Abstract
The morphology of dendrites and the axon determines how a neuron processes and transmits information. Neurite morphology is frequently analyzed by Sholl analysis or by counting the total number of neurites and branch tips. However, the time and resources required to perform such analysis by hand is prohibitive for the processing of large data sets and introduces problems with data auditing and reproducibility. Furthermore, analyses performed by hand or using course-grained morphometric data extraction tools can obscure subtle differences in data sets because they do not store the data in a form that facilitates the application of multiple analytical tools. To address these shortcomings, we have developed a program (titled “Bonfire”) to facilitate digitization of neurite morphology and subsequent Sholl analysis. Our program builds upon other available open-source morphological analysis tools by performing Sholl analysis on subregions of the neuritic arbor, enabling the detection of local level changes in dendrite and axon branching behavior. To validate this new tool, we applied Bonfire analysis to images of hippocampal neurons treated with 25 ng/ml Brain-Derived Neurotrophic Factor (BDNF) and untreated control neurons. Consistent with prior findings, conventional Sholl analysis revealed that global exposure to BDNF increases the number of neuritic intersections proximal to the soma. Bonfire analysis additionally uncovers that BDNF treatment affects both root processes and terminal processes with no effect on intermediate neurites. Taken together, our data suggest that global exposure of hippocampal neurons to BDNF results in a reorganization of neuritic segments within their arbors, but not necessarily a change in their number or length. These findings were only made possible by the neurite-specific Sholl data returned by Bonfire analysis.
Keywords: Sholl Analysis, Neurite, BDNF, Morphology, Computer-assisted, Digitization, Tracing
Introduction
Neuronal morphology is important for determining how action potentials propagate, how information is processed (1-4), and neuronal function (5-8). Neurite branching affects how single neurons integrate synaptic inputs (9,10) and how they communicate as networks (11). Alterations in neuronal morphology and branching patterns have been observed in a wide range of developmental or acquired disorders (12-15) in which it is thought that altered arbor structure plays a role in the pathogenesis of the disorder. Understanding the factors affecting neuronal morphology is, therefore, integral to understanding nervous system health and disease.
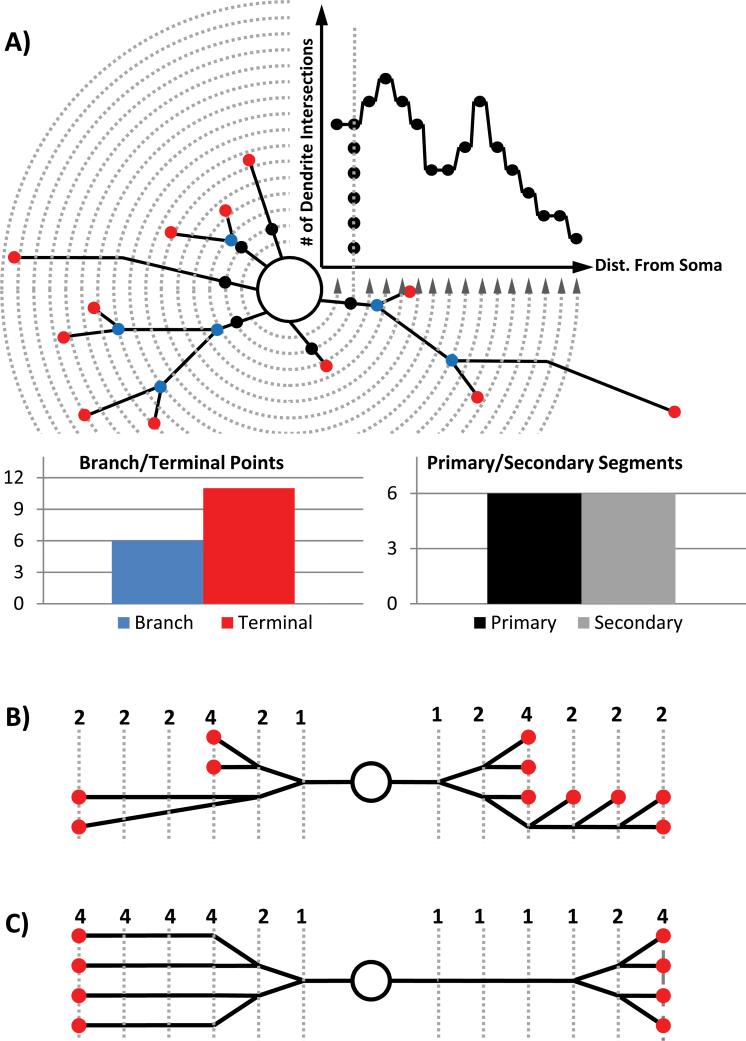
Neuronal morphology is a complex phenomenon to study due to the wide range of metrics which may be quantified (16,17). The need for streamlined methods of acquiring neuronal morphological data has given rise to a growing number of fully-automated and semi-automated tools (18-22). Each has unique strengths and weaknesses associated with removing user control and relying on computers for neurite identification, resulting in a trade-off between speed and reliability. Often, neurite morphology is analyzed by counting the total number of neurites, branch points, tips, or by Sholl analysis (Fig. 1 A) (23). Each of these methods used in isolation falls short of providing a full description of arbor morphology because it is possible to generate the same output from two different input arbors (Fig. 1 B-C) (17). Additionally, the pattern of dendritic and axonal extensions is a result of cytoskeletal assembly and disassembly processes regulated by intracellular and extracellular factors. Because of the highly local role of many of these regulatory processes, final arbor morphology is a product of specific effects acting locally in each neurite. Axons and dendrites, for example, develop distinct morphological behaviors even when exposed to the same global environment (24). The application of these data extraction techniques to whole neuronal arbors destroys the ability to observe local changes. These failures of traditional data-generating tools make it difficult to test specific hypotheses about the biochemical processes driving arborization. This problem is compounded by the time associated with manual quantification, which makes it impossible to practically employ more than a very few quantification techniques, limits data sharing opportunities between laboratories, and creates problems with reproducibility and data auditing (21).
