Abstract
Background
Leaf color variations are common in plants. Herein we describe a natural mutant of barley cultivar Edamai No.6, whs18, whose leaf color showed stable and inheritable stage-green-revertible-albino under field condition.
Methods
Bulked Segregant Analysis (BSA) based on SSR assay and Specific Length Amplified Fragment Sequencing (SLAF-seq) was used to map the candidate gene for this trait.
Results
We found that leaf color of whs18 was green at seedling stage, while the seventh or eighth leaf began to show etiolation, and albino leaves emerged after a short period. The newly emerged leaves began to show stripe white before jointing stage, and normal green leaves emerged gradually. The duration of whs18 with abnormal leaf color lasted for about 3 months, which had some negative impacts on yield-related-traits. Further investigations showed that the variation was associated with changes in chlorophyII content and chloroplast development. Genetic analysis revealed that the trait was controlled by a single recessive nuclear gene, and was designed as HvSGRA in this study. Based on the F2 population derived from Edamai No.9706 and whs18, we initially mapped the HvSGRA gene on the short arm of chromosome 2H using SSR and BSA. GBMS247 on 2HS showed co-segregation with HvSGRA. The genetic distance between the other marker GBM1187 and HvSGRA was 1.2 cM. Further analysis using BSA with SLAF-seq also identified this region as candidate region. Finally, HvSGRA interval was narrowed to 0.4 cM between morex_contig_160447 and morex_contig_92239, which were anchored to two adjacent FP contigs, contig_34437 and contig_46434, respectively. Furthermore, six putative genes with high-confidence in this interval were identified by POPSEQ. Further analysis showed that the substitution from C to A in the third exon of fructokinase-1-like gene generated a premature stop codon in whs18, which may lead to loss function of this gene.
Conclusions
Using SSR and SLAF-seq in conjunction with BSA, we mapped HvSGRA within two adjacent FP contigs of barley. The mutation of fructokinase-1-like gene in whs18 may cause the stage green-revertible albino of barley. The current study lays foundation for hierarchical map-based cloning of HvSGRA and utilizing the gene/trait as a visualized maker in molecular breeding in future.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2015-1) contains supplementary material, which is available to authorized users.
Keywords: Fine mapping, Barley, Stage Green-Revertible Albino, Specific Length Amplified Fragment Sequencing, Bulked Segregant Analysis
Background
Variations in leaf colors are very special in the kingdom of plants, and different kinds of them have been reported [1]. Thus far, more than 80 Chl-deficient mutants have been discovered in rice, and they are referred to as virescent, stripe, albino, chlorina, zebra and yellow variegated according to their diverse phenotypes (http://www.shigen.nig.ac.jp/rice/oryzabase/) [2]. Leaf color mutants have been utilized extensively in theoretical studies during the last several years. For example, leaf color mutants have been used to interpret the development of chloroplasts and metabolism of chlorophyll in plants [3, 4], and to illustrate the pathways of photosynthesis [5], the mechanisms of photomorphism [6], metabolism of phytohormones [7] and molecular mechanisms of disease resistance [8].
Green-revertible albino is a special type of leaf color variation. Leaf color of this type of mutant is albino under certain conditions or at certain developmental stages, while it shows green leaves gradually and thus guarantees the mutant to be able to grow and mature normally [9], such as the rice mutant Qiufeng M [9] and low temperature albino 1 [10]. As a visualized marker, this type of variation has been used successfully in breeding male-sterile lines in rice and enhancing the seed purify of hybrids in plants [11–14]. Furthermore, some of these type of leaf color variations are reported to be controlled by temperature, such as the rice mutant low temperature albino 1 [10] and maize inbred line A661 [15]. They can also be used to illustrate the mechanisms of sense and response of temperature in plant.
The completion of draft genome sequences of several higher plant species has opened an unprecedented opportunity for functional genomics studies in them [14, 16]. In rice, more than 40 green-revertible albino mutants have been identified [1]. Several genes responsible for this type of mutation have also been identified using the strategy of forward genetics. These genes are involved in diverse physiological processes. For example, one base substitution of C to T in the coding region of chloroplast protein synthesis elongation factor Tu could result in the green-revertible albino [17]. The silence of cytochrome P450 gene CYP-2 maybe responsible for the low temperature induced seedling-specific albino [18]. The 5-bp deletion in the coding region of the pentatricopeptide repeat (PPR) gene generated a premature stop codon in rice mutant young seedling albino and caused the seedling-specific albino phenotype [14]. The 45-bp insertion occurred in the first exon of a heme oxygenase gene may lead to the green-yellow phenotype in rice [19]. In maize, “zebra7 (zb7)” showed transverse green/yellow striped leaves in young plants, and map-based-cloning demonstrated that zb7 encoded the lspH protein with a mis-sense mutation in a conserved region [16].
On the other hand, combining classical bulked segregant analysis (BSA) with Next Generation Sequencing (NGS) technology has made gene cloning essentially a single-step computational procedure once a mapping population has been established [20]. Recently developed Specific Length Amplified Fragment Sequencing (SLAF-seq) is an efficient method of large-scale genotyping, which is based on reduced representation library (RRL) [21] and high-throughput sequencing [22]. SLAF-seq has been widely employed in many species [23–31], and was proven to be an effective strategy for construction of high density genetic map [23–27, 29], identification of gene for qualitative trait [28, 30] as well as QTLs for quantitative traits [24, 26, 31].
As the fourth cereal crop of the world, barley (Hordeum vulgare L., 2n = 2H = 14) provides economically important source of human and animal nutrition and underpins the malting and brewing industries. Barley also performs as a genetic model species for Triticeae genomics [32]. The recently released whole genome sequence of barley cv. Morex [33] has opened an unprecedented opportunity for performing functional genomics studies in barley. However, the genetic basis of leaf color variations in barley was largely unexplored.
In the present study, traditional SSR marker assays and SLAF-seq in conjunction with BSA were utilized to fine map a novel stage green-revertible albino gene (HvSGRA) in barley mutant whs18. The mutant was originally isolated from the malting barley variety Edamai No. 6 and showed stable and inheritable character of stage green-revertible albino. The candidate genes were also discussed and analyzed in this study. The current study will provide a basis to clone and utilizing the gene that is responsible for the albino trait in the future.
Results
Phenotypic characterization of the barley stage green-revertible albino mutant “whs18”
“whs18” was a natural mutant of the malting barley variety “Edamai No. 6”. After three years’ observation, we found that leaf color of whs18 showed stage green-revertible albino under the field condition. To be detailed, whs18 had wild-type leaf color at the seedling stage, while the seventh or eighth and later leaves showed etiolation (Fig. 1a), and albino leaves (Fig. 1b) emerged soon. The newly emerged leaves began to show stripe white (Fig. 1b, c) before the jointing stage, and normal green leaves (Fig. 1c) emerged gradually before heading time. Thus, leaf color of whs18 could harbor the stereo model of “albino-stripe white-green” (Fig. 1d) after green leaves appeared again. Leaves with different color at the corresponding stage were called green leaf at the seedling stage (GL), etiolated leaf (EL), albinistic leaf (AL), stripe white (SW), respectively. The whole developmental period with abnormal leaf color generally lasted for about 3 months.
