Abstract
The microbiome is a complex community of Bacteria, Archaea, Eukarya and viruses that infect humans and live in our tissues. It contributes the majority of genetic information to our metagenome, and consequently, to our resistance and susceptibility to diseases, especially common inflammatory diseases, such as type 1 diabetes, ulcerative colitis, and Crohn's disease. Here we discuss how host-gene-microbial interactions are major determinants for the development of these multifactorial chronic disorders and thus, for the relationship between genotype and phenotype. We also explore how genome-wide association studies (GWAS) on autoimmune and inflammatory diseases are uncovering mechanism-based sub-types for these disorders. Applying these emerging concepts will permit a more complete understanding of the etiologies of complex diseases and underpin the development of both next generation animal models and new therapeutic strategies for targeting personalized disease phenotypes.
Recent advances in diverse areas of science and technology make this a unique time to study the genetics and pathogenesis of complex diseases, such type 1 diabetes (T1D) and inflammatory bowel disease (IBD), which includes Crohn's disease (CD) and ulcerative colitis (UC). These distinct diseases are now understood to share important common characteristics and aspects of their disease mechanisms. In all three diseases, the immune system damages tissues: T1D is likely an autoimmune disease, whereas CD and UC are likely caused by inappropriate inflammatory responses to components of our microbiome (see Box 1 for definition of key terms). Many genetic loci regulate the risk for each disease. Although a threshold dose of these susceptibility alleles provides the foundation for developing the disease, these alleles are not sufficient to cause the disease.
Text Box: Definition of terms.
Dysbiosis: Most commonly refers to a disruption in the normal homeostatic and beneficial relationship between microbes and their host, including disruptions in microbial community structure and function. Alterations in microbial community structure, involving Bacteria, Archaea, and/or Eukarya can occur in any body habitat but has been best described in the gut where it has been associated with a number of disease states including, for example, inflammatory bowel disease.
GWAS: Analysis of common alleles (mostly single nucleotide polymorphisms, SNPs) in a population that associates genetic loci with disease susceptibility. These loci contain “candidate” disease genes.
Familial clustering: If a family member is diagnosed with a disease such type 1 diabetes, ulcerative colitis, and Crohn's disease, then the risk of other first degree family members is much greater (perhaps as much as 50-fold for some multifactorial disorders) than that for a person taken at random from the general population. Familial clustering is caused by a combination of inherited genetic variants from the parents to the children and shared environmental factors within the families. Susceptibility variants are being discovered rapidly by genome-wide association studies, but the environmental factors remain unknown although numerous candidates are recognized, most particularly a role for the microbiome and infections.
Metagenetics: Approaching genetic and genomic studies by considering all of the genes in the metagenome as opposed to considering, in isolation, host genes or genes that confer particular properties (e.g., virulence or commensalism) upon an individual microbe. Importantly, the history of microbial inputs into the metagenomic profile of an individual is important for identifying the causes of complex disease, requiring expensive, but essential longitudinal studies, including information from maternal and gestational exposures and phenotypes.
Metagenome: As used here, metagenome is the sum of all genes and genetic elements and their modifications in the somatic and germ cells of a host plus all genes and genetic elements in all microorganisms that live on or in that host at a given time. The metagenome has transient elements (e.g. during infection with a rare pathogen) and more persistent elements (e.g., infection with latent eukaryotic virus; presence of commensal bacteria).
It has been obvious for decades that complex gene-gene and gene-environment interactions govern these diseases, but not surprisingly, untangling this web of interactions has been extremely difficult (Figure 1). Despite the failure to identify single causal agents for each disease, there is strong evidence that microbes contribute to pathogenesis. Furthermore, genome-wide association studies (GWAS), which use large study populations and careful replication of results, have effectively identified many important loci in the host that increase one’s risk for the disease, and these results have fundamentally altered how we conceptualize these diseases (Stappenbeck et al., 2011; Khor et al., 2011; Anderson et al., 2011; Franke et al., 2010; Todd, 2010). Correlation of GWAS data with genome-wide gene expression analyses (eQTLs), in combination with protein-protein interaction data, is greatly assisting the identification of candidate causal genes within these loci (Anderson et al., 2011; Franke et al., 2010; Cotsapas et al., 2011; Rossin et al., 2011; Fehrmann et al., 2011). Recently, numerous approaches have been developed to start defining mechanisms for complex inflammatory diseases by using leads from GWAS and analyses of the microbiome. These promising approaches include: the introduction of mutations in GWAS-identified loci into the mouse genome (Cadwell et al., 2010; Bloom et al., 2011); the creation of induced pluripotent stem cells (iPSCs) from patients and their differentiation into relevant cell types [e.g. (Rashid et al., 2010)]; and, humanized mouse models in which the murine immune system is replaced by transplantation [e.g. (Brehm et al., 2010; Esplugues et al., 2011)] or human microbial communities are transplanted into formerly germ-free mice (Goodman et al., 2011). Currently the great challenges in this field are to (1) understand how both microbiome and GWAS-identified genes contribute to disease; (2) elucidate the molecular mechanisms by which causal genes act during pathogenesis; and (3) validate biomarkers and druggable pathways via genotype-phenotype studies [e.g. (Dendrou et al., 2009; Bloom et al., 2011; Cadwell et al., 2010)].
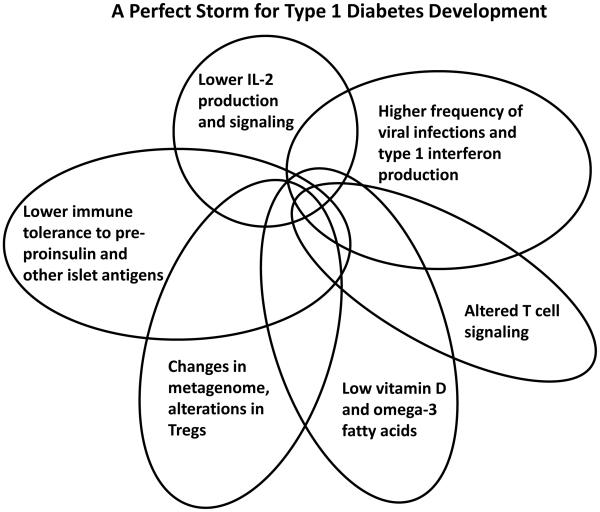
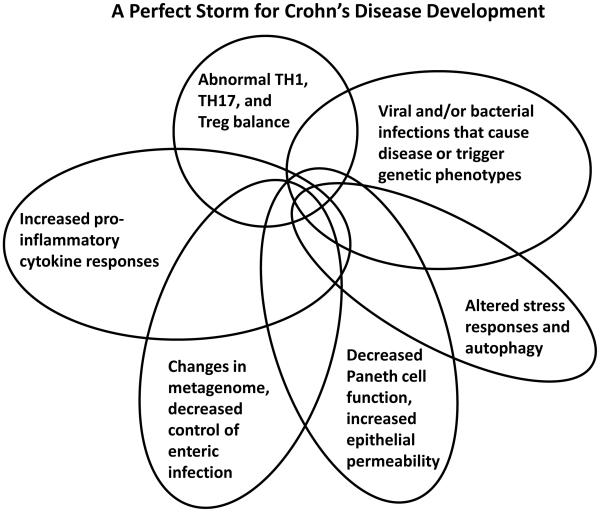
Figure 1. Perfect Storms for Developing Crohn's Disease and Type 1 Diabetes.
A series of overlapping events and phenotypes driven by metagentic and environmental processes that, in sum, contribute to the development and pathogenesis of type 1 diabetes (left) and Crohn’s disease (right).
By peering through the lens of recent studies on CD, UC, and T1D, this review seeks to delineate emerging concepts in research on complex inflammatory diseases and to comment on the implications of these concepts for the interpretation of genetic and pathogenetic data. Two concepts are emphasized and integrated herein: (1) that single disease diagnoses are unlikely to be single phenotypes and may instead be the sum of multiple mechanism-based disease subsets, and (2) that the interactions of individual microorganisms and their genomes with specific host genes or pathways underpin the relationship between genotype and phenotype in these complex diseases. In this view, disease genetics may be combinatorial with different host-gene-microbial interactions, contributing to the pathogenesis of disease in subsets of patients. These two interrelated concepts, therefore, define T1D, CD, and UC as metagenetic (Box 1), rather than simply 'genetic', diseases. These concepts will guide the design and interpretation of future experiments that seek to dissect the pathophysiologic mechanisms underlying a number of complex diseases and to identify more effective approaches for their treatment and prevention.