Figure 1.
Schematic of several classic morphological analysis tools and the digitized process for performing them available through the Bonfire program. A) Schematic of Sholl analysis, branch/terminal point counting and segment counting. B) Two distinct arbors (left and right) that generate identical Sholl curves. C) Two distinct arbors (left and right) that generate identical terminal point and branch counts. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
In this study, we report the release of a unique tool that incorporates two existing morphological analysis platforms, along with custom analytical components, to provide detailed neurite-level morphological data. The program incorporates NeuronJ (18) to acquire spatial information about the position of neuritic segments in space relative to the rest of the cell (Fig. 2 B). It then exports this information to NeuronStudio (25) to allow the user to define structural information about the connectivity between neurites (Fig. 2 C). Custom portions of the program then assign “identities” to each neuritic segment according to its location within an arbor. Assigning an identity to each segment allows the program to perform a series of analyses relating morphological metrics to segmental identity (Fig. 3). The workflow created is intended to streamline data digitization and storage processes while preserving the reliability of user control.
Figure 2.
Schematic of the digitization and analysis process available through the Bonfire program. A) A neuron that has been “skeletonized” as a first step in digitizing its structure. BE) Example graphical output of each step of the Bonfire procedure, as applied to segments of the image shown above in A). Example non-linkage errors (D, red spots) have been left in the figure intentionally to demonstrate Bonfire program error identification. [Color figure can be viewed in the online issue which is available at wileyonlinelibrary.com.]
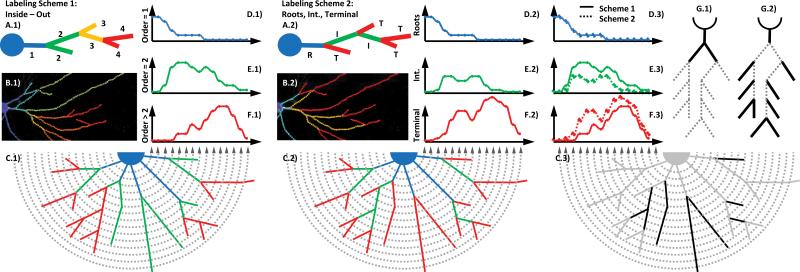
Figure 3.
Two structure-dependent labeling schemes that assign an identity to neurite segments based on their location within their arbors. A) Randomly generated arbor, labeled according to the “inside-out” scheme (I/OA.1) or the “Roots, Intermediate, Terminal” scheme (RITA.2) (17), and color-coded according to order. B) An example neuritic arbor, which has been digitized and color-coded according to branch order using I/O labeling (B.1) and RIT labeling (B.2), respectively. C) Schematized example neuritic arbor, color-coded according to I/O labeling (C.1) and RIT labeling (C.2), respectively, with superimposed Sholl rings. DF) The order-specific Sholl curves resulting from the arbors shown in Fig. 2C. C.3) Example arbor in gray, showing neuritic segments that change groupings between the two labeling schemes (black), accompanied by a schematic showing the relative areas of emphasis for the I/O (G.1) and RIT (G.2) labeling schemes. [Color figure can be viewed in the online issue which is available at wileyonlinelibrary.com.]
We validate the program's ability to detect changes in arbor morphology by applying this analysis to neurons incubated with BDNF, a well-studied extracellular factor that regulates neurite morphology (26-28). Most studies have found that BDNF increases dendrite number close to the soma and reduces distal dendrite number (29,30). BDNF promotes similar increases in proximal axon complexity (31-33). Historically, BDNF has been shown to increase dendrite complexity (34,35) and number (36) by altering the rates at which new dendrite branches are gained and lost (36). The exact mechanisms by which these effects are mediated are still debated and may differ by cell type or location in the same cell (37-40). Bonfire analysis was applied to our experimental data to assess whether this novel analysis detects effects of BDNF on neurites that may have been missed by conventional Sholl analysis.
Materials and Methods
Cell Culture & Imaging
Hippocampal neurons were isolated from E18 rat embryos and cultured as previously described (41). Briefly, embryos were removed by Cesarian section at 18 days gestation and decapitated. The hippocampi were manually dissected under a microscope, and cells were triturated with a fire polished glass pipette tip, counted on a hemocytometer, and plated at a density of ~1800 cells per mm2 on 35 mm petri plates (Corning) coated with 1 mg/ml poly-D-lysine (Sigma-Aldrich). Cultures were maintained in Neurobasal medium containing penicillin, streptomycin, glutamine, and B27 supplement (NB; all purchased from Invitrogen).