Fig. 1.

The dynamic leaf color variation of whs18 under field condition. a Etiolated leaf b Albino leaf c Strip white leaf d Stereo leaf color performance
The impact of temperature on the stage green-revertible albino of whs18
To analyze whether the color variation of whs18 was induced by environment, three different temperature treatments (See methods) were conducted on whs18. After one month’s temperature treatment, we found that both of the seedlings in the incubator (Fig. 2a, left) and under the plastic membrane (Fig. 2b, left) grew much faster than the seedlings grown under normal condition (Fig. 2a and b, right) due to the warmer temperature. Meanwhile, the seedlings grown outside began to show ELs, and showed stage green-revertible albino as expected (Fig. 2a and b, right). However, we didn’t observe any color variation for the seedlings either in the incubator or under the plastic membrane (Fig. 2a and b, left). Another analysis showed that when encountered a period of low temperature, ELs and ALs emerged in all of the seedlings planted at three different dates (Fig. 2c), even though they have developed quite different numbers of leaves (Fig. 2c). The newly emerged leaves also showed green nearly at the same time (Fig. 2d).
Fig. 2.

The phenotype of whs18 under different temperature treatment. a T1: whs18 grown inside or outside of incubator b T2: whs18 grown inside or outside of plastic membrane c T3-1: whs18 sowed at different date d T3-2: whs18 sowed at different date
Effect of stage green-revertible albino on agronomic traits
To investigate the effect of the stage albino on agronomic trait of barley, we measured some yield related traits in Edamai No.6 and whs18. As shown in Table 1, heading date (HD) of whs18 was 8 days’ later than Edamai No.6. Furthermore, stage albino significantly reduced the plant height (PL) and length of the main spike (LS) of barley for about 10 cm and 0.8 cm, respectively. Spike number per plant (SNP) and weight of 1000 grains (WTG) were also decreased significantly by stage albino. While it seemed that the stage green-revertible albino didn’t have any significant effect on grain number of the main spike (GNS).
Table 1.
Comparison of agronomic traits between Edamai No. 6 and whs18 (p = 0.05)
| HD | PL(cm) | SNP | LS(cm) | GNS | WTG(g) | |
|---|---|---|---|---|---|---|
| Edamai No. 6 | Mar.21 | 77.4 | 6.7 | 7.1 | 33.3 | 43.6 |
| whs18 | Mar.29 | 67.0a | 5.3a | 6.3a | 31.2 | 38.3a |
HD Heading Date, PL Plant Height, SNP Spike Number per Plant, LS Length of the main Spike, GNS Grain Number of the main Spike, WTG Weight of 1000 Grains
aIndicated that there was significant difference between Edamai No.6 and whs18 at the level of 0.05
Effect of stage green-revertible albino on photosynthesis relevant traits of barley
Leaf color variations were also associated with abnormal chlorophyll metabolism. We quantified the contents of chlorophylls Edamai No.6 and whs18 at different developmental stages to determine whether the albino phenotype of whs18 resulted from impaired chlorophyll biosynthesis. As expected, the amount of chlorophylls of Edamai No.6 showed no significant change at different stages, while both of chlorophylla and chlorophyllb in whs18 showed a significant dynamic change of decrease to increase along with the leaf color changing from green to albino to green. The total chlorophyll in ALs of whs18 was less than one tenth of total chlorophyll in corresponding leaves of WT plants (Table 2).
Table 2.
Concentration of chlorophylls in Edamai No.6 and whs18
| ChlorophyIIs (mg/g) | GL | EL | AL | SW | FL | |
|---|---|---|---|---|---|---|
| Total chlorophyII | Edamai No.6 | 1.417 ± 0.063 | 1.582 ± 0.131 | 1.340 ± 0.036 | 1.34 ± 0.036 | 1.772 ± 0.072 |
| whs18 | 1.283 ± 0.073a | 0.828 ± 0.094b | 0.091 ± 0.009b | 0.564 ± 0.049b | 1.533 ± 0.060a | |
| ChlorophyIIa | Edamai No.6 | 1.194 ± 0.053 | 1.330 ± 0.168 | 1.071 ± 0.032 | 1.071 ± 0.032 | 1.408 ± 0.065 |
| whs18 | 0.976 ± 0.049a | 0.701 ± 0.082b | 0.085 ± 0.007b | 0.424 ± 0.040b | 1.257 ± 0.056a | |
| ChlorophyIIb | Edamai No.6 | 0.271 ± 0.009 | 0.253 ± 0.068 | 0.268 ± 0.011 | 0.268 ± 0.011 | 0.364 ± 0.011 |
| whs18 | 0.245 ± 0.033 | 0.127 ± 0.069b | 0.006 ± 0.003b | 0.140 ± 0.113b | 0.324 ± 0.075 |
GL Green Leaf at the seedling stage, EL Etiolated Leaf, AL Albinistic Leaf, SW Stripe White leaf, FL Flag Leaf
aIndicated that there was significant difference between Edmai No.6 and whs18 at the level of p = 0.05
bIndicated that there was significant difference between Edmai No.6 and whs18 at the level of p = 0.01
Ultrastructure of chloroplast in EL, ALand flag leaf (FL, normal green) of Edamai No.6 and whs18 was also observed at the corresponding stages. As shown in Fig. 3, stacked grana and stroma thylakoid was clearly observed and arranged regularly in Edamai No.6 at all of the three stages (Fig. 3a, b and c). However, in EL of whs18, there was no clear boundary between chloroplast and the cell wall. Grana could not be stacked normally in EL of whs18, and grana and stroma thylakoid was linear (Fig. 3d) in these ELs. What’s more, ultrastructure of chloroplast was disrupted completely in AL of whs18 (Fig. 3e). We couldn’t observe any structure of chloroplast in the whole scope. Subsequently, ultrastructure of chloroplast in FL of whs18 was intact (Fig. 3f) and similar to that of Edamai No.6 (Fig. 3c).
Fig. 3.

Ultrastructure of chloroplast of Edamai No.6 and whs18 at different developmental stages. a Edamai No.6 at etiolated stage, b Edamai No.6 at albino stage, c Flag leaf of Edamai No.6 d Etiolated leaf of whs18, e Albinistic leaf of whs18, f Flag leaf of whs18
Genetic analysis of the gene (s) controlling the mutant phenotype
To illustrate the genetic characteristics of the gene (s) controlling the phenotype of stage green-revertible albino, whs18 was crossed with two wild-type barley cultivars (Table 3). Our analysis showed that all of the four F1 population consisted of about 100 individuals showed normal phenotype, either use whs18 as the male or female parent. For all of the tested F2 populations, the segregation ratio of green and albino plants fit a 3:1 Chi square test (Table 3), suggesting that the inheritance of the stage green revertible albino in barley follows a simple Mendelian inheritance pattern and behaves as a single recessive trait, and the gene controlling the trait was designed as HvSGRA in this study.