Host Genetic Grist for the Metagenetic Mill
Recently meta-analyses of GWAS of large cohorts of patients of European descent with UC or CD have been performed (Franke et al., 2010; Anderson et al., 2011; Khor et al., 2011). These studies identified 98 loci, and candidate genes within these loci candidate genes, that have a putative role in IBD. Similar studies of T1D identified 53 disease susceptibility loci (Barrett et al., 2009) (www.t1dbase.org). Importantly, many disease susceptibility loci are shared amongst common autoimmune and inflammatory diseases including T1D, Graves’ disease, celiac disease, CD, UC, psoriasis, rheumatoid arthritis, alopecia areata, multiple sclerosis, and systemic lupus erythematosus (Cotsapas et al., 2011; Khor et al., 2011). It is striking that T1D and CD share 13/52 (25%) risk loci outside the human leukocyte antigen (HLA) gene complex, despite the fact that these diseases are neither thought to be related diseases nor reported to be shared within families more often than expected by chance (www.t1dbase.org). Notably, the candidate causal genes in these 13 susceptibility loci regulate immunity. These include (Khor et al., 2011): PTPN22, which is involved in T and B cell signaling; IL10, encoding a powerful cytokine that suppresses inflammatory responses (including in specialized T regulatory cells in the gut) (Maloy and Powrie, 2011); BACH2, which regulates B cell gene expression and possibly IgA production; TAGAP, which is involved in T cell activation; IKZF1, which negatively regulates B cells; IL2RA, which controls T regulatory lymphocyte development and function; GSDMB/GSDMA/ORMDL3, which is involved in stress responses; FUT2, which controls microbial susceptibility (Smyth et al., 2011; Franke et al., 2010; McGovern et al., 2010); and IL27, which suppresses inflammatory responses and regulates IL-10 signaling (Imielinski et al., 2009)(Barrett et al., 2009). This is a remarkable concordance of involved genes for two unrelated diseases, indicating that different diseases can have common mechanistic components and that the immune system is key for both diseases.
However, not withstanding all insights into disease mechanisms that the GWAS approach has already provided, the inheritance and the strong clustering of these multifactorial diseases within families (Box 1), which encompasses both inherited genetic variants and intrafamilial environmental factors, remains only partially explained. Assuming a simple statistical model of gene interaction (Clayton, 2009), the numerous identified loci account for not more that 25% of the familial clustering of CD and UC (Anderson et al., 2011; Franke et al., 2010). This contrasts with T1D, in which the HLA effect is uniquely large and, together with 52 non-HLA loci, can account for almost all of the familial clustering (Clayton, 2009). For T1D, the massive effect of the HLA region, owing to functional polymorphisms in the HLA class II and class I genes contributes almost 50% of familial clustering on its own (Clayton, 2009)(Todd, 2010). There are, however, probably hundreds of non-HLA loci affecting the risk of CD, UC, and T1D risk that remain unmapped owing, to their very small effect sizes (Barrett et al., 2009; Anderson et al., 2011; Franke et al., 2010). These putative loci will be difficult to map unless they contain rare mutations of higher penetrance, an occurrence that is just beginning to yield informative findings (Nejentsev et al., 2009; Rivas et al., 2011) and holds continued promise with the rapid use of high throughput next generation sequencing.
In humans, the HLA locus contains a large number of genes encoding the Major Histocompatibility Complex (MHC) molecules (which are responsible for presenting antigens to cells of the immune system), along with a number of other genes that modulate immune responses. The remarkable contribution of HLA variations (Todd, 2010) to T1D risk is an unusual feature of a common disease. Nevertheless, HLA genotypes that greatly predispose individuals to T1D are not sufficient to cause the disease because only ~5% of high risk HLA carriers develop T1D. HLA are expressed by antigen presenting cells (APCs), such as macrophages, B lymphocytes, and dendritic cells (DCs). DCs are highly potent APCs that reside in the pancreas and its islets (i.e., collections of insulin-producing beta cells and other endocrine cells) and could initiate the autoimmune destruction of beta cells by T cells (Calderon et al., 2011a; Calderon et al., 2011b). Interestingly, the pancreatic lymph nodes, where DC priming of T cells for the induction of T1D may occur, also drain to parts of the intestine, providing a site where the microbiome might influence the genesis of T1D (Turley et al., 2005; Wen et al., 2008). Because the insulin gene is one of the strongest non-HLA T1D susceptibility loci in the genome (Todd, 2010) (www.t1dbase.org), insulin and its precursors are likely primary autoantigens. These very strong associations with HLA and this autoantigen gene are not a feature of CD or UC, in which no particular antigen is known to be targeted, and hence their classification as inflammatory rather than autoimmune diseases. GWAS point to several other immunologic components of T1D etiology, including IL-2 production and receptor signaling [IL-2 gene, IL-2 receptors IL2RA (CD25) and IL2RB (CD132) (Todd, 2010)], immune tolerance and T cell receptor signaling [PTPN2 (Long et al., 2011), PTPN22 (Arechiga et al., 2009; Bottini et al., 2006)], and recently, the immune response to viral infections and the type 1 interferon responses [IFIH1 (encoding MDA5), GPR183 (EBI2) (Heinig et al., 2010), TLR7 and TLR8, and FUT2 (Smyth et al., 2011)].
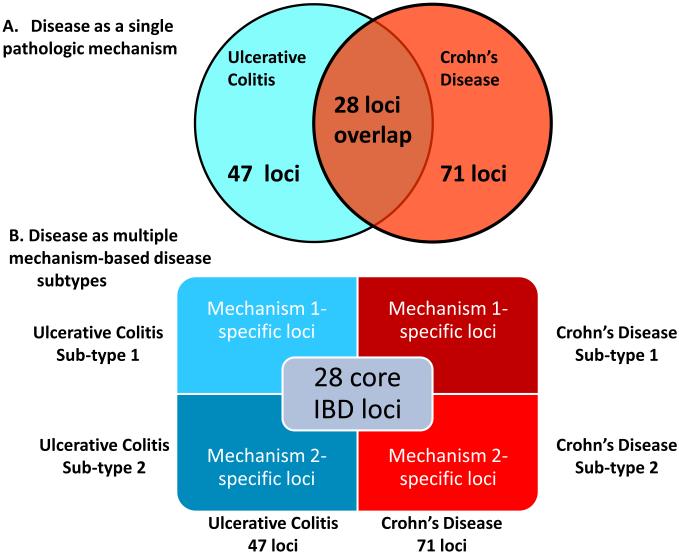
Twenty-eight loci (28/71, 39%) of CD risk loci are shared with UC, indicating that a set of core mechanisms participate in these diseases (Figure 2) (Khor et al., 2011). These diseases genes implicate numerous processes in both CD and UC, including T cell differentiation and function, autophagy, endoplasmic reticulum stress, oxidative stress, and mucosal immune defenses, amongst others. There are important gene-gene and pathway-pathway interactions within this core set of processes. For example, the CD risk gene NOD2 links to autophagy through interactions between the NOD2 protein and with Atg16L1, induction of pro-inflammatory cytokines, control of bacterial infection, and sensing of pathogen associated molecular patterns (Levine et al., 2011). Particularly notable are pathways involving the cytokines IL-23 and IL-12, which regulate the development of TH1 and TH17 CD4 T cells, and IL-10, which is essential in the function of certain regulatory T cells (Tregs) via its anti-inflammatory activity. Rare mutations in genes encoding IL-10 receptors confer susceptibility to early-onset IBD (Glocker et al., 2009). These genetic clues point to a key role for regulating the balance between pro-and anti-inflammatory T cells in CD and UC. The regulation of T cell differentiation is also a key target for host-gene-microbial interactions, as discussed below.
Figure 2. Refining the Relationship between Genotype and Phenotype in Complex Inflammatory Diseases.
(a) Traditionally a disease is considered as a single phenotype, with genes or loci conferring risk to two diseases shown as overlapping in a Venn diagram. (b) We propose a new view of the genotype-phenotype relationship in which different sets of loci are responsible for mechanistically distinct subtypes of diseases, and the sum of these subtypes constitutes the overall diagnosis. Here two disease subtypes are indicated for simplicity, but many such sub-types may exist, and sets of overlapping risk loci may be associated with these multiple mechanistically distinct disease phenotypes.
