDAC-dependent manner (Figure 1). Future studies will need to address the chromatin context by which these distinct DNA methylation-dependent splicing mechanisms occur.
Histone Post-translational Modifications
In contrast to the studies that address genomic regions implicated in transcriptional initiation, the role of specific histone modifications in intragenic regions is not well understood. Although a myriad of intragenic histone post-translational modifications have been identified, we will focus on histone lysine acetylation and methylation.
Intragenic Histone Acetylation
High throughput studies on genomic localization of histone acetyltransferases (HATs) and HDACs have shown that some HATs and HDACs are co-enriched at the promoter and intragenic regions of transcriptionally active genes in mammals50. Consistent with this observation, various acetylated histone residues, such as H3K14 acetylation (H3K14Ac), are enriched in the transcribed regions 51, 52. These observations are relevant based on the fact that HATs that can catalyze intragenic histone acetylation also play a critical role in vessel formation. For instance, PCAF, a HAT, contributes to arteriogenesis, the remodeling of pre-existing collateral arterioles into larger arteries53. Furthermore, mice that are deficient in HBO1, a prominent H3K14 HAT, are embryonically lethal and show defects in embryonic vascular remodeling52. Future studies will have to investigate if other HATs and HDACs that catalyze intragenic histone acetylation also play a prominent role in the vasculature and are involved in EC biology.
Mechanistically, the observation that HATs and HDACs are enriched in intragenic regions suggest that dynamic histone acetylation in the intragenic region is important for transcription elongation. In agreement, hyperacetylated histone residues, such as AcH3K14, can co-enrich with Brd2 and Brd3 at intragenic regions in mammals. These bromodomain-containing proteins can bind these acetylated histone residues and facilitate transcription through chromatin in vitro54. This is consistent with an elongating RNA polymerase II (Pol II) transcription complex evicting histones as the DNA template is made accessible for Watson-Crick base pairing of DNA with RNA substrates (Figure 1).
Moreover, yeast studies demonstrate that dynamic intragenic histone acetylation maintains the delicate balance of efficient transcription elongation without cryptic initiation of transcription in intragenic regions. The loss of HATs Gcn5 and Elp3 in yeast result in histone H3 hypoacetylation in the intragenic region of genes and is associated with their transcriptional inhibition55. Overall inhibition of transcription by Gcn5 deficiency is due to defects in nucleosomal eviction during transcription elongation56. In contrast, mutations of HDACs, such as the Rpd3S HDAC complex, result in increased histone acetylation and intragenic cryptic transcript initiation57. Future studies will be needed to better understand the functional role and relevant protein complexes implicated in dynamic intragenic histone acetylation in mammals.
Intragenic Histone methylation
Intragenic histone methylation also plays a prominent role in transcriptional elongation. One of the most prominent modifications is H3K36 trimethylation (H3K36me3), which is catalyzed by SET2. SET2-dependent H3K36me3 suppresses spurious intragenic transcription by recruiting the Rpd3S complex and preventing the incorporation of newly acetylated histones over transcribed intragenic regions (ORFs) in yeast57, 58. The functional role of H3K36me3 is conserved in humans and, in part, mediated by FACT-dependent histone H2B exchange59. Importantly, mice that are deficient for HYPB/Set2D, the H3K36 methyltransferase, are embryonically lethal and show defects in embryonic vascular remodeling suggesting that H3K36me3 may play a prominent role in vascular biology60.
The function of other intragenic histone methylation-based modifications is currently being investigated. Of note, H3K27me3, which commonly occurs at the promoter, is enriched at the intragenic regions of some target genes61, 62. In-depth analysis in neural stem cells have shown that intragenic region of some TGF-β-inducible genes are enriched for H3K27me361, 62. Importantly, TGF-β induces the recruitment of JmjD3 (KDM6B), a H3K27me3 demethylase, to catalyze histone demethylation and promote transcriptional elongation. TGF-β signaling plays a critical role in developmental vessel formation and mutations in the regulators of TGF-β signaling cause diseases with endothelial pathologies, such as Hereditary Hemorrhagic Telangiectasia63. Thus, critical TGF-β inducible genes involved in vessel development and disease might also be regulated by intragenic H3K27me3 in ECs. Consistent with this possibility, previous JmjD3-deficient mouse ESCs show compromised cardiovascular differentiation64. Future studies will have to address the role of intragenic H3K27me3 in ECs and define how intragenic H3K27me3 represses transcriptional elongation.