Table 3.
Genetic analysis of the albino mutant
| Cross | No. of normal plant | No. of mutant plant | Expected ratio | Calculated ratio |
|---|---|---|---|---|
| χ 2 (0.05,1) = 3.84 | ||||
| Edamai No.9706 × whs18 | 397 | 117 | 3:1 | 1.37 |
| Edamai No.934 × whs18 | 266 | 94 | 3:1 | 0.24 |
| whs18 × Edamai No.9706 | 221 | 65 | 3:1 | 0.79 |
| whs18 × Edamai No.934 | 164 | 51 | 3:1 | 0.19 |
Preliminary mapping of the HvSGRA gene
To map the HvSGRA gene, an F2 mapping population was generated from the cross between Edamai No.9706 and whs18. More than 400 SSR primer-pairs distributing on the seven barley chromosomes were used to assay the two parents. About one-third of them showed polymorphism between the two parents. Further BSA analysis indicated that the HvSGRA gene was on chromosome 2H of barley. Thus SSR markers on 2H were screened in the F2 population consisting of 135 albino individuals, and the HvSGRA gene was mapped with two SSR markers GBMS247 and GBM1187 within a genetic distance of 1.2 cM (Fig. 4a). Further analysis anchored the two markers on morex_contig_38923 and morex_contig_40266 (Fig. 4c), respectively. The two contigs (morex_contig_38923 and morex_contig_40266) were genetically positioned at 2.2622 and 7.4363 cM of barley 2H chromosome, respectively.
Fig. 4.

Mapping of HvSGRA gene. (a) Primary mapping of HvSGRA (b) Fine mapping of HvSGRA (c) Physical mapping of HvSGRA
SLAF-seq was conducted on the two parents and two bulks with 50 normal or mutant individuals from the F2 population, respectively. The SLAF-seq data analysis generated 1.39 Gb, 1.39 Gb, 3.59 Gb and 3.54 Gb data for Edamai No.9706, whs18, dominant bulk and recessive bulk, respectively. A total of 100,926 SLAFs were generated, including 10,278 SNPs, 467 repeat, and the other 87,569 showed no polymorphism between the two parents. 59 Diff-markers that are likely associated with the trait were identified after statistic analysis. A total of 38 of them could be anchored to the morex_contigs (Additional file 1). Then five regions with three or more Diff-markers were considered for further analysis, including 2.1955 to 7.4363 cM and 52.4788 to 58.0524 cM on the short arm of chromosome 2H, 49.5042 to 56.1615 cM on the short arm of 6H, 113.2082 to 119.0587 cM on the long arm of 6H, and 104.8159 to 110.2691 cM on the long arm of 7H (Additional file 1). Number of Diff_marker of each region was six, four, five, four and four, respectively (Additional file 1).
Fine mapping of the HvSGRA gene
The result of the preliminary mapping using SSR and SLAF-seq suggested that the HvSGRA gene was in the interval of 2.2622 to 7.4363 cM on 2H of barley. In order to further fine map the HvSGRA gene, the closest linked SSR marker GBMS247 and GBM1187 were used to screen a larger F2 population consisting of 693 albinos, and totally 19 recombinants were identified. Two other SSR markers (Bmac0134 and Awbms0062) between GBMS247 and GBM1187 were scored on the panel of 19 recombinants for the HvSGRA interval. The candidate gene was finally located between Bmac0134 and Awbms0062 (Fig. 4b). The two SSR markers were anchored to morex_contig_42056 and morex_contig_92239 (Fig. 4c), respectively.
For our further analysis, several primer-pairs based on eight morex-contigs nearby morex_contig_42056 and morex_contig_92239 were designed and used to generate new molecular markers potentially linked to HvSGRA gene. Five primer-pairs of them showed polymorphism between the two parents (Additional file 2), including MC7646, MC67200, MC160447, MC1592730 and MC138067. As shown in Fig. 5, HvSGRA showed cosegregation with MC1592730 and MC138067 in the larger population consisting of 693 albino individuals. Finally, the HvSGRA was mapped between morex_contig_160447 and morex_contig_92239, with the genetic distance being 0.2 cM and 0.2 cM (Fig. 4b and c), respectively. They were anchored to two adjacent FP_contigs of barley, contig_34437 and contig_46434 (Fig. 4c), respectively.
Fig. 5.
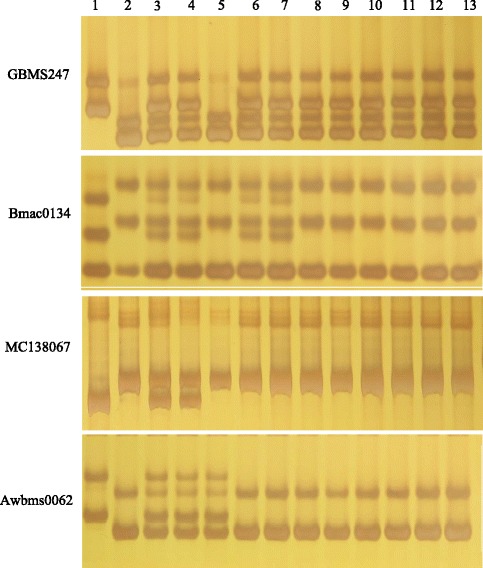
Genotype of the two parents and lines from the derived F2 population. Lane1: Edamai No.9706, Lane2:whs18, Lane3-4: Wild-type lines, L5-13: Mutant Lines
Candidate gene analysis of HvSGRA
Our further analysis showed that there were 27 and 33 morex-contigs on contig_34437 and contig_46434, respectively. Six putative genes with high confidence on these morex_contigs were extracted from the most current POPSEQ barley map [34]. They included cytochrome P450, fructokinase-1-like (FLN), Laccase and unknown genes (Table 4).
Table 4.
Putative genes in the candidate region
| Morex_contig | FP_contig | HC_genes_CDS | Gene annotation |
|---|---|---|---|
| morex_contig_160711 | contig_34437 | AK249799.1 | fructokinase-1-like (partial sequence) |
| morex_contig_244330 | contig_34437 | AK372445 | cytochrome p450 (partial sequence) |
| morex_contig_67200 | contig_34437 | MLOC_75134.1 | cytochrome p450 (full length) |
| morex_contig_1592730 | contig_46434 | AK356172 | uncharacterized protein (partial sequence) |
| morex_contig_6591 | contig_46434 | AK369630 | cbs domain containing protein (partial sequence) |
| morex_contig_138067 | contig_46434 | MLOC_6767.1 | Laccase (full length) |
All of these six genes were cloned and sequenced from the wild-type Edamai No. 6 and the mutant whs18. Our final comparison between them showed that there were totally14 stable SNPs in the third exon of fructokinase-1-like gene HvFLN1 (Genbank: AK249799) between Edamai No.6 and whs18 (Fig. 6) both on DNA and cDNA levels. Most importantly, one of these SNPs was a substitution from C to A in whs18. The substitution generated a premature stop codon of TAA in whs18 (Fig. 6). The coding region of the wild-type HvFLN1 was 1734 bp in length and the substitution lead to the missing of 330 animo acids in C terminal of HvFLN1 in whs18. Thus, the mature product of HvFLN1 in whs18 was lack of the ATP binding site of the kinase, which may lead to loss function of this gene in whs18. The coding regions of the other five genes were exactly the same between Edamai No. 6 and whs18. Our further analysis showed that the putative amino acid sequence encoded by HvFLN1 showed 27 and 41 % similarity to AtFLN1 (Genbank: NP_190977) and AtFLN2 (Genbank: NP_177080) of Arabidopsis, respectively (Fig. 7).