At 5 days in vitro (DIV5), cells were transfected with cDNA encoding GFP using Effectene (Qiagen), as previously described (42-45). The low efficiency of this transfection technique in this cell system allows the easy identification of the processes associated with single neurons. In the BDNF treatment groups, treatment occurred from DIV7-10, during which regular NB was replaced with NB containing 25 ng/ml BDNF. This BDNF concentration does not stimulate p75 or other Trk (A and C) receptors, (46). Cultures were fixed in 4% paraformadehyde on DIV10 and immunostained with rat anti-GFP (a gift from Dr. Shu-Chan Hsu, Rutgers University) and MAP-2 (Sigma-Aldrich). Neurons were imaged in the GFP channel at 200× using an Olympus Optical IX50 microscope with a Cooke Sensicam CCD cooled camera, fluorescence imaging system, and ImagePro software (MediaCybernetics). Images were acquired in 8-bit TIFF format, measuring 512 × 640 pixels.
Program Mechanics & Usage
The Bonfire program is a series of custom scripts written in MATLAB (MathWorks). These scripts, as well as instructions on its use and sample data, are available in the supplemental online material accompanying this article. Neuronal morphology was digitized in three stages based on the initial images. In the first stage, the semi-automated tools available through the NeuronJ plugin (18) to ImageJ (NIH, Bethesda MD) were used to define positions of all neurites (Fig. 2 A-B). The data for each neurite are exported using NeuronJ in the form of a series of nodes with defined positions in the X-Y plane, where nodes belonging to an individual neurite segment are linked by association. These tools allow the user to guide the tracing of each neuritic segment with course resolution by hand but reduce tracing time by using a curve-fitting algorithm to fine-tune the exact location of the nodes defining the neurite position.
In the second stage, portions of the Bonfire program were used to convert the strings of nodes provided by NeuronJ into SWC format (47) for further manipulation. NeuronStudio (25) is then used to define the pattern of connectivity between neurite segments (Fig. 2 C). The transformation of the data into SWC format allows for the linkage of the simple strings of nodes defined previously into more complex branching structures. After linking is complete, another component of the Bonfire program checks the resulting structure for errors and non-linkages (Fig. 2 D), based on the assumption that each neuritic segment may only give rise to two or less daughter segments (48). This assumption is made to facilitate future integration with theoretical growth models (49) and does not result in significant loss of data (50). These two steps fully determine the structure of each cell's neuritic arbor in 2-dimensional space and encode it in a digital format.
Using these digitized neuritic arbors, a second component of the Bonfire program was then used to perform process identification and extract the following metrics: number of primary neurites, number of secondary neurites, number of branch points per cell, number of terminal neurite tips per cell, and Sholl analysis performed with a 3.0 μm ring interval (Fig. 2 E). Sholl analysis is performed conceptually by drawing concentric circles around the cell body at incrementally increasing radii and counting the number of times each circle crosses a neuritic segment (shown counting around the circle counterclockwise for demonstration in Fig. 1 A). The number of intersections is graphed as a function of radial distance from the cell body (Fig. 1 A upper right quadrant) to give a quantitative representation of how neurite density varies spatially.
Because the location and linkage pattern of neurites is user-defined using external tools, the algorithms associated with data extraction are geometric in nature and do not depend on conceptually complex image analysis. For example, the algorithm for Sholl analysis is based on the assumption that if a neuritic segment starting at one node (node Nn with Cartesian coordinates Xn,Yn) lies inside of a soma-centric circle with radius r (Cr), and its daughter node (node Nn+1 with Cartesian coordinates with coordinates Xn+1,Yn+1) lies outside of radius r, then that neurite must cross the circle with radius r. Cr is also intersected by this neuritic segment if the reverse is true. Therefore, Cr is intersected by a neuritic segment when the criteria outlined in Equation 1 are met, as follows:
| Eq. 1 |
The above holds true regardless of the spacing between nodes defining a neurite path, and so can be used for all mother-daughter node pairs. To return the cumulative Sholl curve, this same check is performed between every mother-daughter node pair and every circle, and the results are summed by circle. Because every node is checked for the existence of a mother node, this reliably returns the Sholl information for the entire arbor. Furthermore, because every node-pair can be tagged with a structure-based identity, it is possible to later tabulate which identity-classes intersect with specific circles. Additional descriptions of the algorithms involved in data management can be found as comments in the MATLAB code accompanying this article. Afterward, data were transferred to Excel to facilitate statistical analysis. The experimenter was blinded to experimental conditions during all data analysis.
Branch Identity-specific Data Analysis
Two labeling schemes were used to assign structure-based identity to segments, which allow us to analyze subregions of the arbor with varying degrees of specificity. Neuritic segments, or branches, are the uninterrupted stretches of neurite starting at one branch point, or starting at the cell body in the case of root segments, and ending when the neurite terminates or at the next branch point. These segments can be grouped together in different ways. In the most frequently used convention, these processes are assigned a number, or branch order. In this convention, termed “inside-out” labeling (I/O) here, branch order starts at 1 with any branch initiating at the soma and increases by 1 with each branch point reached, moving from the soma to the branch tips (Fig. 3 A.1 – F.1) (17). The second convention is the “roots, intermediate, terminal” (RIT) convention (Fig. 3 A.2 – F.2), in which any neurite originating in the soma is a root segment, any neurite with no daughter neurites is a terminal segment, and any neurite not a root or a terminal is an intermediate segment (17).