Mechanism-based Disease Sub-types
GWAS have revealed a wealth of genes potentially involved in T1D, UC, and CD, but no single gene or set of genes is prognostic. How can we interpret this observation? Here, we argue for an important contributor to this observation- the concept that 'diagnosis' does not equal 'single phenotype'. Without a distinct phenotype, genetic results are often difficult to interpret. This basic principle comes into sharp focus as one considers current genetic and pathogenesis studies of CD, UC and T1D.
Why is a diagnosed 'disease' an imprecise phenotype? It is not because patients have been misdiagnosed - the diagnosis of UC or CD or T1D has stood the test of time to predict patient prognosis. However, we believe that there are many pathways to the same diagnosis. A diagnosis may be 'clinically' precise but 'mechanistically' imprecise. Thus, clinical diagnoses are poor phenotypes for genetic studies unless a single mechanism is responsible for the diagnosis, as in the case of a rare gene mutation in a monogenic disease. The complexity of GWAS results is consistent with the existence of multiple disease subtypes within T1D, UC, or CD, each based on the specific mechanism (Figure 2). Support for this idea comes from the observation that subsets of IBD patients respond differentially to mechanistically distinct interventions (Melmed and Targan, 2010).
Why do diagnostic categories group different mechanistic processes under the same moniker? Over many decades, pathologists have lumped patients with similar but non-identical clinical and pathological signs and symptoms into diagnostic categories that predict outcome and complications. Indeed, this has enormous value clinically, but it emphasizes similarities between patients in outcome rather than differences in pathways that lead to a common endpoint. Complex diseases are diagnosed by summing up multiple factors that may be causes or mere consequences of the disease process. Disease 'diagnosis' does not require the presence in the tissue of all of the abnormalities that may be 'classically' seen in a given disease (Gianani et al., 2010; Odze, 2003). For example, at the polar extreme, CD is easily distinguished from UC by its classical ileal involvement (i.e., involvement of tissue at the end of the small intestine), fissures, granulomas, transmural inflammation (i.e., inflammation through the entire intestinal wall), fat wrapping of the intestine, patchy pathology, skip lesions, and patient presentation with bowel strictures or percutaneous fistulae. However, like UC, CD can be restricted to the colon, and the inflammatory infiltrates of CD and UC overlap. UC can be patchy, and the patient presentations of the two diseases can overlap extensively. Similarly, the genetics, pathology, and pathogenesis of IBD may differ between young and old patients with the same diagnosis (Imielinski et al., 2009; Odze, 2003). Even when all classical aspects of a disease are present, the mechanism responsible for the pathology observed may differ from one person to another. Based on these considerations, it is no surprise that the genetics of T1D, CD, and UC are complex because different phenotypes may have been grouped into a single analysis.
This putative mechanistic heterogeneity is reflected in sometimes subtle, but quantifiable, characteristics of the disease process and pathology. Taking such differences into account can be used to identify disease subtypes that are more recognizable as molecularly-defined pathological conditions and that more closely relate to specific pathogenetic mechanisms underpinned by distinct sets of genetic risk loci and that (Figure 3). For example, variations in the ATG16L1 gene (i.e., hypomorphic expression in the mouse and homozygosity for the T300A variant in humans) result in abnormalities in Paneth cell granules and secretion (Cadwell et al., 2008; Cadwell et al., 2010). Paneth cells are innate immune epithelial cells positioned at the base of small intestinal crypts, where they secrete antimicrobial peptides and other factors that help shape the configuration of the intestine’s bacterial community. Abnormalities in Paneth cells are observed in the subset of CD patients homozygous for the T300A allele, thus defining a pathologic subtype of CD (Figure 2B). If one used criteria including Paneth cell abnormalities in CD diagnosis, the frequency of the ATG16L1 T300A allele would be higher in patients with the 'Paneth cell-sub-type' of CD than in the CD population as a whole (Figure 3). If multiple risk loci contribute to such Paneth cell changes, one might be able to detect gene-gene interactions in this subset of patients compared to other subsets.
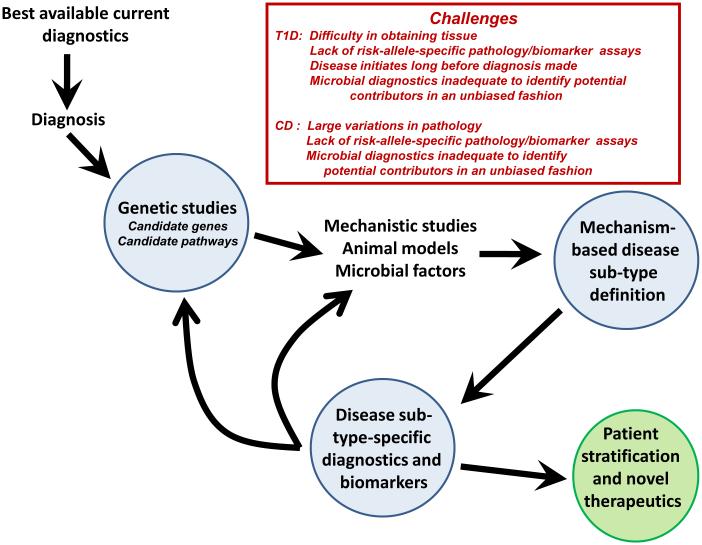
Figure 3. The Iterative Redefinition of Mechanism-based Disease Subtypes.
Here we present a conceptual work flow for breaking a broad disease diagnosis into its component sub-types by the iterative application of genetics and mechanistic studies. One output would be therapeutics based on disease sub-type and patient stratification into groups more likely to respond to a given therapy or preventive strategy. The box shows specific challenges for this process for type 1 diabetes and Crohn’s disease.
A similar situation exists in T1D. Biopsy specimens of the pancreas are virtually impossible to obtain. Therefore, T1D is defined clinically by the downstream consequences of destroying the insulin-secreting β-cells of the pancreatic islets, namely, high blood glucose and absolute insulin dependence, rather than by the mechanisms for their destruction. It is, therefore, possible that several different pathologic processes result in this disease. T1D patients diagnosed under age 10 years frequently exhibit islet inflammation or insulitis, whereas patients diagnosed over age 10 years exhibit insulitis less frequently. More recently, this histopathological heterogeneneity has become even more evident (Gianani et al., 2010), thanks to the Juvenile Diabetes Research Foundation nPOD project (www.jdrfnpod.org). As for CD, the diagnosis of T1D may reflect the presence of more than one pathogenetic mechanism and thus, represent more than one disease subtype, although in T1D the HLA effect are an essential common pathway.
The concept that disease diagnoses include mechanism-based disease subtypes has many implications for interpreting human genetic studies and for understanding the relationship between the microbiome and genetic susceptibility, as discussed below. Including disease subtypes within a single diagnosis would decrease the power to define causal alleles and to detect gene-gene interactions that contribute to a single disease sub-type. In this view, the difficulty of interpreting how multiple small genetic effects sum to predispose an individual to a clinical diagnosis may partly reflect insufficient precision in selection of specific phenotypes to study.
It is important to recognize that it is the power and informativeness of GWAS themselves that drive the concept of mechanism-based disease subtypes (Figure 2). In the absence of candidate genetic mechanisms for defining disease subtypes, there is limited clinical utility in focusing on low frequency characteristics or subtypes within a larger diagnostic category which predicts patient outcome. We, therefore, argue for iterative high-precision phenotyping of patients into mechanism-based subtypes in future studies; this will allow more accurate interpretation of genetic, pathogenesis, outcome, and therapeutic studies (Figure 3). Such definitions must be iteratively re-assessed because risk alleles are defined and disease mechanisms are delineated so that the field is not limited by inflexible definitions of disease that may obscure mechanistic heterogeneity. This type of approach is a necessary presage to so-called stratified or personalized medicine. The genetic and pathological complexity of T1D, CD, and UC is particularly well suited for testing whether iteratively redefining disease diagnoses can enhance the value of genetic and pathogenesis studies. Importantly, precision in disease categorization would make defining the impact of host-gene-microbial interactions on diseases processes more robust.