Fig. 6.

Sequence comparison of the third extron of HvFLN1 gene between Edamai No.6 and whs18. The red * indicated the SNP that generated the premature stop codon in whs18
Fig. 7.

Homology analysis of fructokinase and fructokinase-like proteins in plant. “red circle symbol” indicated the FLNs proteins, “red triangle symbol” indicated the FRKs proteins
Discussion
Hundreds of mutants with altered leaf color have been previously described in plants, especially in Arabidopsis (http://www.arabidopsis.org) and rice (http://rmd.ncpgr.cn/) including green-revertible albino mutants. However, few were described in barley. Cytoplasmic line 2 (CL2) was a cytoplasmically inherited chlorophyII-deficient mutant of barley. At emergence, the CL2 seedlings-phenotype varied from a grainy light green to an albino color, and gradually greened during the following days [35]. In this study, whs18, a natural leaf color mutant of barley cultivar Edamai No.6, was characterized and analyzed. Leaf color of whs18 was not only different from the wild type, but also was different at different developmental stages (Fig. 1), including GL, EL, ALand SW. This phenomenon was called stage green-revertible albino in rice [9, 13, 17, 19, 36]. There has been no report about this type of leaf color variation in barley. Characterization of the mutant and fine mapping of the gene will pave the way for map-based cloning of the gene and unraveling the molecular mechanisms of the variation. The mutant will also be a new gene resource for barley molecular breeding in future.
Several leaf color variations have been reported to be induced by low temperature, such as the maize inbred line A661I, exposure of whose seedlings to low temperatures during early leaf biogenesis led to chlorophyll losses [15]. Rice mutant low temperature albino 1 (Ita 1) showed albino leaves before 4-leaf stage when grown under temperature lower than 20 °C [10]. Another rice mutant mr21 showed yellow leaf color under the temperature lower than 25 °C and turned green with an increase in temperature [37]. However, in the barley Cytoplasmic line 2 mentioned above, higher temperatures during seed formation were negatively associated with pigment content in the seedlings, while higher temperature during the growth of its seedlings had an opposite effect on pigment content [35]. The abnormal leaf color of the rice mutant hfa-1 seemed to be induced by warm temperature too. Leaf color of hfa-1 was albino when the temperature was between 25-30 °C, while it was yellow under the temperature of 15-20 °C [36]. To analyze the interaction between the color variation of whs18 and environment, whs18 was subjected to three different treatments. As shown in Fig. 2, both of the seedlings in the incubator and plastic membrane showed normal green leaves, while the seedlings grown outside showed abnormal leaf color after one month later (Fig. 2a and b). Furthermore, seedlings sowing at different dates showed etiolation nearly at the same time (Fig. 2c and d), suggesting that the abnormal leaf color of whs18 was mainly induced by low temperature. Actually, to analyze the precise shift temperature of the leaf color variation in whs18, the 10-day-old seedlings of whs18 have been treated at the temperature of 10/0 °C, 10/4, 15/8 and 20/13 °C (day/night, 12/12 h) for one month, respectively. However, we didn’t observe any color variation under those controlled conditions. On the other hand, the maximum and minimum temperatures were also recorded from mid-November (10-day-old seedling) to the end of December (albino leave emerged) in 2014. We found that the minimum temperature of 10 days was 0 °C and 7 days was lower than 0 °C during this time (Data not shown), suggesting that the temperature under the ice-point may be crucial for the leaf color variation of whs18.
As we all known, most of the leaf color variation couldn’t survive under normal condition, especially these albino mutants. Stage green-revertible albino mutant was a special kind of leaf color mutant. This type of mutant could develop normal green leaves after the abnormal ones, which enabled it to flower and mature normally. For example, the green-revertible albino in rice Qiufeng M lead to the significant reduction of neck length and 1000-grain weight at 1 % level, while the other agronomic characters investigated showed no significant difference between the mutant and the wild-type [9]. As compared to Edamai No.6, the abnormal vegetative growth in whs18 lead to the reduction of plant height and other yield related traits, including SNP, LS, GNS and WTG, and the HD was about eight days later in whs18 (Table 1). It was probably because that the duration of whs18 with abnormal leaf color lasted for about three months, which was nearly half of the whole growing season of barley in Wuhan city. The abnormal chlorophyll metabolism in these leaves (Table 2, Fig. 3) influenced the photosynthesis of these leaves, and then played some negative impact on these agronomic traits investigated.
Most of the leaf color variations were controlled by one single recessive nuclear gene, and a few of them were also reported to be controlled by cytogene, such as the stage albinism line of winter wheat FA85 [38]. While the abnormal leaf color of Mt135 was controlled by nucleo-cytoplasmic gene interaction [2]. Our genetic analysis suggested that the stage green-revertible albino of whs18 was a qualitative trait that controlled by one single recessive nuclear gene (Table 3) , which allowed us to hunt for this gene directly using the strategy of BSA based on molecular markers. We preliminarily mapped the HvSGRA gene on chromosome 2H of barley using traditional BSA method.
Recently, a large amount of data from NGS and modern tools enable us to extract putative genes in certain regions without a precise genomic sequence in barley. The strategy of mapping-by-sequencing has accelerated the forward genetics significantly in barley [39]. Using the approach of mapping-by-sequencing applied on the whole-exome capture data, HvPHYTOCHROME C was identified as a candidate gene for the early maturity locus modulating the circadian clock and photoperiodic flowering in barley [40]. We assumed to conduct SLAF-seq to develop closer SNP markers linked to the gene. Five regions were identified by SLAF-seq and BSA, including the interval identified by traditional BSA (Additional file 1). Xia et al. [30] also identified three independent candidate regions on chr3 of maize for a qualitative trait using the strategy of BSA based on SLAF-seq, with only three Diff_markers for each region. Actually, SSR markers in the other four regions identified by SLAF-seq (Additional file 1) were also employed in our preliminary mapping analysis. These markers showed no polymorphism either between the two parents or the two pools, and they were excluded from our further analysis. Thus the overlapped region identified by SSR and SLAF-seq was considered the candidate region. On the other hand, marker density (Six Diff_markers) in the HvSGRA interval (2.1955 to 7.4363 cM on the short arm of chromosome 2H) identified by SLAF-seq may be insufficient to pin the candidate gene in barley (Additional file 1). Similar phenomenon was also observed in cotton [28]. And our further map-based cloning based on the physical map of barley narrowed down the HvSGRA interval to two morex_contigs on two adjacent FP_contigs of barley, contig_46434 and contig_34437 (Fig. 4).