Having neurites assigned segmental identities in I/O and RIT labeling allows the performance of more traditional forms of analysis on specific subregions of the arbor. For example, Figure 3 A.1 – F.1 shows Sholl analysis performed on neurite segments that have been consolidated into three separate groups based on their structural identity, resulting in three separate Sholl curves for the same cell. This technique is performed the same way as standard Sholl analysis but uses three possible groupings of segments. In the example shown in Figure 3, the first grouping contains only primary segments, and therefore, only the intersections of primary segments with the Sholl rings are tallied in the generation of the Sholl curve for that group (Fig. 3 D.1). In the second group, only secondary processes are counted, and in the third group, all segments with order ≥ 3 are counted, generating the Sholl curves shown in Figure 3 E.1 – F.1, respectively. A similar analysis can be performed using the RIT labeling scheme in which only the Sholl intersections with root segments (Fig. 3 D.2), intermediate segments (Fig. 3 E.2), or terminal segments (Fig. 3 F.2) are counted. Note that the sum of all the component Sholl curves adds up to the total Sholl curve for the cell, and therefore, is the same in both of these cases.
Results
Global analysis
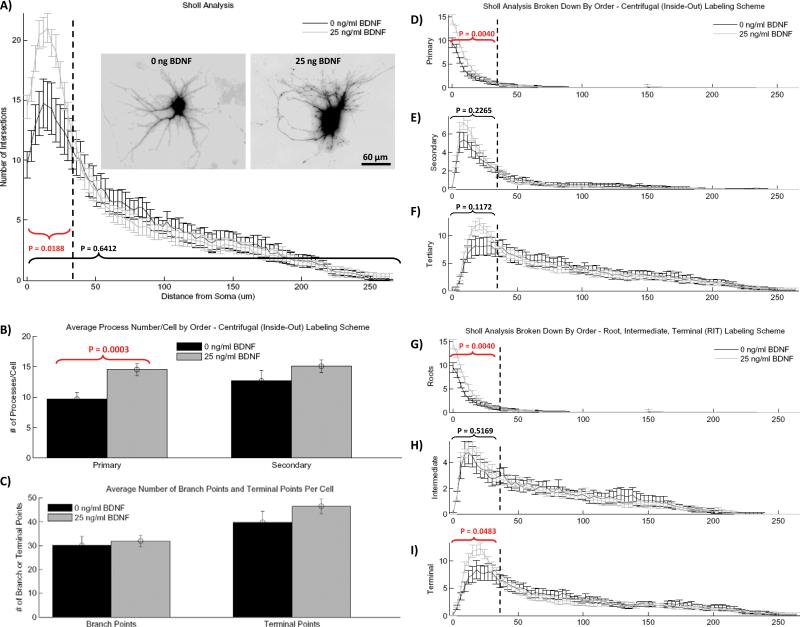
As seen by other groups (31), global exposure to BDNF in vitro increases proximal neurites within the first 35 μm of the soma but has no effect on distal neurites (Fig. 4 A). Quantification of the number of primary and secondary neurites shows that BNDF treatment causes a statistically significant increase in the number of primary extensions, and while there is a trend of an increasing number of secondary neurites it does not reach statistical significance (Fig. 4 B). In addition, we analyzed the effect of global BDNF administration on the average number of branch points and number of terminal branches per cell (Fig. 4 C), which showed no change between conditions. Similarly, global BDNF exposure causes no significant change in average process length or total arbor length (data not shown). While the specifics of BDNF treatment vary based on the type of neuron and system (in vitro vs. in vivo) (29-33), these results are broadly consistent with the general scientific consensus that BDNF treatment increases arbor “complexity.”
Figure 4.
Results of the Bonfire analysis for hippocampal neurons treated with 25 ng/ml BDNF and untreated control neurons. A) Total Sholl curves with example inverted GFP images (inset). B) Average number of primary and secondary processes per cell. C) Average number of branch points and terminal points per cell. DF) Segment identity-specific Sholl analysis according to the I/O labeling scheme, where segments have been grouped as either primary (D), secondary (E) or tertiary and greater (F). GI) Segment identity-specific Sholl analysis according to RIT labeling scheme, where segments have been grouped as either root segments (G), intermediate segments (H), or terminal segments (I). All error bars represent the standard error of the mean (SEM). Statistical analysis of dendrite number was performed on the total number of Sholl intersections in the bracketed regions using two-tailed unpaired t test with Welch correction (n = 18 for 0 ng/ml BDNF condition, n = 24 for 25 ng/ml condition). [Color figure can be viewed in the online issue which is available at wileyonlinelibrary.com.]
Local analysis
A more detailed picture of BDNF-induced morphological changes is created by the local-level Sholl analysis. RIT-based Sholl analysis identifies that the increased number of proximal Sholl intersections is due to two effects: 1) there is an increase in the number of primary neurites (Fig. 4 B), which causes an increase in the number of Sholl intersections with primary neurites (Fig. 4 D and G), and 2) there is an increase in the number of Sholl intersections with terminal neurites (Fig. 4 J). However, when the I/O labeling scheme is used (Fig. 4 D-F), only primary neurites appear to change significantly between conditions. This is because the neurite sub-type responsible for much of the change (terminal neurites) is split between the second two classifications using the I/O labeling scheme, making a statistically significant change impossible to detect. Taken together, these two schemes indicate that the increase in proximal Sholl intersections is caused by increased primary sprouting as well as increased presence of low-order branches that terminate rather than branch into daughter segments.