Therapeutic potential of Targeting Epigenetic Modifiers
Alterations in the activities of critical epigenetic regulators might be integral in the therapeutic approaches of CVDs. Such therapeutic approaches include the differentiation of inducible pluripotent stem cells (iPSCs) to endothelial cell (EC)-like cells for treating CVDs and the manipulation of CVD-associated lncRNAs, such as ANRIL.
Epigenetics in cellular reprogramming and its application to CVD
The advent of inducible pluripotent stem cells (iPSCs) offers a promising potential for cell-based therapies of CVD. Briefly, iPSCs are somatic cells that are reprogrammed to an embryonic stem (ES) cell-like state. Although they were initially established by the combined ectopic expression of Oct3/4, Sox2, Klf4, c-Myc, it was later discovered that different cocktails of trans factors could facilitate this process65. The use of iPSC-differentiated EC-like cells have been shown to contribute to vascular repair in various vascular injury models, such as hind limb ischemia66, 67. These studies are promising, given that patient derived iPSC-differentiated EC-like cells may offer treatment options for critical limb ischemia through the process of modifying blood vessel formation (Figure 2).
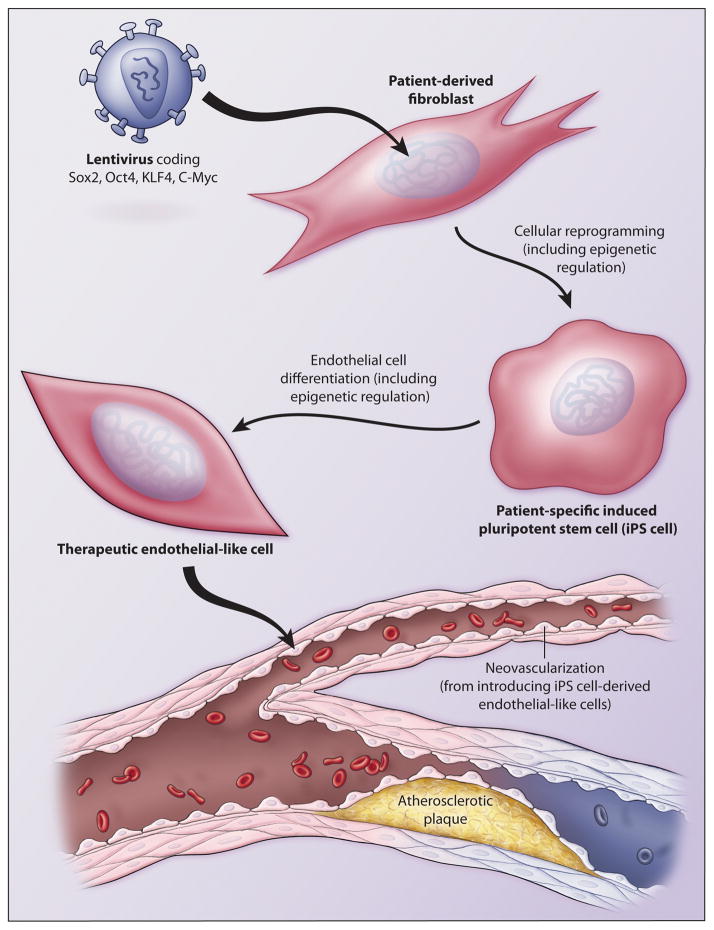
Figure 2.
Inducible pluripotent stem cell (iPSC)-based therapies for cardiovascular disease (CVD). Patient-derived fibroblasts can be reprogrammed into iPSCs and differentiated into endothelial (EC)-like cells for cell-based therapies of CVD. The processes of cellular reprogramming and endothelial differentiation involve dynamic changes in the epigenetic landscape.