The recent release of the barley draft genome assembly [33] provides a resource for pursuing the SGRA gene, however, the precise genomic sequence is still not available. Synteny-based isolation of genes conserved across cereal species is a common and effective way in map-based cloning of barley genes [41, 42]. Whereas, our in silico comparison showed that only 18 of the 60 morex_contigs on FP_contig_46464 and FP_contig_34437 could be anchored to the genome of Brachypodium distachyon. Nine and seven of these 18 morex_contigs were distributing on Bd1 and Bd3, respectively (Data not shown). Furthermore, as for the two morex_contigs surrounded the HvSGRA gene, morex_contig_160447 was anchored to Bd5 of Brachypodium distachyon, and we couldn’t find any homolog contigs for morex_contig_92239. The similar result was also observed in rice. This hindered us from getting a precise physical order of these morex_contigs in the chromosome region containing the HvSGRA gene. So all of the genes in FP_contig_46434 and FP_contig_34437 with high confidence were extracted from the POPSEQ barley map [34]. Finally, six genes were identified as candidate gene for HvSGRA (Table 4), including four genes with known function, namely cytochrome P450, fructokinase-1-like (FLN1), Laccase. Out of these genes, cytochrome P450 [18] and fructokinase (FRK) [43, 44] have been reported to be associated with albino in plant. Our further analysis showed that one base’s substation of C to A in the third exon of HvFLNs gene generated a premature stop codon in whs18 (Fig. 6a). The lack of ATP binding site of HvFLN1 in whs18, which is essential for the kinase activity, may lead to loss of function of this gene. FRK is a kind of kinase that primarily phosphorylates fructose in carbohydrate metabolism, which is involved in many developmental and physiological processes of plant [45]. Except for FRKs, there is still a kind of members called FLNs in plants (Fig. 7). These FLNs were distinctly grouped for a new branch during the homology analysis (Fig. 7), suggesting that they may play different roles in plant as compared to FRKs. However, these FLNs were quite conserved among different species, especially the C terminal of these proteins. The HvFLN1 identified in our study showed high level of similarity to AtFLN1 and AtFLN2 of Arabidopsis, respectively (Fig. 7). The most recent investigations showed that the mutation of AtFLN1 or AtFLN2 in Arabidopsis displayed a pale-yellow or albino cotyledons phenotype and were seedling lethal on MS medium without sucrose, and only could survive on sucrose-containing medium [43, 44]. The high similarity between the barley gene and its counterparts of Arabidopsis suggested that they may play a similar role in developmental and physiological processes of plant. And further functional validation of the candidate gene, such as knock out or functional complementation, was also needed to confirm the role of HvFLN1 in the green-revertible albino phenotype of barley. Illustrating the molecular mechanisms of this interesting phenomenon will also help us to understand it well, which will be helpful for further use of the gene and the trait in future.
Conclusions
SSR assay and SLAF-seq in conjunction with BSA were conducted to map a gene controlling the stage green-revertible albino in barley. The HvSGRA gene was mapped between two adjacent FP contigs of barley. Substitution of C to A in the third exon of fructokinase-1-like gene in whs18 may be responsible for the stage green-revertible trait of barley. The current study will lay the foundation for hierarchical map-based cloning of the HvSGRA gene and utilizing the gene or the trait in the molecular breeding of barley in the future.
Methods
Measurement of agronomic traits of whs18 and Edamai No.6
“whs18” was initially isolated from the elite malting barley cultivar Edamai No.6 as a spontaneous mutant in the field. Leaf color of whs18 showed stage green-revertible albino. Edamai No.6 and whs18 were planted in the field, and the management of the filed experiments was in accordance with local standard practices. At maturity, agronomic traits including plant height (PL), heading date (HD), spike number per plant (SNP), length of the main spike (LS), grain number of the main spike (GNS) and weight of 1000 grains (WTG) were measured for ten plants of Edamai No.6 and whs18.
Measurement of concentration of chlorophyll
Concentration of chlorophylls in leaves of whs18 with different colors were measured [46], and also that in corresponding leaves of Edamai No.6. In brief, healthy and fresh leaves with different color were collected at different stages, namely green leaves (GL) at seedling stage, etiolated leaves (EL) at etiolation stage, albinistic leaves (AL) at albino stage and flag leaves (FL) at heading stage, and cut into sections of 1 mm to 2 mm. Half of one gram of each sample was transferred into a mortar with 25 ml 95 % ethanol and grounded until the leaves turned into white. The tissue was transferred into 50 ml volumetric flask and brought up to volume by 95 % ethanol. The extraction solution was used to measure the absorbance values under 663 nm and 645 nm using a spectrophotometer with 95 % ethanol as control. Each sample was assayed with three biological replicates, and content of chlorophyll was calculated according to the following equations:
CT was the content of total chlorophyll with unit of mg/g fresh weight. A663 and A645 were the absorbance at 663 nm and 645 nm, respectively. V is the final volume of the extraction solution (mL), and m is the initial weight of leaves (g).
Transmission electron microscopy (TEM) assay
Corresponding leaves of Edamai No.6 and whs18 at three of the five developmental stages were sampled for TEM assay [10], including etiolated leaves, albinistic leaves and flag leaves. For detailed, leaves were cut into sections of 1 mm × 1 mm and fixed in 2.5 % glutaraldehyde in 0.1 mol/L phosphate buffer (PBS, pH 7.2) at 4 °C for 3 days, following by washing with 1 mol/L PBS for three times, 15 min per time. Samples were fixed in 1 % osmium tetroxide in 0.1 mol/L PBS for about 2.5 h until it turned into black, followed by washing with 1 mol/L PBS as before, then dehydrated with a gradient acetone series (30-50 %-70-80 %-90-100 %), 30 min for each concentration, and embedded in Polybed 812 (Epon) resin. Ultrathin sections were obtained with an ultramicrotome, mounted on grids, stained by lead citrate and uranyl acetate for 10 and 30 min, respectively. The final dried ultrathin sections were observed and photographed using a transmission electron microscope H-7650 (Hitachi, Japan) according to the protocol provided by the manufacture.
Temperature treatment
To characterize whether the stage green-revertible albino of whs18 was induced by low temperature, the 10-day-old seedlings of whs18 grown in pots were transferred into the incubator with 18/10°Cday-night temperature (T2), or 10-day-old seedlings of whs18 grown in the field were covered with two layers of plastic membrane (T4). Leaf color of them was compared with the corresponding seedlings grown under the normal condition, which were referred to T1 and T3 for the pots and the field, respectively. whs18 was also sowed at three different dates with 10 days’ interval in 2012 and 2013, including normal sowing date (Nov.7th) and 10 days’ earlier (Oct.28th) or later than (Nov.17th) the normal sowing date, and the leaf color was also observed at different stages.