Discussion
Morphological Analysis
The data generated during the study of neuroscience are as variable as the subject itself, including genetic, proteomic, morphological, electrophysiological, histological, and behavioral data. Even within the subtype of morphological data, there is no consensus on which metrics are most significant, and there are frequently multiple means of capturing, analyzing, and storing the data (22). Consequently, every laboratory accumulates a unique set of tools according to their means and needs. Recently, researchers have turned increasingly to the field of neuroinformatics, which aims to create tools and standards to help integrate information across research platforms and laboratories (51). In an environment where such diversity is required for its continued productivity, the development of tools meant to bridge existing platforms can be more effective than the development of tools which aim to vertically integrate all steps in analysis in one platform.
Digitizing neuronal morphology generally consists of 6 steps: 1) image acquisition, 2) skeletonization, 3) generation of meta-data, such as arbor structure, 4) quantification, 5) analysis and interpretation, and finally, 6) data storage. Numerous software packages have been developed to assist neuroscientists in these various steps, including automated (19,21,52) and hand-assisted (18) skeletonization tools, neurite linking tools (25,53,54), automated data-analysis tools (20,55), and database tools (56,57). One notable absence from the collection of available open-source tools is a method for deriving Sholl profiles, which interfaces with the other tools used to digitize neuronal morphology. Current Sholl tools available for download operate by simple image thresholding (58), are entirely user-driven (53,59), or provide limited and highly specific output (25). While commercial alternatives exist, they are frequently cost-prohibitive and cannot be altered to meet unique demands.
The Bonfire program integrates with two existing tools used in neuronal digitization (NeuronJ and NeuronStudio) and provides a means of extracting Sholl profiles from digitized neurons stored in standard SWC format. Furthermore, the linkage of an automated, structure-based labeling system with an automated Sholl analysis algorithm creates a powerful new method for quantifying highly specific changes occurring in dendrites and axons following genetic or pharmacologic manipulations. The reason for having multiple methods of segmenting the data is that each method only focuses on a small region of the neuritic arbor, making it better suited to identify effects in that region. The I/O scheme focuses on primary and secondary neurites at the expense of higher order neurites (Fig 3 G.1, D.3 – F.3). Such a scheme may uncover morphological effects caused by factors acting at the cell body but may miss changes preferentially affecting only the more terminal regions of the arbor. Conversely, the RIT scheme captures effects of factors at terminal segments but causes a loss of resolution in the intermediate segments since they are grouped together (Fig. 3 G.2, D.3 – F.3). The RIT scheme is best for identifying factors that affect the stability or creation of terminal segments, but this scheme misses effects that occur close to the cell body. The difference in regions of focus between the two schemes is generated by their accounting for the same set of processes differently (Fig. 3 C.3). This identity-specific information reveals trends in neurite patterning, which were previously obscured by global-level analyses.
Biological Findings
Primary segment numbers confirm that BDNF exposure increases primary neurite number. However, the absence of a significant change in the number of branch points or terminal points and the lack of a change in the average segment length implies that the effect of BDNF is not due to overall increased neurite branching. Taken together with the absence of a change in the total arbor length, these findings, which represent an entirely new function for BDNF, suggest that global exposure of hippocampal neurons to BDNF in vitro results in a reorganization of neuritic segments within their arbors but not necessarily a change in overall neurite number or length.
Data extracted using Bonfire analysis provide a more detailed view of these morphological changes, and for the first time, tie the effects of BDNF to specific regions of the arbor. The importance of having the multiple methods of data segmentation provided by the two labeling schemes is clarified by the fact that the analysis identified not only a global effect of bath application of BDNF but also that this global effect is predominantly driven by a change in two specific arbor subregions. These details about the local nature of morphological changes have not been identified using conventional methods (43,59,60).
Future Directions
One of the most exciting opportunities opened by the generation of morphological data containing local-level detail on arbor structure is the ability to fuel mathematical exploration of the molecular processes locally guiding arborization (61-65). Much work has been done deriving generative models of neuronal morphologies (66-69). These models are based on observation, but their mechanics are meant to represent biological processes driving cellular morphology (61-63,70). Providing these models with a more detailed source of experimental data would improve mathematicians’ ability to generate and test specific hypotheses about biochemical regulatory networks. As mentioned previously, final arbor structure is a result of balanced cytoskeletal assembly and disassembly processes which are regulated by local and global factors. Detailed morphological analysis of specific regions of the dendritic and axonal arbors may provide a convenient window into the regulation of neuronal structure by locally active factors. The result would be a closer integration of mathematical modeling with experimental science.
For example, the mathematical interpretation of Sholl data can be performed using multiple methods (65). The selection of the most informative one may be dependent on cell type, or even the process type within one cell, implying that the biological drivers of neurite branching and growth may change based on context. However, even using multiple mathematical approaches, interpretation of standard Sholl analysis is not straightforward. An observed increase in Sholl intersections may be due to a variety of changes in branching behavior, including increased sprouting from the soma, more rapid neurite bifurcation, delayed termination, or even neurite extension from the periphery back toward the soma. It is therefore necessary to determine the contribution of each process type to the overall Sholl curve if such information is to be instructive of the underlying biological processes.