A major challenge in applying patient derived iPSC-differentiated ECs for treating CVDs is the low efficiency (<3%) in iPSC reprogramming65. The low efficiency reflects the required, dramatic changes in gene expression that are dependent in a major way on profound changes in their epigenetic landscape. In a comprehensive analysis of epigenetic changes during cellular reprogramming, chromatin was found to be more “open” during the early reprogramming phase and it subsequently returns to its original stem-cell like state based on overall changes in levels of the repressive H3K27me3 modification at genomic loci68. Genes that are expressed during the reprogramming process are associated with pluripotency and their expression is correlated with a decrease in H3K27me3 and the presence of H3K4me3 at their proximal promoters. In contrast, increased H3K27me3 enrichment occurs at genes that are associated with development and cell fate determination. During the later phases of iPSC reprogramming, DNA methylation patterns are also altered and associated with repression of gene expression.
Consistent with the importance of altering the epigenetic landscape, inhibition of epigenetic modifiers, such as DNA methyltransferases (DNMT) and histone deacetylases (HDACs), improve the efficiency of iPSC reprogramming69, 70. A possible reason for this improvement is that such manipulation helps transition partially reprogrammed cells to more fully totipotent iPSCs. Indeed, pharmacological inhibition of epigenetic modifiers have been used to direct the transition of partially reprogrammed cells to more totipotent iPSCs69, 70.
Little is known about the effects of altering epigenetic modifier activity on the differentiation of iPSCs to EC-like cells even though significant progress has been made in generating EC-like cells efficiently via iPSC differentiation to cord blood endothelial colony forming-like cells66. However, studies have shown that TGFβ, Wnt, and VEGF signaling play a prominent role in differentiating iPSCs to EC-like cells71. Future studies may identify critical epigenetic modifiers that improve the efficiency of EC differentiation. Furthermore, it is essential that we understand the epigenetic modifications that are prevalent at EC-enriched genes and how exogenous stimuli and developmental processes, especially TGFβ, Wnt, and VEGF signaling, set up these unique epigenetic signatures.
The lncRNA ANRIL and its role in CVD
Intriguingly, lncRNA might reflect the interaction between epigenetic pathways and CVD genetic determinants. In CAD, for instance, many of the associated genetic variants occur in non-protein coding regions 1. Although this can reflect the contribution of distal regulatory elements for protein-coding genes, it can alternatively suggest the actions of long noncoding RNAs (lncRNAs). The latter possibility is exemplified by the 9p21 chromosome locus, which contains the strongest genetic risk factor for CAD1.
The 9p21 risk variants are single nucleotide polymorphisms (SNP) commonly observed in GWAS studies on CAD. These variants are associated with CAD-related outcomes, including myocardial infarction, and are independent of covariates, such as lipids and hypertension1. Strikingly, the top SNP risk allele in the locus increases CAD risk by 36% per copy1. Aside from CAD, the 9p21 variants are associated with other vascular phenotypes, including stroke1.
These 9p21 risk variants map to a long noncoding RNA gene known as antisense non-coding RNA at the INK4 locus (ANRIL). The closest flanking protein coding genes are CDKN2A and CDKN2B, which are about 100 kb away and encode p16INK4A/p14ARF and p15INK4B respectively. Importantly, some of these genetic variants are associated with increased ANRIL expression in cancer and vascular tissue samples72, 73. Increased ANRIL expression is observed to inhibit apoptosis, increase cell proliferation and enhance cell adhesion, which might support its pro-atherogenic function73. Importantly, ANRIL regulates the expression of genes associated with these processes 73. Mechanistically, ANRIL can associate with polycomb repressive complex 2 (PRC2), which catalyzes histone H3 lysine 27 trimethylation (H3K27me3), to repress target protein-coding genes, including p15INK4B and p2172–74. ANRIL can also recruit PRC2 to repress KLF2, a laminar flow-inducible transcription factor that is important for maintaining a healthy endothelial cell (EC) phenotype, suggesting that ANRIL can potentially act similarly in CVD72.