Genetic analysis of the gene controlling the stage green-revertible albino of barley
Genetic characterization of the gene (s) controlling the stage green-revertible albino of whs18 was analyzed based on the phenotype of F1 hybrid and F2 population derived from four crosses, including Edamai No.9706 × whs18, Edamai No.934 × whs18, whs18 × Edamai No.9706 and whs18 × Edamai No.934. Chi-squared test was used to determine the suitability of observed data with expected segregation ratios.
SSR-based genotyping and bulked segregant analysis
To determine the position of the stage green-revertible albino gene sgra in “whs18” on genetic map, an F2 population derived from barley cultivar Edamai No.9706 and whs18 was used. Traditional BSA strategy was conducted to prepare genomic DNA pools for analysis. To be detailed, genomic DNA of 10 green lines and 10 albino lines from the F2 population along with the two parents was extracted using the modified CTAB method [47]. Equal amounts of DNA from the 10 green or albino lines were pooled together to construct the dominant or recessive bulks, respectively. More than 400 known SSR markers from GrainGenes (http://wheat.pw.usda.gov/GG2/index.shtml) distributing on the seven chromosomes of barley were screened in the parents and the two bulks. PCR reaction and the polyacrylamide gel analysis were conducted to analyze the genotype of the samples according to Liu et al. [48]. The probably polymorphic markers that associated with the trait were used to genotype the 135 homozygous recessive individuals from the F2 population firstly. Linkage between molecular markers and the SGRA gene was analyzed using Mapmaker 3.0b [49] with an LOD score of 3.0 as the threshold. The genetic map was drawn with the software Mapdraw V2.1 [50]. Closest linked markers were used as queries to search in the database of assembly_WGS Morex on IBSC [33] using BLASTN and were anchored to the genome of barley.
SNP-based genotyping and bulked segregant analysis
To narrow down the candidate gene interval, SLAF-seq [22] was conducted with Illumina Genome Analyzer II (Illumina Inc., San Diego, CA, USA) to develop SNP markers that probably linked to the gene. Specifically, genomic DNA of 50 normal green and 50 albino plants from the F2 population along with the two parents was extracted as mentioned above. Equal amounts of DNA from the 50 normal green or albino plants were pooled together to construct the dominant or recessive bulks, respectively. The totally four samples were subjected to SLAF-seq and SNP-index analysis. The procedure was performed as described by Sun et al. [22]. Data analysis and association analysis was the same as Xia et al. and Chen et al. [28, 30]. Diff-markers likely associated with the trait were anchored to the genome of Morex (Data version: assembly3_WGSMorex) [33] using blat [51]. The chromosome interval with five or more consecutive Diff-markers was considered for further analysis.
Fine mapping of the HvSGRA gene
The closest markers linked to the candidate gene were screened in a larger F2 population consisting of 693 homozygous recessive individuals derived from Edamai No. 9706 and whs18. More SSR markers between or nearby the closest markers were genotyped in the resulting recombinants to narrow down the interval containing the candidate gene, which had been marker-selected from the larger population. The markers were anchored to morex_contigs as mentioned above. Morex-contigs nearby them were picked up and used to generate new molecular markers potentially linked to HvSGRA gene. Primers were designed using the DNAMAN software.
Physical mapping of the HvSGRA gene and candidate gene analysis
The closest markers linked to the candidate gene were anchored to the barley physical map by doing blast with BAC end sequences and fully sequenced BACs [33], and the putative FingerPrinted contigs (FP contig version: fpc_10) containing HvSGRA gene were identified. Then morex contigs on the FP contigs were extracted from IBSC, and putative genes with high confidence on these morex_contigs were extracted from the most current POPSEQ barley map [34]. Genes were annotated after running blastn search against the database of HC_genes_CDS_Seq of barley [33].
As shown in Table 4, some morex_contigs only contained partial sequence of the genes, so the full genomic sequence of these genes were first cloned in silico. Several overlapped primers were designed based on the sequence of the Morex-contigs and the candidate genes using DNAMAN5.0 software. The specific PCR product from DNA and cDNA of the wild-type Edamai No.6 and the mutant whs18 was sequenced and compared.
Amino acid sequences of FRKs or FLNs from other plant species were downloaded from NCBI (www.ncbi.nlm.nih.gov). The deduced amino acid sequence of HvFLN and FRKs or FLNs from other plant species were aligned and compared using the software MEGA 5.1.
Acknowledgements
We thank Doctor Liu Gang at China Seed Group Company Limited for his assistance in the data analysis of SSR assay and BSA analysis. We thank Dr. Liangliang Gao (University of Minnesota) for reading this manuscript and providing suggestions to improve it. This work was financially supported by National Natural Science Foundation of China (31201211), Natural Science Foundation of Hubei Province (2015CFA108) and China Agriculture Research System (CARS-05).
Abbreviations
- SGRA
Stage Green-Revertible Albino Gene
- BSA
Bulked Segregant Analysis
- SLAF-seq
Specific Length Amplified Fragment Sequencing
- GL
Green leaf at the seedling stage
- EL
Etiolated leaf
- AL
Albinistic leaf
- SW
Stripe White leaf
- FL
Flag leaf
- PL
Plant height
- HD
Heading date
- SNP
Spike number per plant
- LS
Length of the main spike
- GNS
Grain number of the main spike
- WTG
Weight of one thousand grains
- NGS
Next Generation Sequencing
- FRK
Fructokinase
- FLN
Fructokinase-like protein
Additional files
Comparison between the markers identified by SLAF-seq and SSR assay. (XLSX 13 kb)
Information of the HvSGRA associated markers. (XLSX 10 kb)
Footnotes
Competing interests
All of the authors declare that they have no competing interests.
Authors’ contributions
DDQ carried out the whole experiment and drafted the manuscript, JD and STG carried out the field experiment, GGG carried out the data analysis of the SLAF-seq, FCX participated in the measurement of concentration of chlorophyll, QX and YXX helped in DNA extraction and MFL designed the experiment. All authors have read and approved the final manuscript.
Contributor Information
Dandan Qin, Email: hnqdd@163.com.
Jing Dong, Email: dongjingsir061@163.com.
Fuchao Xu, Email: xfc2008xfc@163.com.
Ganggang Guo, Email: guoganggang@caas.cn.
Shuangtao Ge, Email: gst419@163.com.
Qing Xu, Email: 395045924@qq.com.
Yuxin Xu, Email: xyx980223@icloud.com.
Meifang Li, Email: limeifang100@126.com.