Host-gene-microbial Interactions in Metagenetics
Metazoan organisms are complex communities that include a core organism in combination with a veritable zoo of other organisms that live on or in the body – our microbiome. The microbiome includes eukaryotic viruses, Eukarya, bacteria viruses, Bacteria, Archaea, and, for many, helminths (Virgin et al., 2009; Kau et al., 2011; Garrett et al., 2010b; Spor et al., 2011). The importance of understanding the microbiome has been repeatedly emphasized, giving rise to a large number of international human microbiome projects (e.g. https://commonfund.nih.gov/hmp/, http://www.metahit.eu/) that have focused initially on the bacterial component of the microbiome. The host plus non-host genes of this polyglot and interactive community constitute our metagenome (Box 1). A critical emerging concept is that bacterial and viral interactions in the pathogenesis of inflammatory disease occur in a host gene-specific fashion [see below, (Virgin et al., 2009; Cadwell et al., 2010; Bloom et al., 2011; Elinav et al., 2011)]. Understanding the metagenome is, therefore, highly relevant to understanding T1D, UC, CD, and other common multifactorial diseases.
Intestinal bacteria play a role in driving IBD, and emerging data support a similar view for T1D (Wen et al., 2008; Giongo et al., 2011; Roesch et al., 2009). The evidence that bacteria plays a role in IBD includes two major observations: that surgical diversion of the fecal stream ameliorates inflammation (Sartor, 2008) and that antibiotics help some patients. In mouse models of colitis viruses, bacteria, or both acting together can contribute to the pathology via signaling through innate immune sensors and regulation of pro- and anti-inflammatory cytokines (Levine et al., 2011; Maloy and Powrie, 2011; Khor et al., 2011). For many years, enterovirus infection has been associated with T1D [e.g. (Yeung et al., 2011; Oikarinen et al., 2011; Stene et al., 2010)], and the major sensor for enterovirus RNA is the T1D susceptibility gene IFIH1, encoding MDA5 (Nejentsev et al., 2009; McCartney et al., 2011). The mechanisms for these associations between components of the microbiome and T1D, CD, or UC have proven elusive. The lack of integration among scientific disciplines and among training programs, together with limitations in technology, has substantially limited the understanding of metagenomic contributions to disease.
Adult mammals are permanently infected by many viruses, and they are populated by large site-specific bacterial and phage communities without overt negative effects (Virgin et al., 2009; Foxman and Iwasaki, 2011; Spor et al., 2011). Thus, the bacterial microbiota (and their phages) and the eukaryotic virome are two major (but not the only) contributors to the metagenome. The intestinal microbiome plays a critical role in mammalian physiology by synthesizing vitamins and harvesting energy from food (Spor et al., 2011; Kau et al., 2011). Further, the normal function of the innate immune system, which is critically involved in the pathogenesis T1D, UC, and CD, is regulated by both chronic viral infections and resident bacterial communities (Barton et al., 2007; White et al., 2010; Virgin et al., 2009; Spor et al., 2011; Kau et al., 2011). The microbiome and metagenome varies from person to person based on host genetics, diet, exposure, geography (including westernization as approximated by the gross national product of a country), socioeconomic status, mode of delivery, and gestational age at birth, breast feeding, antibiotic use, and additional factors (Virgin et al., 2009; Benson et al., 2010; Spor et al., 2011; Kau et al., 2011; Penders et al., 2006). Such variations could certainly provide variable environmental inputs that contribute to the incidence of T1D, UC, and CD, within the genetic foundation revealed by GWAS (Bach, 2002; Vehik and Dabelea, 2011; Ehlers and Kaufmann, 2010).
There are extensive interactions between host and non-host genes within the metagenome, and bacteria and eukaryotic viruses alter our physiology and fitness (Spor et al., 2011; Virgin et al., 2009; Hansen et al., 2010). These genetic interactions within the metagenome create a complex and poorly understood host-gene-microbial interaction matrix that can define phenotype. Host genes, such as those involved in innate and adaptive immunity (e.g., NOD2, NLRP6, HLA, TLR2, and MYD88) shape the bacterial microbiota (Spor et al., 2011; Elinav et al., 2011; Wen et al., 2008). Forward genetic screens in mice suggest that resistance to individual viruses involves hundreds of genes (Virgin et al., 2009), making it likely that many host genes regulate the microbiome (and thus the metagenome). Importantly, key interactions between members of the microbiome have been and are increasingly being reported. For example, murine norovirus can trigger an intestinal inflammatory process in mice with a mutation in Atg16L1. This process can be treated with antibiotics and is thus presumed to be bacteria-dependent (Cadwell et al., 2010) (Figure 4). The existence of such interactions indicates that it will be important to consider that the disease contributions of microbiome members (e.g., helminths and bacteria) are potentially dependent on each other.
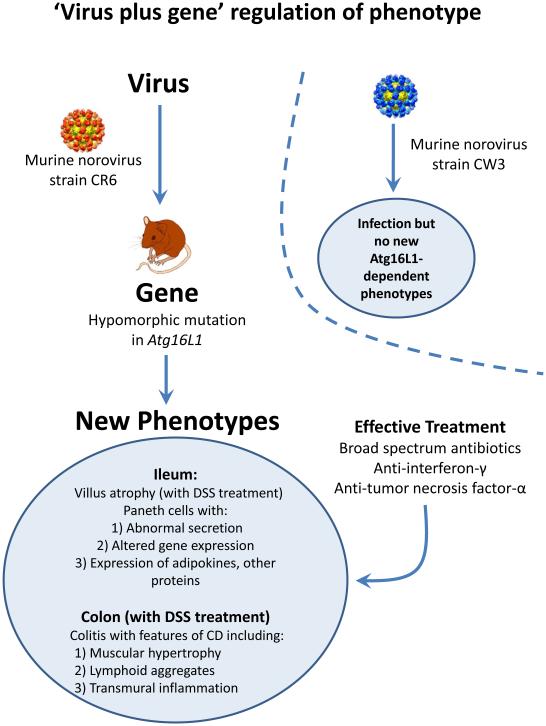
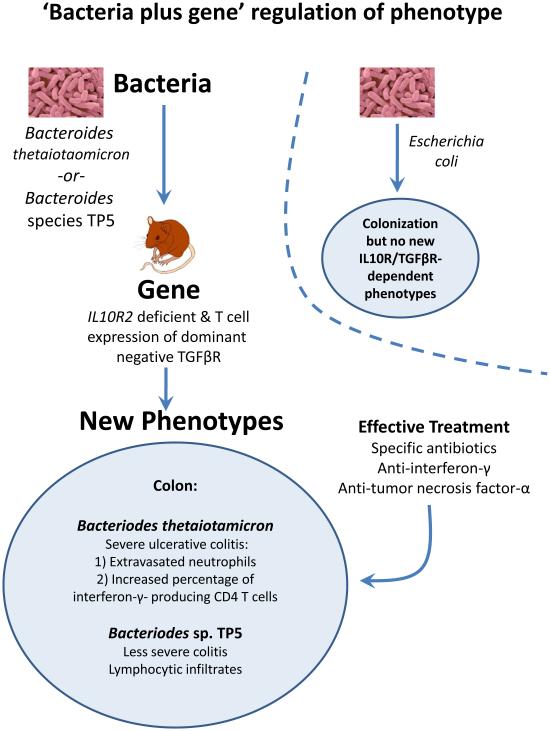
Figure 4. Microbe Plus Gene Interactions Determine Inflammatory Bowel Disease Phenotypes.
(a) Two recent studies analyzed the capacity of two different strains of murine norovirus, MNV strain CR6 versus MNV strain CW3, to trigger phenotypes when orally inoculated into mice with a mutation in the Crohn's disease risk gene Atg16L1 (Cadwell et al., 2010; Cadwell et al., 2008). This mutation results in decreased expression of Atg16L1 protein (hypomorphic, Atg16L1HM). Even though MNV CW3 and MNV CR6 are closely related, they have different effects on intestinal pathology in Atg16L1HM mice. Some of these interactions are observed only when mice are fed the chemical dextran sodium sulfate (DSS). (b) Two other studies analyzed the capacity of two different species of Bacteroides to trigger phenotypes in combination with mutations in the IL-10 receptor and T cell expression of a dominant negative form of the TGFβ-receptor (dnKO mice) (Bloom et al., 2011; Kang et al., 2008). dnKO mice are cured of their spontaneous colitis by treatment with antibiotics, but oral feeding of 'cured' mice with fecal contents or specific bacteria re-induce disease. Even though Bacteroides thetaiotaomicron and Bacteroides sp. TP5 are closely related, they induce different forms of inflammation when fed to antibiotic-cured dnKO mice. In the same studies dysbiosis (Box 1) with increases in the numbers of Enterobacteriaceae was noted in dnKO mice prior to curing the mice with antibiotics. However, E. coli inoculation did not trigger the pathologies seen with either Bacteroides species.