Additional studies on ANRIL will need to be conducted in relevant cell types of CVD, especially ECs. Furthermore, the mechanism by which ANRIL enhances CVD risk also remains elusive and requires further investigation. Since the ANRIL lncRNA can recruit epigenetic modifiers to regulate a distinct set of genes, this may suggest that manipulation of lncRNA expression can be used to modulate the expression of specific genes such as those relevant to CVD.
Conclusions
With the use of genome-wide approaches for probing the epigenome, the catalogue of pathological epigenetic changes that occur in CVDs grow ever larger. The current challenge is to understand how these epigenetic changes mechanistically alter gene expression and to identify the regulators responsible for these changes. Thus, it is important to have a greater understanding of various mechanisms of epigenetic regulation, including those mediated by intragenic epigenetic modifications and lncRNAs. Consistent with the importance of intragenic epigenetic modifications, most atherosclerosis-specific differentially methylated CpGs are mapped to intragenic regions40. Also, enzymes that catalyze these modifications play important roles in vascular biology52, 53, 60. It is important to note that the studies on intragenic epigenetic modifications and their epigenetic modifiers focus on protein-coding genes. It is unknown if similar mechanisms are involved in regulating non-protein coding genes and if lncRNAs play a role in regulating these modifications. Greater understanding of intragenic epigenetic modifications and lncRNAs may result in the identification of novel targets for pharmacological inhibition in CVD and improve the efficacy of cell-based therapies of CVDs.
Significance.
The emergence of whole genome approaches to investigating the role of epigenetic pathways has provided a platform for gaining further insight into under-appreciated aspects of gene regulation, including intragenic epigenetic modifications and long non-coding RNAs These facets of gene regulation contribute to the function of the vascular endothelium and the pathological changes that occur in CVDs. Greater understanding of epigenetic pathways in the vascular endothelium provides the exciting potential to improve diagnostic and therapeutic approaches for treating CVDs, especially cell-based therapies.
Acknowledgments
Sources of Funding
M.S.Y. is a recipient of a CIHR Frederick Banting and Charles Best Canada graduate scholarship. This work was supported by grant P01 HL076540-06A1 from NHLBI/NIH and CIHR operating grant MOP-142307 to P.A. Marsden.
Nonstandard Abbreviations and Acronyms
- lncRNA
long noncoding RNA
- iPSC
inducible pluripotent stem cells
- EC
endothelial cells
- DNMT
DNA methyltransferase
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- Pol II
RNA polymerase II
- SNP
Single Nucleotide Polymorphism
Footnotes
All authors, M.S.Y. and P.A.M, have contributed to the writing of this article and have approved it.
Disclosures
None.
References
- 1.Abraham G, Bhalala OG, de Bakker PI, Ripatti S, Inouye M. Towards a molecular systems model of coronary artery disease. Curr Cardiol Rep. 2014;16:488. doi: 10.1007/s11886-014-0488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol (1985) 2010;109:916–926. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 3.Turgeon PJ, Sukumar AN, Marsden PA. Epigenetics of Cardiovascular Disease - A New “Beat” in Coronary Artery Disease. Med Epigenet. 2014;2:37–52. doi: 10.1159/000360766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 5.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Gu TP, Weber AR, Shen JZ, Li BZ, Xie ZG, Yin R, Guo F, Liu X, Tang F, Wang H, Schar P, Xu GL. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015;43:3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang YZ, Manduchi E, Stoeckert CJ, Jr, Davies PF. Arterial endothelial methylome: differential DNA methylation in athero-susceptible disturbed flow regions in vivo. BMC Genomics. 2015;16:506. doi: 10.1186/s12864-015-1656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang YZ, Jimenez JM, Ou K, McCormick ME, Zhang LD, Davies PF. Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-Like Factor 4 promoter in vitro and in vivo. Circ Res. 2014;115:32–43. doi: 10.1161/CIRCRESAHA.115.303883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 11.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, Blum R, Bowman C, Hu D, Shilatifard A, Shen S, Dynlacht BD. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci. 2014;71:379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valdes-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 2012;22:307–321. doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deroanne CF, Bonjean K, Servotte S, Devy L, Colige A, Clausse N, Blacher S, Verdin E, Foidart JM, Nusgens BV, Castronovo V. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene. 2002;21:427–436. doi: 10.1038/sj.onc.1205108. [DOI] [PubMed] [Google Scholar]