References
- 1.Dong H-l, Z-j F, Wei L, Wu X-Z, Zhou P. Research Advances on Green Revertible Albino Mutants of Rice Leaves in Seedling Stage. Hubei Agric Sci. 2012;51:5241–5247. [Google Scholar]
- 2.Zhao H-B, Guo H-J, Zhao L-S, Gu J-Y, Zhao S-R, Li J-H, Liu L-X. Agronomic Traits and Photosynthetic Characteristics of Chlorophyll-Deficient Wheat Mutant Induced by Spaceflight Environment. Acta Agron Sin. 2011;37:119–126. [Google Scholar]
- 3.Leon P, Arroyo A, Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49(1):453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- 4.Von Wettstein D, Gough S, Kannangara CG. Chlorophyll Biosynthesis. Plant Cell. 1995;7(7):1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fambrini M, Castagna A, Vecchia FD, Degl’Innocenti E, Ranieri A, Vernieri P, Pardossi A, Guidi L, Rascio N, Pugliesi C. Characterization of a pigment-deficient mutant of sunflower (Helianthus annuus L.) with abnormal chloroplast biogenesis, reduced PS II activity and low endogenous level of abscisic acid. Plant Sci. 2004;167(1):79–89. doi: 10.1016/j.plantsci.2004.03.002. [DOI] [Google Scholar]
- 6.Parks BM, Quail PH. Phytochrome-Deficient hy1 and hy2 Long Hypocotyl Mutants of Arabidopsis Are Defective in Phytochrome Chromophore Biosynthesis. Plant Cell. 1991;3(11):1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal GK, Blaxter ML, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC Gene. Plant Physiol. 2001;125(3):1248–1257. doi: 10.1104/pp.125.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh UP, Prithiviraj B, Sarma BK. Development of Erysiphe pisi (Powdery Mildew) on Normal and Albino Mutants of Pea (Pisum sativum L.) Entwicklung von Erysiphe pisi (Echter Mehltau) an normalen Pflanzen und an Albinomutanten der Erbse (Pisum sativum L.) J Phytopathol. 2000;148(11–12):591–595. [Google Scholar]
- 9.Chen T, Zhang Y, Zhao L, Zhu Z, Lin J, Zhang S, Wang C. Physiological Character and Gene Mapping in a New Green-revertible Albino Mutant in Rice. J Genet Genomics. 2007;34(4):331–338. doi: 10.1016/S1673-8527(07)60035-6. [DOI] [PubMed] [Google Scholar]
- 10.Peng Y, Zhang Y, Lv J, Zhang J, Li P, Shi X, Wang Y, Honglei Z, He Z, Teng S. Characterization and Fine Mapping of a Novel Rice Albino Mutant low temperature albino 1. J Genet Genomics. 2012;39:385–396. doi: 10.1016/j.jgg.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, Wang ML, Zhang YZ, Du LF, Pan T. A chlorophyll-reduced seedling mutant in oilseed rape, Brassica napus, for utilization in F1 hybrid production. Plant Breed. 2000;119(2):131–135. doi: 10.1046/j.1439-0523.2000.00453.x. [DOI] [Google Scholar]
- 12.Shen S-q, Q-y S, Wu BJ-s, D-x CH-r, Y-w X. Development of a Greenable Leaf Color Mutant Baifeng A and Its Application in Hybrid Rice Production. Chinese Rice Science. 2004;18:34–38. [Google Scholar]
- 13.Wu W, Liu X, Shu X-l, Shu Q-Y, Xia Y-w, Wu D-X. Two-line Hybrid Rice Male Sterile Line “BHR111S” with a Marker of Green-revertible Albino Leaves. J Nucl Agric Sci. 2006;20:103–105. [Google Scholar]
- 14.Su N, Hu ML, Wu DX, Wu FQ, Fei GL, Lan Y, Chen XL, Shu XL, Zhang X, Guo XP, et al. Disruption of a Rice Pentatricopeptide Repeat Protein Causes a Seedling-Specific Albino Phenotype and Its Utilization to Enhance Seed Purity in Hybrid Rice Production. Plant Physiol. 2012;159(1):227–238. doi: 10.1104/pp.112.195081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez VM, Velasco P, Garrido JL, Revilla P, Ordás A, Butrón A. Genetic regulation of cold-induced albinism in the maize inbred line A661. J Exp Bot. 2013;64(12):3657–3667. doi: 10.1093/jxb/ert189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu XM, Hu XJ, Zhao YZ, Song WB, Zhang M, Chen ZL, Chen W, Dong YB, Wang ZH, Lai JS. Map-Based Cloning of zb7 Encoding an IPP and DMAPP Synthase in the MEP Pathway of Maize. Mol Plant. 2012;5(5):1100–1112. doi: 10.1093/mp/sss038. [DOI] [PubMed] [Google Scholar]
- 17.Chen T, Zhang Y, Zhao L, Zhu Z, Lin J, Zhang S, Wang C. Fine mapping and candidate gene analysis of a green-revertible albino gene gra(t) in rice. J Genet Genomics. 2009;36(2):117–123. doi: 10.1016/S1673-8527(08)60098-3. [DOI] [PubMed] [Google Scholar]
- 18.Xia J-C, Wang Y-P, Ma B-T, Yin Z-Q, Hao M, Kong D-W, Li S-G. Ultrastructure and Gene Mapping of the Albino Mutant al12 in Rice (Oryza sativa L.) Acta Genet Sin. 2006;33(12):1112–1119. doi: 10.1016/S0379-4172(06)60149-2. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Wang Y, Chai J, Wang L, Wang C, Long W, Wang D, Wang Y, Zheng M, Peng C, et al. Green-revertible Chlorina 1 (grc1) is required for the biosynthesis of chlorophyll and the early development of chloroplasts in rice. J Plant Biol. 2013;56:326–335. doi: 10.1007/s12374-013-0180-9. [DOI] [Google Scholar]
- 20.Liu WY, Kang JH, Jeong HS, Choi HJ, Yang HB, Kim KT, Choi D, Choi G, Jahn M, Kang BC. Combined use of bulked segregant analysis and microarrays reveals SNP markers pinpointing a major QTL for resistance to Phytophthora capsici in pepper. Theor Appl Genet. 2014;127:2503–2513. doi: 10.1007/s00122-014-2394-8. [DOI] [PubMed] [Google Scholar]
- 21.Van Tassell CP, Smith TPL, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Meth. 2008;5(3):247–252. doi: 10.1038/nmeth.1185. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H, et al. SLAF-seq: An Efficient Method of Large-Scale De novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE. 2013;8(3):e58700. doi: 10.1371/journal.pone.0058700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Wang L, Xin H, Li D, Ma C, Ding X, Hong W, Zhang X. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013;13:141. doi: 10.1186/1471-2229-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Tian L, Zhang J, Huang L, Han F, Yan S, Wang L, Zheng H, Sun J. Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max. BMC Genomics. 2014;15(1):1086. doi: 10.1186/1471-2164-15-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Z, Huang L, Zhu R, Xin D, Liu C, Han X, Jiang H, Hong W, Hu G, Zheng H, et al. A High-Density Genetic Map for Soybean Based on Specific Length Amplified Fragment Sequencing. PLoS ONE. 2014;9(8):e104871. doi: 10.1371/journal.pone.0104871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Q, Wang Y, Qin X, Zhang Y, Zhang Z, Wang J, Li J, Lou Q, Chen J. An SNP-based saturated genetic map and QTL analysis of fruit-related traits in cucumber using specific-length amplified fragment (SLAF) sequencing. BMC Genomics. 2014;15(1):1158. doi: 10.1186/1471-2164-15-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai C, Cheng F-Y, Wu J, Zhong Y, Liu G. The First High-Density Genetic Map Construction in Tree Peony (Paeonia Sect. Moutan) using Genotyping by Specific-Locus Amplified Fragment Sequencing. PLoS ONE. 2015;10(5):e0128584. doi: 10.1371/journal.pone.0128584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Yao J, Chu L, Yuan Z, Li Y, Zhang Y. Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor Appl Genet. 2015;128(3):539–547. doi: 10.1007/s00122-014-2452-2. [DOI] [PubMed] [Google Scholar]
