Abstract
The Syrian golden hamster is an attractive animal for research on infectious diseases and other diseases. We report here the sequencing, assembly, and annotation of the Syrian hamster transcriptome. We include transcripts from ten pooled tissues from a naïve hamster and one stimulated with lipopolysaccharide. Our data set identified 42,707 non-redundant transcripts, representing 34,191 unique genes. Based on the transcriptome data, we generated a custom microarray and used this new platform to investigate the transcriptional response in the Syrian hamster liver following intravenous adenovirus type 5 (Ad5) infection. We found that Ad5 infection caused a massive change in regulation of liver transcripts, with robust up-regulation of genes involved in the antiviral response, indicating that the innate immune response functions in the host defense against Ad5 infection of the liver. The data and novel platforms developed in this study will facilitate further development of this important animal model.
Keywords: Adenovirus, Transcriptome, Syrian hamster, Microarray, Liver, Antiviral response, Innate immunity
Highlights
-
•
Syrian hamster transcriptome; 42,707 transcripts representing 34,191 unique genes
-
•
Syrian hamster custom expression microarray platform
-
•
Ad5 intravenous infection of the Syrian hamster liver
-
•
Ad5 upregulation of hamster liver genes involved in innate antiviral response.
Introduction
The Syrian golden hamster has been used broadly in biomedical research for decades, especially as a model for cancer development. More recently, it has been served as models in research on diabetes (Bhathena et al., 2011), atherosclerosis (Dillard et al., 2010, Jove et al., 2013), and neural plasticity (Staffend and Meisel, 2012). Also, the Syrian hamster is frequently employed in studies of infectious disease, as it is highly susceptible to infection with a wide range of viruses, bacteria, and parasites (Espitia et al., 2010, Zivcec et al., 2011). The Syrian hamster has been used to study emerging and highly pathogenic RNA viral infections and virus-induced diseases, including flaviviruses (West Nile virus, Japanese encephalitis virus, St. Louis encephalitis virus, Yellow fever virus), alphaviruses (Venezuelan-, Western-, Eastern equine encephalitis virus), henipavirus (Nipah virus), arenaviruses (Pirital virus, Pichinde virus ), bunyaviruses (Rift valley fever virus, Punta Toro virus, Andes virus), filoviruses (Ebola virus, Marburg virus), and SARS-coronavirus (Ebihara et al., 2013, Gowen and Holbrook, 2008, Holbrook and Gowen, 2008, Nakayama and Saijo, 2013, Roberts et al., 2005). Further, the Syrian hamster has proved to be a useful model to evaluate vaccines and anti-viral drugs against both RNA and DNA viruses (Julander et al., 2007, Julander et al., 2011, Monath et al., 2010, Morrey et al., 2004, Morrey et al., 2008, Toth et al., 2008, Wold and Toth, 2012).
Still further, the Syrian hamster has served as a model to study human adenovirus (Ad) pathogenesis and to evaluate oncolytic Ad vectors in cancer gene therapy (Bortolanza et al., 2007, Cerullo et al., 2012, Dhar et al., 2009a, Dhar et al., 2009b, Dhar et al., 2012, Dhar et al., 2014, Lichtenstein et al., 2009, Spencer et al., 2009, Thomas et al., 2006, Thomas et al., 2007, Thomas et al., 2008, Toth et al., 2007, Wold and Toth, 2012, Ying et al., 2009, Young et al., 2013a, Young et al., 2013b). Human Ads are double-stranded DNA viruses with a non-enveloped icosahedral capsid. There are approximately 60 types (previously referred to as serotypes) that are classified into seven species from A to G (Berk, 2013). In immunocompetent healthy individuals, most Ad infections are generally mild and self-limiting, resulting in infections in the upper respiratory tract, gastrointestinal, and urinary tracts (Wold and Ison, 2013). Some serotypes can cause serious ocular illness such as epidemic keratoconjunctinitis. In immunocompromised individuals, especially in pediatric hematopoietic stem cell transplant patients, Ads can cause severe disseminated multi-organ infections which are sometimes lethal (Echavarria, 2008, Ison, 2006, Lion, 2014, Matthes-Martin et al., 2012, Matthes-Martin et al., 2013, Sandkovsky et al., 2014, Stercz et al., 2012, Wold and Ison, 2013).
Syrian hamsters are permissive for human Ads (reviewed in Wold and Toth (2012)). Ad5 infection following intravenous (i.v.) administration results in the hamster׳s body weight loss, elevated serum liver enzyme levels, and virus replication in the liver, lungs, and other organs (Lichtenstein et al., 2009; Ying et al., 2009). When the hamster is immunosuppressed using high-dose cyclophosphamide, Ad5 replication continues for extended periods in multiple organs, especially the liver, resulting in increased mortality (Thomas et al., 2008, Toth et al., 2008). This extended replication in hamsters recapitulates human Ad infection in immunocompromised patients.