Metagenetic influences on disease could occur in various ways. Familial disease clustering may reflect intrafamilial behavioral and dietary factors that define the metagenome. A major influence on the human metagenome may be vertical transmission of the maternal microbiome. In the controlled environment of mouse colonies, the bacterial microbiota is clearly maternally inherited. Furthermore, this microbiome can have profound pathological effects in mice carrying specific mutations, such that studies of host-gene functions must now consider contributions of the metagenome (see below).
The situation in humans is more complex (Hansen et al., 2011; Benson et al., 2010), although the although the importance of early environmental exposures has been well documented, including studies with mono- and dizygotic twins (Turnbaugh et al., 2009). Children delivered vaginally initially acquire a distinctly different intestinal bacterial microbiota than those born by caesarian section (Penders et al., 2006; Dominguez-Bello et al., 2010). In a meta-analysis, delivery by caesarean section increases the risk of T1D by 20% (Cardwell et al., 2008). An association with increased risk for T1D has also been reported for higher birth weight (Cardwell et al., 2010), and early infant diet (Pfluger et al., 2010) Figure 5). Furthermore, changing microbial exposures and infections likely has a major influence on the incidence of other diseases, as well. The dramatic rise in the incidence of allergy, asthma, and T1D in the last 60 years correlates with vast improvements in healthcare and sanitation (Bach, 2002; Vehik and Dabelea, 2011; Ehlers and Kaufmann, 2010). For example, severe rhinovirus infection before age 3 years, coupled to an asthma-predisposing inherited host genome, has been associated with increased risk of asthma (Foxman and Iwasaki, 2011). Thus, the metagenome could contribute to disease susceptibility and potentially explain a proportion of the familial clustering, the so-called “missing heritability” of multifactorial diseases.
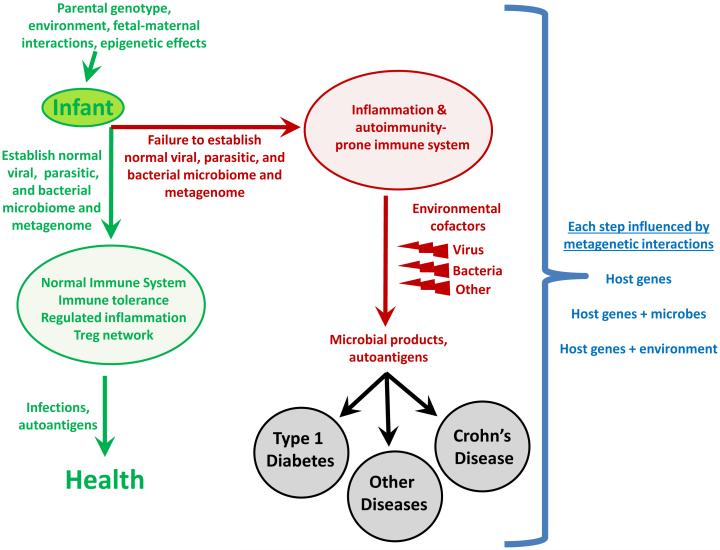
Figure 5. A Metagenetic View of Developing Normal and Pathological Immune Responses.
This flow-chart depicts stages in the development of normal immune responses or auto-immune and inflammatory diseases at which metagenetic interactions (i.e., gene-gene and gene-microbe interactions) might play a determining role. 'Microbial products' refers to molecules that interact with host innate immune sensors and initiate inflammation.
Metagenetic Effects on Immunity and Autoimmunity
GWAS point to a fundamental role for the immune system in the pathogenesis of T1D, UC, and CD. An emerging concept is that bacterial and viral interactions contribute to both normal immune physiologies and abnormal pathologic responses that occur in a host gene-specific fashion (Virgin et al., 2009; Cadwell et al., 2010; Bloom et al., 2011; Garrett et al., 2007; Elinav et al., 2011). The microbiome has significant effects on the development of the immune system (Lee et al., 2011; Mazmanian et al., 2008; Sartor, 2008; Ivanov et al., 2009) and on physiology, including susceptibilities to obesity and the metabolic syndrome (Kau et al., 2011). Serum IgE responses to antigenic challenge are lower in mice colonized with Clostridium, confirming that bacteria can have profound effects on systemic immune responses involved in allergy (Atarashi et al., 2011). CNS inflammation induced by autoantigen is limited in germ-free mice, but it can be restored by colonization with specific bacteria(Lee et al., 2011). In the NOD mouse model, autoimmune diabetes is regulated by a MyD88-dependent interaction of intestinal microbes with the innate immune system (Wen et al., 2008). Germ-free K/BxN T cell receptor transgenic mice are resistant to arthritis caused by autoantibodies to the self antigen glucose-6-phosphate isomerase. When these animals are colonized with segmented filamentous bacteria (SFB), it regenerates TH17 responses in the small intestine, autoantibody production, and arthritis (Wu et al., 2010). Furthermore, the normal intestinal microbiome is essential for effective resistance to oral inoculation with Toxoplasma gondii and for generating appropriate CD8+ T cell responses to influenza (Ichinohe et al., 2011; Benson et al., 2009).
The mechanisms responsible for these observations are under intensive investigation. One recent study shows that intestinal bacteria induce Tregs in an antigen-specific and T cell receptor-dependent fashion (Lathrop et al., 2011). This is a key observation because it provides a mechanism, in addition to thymic exposure to self antigens, for how regulatory responses can be generated to blunt inflammation. Given the continuous presence of the stimulating antigens for these Tregs in the normal intestinal microbiome, such cells could have profound effects on both intestinal and systemic immune responses, including those responses to self antigens. This is particularly important because it has been reported that the intestine is where T lymphocytes migrate to accept differentiation signals regulating autoimmune responses (Esplugues et al., 2011). It was also shown that injection of Staphylococcus aureus or its superantigen S. aureus enterotoxin B (SEB) was able to induce these intestinal regulatory TH17 cells, which is consistent with SEB injection being immune tolerogenic (Esplugues et al., 2011). These studies suggest that variation in the metagenome between individual humans, between mice in different research facilities, or even between animals from different cages within the same facility, could have profound effects on many aspects of the immune response. This concept has key implications for the interpretation of mouse studies. The microbiome is maternally inherited in mice, but it can differ among research facilities; there may even be significant microenvironmental variation between cages of mice or between mice born of different dams. Given that the microbiome influences immunity so extensively, experiments must control for these factors. Currently, this is neither consistently performed nor required by peer reviewers.
Host-gene-metagenome Interactions in UC and CD
Correlations between communities of intestinal bacteria and CD or UC have led to the concept of dysbiosis (Box 1), as a contributor to these diseases [e.g. (Sartor, 2008)]. This important hypothesis emphasizes the potential role that changes in the bacterial microbiota have on disease. However, now this hypothesis needs to expand and include both non-bacterial components of the metagenome and highly specific interactions between individual bacteria or viruses and host genes, which have recently been identified as contributors to disease pathogenesis. The relative contribution of dysbiosis versus the contribution of single organisms within the microbiome to the etiology of complex inflammatory diseases is unresolved. A confounding element has been the reliance on antibiotic treatment to assess bacteria as causes for intestinal disease. Because antibiotics can treat enteric inflammatory disease triggered by viruses (Figure 4) (Cadwell et al., 2010), a broader approach- including proof that specific bacteria or viruses are both necessary and sufficient for a phenotype- will be required to understand metagenetics of disease. Specific risk alleles for CD or UC could affect IBD by altering bacterial populations or individual bacterial types (Maloy and Powrie, 2011; Garrett et al., 2010b; Spor et al., 2011). Data from numerous mouse models of transmissible colitis confirm this point and are discussed below. The complexity of these reciprocal interactions between host and non-host genes within the metagenome underlines the critical need for new concepts and methodologies in computational and systems biology, which can deal with individual host-gene-microbial interactions in the broader context of the metagenome.