Conclusions
The Bonfire program provides a useful bridge to integrate several existing digitization tools, makes the data available in a variety of formats (.swc morphological descriptions of individual neurons, .xls summary reports, and graphical analysis provided within MATLAB), and assists in the generation of morphological databases where information is stored concisely and can be revisited with unique analyses or audited for correctness as the need arises without having to re-digitize the data. The Bonfire program capitalizes on the strengths of previously developed tracing programs to reduce the time required for high-fidelity tracing, while preserving the flexibility of laboratories to generate digitized neuronal files using their own preferred method. Though the program currently integrates most effectively with NeuronJ and NeuronStudio, it operates on standard SWC encoded neuronal information, which can be created using a variety of methods. Furthermore, the MATLAB code is readily available for modification, enabling users to modify or expand on the analysis to meet their own needs. In making this tool freely available, we hope to increase the accessibility of high-quality morphological analysis tools to the scientific public and to establish a precedent for building and sharing open-source tools for improved morphological analysis of neurons.
Supplementary Material
Footnotes
Acknowledgements: This work was supported in part by a Busch Biomedical Grant, NSF grant IBN-0548543, NSF grant IBN-0919747, March of Dimes Foundation Grant 1-FY04-107, March of Dimes Foundation Grant 1-FY08-464 (to B.L.F). C.G.L. was supported by NIH Biotechnology Training Grant T32 GM008339-20 and NJ Commission on Spinal Cord Research Predoctoral Fellowship 08-2941-SCR-E-0. E.S.S was supported in part by NIH training grant 5 T32 MH019957-10 and T32 GM00839. M.L.P. was supported by University and Louis Bevier Graduate and Dissertation Fellowship. M.L.P. was supported in part by the IGERT Program on Integratively Engineered Biointerfaces at Rutgers: NSF Grant DGE-0333196. We would like to thank Jose Fernandez for his advice and contribution to this work.
Bibliography
- 1.Eilers J, Konnerth A. Dendritic signal integration. Curr Opin Neurobiol. 1997;7(3):385–90. doi: 10.1016/s0959-4388(97)80067-0. [DOI] [PubMed] [Google Scholar]
- 2.Hausser M, Spruston N, Stuart GJ. Diversity and dynamics of dendritic signaling. Science. 2000;290(5492):739–44. doi: 10.1126/science.290.5492.739. [DOI] [PubMed] [Google Scholar]
- 3.Schaefer AT, Larkum ME, Sakmann B, Roth A. Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. J Neurophysiol. 2003;89(6):3143–54. doi: 10.1152/jn.00046.2003. [DOI] [PubMed] [Google Scholar]
- 4.Vetter P, Roth A, Hausser M. Propagation of action potentials in dendrites depends on dendritic morphology. J Neurophysiol. 2001;85(2):926–37. doi: 10.1152/jn.2001.85.2.926. [DOI] [PubMed] [Google Scholar]
- 5.Elston GN. Pyramidal cells of the frontal lobe: all the more spinous to think with. J Neurosci. 2000;20(18):RC95. doi: 10.1523/JNEUROSCI.20-18-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch C, Segev I. The role of single neurons in information processing. Nat Neurosci. 2000;3(Suppl):1171–7. doi: 10.1038/81444. [DOI] [PubMed] [Google Scholar]
- 7.Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron. 2001;29(3):779–96. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 8.Stuart G, Spruston N, Hèausser M. Dendrites. xxi. Oxford University Press; New York ; Oxford: 1999. p. 376. [Google Scholar]
- 9.Vetter P, Roth A, Hausser M. Propagation of action potentials in dendrites depends on dendritic morphology. Journal of Neurophysiology. 2001;85(2):926–937. doi: 10.1152/jn.2001.85.2.926. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer AT, Larkum ME, Sakmann B, Roth A. Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. Journal of Neurophysiology. 2003;89(6):3143–3154. doi: 10.1152/jn.00046.2003. [DOI] [PubMed] [Google Scholar]
- 11.Brette R, Rudolph M, Carnevale T, Hines M, Beeman D, Bower JM, Diesmann M, Morrison A, Goodman PH, Harris FC. Simulation of networks of spiking neurons: A review of tools and strategies. Journal of Computational Neuroscience. 2007;23(3):349–398. doi: 10.1007/s10827-007-0038-6. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arendt T, Zvegintseva HG, Leontovich TA. Dendritic Changes in the Basal Nucleus of Meynert and in the Diagonal Band Nucleus in Alzheimers-Disease - a Quantitative Golgi Investigation. Neuroscience. 1986;19(4):1265–1278. doi: 10.1016/0306-4522(86)90141-7. [DOI] [PubMed] [Google Scholar]
- 13.Harrison PJ. The neuropathology of schizophrenia - A critical review of the data and their interpretation. Brain. 1999;122:593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cerebral Cortex. 2000;10(10):981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 15.Lewis DA, Glantz LA, Pierri JN, Sweet RA. Altered cortical glutamate neurotransmission in schizophrenia - Evidence from morphological studies of pyramidal neurons. Glutamate and Disorders of Cognition and Motivation. 2003;1003:102–112. doi: 10.1196/annals.1300.007. [DOI] [PubMed] [Google Scholar]
- 16.Dragunow M. Opinion - High-content analysis in neuroscience. Nature Reviews Neuroscience. 2008;9(10):779–788. doi: 10.1038/nrn2492. [DOI] [PubMed] [Google Scholar]