- 18.Jin G, Bausch D, Knightly T, Liu Z, Li Y, Liu B, Lu J, Chong W, Velmahos GC, Alam HB. Histone deacetylase inhibitors enhance endothelial cell sprouting angiogenesis in vitro. Surgery. 2011;150:429–435. doi: 10.1016/j.surg.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaluza D, Kroll J, Gesierich S, Manavski Y, Boeckel JN, Doebele C, Zelent A, Rossig L, Zeiher AM, Augustin HG, Urbich C, Dimmeler S. Histone deacetylase 9 promotes angiogenesis by targeting the antiangiogenic microRNA-17-92 cluster in endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:533–543. doi: 10.1161/ATVBAHA.112.300415. [DOI] [PubMed] [Google Scholar]
- 20.Rossig L, Urbich C, Bruhl T, et al. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Q, Zeng L, Zhang Z, Margariti A, Ali ZA, Channon KM, Xu Q, Hu Y. Sca-1+ progenitors derived from embryonic stem cells differentiate into endothelial cells capable of vascular repair after arterial injury. Arterioscler Thromb Vasc Biol. 2006;26:2244–2251. doi: 10.1161/01.ATV.0000240251.50215.50. [DOI] [PubMed] [Google Scholar]
- 22.Lee DY, Lee CI, Lin TE, Lim SH, Zhou J, Tseng YC, Chien S, Chiu JJ. Role of histone deacetylases in transcription factor regulation and cell cycle modulation in endothelial cells in response to disturbed flow. Proc Natl Acad Sci U S A. 2012;109:1967–1972. doi: 10.1073/pnas.1121214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zampetaki A, Zeng L, Margariti A, Xiao Q, Li H, Zhang Z, Pepe AE, Wang G, Habi O, deFalco E, Cockerill G, Mason JC, Hu Y, Xu Q. Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation. 2010;121:132–142. doi: 10.1161/CIRCULATIONAHA.109.890491. [DOI] [PubMed] [Google Scholar]
- 24.Pandey D, Sikka G, Bergman Y, Kim JH, Ryoo S, Romer L, Berkowitz D. Transcriptional regulation of endothelial arginase 2 by histone deacetylase 2. Arterioscler Thromb Vasc Biol. 34:1556–1566. doi: 10.1161/ATVBAHA.114.303685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. 2015 doi: 10.1093/eurheartj/ehv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein S, Schafer N, Breitenstein A, Besler C, Winnik S, Lohmann C, Heinrich K, Brokopp CE, Handschin C, Landmesser U, Tanner FC, Luscher TF, Matter CM. SIRT1 reduces endothelial activation without affecting vascular function in ApoE−/− mice. Aging (Albany NY) 2010;2:353–360. doi: 10.18632/aging.100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JY. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci U S A. 2010;107:10268–10273. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmos Y, Sanchez-Gomez FJ, Wild B, Garcia-Quintans N, Cabezudo S, Lamas S, Monsalve M. SirT1 regulation of antioxidant genes is dependent on the formation of a FoxO3a/PGC-1alpha complex. Antioxid Redox Signal. 2013;19:1507–1521. doi: 10.1089/ars.2012.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan YZ, Gao P, Zhou S, Zhang ZQ, Hao DL, Lian LS, Li YJ, Chen HZ, Liu DP. SIRT1-mediated epigenetic downregulation of plasminogen activator inhibitor-1 prevents vascular endothelial replicative senescence. Aging Cell. 2014;13:890–899. doi: 10.1111/acel.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Kim CW, Simmons RD, Jo H. Role of flow-sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero-miRs. Arterioscler Thromb Vasc Biol. 2014;34:2206–2216. doi: 10.1161/ATVBAHA.114.303425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 32.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 33.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B, Jiang Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 5:e1506. doi: 10.1038/cddis.2014.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu VW, Huang PL. Cardiovascular roles of nitric oxide: a review of insights from nitric oxide synthase gene disrupted mice. Cardiovasc Res. 2008;77:19–29. doi: 10.1016/j.cardiores.