IBD in humans and mice is associated with alterations in the balance between TH1, TH17, and Treg cells, and this balance is dependent on the metagenome (Garrett et al., 2010b; Maloy and Powrie, 2011). The relevance of these studies to human CD and UC is strongly supported by the identification of genes regulating these pathways in GWAS on IBD (see above). The role of specific bacteria and helminths in regulating these T cell responses in both the small and large intestine is highly relevant to understanding the genetics and pathogenesis of IBD. Polysaccharide A synthesized by the common colonic commensal Bacteroides fragilis induces Tregs that secrete IL-10 and inhibit intestinal inflammation (Round and Mazmanian, 2010; Mazmanian et al., 2008). Similarly, a protein antigen secreted by the intestinal helminth Heligmosomoides polygyrus induces Foxp3+ Treg cells in vitro and in vivo in mice (Grainger et al., 2010). Furthermore, enteric carriage of a community of Clostridium species induces IL-10 secreting Foxp3+ Tregs in the colon, likely via induction of TGFβ (Atarashi et al., 2011). These findings are interesting in light of the ubiquity of Bacteroides and Clostridia as commensal organisms in human and mouse, and differences in human carriage of helminths across the world.
In mice, the presence of distant relatives of Clostridia, called segmented filamentous bacteria (SFB), drive resistance to the enteric pathogen Citrobacter rodentium and the induction of CD4+ TH17 cells in the lamina propria of the small intestine (Ivanov et al., 2009; Gaboriau-Routhiau et al., 2009). The discovery that SFB influence CD4+ T cell differentiation was based on investigators noticing differences in intestinal TH17 cell numbers between mice of the same strain purchased from different vendors, followed by the demonstration that co-housing of these mice resulted in induction of TH17 cells (Ivanov et al., 2009). SFB are highly evolved for their commensal relationship with the mouse intestine (Sczesnak et al., 2011). Similar organisms have not yet been reported in humans, but it seems likely that similarly co-evolved organisms will play a role in human intestinal biology and immunoregulation. The discovery of the role for SFB in CD4+ T cell responses is similar to the discovery of a virus-plus-gene trigger for an intestinal disease in mice with similar symptoms as CD (Cadwell et al., 2010). This finding occurred by comparing intestinal phenotypes in one strain of mice bred in two different facilities. Both of these findings underline the critical importance of directly analyzing the contributions of the entire microbiome, rather than individual components, in animal models of diseases.
Transmissible Colitis and Host-gene-metagenome Interactions
Recent studies have made the striking observation that genetically determined colitis is transmissible, revealing a key role for host genes in defining the microbiome and for metagenomic contributions to enteric disease. Mice lacking both Rag2 and the transcription factor T-bet develop colitis that can be transmitted from a mutant mother to wild-type fosterling mice (Garrett et al., 2007; Garrett et al., 2010a). Although there are expansions of specific bacterial types in these mice, including Klebsiella pneumoniae and Proteus mirabilis, another cofactor, in addition to these bacteria, is required to generate the colitis phenotype. This cofactor is not yet identified. Similarly, mice deficient in NLRP6, caspase-1, IL-18, or ASC (all proteins that regulate the expression of pro-inflammatory cytokines such as IL-18) develop colitis that is transmissible to co-housed wild-type mice (Elinav et al., 2011).
Recent studies in another mouse model of transmissible colitis, which has similarities to UC provide an example of the specificity of host-gene-bacterial relationships and IBD (Figure 4) (Bloom et al., 2011; Kang et al., 2008). Mice lacking the IL-10 receptor and expressing a dominant negative form of the TGFβ receptor in T lymphocytes develop IFNγ- and TNFα-dependent colitis (Kang et al., 2008). The disease is cured by antibiotic treatment and re-induced by co-housing diseased and cured animals or by simply feeding cured mice the common commensal bacteria Bacteroides thetaiotaomicron (B. theta)(Bloom et al., 2011). In the same mice, the related Bacteroides sp. TP5 induced a lymphocytic inflammatory infiltrate different from that induced by B. theta, indicating the remarkable specificity of host-gene-bacterial interactions (Figure 4). The authors noted dysbiosis in diseased animals with increased numbers of Enterobacteriaceae, but these bacteria did not induce disease despite being present in higher numbers in sick mice. This study shows that a single bacterial type can cause IBD-like pathology in the proper genetic setting, a bacteria-plus-gene interaction that triggers intestinal inflammation. Importantly, the observation that two closely related bacteria induce different pathologies in the same genetically susceptible host provides support for the concept that genes present in the non-host metagenome can determine a host phenotype.
A similar observation, in this case of a virus-plus-gene interaction that triggers IBD-like pathology, has been described in mice mutant for the CD risk gene Atg16L1 (Figure 4) (Cadwell et al., 2008; Cadwell et al., 2010). Abnormal Paneth cells were observed in umans carrying the ATG16L1 T300A allele and mice hypomorphic for expression of Atg16L1 raised in a conventional clean barrier (Cadwell et al., 2008). Importantly, the phenotype of the mice varied between different facilities and could be induced in mutant mice, but not wild type mice, by inoculation with a specific strain of murine norovirus (Karst et al., 2003; Thackray et al., 2007). When these mice were challenged with dextran sodium sulfate (DSS) they developed inflammatory phenotypes specific to the combination of Atg16L1 mutation and an individual norovirus strain (Figure 4). Virus-triggered pathologies could be treated by blocking TNFα or IFNγ or treatment with antibiotics. Interestingly, infection with murine norovirus enhances signaling through Nod1 and Nod2 via the induction of type 1 IFN, potentially providing a direct link between enteric viral infection and NOD signaling pathways implicated in IBD risk (Kim et al., 2011). These data raise the possibility that patterns of viral infection and specific components of the bacterial metagenome act together to influence the penetrance of UC and CD susceptibility risk alleles in humans. Furthermore, these data show that closely related viruses can have quite different effects on the phenotype of a host genetically prone to a disease process. This finding further supports the concept that genes in the non-host metagenome can determine host phenotypes.
Host-gene-metagenome Interactions in T1D
For T1D, recent observations fit with a 'perfect storm' scenario in which numerous events combine to increase susceptibility to disease development in early childhood (Figure 1). These events include susceptibility alleles in HLA class II genes and INS that cause increased autoreactivity against insulin, its precursors, and other islet antigens; lowered IL-2, IL-10, and IL-27 production and signaling; altered T cell receptor signaling and regulation (via for example susceptibility alleles in PTPN2, PTPN22, CTLA4, IL2RA); and increased type 1 IFN production and responsiveness (Todd, 2010; Robinson et al., 2011; Bluestone et al., 2010). The ‘perfect T1D storm’ is generated when these factors combined with a permissive, modern environment of widespread vitamin D deficiency (Cooper et al., 2011) and other still unidentified environmental factors (Figure 5). In particular, the T1D susceptibility genes and candidates IFIH1 (Nejentsev et al., 2009), GPR183 (EBI2)(Heinig et al., 2010) TLR7, TLR8 (Barrett et al., 2009), and FUT2 (Smyth et al., 2011), strongly suggest an etiological role for virus-induced, type 1 interferon production. A common knockout mutation of FUT2 in several populations causes the non-secretor status (i.e., a lack of shedding of the A and B blood group antigens into saliva and intestinal secretions). This T1D-predisposing FUT2 genotype is also associated with increased risk of CD (McGovern et al., 2010; Franke et al., 2010), providing another direct mechanistic link between these two diseases and microbial infections. The FUT2 non-secretor genotype is associated with resistance to certain strains of norovirus and Helicobacter pylori (Smyth et al., 2011). Investigations of the mechanisms involved in the FUT2 associations with chronic and infectious disease are urgently required, as is the case for many of the newly-identified GWAS candidate genes.