- 17.Uylings HBM, van Pelt J. Measures for quantifying dendritic arborizations. Network-Computation in Neural Systems. 2002;13(3):397–414. [PubMed] [Google Scholar]
- 18.Meijering E, Jacob M, Sarria JCF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry Part A. 2004;58A(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 19.Pool M, Thiemann J, Bar-Or A, Fournier AE. NeuriteTracer: A novel ImageJ plugin for automated quantification of neurite outgrowth. Journal of Neuroscience Methods. 2008;168(1):134–139. doi: 10.1016/j.jneumeth.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Popko J, Fernandes A, Brites D, Lanier LM. Automated analysis of NeuronJ tracing data. Cytometry A. 2009;75A(4):371–6. doi: 10.1002/cyto.a.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallotton P, Lagerstrom R, Sun C, Buckley M, Wang DD, De Silva M, Tan SS, Gunnersen JA. Automated analysis of neurite branching in cultured cortical neurons using HCA-Vision. Cytometry Part A. 2007;71A(10):889–895. doi: 10.1002/cyto.a.20462. [DOI] [PubMed] [Google Scholar]
- 22.Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136(3):661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- 23.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. [PMC free article] [PubMed] [Google Scholar]
- 24.Georges PC, Hadzimichalis NM, Sweet ES, Firestein BL. The Yin-Yang of Dendrite Morphology: Unity of Actin and Microtubules. Molecular Neurobiology. 2008;38(3):270–284. doi: 10.1007/s12035-008-8046-8. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nature Protocols. 2006;1(4):2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- 26.Lykissas MG, Batistatou AK, Charalabopoulos KA, Beris AE. The role of neurotrophins in axonal growth, guidance, and regeneration. Current Neurovascular Research. 2007;4(2):143–151. doi: 10.2174/156720207780637216. [DOI] [PubMed] [Google Scholar]
- 27.McAllister AK. Neurotrophins and neuronal differentiation in the central nervous system. Cellular and Molecular Life Sciences. 2001;58(8):1054–1060. doi: 10.1007/PL00000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitford KL, Dijkhuizen P, Polleux F, Ghosh A. Molecular control of cortical dendrite development. Annual Review of Neuroscience. 2002;25:127–149. doi: 10.1146/annurev.neuro.25.112701.142932. [DOI] [PubMed] [Google Scholar]
- 29.Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. Journal of Neuroscience. 2003;23(17):6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolwani RJ, Buckmaster PS, Varma S, Cosgaya JM, Wu Y, Suri C, Shooter EM. BDNF overexpression increases dendrite complexity in hippocampal dentate gyrus. Neuroscience. 2002;114(3):795–805. doi: 10.1016/s0306-4522(02)00301-9. [DOI] [PubMed] [Google Scholar]
- 31.Danzer SC, Crooks KRC, Lo DC, McNamara JO. Increased expression of brain-derived neurotrophic factor induces formation of basal dendrites and axonal branching in dentate granule cells in hippocampal explant cultures. Journal of Neuroscience. 2002;22(22):9754–9763. doi: 10.1523/JNEUROSCI.22-22-09754.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lom B, Cogen J, Sanchez AL, Vu T, Cohen-Cory S. Local and target-derived brain-derived neurotrophic factor exert opposing effects on the dendritic arborization of retinal ganglion cells in vivo. Journal of Neuroscience. 2002;22(17):7639–7649. doi: 10.1523/JNEUROSCI.22-17-07639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel MN, McNamara JO. SELECTIVE ENHANCEMENT OF AXONAL BRANCHING OF CULTURED DENTATE GYRUS NEURONS BY NEUROTROPHIC FACTORS. Neuroscience. 1995;69(3):763–770. doi: 10.1016/0306-4522(95)00281-m. [DOI] [PubMed] [Google Scholar]
- 34.Baker RE, Dijkhuizen PA, Van Pelt J, Verhaagen J. Growth of pyramidal, but not nonpyramidal, dendrites in long-term organotypic explants of neonatal rat neocortex chronically exposed to neurotrophin-3. Eur J Neurosci. 1998;10(3):1037–44. doi: 10.1046/j.1460-9568.1998.00118.x. [DOI] [PubMed] [Google Scholar]
- 35.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15(4):791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 36.Horch HW, Kruttgen A, Portbury SD, Katz LC. Destabilization of cortical dendrites and spines by BDNF. Neuron. 1999;23(2):353–64. doi: 10.1016/s0896-6273(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 37.Dijkhuizen PA, Ghosh A. BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J Neurobiol. 2005;62(2):278–88. doi: 10.1002/neu.20100. [DOI] [PubMed] [Google Scholar]
- 38.Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3'-kinase-Akt-mammalian target of rapamycin pathway. J Neurosci. 2005;25(49):11300–12. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci. 2005;25(49):11288–99. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24(33):7366–77. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AED, Bredt DS. Cypin: A cytosolic regulator of PSD-95 postsynaptic targeting. Neuron. 1999;24(3):659–672. doi: 10.1016/s0896-6273(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 42.Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL. Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci. 2004;7(2):145–52. doi: 10.1038/nn1179. [DOI] [PubMed] [Google Scholar]