- 29.Ma J-Q, Huang L, Ma C-L, Jin J-Q, Li C-F, Wang R-K, Zheng H-K, Yao M-Z, Chen L. Large-Scale SNP Discovery and Genotyping for Constructing a High-Density Genetic Map of Tea Plant Using Specific-Locus Amplified Fragment Sequencing (SLAF-seq) PLoS ONE. 2015;10(6):e0128798. doi: 10.1371/journal.pone.0128798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia C, Chen L-l, Rong T-z, Li R, Xiang Y, Wang P, Liu C-h, Dong X-q, Liu B, Zhao D, et al. Identification of a new maize inflorescence meristem mutant and association analysis using SLAF-seq method. Euphytica. 2015;202(1):35–44. doi: 10.1007/s10681-014-1202-5. [DOI] [Google Scholar]
- 31.Xu F, Sun X, Chen Y, Huang Y, Tong C, Bao J. Rapid Identification of Major QTLs Associated with Rice Grain Weight and Their Utilization. PLoS ONE. 2015;10(3):e0122206. doi: 10.1371/journal.pone.0122206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ariyadasa R, Mascher M, Nussbaumer T, Schulte D, Frenkel Z, Poursarebani N, Zhou R, Steuernagel B, Gundlach H, Taudien S, et al. A Sequence-Ready Physical Map of Barley Anchored Genetically by Two Million Single-Nucleotide Polymorphisms. Plant Physiol. 2014;164(1):412–423. doi: 10.1104/pp.113.228213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Barley Genome Sequencing C. Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 34.Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH, et al. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ) Plant J. 2013;76(4):718–727. doi: 10.1111/tpj.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prina AR, Arias MC, Lainez V, Landau A, Maldonado S. A cytoplasmically inherited mutant controlling early chloroplast development in barley seedlings. Theor Appl Genet. 2003;107:1410–1418. doi: 10.1007/s00122-003-1391-0. [DOI] [PubMed] [Google Scholar]
- 36.Guo T, Huang X, Huang Y-X, Liu Y-Z, Zhang J-G, Chen Z-Q, Wang H. Characterizations of a Mutant Gene hw-1(t)for Green-revertible Albino, High Tillering and Dwarf in Rice(Oryza sativa L.) Acta Agron Sin. 2012;38:23–35. doi: 10.3724/SP.J.1006.2012.00023. [DOI] [Google Scholar]
- 37.Chen J, Zhao J, Liu X, Li C, Lin D, Dong Y, Ye S, Zhang X. Genetic Analysis and Molecular Mapping of a New Thermosensitive Leaf-color Mutant in Oryza sativa. Chin Bull Bot. 2010;45(4):419–425. [Google Scholar]
- 38.D-y H, Xu H, Du G-y, Lin J-t, Duan M, Guo A-g. Proteome analysis of chloroplast proteins in stage albinism line of winter wheat (triticum aestivum) FA85. BMB Rep. 2009;42:450–455. doi: 10.5483/BMBRep.2009.42.7.450. [DOI] [PubMed] [Google Scholar]
- 39.Mascher M, Jost M, Kuon J-E, Himmelbach A, Aszfalg A, Beier S, Scholz U, Graner A, Stein N. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 2014;15(6):R78. doi: 10.1186/gb-2014-15-6-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pankin A, Campoli C, Dong X, Kilian B, Sharma R, Himmelbach A, Saini R, Davis SJ, Stein N, Schneeberger K, et al. Mapping-by-Sequencing Identifies HvPHYTOCHROME C as a Candidate Gene for the early maturity 5 Locus Modulating the Circadian Clock and Photoperiodic Flowering in Barley. Genetics. 2014;198(1):383–396. doi: 10.1534/genetics.114.165613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottwald S, Stein N, Börner A, Sasaki T, Graner A. The gibberellic-acid insensitive dwarfing gene sdw3 of barley is located on chromosome 2HS in a region that shows high colinearity with rice chromosome 7 L. Mol Genet Genomics. 2004;271:426–436. doi: 10.1007/s00438-004-0993-9. [DOI] [PubMed] [Google Scholar]
- 42.Li C, Ni P, Francki M, Hunter A, Zhang Y, Schibeci D, Li H, Tarr A, Wang J, Cakir M, et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct Integr Genomics. 2004;4:84–93. doi: 10.1007/s10142-004-0104-3. [DOI] [PubMed] [Google Scholar]
- 43.Arsova B, Hoja U, Wimmelbacher M, Greiner E, Üstün Ş, Melzer M, Petersen K, Lein W, Börnkeb F. Plastidial Thioredoxin z Interacts with Two Fructokinase-Like Proteins in a Thiol-Dependent Manner: Evidence for an Essential Role in Chloroplast Development in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2010;22(5):1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C, Yu QB, Lv RH, Yin QQ, Chen GY, Xu L, Yang ZN. The Reduced Plastid-Encoded Polymerase-Dependent Plastid Gene Expression Leads to the Delayed Greening of the Arabidopsis fln2 Mutant. PLoS ONE. 2013;8(9):e73092. doi: 10.1371/journal.pone.0073092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granot D, Kelly G, Stein O, Vid-Schwartz R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J Exp Bot. 2014;65(3):809–819. doi: 10.1093/jxb/ert400. [DOI] [PubMed] [Google Scholar]
- 46.Cui Q, Li X, Cui S. Measurement of Concentration of chlorophyll in wheat leaves using spectrophotometer. J Anhui Agric Sci. 2006;34(10):2063. [Google Scholar]
- 47.Zhang X, Wang L, Shou L. Modified CTAB Method for Extracting Genomic DNA from Wheat Leaf. Agric Sci Technol. 2013;14(7):946–949. [Google Scholar]
- 48.Liu G, Jia L, Lu L, Qin D, Zhang J, Guan P, Ni Z, Yao Y, Sun Q, Peng H. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet. 2014;127(11):2415–2432. doi: 10.1007/s00122-014-2387-7. [DOI] [PubMed] [Google Scholar]
- 49.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–81. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 50.Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic lingage data. Hereditas. 2003;25:317–321. [PubMed] [Google Scholar]
- 51.Kent WJ. BLAT-The BLAST-Like Alignment Tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202.ArticlepublishedonlinebeforeMarch2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