Defining the Metagenome Now and in the Future
Technologies for analyzing human loci involved in complex diseases have, until recently, outstripped technologies for analyzing the metagenome. For example, SNP-based GWAS cover the entire human genome, although at low resolution, while most common tools and methods applied to the non-host metagenome focus on only one component, such as a particular bacteria, viruses, or phage. The non-host metagenome is so complex that researchers have focused on DNA sequencing, even though many organisms relevant to disease - including enteroviruses that have been linked to T1D and viruses that cause intestinal disease- have RNA genomes. Although our knowledge of the human gut metagenome is in its infancy, this metagenome can now be explored in detail by deep, next-generation sequencing of both RNA and DNA, and then stratified by host genotype, disease-risk, or disease-status. Investigators are increasingly using shotgun sequencing of RNA + DNA, which theoretically can detect any organism (e.g. Finkbeiner et al., 2008). However, studies to date have often relied on the DNA sequencing of 16S rRNA genes of bacteria. This standard and reliable method has identified dysbiosis in IBD and T1D (Wen et al., 2008; Roesch et al., 2009; Giongo et al., 2011; Sartor, 2008; Garrett et al., 2010b). Whether these changes are causal or secondary to disease is unclear.
An outstanding example of consequences of relying on the analysis of only a subset of the metagenome is the recent appreciation that bacterial phage viruses are a major and dynamic part of the intestinal microbiome (Reyes et al., 2010). This adds an enteric bacterial 'virome' to the eukaryotic virome that lives in our tissues (Virgin et al., 2009). Bacteria are not the only cells, in addition to host cells, that can be infected by viruses with consequent changes in biology. For example, an RNA virus infects the eukaryotic pathogen Leishmania and regulates the host inflammatory responses during parasite infection (Ives et al., 2011). Thus, like bacteria and their phages, all Eukarya in the microbiome are candidates for viral infection that might alter biological processes.
The tools to detect and quantify the entire non-host metagenome at a reasonable cost will undoubtedly develop rapidly as metagenomic sequencing technologies and computational approaches to phylogeny and microbe detection are developed and applied. Similarly, sequencing the entire host genome is becoming more cost efficient and practical. This wealth of data will set the stage for metagenetics, but meaningful and robust analyses of the complex interactions within the metagenome will require new computational tools and new conceptualizations of gene-gene and gene-microbe interactions.
Conclusion: The Metagenetics of Mechanism-based Disease Subtypes
Here we have argued that two factors need to be considered as key contributors to the genetics and pathogenesis of complex inflammatory diseases, such as T1D, CD, and UC: specific host-gene-microbial interactions and the mechanistic heterogeneity of phenotypes that constitute complex diseases. Although we have used the lens of T1D, CD, and UC research to support these concepts, it is clear that these ideas may apply to a broader array of diseases, as well. The striking effects of the microbiome on systemic immunity and on diseases that affect both visceral and mucosal tissues suggest that any physiologic process may be altered by the microbiome and gene-specific interactions of the microbiome with the host. At a minimum, the diverse diseases that have been revealed by GWAS to share risk alleles are strong candidates for considering the metagenome, rather than only the host genome, as contributing to health or disease.
The concepts of mechanism-define disease subtypes and host-gene-microbial interactions cooperate in important ways. For example, if the single diagnosis of CD or T1D includes multiple mechanistic phenotypes (Figure 2, 3), a specific host-gene-microbial interaction (Figure 4) might contribute to only one of these phenotypes. In this setting, the impact of interactions between genes in the metagenome, of either microbial or host origin, would be obscured. This could, for example obscure the role of a single microbe in causing one mechanism-based disease subtype rather than causing all cases of a disease. Failure to identify such an agent would prevent the use of approaches that treat or vaccinate against the agent (Figure 2, 5). It is logical and anticipated that stratifying patients for treatment with pathway-specific drugs will improve outcomes and success of Phase II and III clinical trials (Figure 3). This paradigm is highly effective and increasing used in the treatment of cancer, but it also seems likely to benefit those with germ line-based predisposition to disease as well.
Deconvoluting the complex matrix of interactions within the metagenome that contribute to disease will require more complete analyses of the metagenome. It also requires an iterative re-definition of disease subtypes using markers that distinguish between patients based on the mechanism responsible for injury rather than the presence of tissue injury per se. This ambitious goal is daunting to consider, but data discussed herein from human studies, animal studies, and analyses of the microbiome lead us to the inescapable conclusion that complex interactions within the metagenome control phenotypes. We must face this complexity head-on to solve the puzzle of the etiology and pathogenesis of complex diseases.
We, therefore, argue for the inclusion of the metagenome in human genetic studies for these diseases. We view complex diseases as 'metagenetic,' reflecting the contributions of both host and non-host genes within the metagenome. The non-host genes in the metagenome that are relevant to a disease might be viral, bacterial, or derived from additional members of the microbiome, which are still largely uncharacterized. Parasites likely play a critical role in some populations. These metagenetic interactions probably contribute to the development of disease at two levels (Figure 5). First, we envision the normal immune system developing via harmonious relationships within the metagenome. For example, the level of innate immunity in mice is regulated by chronic herpesvirus infection (Barton et al., 2007; White et al., 2010), and therefore acquisition of a specific chronic virus might predispose the host to either helpful or harmful responses to other components of the microbiome. It will be important to develop quantitative and robust ways to indentify such a 'normal' immune system. Second, once a poorly balanced immune system is generated, host gene interactions, with either other host genes or the non-host metagenome, likely synergize to generate inappropriate levels of inflammation in response to microbial products (e.g. CD and UC) or to set the stage for development of HLA-dependent autoimmunity (T1D). Understanding this level of biological complexity will require the involvement of statisticians, computational biologists, geneticists, pathogenesis experts, virologists, bacteriologists, and parasitologists in an integrated fashion to identify mechanistically important interactions. Such an integrated approach can then perhaps make sense of the metagenetics of complex diseases, to the advantage of us all.
Acknowledgements
The authors would like to acknowledge helpful conversations with Thad Stappenbeck, Ramnik Xavier, Emil Unanue, Jeff Gordon, Balfour Sartor, Adolfo Garcia-Sastre, and Dermot McGovern into this review. We thank Tom Smith for providing the images of noroviruses used in Figure 4. HWV is supported by the NCI, NIAID, NCRR, the Crohn's and Colitis Foundation of America, and the Broad Medical Foundation. JAT is supported by the NIDDK, NIHR, the Wellcome Trust, and the Juvenile Diabetes Research Foundation International.