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Won D, Zhu SN, Chen M, Teichert AM, Fish JE, Matouk CC, Bonert M, Ojha M, Marsden PA, Cybulsky MI. Relative reduction of endothelial nitric-oxide synthase expression and transcription in atherosclerosis-prone regions of the mouse aorta and in an in vitro model of disturbed flow. Am J Pathol. 2007;171:1691–1704. doi: 10.2353/ajpath.2007.060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaina S, Heyn H, Carmona FJ, Varol N, Sayols S, Condom E, Ramirez-Ruz J, Gomez A, Goncalves I, Moran S, Esteller M. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 41.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci U S A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 44.Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2013;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Yu NK, Kaang BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med. 2015;47:e166. doi: 10.1038/emm.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurano MT, Wang H, John S, Shafer A, Canfield T, Lee K, Stamatoyannopoulos JA. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015;12:1184–1195. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bastiaansen AJ, Ewing MM, de Boer HC, van der Pouw Kraan TC, de Vries MR, Peters EA, Welten SM, Arens R, Moore SM, Faber JE, Jukema JW, Hamming JF, Nossent AY, Quax PH. Lysine acetyltransferase PCAF is a key regulator of arteriogenesis. Arterioscler Thromb Vasc Biol. 2013;33:1902–1910. doi: 10.1161/ATVBAHA.113.301579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kristjuhan A, Walker J, Suka N, Grunstein M, Roberts D, Cairns BR, Svejstrup JQ. Transcriptional inhibition of genes with severe histone h3 hypoacetylation in the coding region. Mol Cell. 2002;10:925–933. doi: 10.1016/s1097-2765(02)00647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 57.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript Elongation by RNA Polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 58.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 59.Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41:2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu M, Sun XJ, Zhang YL, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci U S A. 2010;107:2956–2961. doi: 10.1073/pnas.0915033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Estaras C, Fueyo R, Akizu N, Beltran S, Martinez-Balbas MA. RNA polymerase II progression through H3K27me3-enriched gene bodies requires JMJD3 histone demethylase. Mol Biol Cell. 2013;24:351–360. doi: 10.1091/mbc.E12-07-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakobsson L, van Meeteren LA. Transforming growth factor beta family members in regulation of vascular function: in the light of vascular conditional knockouts. Exp Cell Res. 2013;319:1264–1270. doi: 10.1016/j.yexcr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 64.Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- 65.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prasain N, Lee MR, Vemula S, et al. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat Biotechnol. 2014;32:1151–1157. doi: 10.1038/nbt.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto T, Shibata R, Ishii M, Kanemura N, Kito T, Suzuki H, Miyake H, Maeda K, Tanigawa T, Ouchi N, Murohara T. Therapeutic reendothelialization by induced pluripotent stem cells after vascular injury--brief report. Arterioscler Thromb Vasc Biol. 2013;33:2218–2221. doi: 10.1161/ATVBAHA.113.301313. [DOI] [PubMed] [Google Scholar]
- 68.Hussein SM, Puri MC, Tonge PD, et al. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- 69.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tonge PD, Corso AJ, Monetti C, et al. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–197. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- 71.Wu YT, Yu IS, Tsai KJ, Shih CY, Hwang SM, Su IJ, Chiang PM. Defining minimum essential factors to derive highly pure human endothelial cells from iPS/ES cells in an animal substance-free system. Sci Rep. 2015;5:9718. doi: 10.1038/srep09718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH, Shu YQ. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2014;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 73.Holdt LM, Hoffmann S, Sass K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]