- 43.Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. Journal of Neuroscience. 2006;26(40):10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrel D, Du YZ, Komlos D, Hadzimichalis NM, Kwon M, Wang B, Brzustowicz LM, Firestein BL. NOS1AP Regulates Dendrite Patterning of Hippocampal Neurons through a Carboxypeptidase E-Mediated Pathway. Journal of Neuroscience. 2009;29(25):8248–8258. doi: 10.1523/JNEUROSCI.5287-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez JR, Welsh WJ, Firestein BL. Structural characterization of the zinc binding domain in cytosolic PSD-95 interactor [cypin]: Role of zinc binding in guanine deamination and dendrite branching. Proteins-Structure Function and Bioinformatics. 2008;70(3):873–881. doi: 10.1002/prot.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Tebar A, Dechant G, Barde YA. Binding of brain-derived neurotrophic factor to the nerve growth factor receptor. Neuron. 1990;4(4):487–92. doi: 10.1016/0896-6273(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 47.Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. Journal of Neuroscience Methods. 1998;84(1-2):49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- 48.Verwer RWH, Vanpelt J. ANALYSIS OF BINARY-TREES WHEN OCCASIONAL MULTIFURCATIONS CAN BE CONSIDERED AS AGGREGATES OF BIFURCATIONS. Bulletin of Mathematical Biology. 1990;52(5):629–641. doi: 10.1007/BF02462102. [DOI] [PubMed] [Google Scholar]
- 49.Vanpelt J, Verwer RWH. GROWTH-MODELS (INCLUDING TERMINAL AND SEGMENTAL BRANCHING) FOR TOPOLOGICAL BINARY-TREES. Bulletin of Mathematical Biology. 1985;47(3):323–336. doi: 10.1007/BF02459919. [DOI] [PubMed] [Google Scholar]
- 50.Verwer RWH, Vanpelt J. TOPOLOGICAL ANALYSIS OF BINARY-TREE STRUCTURES WHEN OCCASIONAL MULTIFURCATIONS OCCUR. Bulletin of Mathematical Biology. 1985;47(2):305–316. [Google Scholar]
- 51.Bjaalie JG, Grillner S. Global neuroinformatics: The international neuroinformatics coordinating facility. Journal of Neuroscience. 2007;27(14):3613–3615. doi: 10.1523/JNEUROSCI.0558-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu WM, Lee HK, Hariharan S, Bu WY, Ahmed S. Quantitative Neurite Outgrowth Measurement Based on Image Segmentation with Topological Dependence. Cytometry Part A. 2009;75A(4):289–297. doi: 10.1002/cyto.a.20664. [DOI] [PubMed] [Google Scholar]
- 53.D'Alessandro G. Neuron_Morpho plugin Homepage. 2007 [Google Scholar]
- 54.Myatt DR, Nasuto SJ. Three-Dimensional Reconstruction of Neurons with Neuromantic. AISB Quarterly. 125:1–2. [Google Scholar]
- 55.Scorcioni R, Polavaram S, Ascoli GA. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat Protoc. 2008;3(5):866–76. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ascoli GA, Donohue DE, Halavi M. NeuroMorpho.Org: A central resource for neuronal morphologies. Journal of Neuroscience. 2007;27(35):9247–9251. doi: 10.1523/JNEUROSCI.2055-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ascoli GA, Krichmar JL, Nasuto SJ, Senft SL. Generation, description and storage of dendritic morphology data. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2001;356(1412):1131–1145. doi: 10.1098/rstb.2001.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghosh A. Sholl Analysis Plugin (v1.0) [Google Scholar]
- 59.Gutierrez H, Davies AM. A fast and accurate procedure for deriving the Sholl profile in quantitative studies of neuronal morphology. Journal of Neuroscience Methods. 2007;163(1):24–30. doi: 10.1016/j.jneumeth.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Chen HX, Firestein BL. RhoA regulates dendrite branching in hippocampal neurons by decreasing cypin protein levels. Journal of Neuroscience. 2007;27(31):8378–8386. doi: 10.1523/JNEUROSCI.0872-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samsonovich AV, Ascoli GA. Statistical determinants of dendritic morphology in hippocampal pyramidal neurons: A hidden Markov model. Hippocampus. 2005;15(2):166–183. doi: 10.1002/hipo.20041. [DOI] [PubMed] [Google Scholar]
- 62.vanPelt J, Dityatev AE, Uylings HBM. Natural variability in the number of dendritic segments: Model-based inferences about branching during neurite outgrowth. Journal of Comparative Neurology. 1997;387(3):325–340. [PubMed] [Google Scholar]
- 63.Sugimura K, Shimono K, Uemura T, Mochizuka A. Self-organizing mechanism for development of space-filling neuronal dendrites. Plos Computational Biology. 2007;3(11):2143–2154. doi: 10.1371/journal.pcbi.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current Opinion in Cell Biology. 2003;15(2):221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 65.Milosevic NT, Ristanovic D. The Sholl analysis of neuronal cell images: Semi-log or loglog method? Journal of Theoretical Biology. 2007;245(1):130–140. doi: 10.1016/j.jtbi.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 66.Ascoli GA. Neuroanatomical algorithms for dendritic modelling. Network-Computation in Neural Systems. 2002;13(3):247–260. [PubMed] [Google Scholar]
- 67.Donohue DE, Ascoli GA. A comparative computer simulation of dendritic morphology. Plos Computational Biology. 2008;4(6) doi: 10.1371/journal.pcbi.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scorcioni R, Ascoli GA. Algorithmic reconstruction of complete axonal arborizations in rat hippocampal neurons. Neurocomputing. 2005;65:15–22. [Google Scholar]
- 69.van Pelt J, Schierwagen A. Morphological analysis and modeling of neuronal dendrites. Mathematical Biosciences. 2004;188:147–155. doi: 10.1016/j.mbs.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Cuntz H, Borst A, Segev I. Optimization principles of dendritic structure. Theor Biol Med Model. 2007;4:21. doi: 10.1186/1742-4682-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.