Reference List
- Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, de VM, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, ANDERSEN V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechiga AF, Habib T, He Y, Zhang X, Zhang ZY, Funk A, Buckner JH. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl. J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HW. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host. Microbe. 2009;6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr., Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464:1293–1300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin. Immunol. 2006;18:207–213. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Brehm MA, Bortell R, diIorio P, Leif J, Laning J, Cuthbert A, Yang C, Herlihy M, Burzenski L, Gott B, Foreman O, Powers AC, Greiner DL, Shultz LD. Human immune system development and rejection of human islet allografts in spontaneously diabetic NOD-Rag1null IL2rgammanull Ins2Akita mice. Diabetes. 2010;59:2265–2270. doi: 10.2337/db10-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon B, Carrero JA, Miller MJ, Unanue ER. Cellular and molecular events in the localization of diabetogenic T cells to islets of Langerhans. Proc. Natl. Acad. Sci. U. S. A. 2011a;108:1561–1566. doi: 10.1073/pnas.1018973108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon B, Carrero JA, Miller MJ, Unanue ER. Entry of diabetogenic T cells into islets induces changes that lead to amplification of the cellular response. Proc. Natl. Acad. Sci. U. S. A. 2011b;108:1567–1572. doi: 10.1073/pnas.1018975108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell CR, Stene LC, Joner G, Cinek O, Svensson J, Goldacre MJ, Parslow RC, Pozzilli P, Brigis G, Stoyanov D, Urbonaite B, Sipetic S, Schober E, Ionescu-Tirgoviste C, Devoti G, de Beaufort CE, Buschard K, Patterson CC. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- Cardwell CR, Stene LC, Joner G, Davis EA, Cinek O, Rosenbauer J, Ludvigsson J, Castell C, Svensson J, Goldacre MJ, Waldhoer T, Polanska J, Gimeno SG, Chuang LM, Parslow RC, Wadsworth EJ, Chetwynd A, Pozzilli P, Brigis G, Urbonaite B, Sipetic S, Schober E, Ionescu-Tirgoviste C, de Beaufort CE, Stoyanov D, Buschard K, Patterson CC. Birthweight and the risk of childhood-onset type 1 diabetes: a meta-analysis of observational studies using individual patient data. Diabetologia. 2010;53:641–651. doi: 10.1007/s00125-009-1648-5. [DOI] [PubMed] [Google Scholar]
- Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5:e1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, Greissl C, Ramos-Lopez E, Hypponen E, Dunger DB, Spector TD, Ouwehand WH, Wang TJ, Badenhoop K, Todd JA. Inherited variation in vitamin D genes is associated with predisposition to autoimmune disease type 1 diabetes. Diabetes. 2011;60:1624–1631. doi: 10.2337/db10-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abecasis G, Barrett JC, Behrens T, Cho J, De Jager PL, Elder JT, Graham RR, Gregersen P, Klareskog L, Siminovich K, vanHeel D, Wijmenga C, Worthington J, Todd JA, Hafler DA, Rich SS, Daly JJ. Pervasive Sharing of Genetic Effects in Autoimmune Disease. PLoS Genetics. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD, Nutland S, Coleman G, Himsworth M, Hardy M, Burren O, Healy B, Walker NM, Koch K, Ouwehand WH, Bradley JR, Wareham NJ, Todd JA, Wicker LS. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers S, Kaufmann SH. Infection, inflammation, and chronic diseases: consequences of a modern lifestyle. Trends. Immunol. 2010;31:184–190. doi: 10.1016/j.it.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O'Connor W, Rongvaux A, Van RN, Haberman AM, Iwakura Y, Kuchroo VK, Kolls JK, Bluestone JA, Herold KC, Flavell RA. Control of T(H)17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RSN, Jensen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJM, Smolonska A, Weersma RK, Hofstra RMW, Buurman WA, Rensen S, Wolfs MGM, Platteel M, Zhernakova A, Elbers CC, Festen EM, Trynka G, Hofker MH, Saris CGJ, Ophoff RA, van den Berg LH, van Heel DA, Wijimenga C, te Meerman GJ, Franke L. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genetics. 2011 doi: 10.1371/journal.pgen.1002197. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat. Rev. Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, de VM, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van GA, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De PM, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010a;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010b;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianani R, Campbell-Thompson M, Sarkar SA, Wasserfall C, Pugliese A, Solis JM, Kent SC, Hering BJ, West E, Steck A, Bonner-Weir S, Atkinson MA, Coppieters K, von HM, Eisenbarth GS. Dimorphic histopathology of long-standing childhood-onset diabetes. Diabetologia. 2010;53:690–698. doi: 10.1007/s00125-009-1642-y. [DOI] [PubMed] [Google Scholar]
- Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Toward defining the autoimmune microbiome for type 1 diabetes. ISME. J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl. J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger JR, Smith KA, Hewitson JP, McSorley HJ, Harcus Y, Filbey KJ, Finney CA, Greenwood EJ, Knox DP, Wilson MS, Belkaid Y, Rudensky AY, Maizels RM. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. J Exp. Med. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. U. S. A. 2011;108 Suppl 1:4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Gulati A, Sartor RB. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr. Opin. Gastroenterol. 2010;26:564–571. doi: 10.1097/MOG.0b013e32833f1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, Li Y, Sarwar R, Langley SR, Bauerfeind A, Hummel O, Lee YA, Paskas S, Rintisch C, Saar K, Cooper J, Buchan R, Gray EE, Cyster JG, Erdmann J, Hengstenberg C, Maouche S, Ouwehand WH, Rice CM, Samani NJ, Schunkert H, Goodall AH, Schulz H, Roider HG, Vingron M, Blankenberg S, Munzel T, Zeller T, Szymczak S, Ziegler A, Tiret L, Smyth DJ, Pravenec M, Aitman TJ, Cambien F, Clayton D, Todd JA, Hubner N, Cook SA. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van GA, Zelenika D, Franchimont D, Hugot JP, de VM, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, Levine A, Piccoli D, Van LJ, Cucchiara S, Monos DS, Guthery SL, Denson L, Wilson DC, Grant SF, Daly M, Silverberg MS, Satsangi J, Hakonarson H. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, Schutz F, Zangger H, Revaz-Breton M, Lye LF, Hickerson SM, Beverley SM, cha-Orbea H, Launois P, Fasel N, Masina S. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331:775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SS, Bloom SM, Norian LA, Geske MJ, Flavell RA, Stappenbeck TS, Allen PM. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H, Akira S, Wobus C, Nunez G. Viral infection augments nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9:496–507. doi: 10.1016/j.chom.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck T, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011 doi: 10.1038/nature10434. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SA, Cerosaletti K, Wan JY, Ho JC, Tatum M, Wei S, Shilling HG, Buckner JH. An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4(+) T cells. Genes. Immun. 2011;12:116–125. doi: 10.1038/gene.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Vermi W, Lonardi S, Rossini C, Otero K, Calderon B, Gilfillan S, Diamond MS, Unanue ER, Colonna M. RNA sensor-induced type I IFN prevents diabetes caused by a beta cell-tropic virus in mice. J Clin Invest. 2011;121:1497–1507. doi: 10.1172/JCI44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C, Dutridge D, Fleshner P, Shih DQ, Melmed G, Mengesha E, King L, Pressman S, Haritunians T, Guo X, Targan SR, Rotter JI. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum. Mol. Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melmed GY, Targan SR. Future biologic targets for IBD: potentials and pitfalls. Nat. Rev. Gastroenterol. Hepatol. 2010;7:110–117. doi: 10.1038/nrgastro.2009.218. [DOI] [PubMed] [Google Scholar]
- Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odze R. Diagnostic problems and advances in inflammatory bowel disease. Mod. Pathol. 2003;16:347–358. doi: 10.1097/01.MP.0000064746.82024.D1. [DOI] [PubMed] [Google Scholar]
- Oikarinen S, Martiskainen M, Tauriainen S, Huhtala H, Ilonen J, Veijola R, Simell O, Knip M, Hyoty H. Enterovirus RNA in blood is linked to the development of type 1 diabetes. Diabetes. 2011;60:276–279. doi: 10.2337/db10-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Pfluger M, Winkler C, Hummel S, Ziegler AG. Early infant diet in children at high risk for type 1 diabetes. Horm. Metab. Res. 2010;42:143–148. doi: 10.1055/s-0029-1241830. [DOI] [PubMed] [Google Scholar]
- Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, hrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, Latiano A, Goyette P, Green T, Halfvarson J, Haritunians T, Korn JM, Kuruvilla F, Lagace C, Neale B, Lo KS, Schumm P, Torkvist L, NIDDK IBD Genetics Consortium,I.I.B.D.G.C. Dubinsky M, Brant SR, Silverberg M, Duerr RH, Altshuler D, Gabriel S, Lettre G, Franke A, D'Amato M, Mcgovern DPB, Cho JH, Rious JD, Xavier RJ, Daly MJ. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature Genetics. 2011 doi: 10.1038/ng.952. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO, Drevlow BE, Zaacks LS, Grober JS, Cohen LM, Kirou KA, Crow MK, Jolly M, Niewold TB. Autoimmune Disease Risk Variant of IFIH1 Is Associated with Increased Sensitivity to IFN-{alpha} and Serologic Autoimmunity in Lupus Patients. J Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME. J. 2009;3:536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastro. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosion JF, Huttenhower C, Littman DR, Ivanov II. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host and Microbe. 2011 doi: 10.1016/j.chom.2011.08.005. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Howson JMM, Clarke P, Downes K, Mistry T, Stevens H, Walker NM, Todd JA. FUT2 non-secretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011 doi: 10.2337/db11-0638. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Rioux JD, Mizoguchi A, Saitoh T, Huett A, rfeuille-Michaud A, Wileman T, Mizushima N, Carding S, Akira S, Parkes M, Xavier RJ. Crohn disease: a current perspective on genetics, autophagy and immunity. Autophagy. 2011;7:355–374. doi: 10.4161/auto.7.4.13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stene LC, Oikarinen S, Hyoty H, Barriga KJ, Norris JM, Klingensmith G, Hutton JC, Erlich HA, Eisenbarth GS, Rewers M. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY) Diabetes. 2010;59:3174–3180. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, Kelley ST, Virgin HW. Murine Noroviruses Comprising a Single Genogroup Exhibit Biological Diversity despite Limited Sequence Divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehik K, Dabelea D. The changing epidemiology of type 1 diabetes: why is it going through the roof? Diabetes Metab. Res Rev. 2011;27:3–13. doi: 10.1002/dmrr.1141. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, Ley TJ, Virgin HW, Fehniger TA. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–4383. doi: 10.1182/blood-2009-09-245464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]