A major limitation for using the Syrian hamster as an animal model for infectious disease is the lack of information on the hamster genome and its transcription. The Syrian hamster genome has just been sequenced and released but has not been annotated (NCBI Bio Project 77669). The transcriptome of Chinese hamster ovary and Syrian baby hamster kidney cell lines as well as hamster brain and liver tissue have been analyzed (Johnson et al., 2014, Schmucki et al., 2013, Vishwanathan et al., 2015). More recently, the Syrian hamster transcriptome with additional tissues from brain, lung, spleen, kidney, heart, and liver has been published (Tchitchek et al., 2014), but the complete global transcriptome representing whole hamster transcripts, especially genes engaging in immune responses, has not been documented. Quantitative reverse transcriptase PCR (qRT-PCR) to detect and quantify some cytokines and chemokines has been published, but to our knowledge, microarrays specific to global hamster genes have not been developed due to the insufficiency of genetic data.
We report here the sequencing, assembly, and annotation of the Syrian hamster whole transcriptome, together with the generation of hamster microarrays. By using these new platforms, we describe the first global transcriptional analysis of gene expression in the liver of Ad5-infected Syrian hamsters. Our results provide unique insights into acute Ad5 infection of the liver, an organ often infected in disseminated Ad disease in humans.
Results
Hamster transcriptome sequencing and annotation
Our goal was to produce a comprehensive catalog of transcripts expressed in the adult hamster. To this end, we isolated RNA from 10 organs from a naïve animal and a hamster treated with LPS. The organs were chosen because they are the targets of many viruses (e.g. liver) or these organs are complex (e.g. brain) in that they are composed of many cells types and thus express many genes. LPS treatment was included to increase the transcription of genes involved in immune responses, as would be seen in microbiological infections. Pooled RNA was subjected to library construction for deep sequencing.
Illumina HiSeq sequencing produced 5.1×1010 raw 100 bp paired-end reads ( Table 1). Raw Illumina RNA-Seq data were adapter trimmed, validated, and filtered based on quality. Sequences that passed validation and quality checks were assembled via the de novo RNA-seq assembler Trinity (Grabherr et al., 2011, Haas et al., 2013). Trinity assembled contigs were validated via ESTscan and custom software, resulting in a set of 49,176 high-confidence transcripts. These high-confidence transcripts denote the assembled transcriptome. Of the 49,176 transcripts in the assembled transcriptome, 44,705 sequences (91%) had at least one BLASTP hit, and 30,956 (63%) had at least one InterProScan hit. The raw reads, as well as the assembled transcripts, have been deposited in GenBank under BioProject ID PRJNA285471.
Table 1.
Analysis of transcriptome by RNA-seq.
| A. Tissue sampled | B. Transcriptome | |
|---|---|---|
| Blood | Total raw read (100bp paired end) | 5.1×1010 |
| Bone Marrow | Total number of validated | 49,176 |
| Brain | Transcripts | |
| Kidney | Total number of unique transcripts | 42,707 |
| Liver | Total number of unique genes | 34,191 |
| Cervical lymph node | ||
| Lung | ||
| Muscle | ||
| Spleen | ||
| Thymus | ||
A. Each tissue was sampled from two hamsters; one treated with LPS and the second was untreated. B. Pooled RNA from both hamsters was sequenced on Illumina HiSeq platform. Summary statistics for raw reads, number of validated transcripts, unique transcript (eliminating transcripts with alternative start site or 5′ end) and total number of genes (based on BLASTP to rat genome).
Generation of custom hamster microarray and analysis
To assess the genome-wide response to Ad5 infections of the liver, two groups of hamsters were administered i.v. with either 1010 pfu of Ad5 or PBS (mock-infection). Ad5 replicates preferentially in the liver of Syrian hamsters following i.v. administration (Ying et al., 2009). Liver RNAs were extracted 18 h post-infection (p.i.) and gene expression was analyzed by a custom designed microarray. Two biological replicates (independent infections) were performed and each replicate was analyzed in triplicate (Ad5-infected was labeled with Cy3 and mock was labeled with Cy5). A dye-swap experiment was also performed to correct for any dye bias, for a total of four technical replicates per biological replicate (one slide). The data were analyzed using an ANOVA model as previously described (Churchill, 2004, Kiesel et al., 2007).
A total of 7110 genes in the liver were differentially expressed with a p≤0.01. Of these, 3788 were upregulated and 3322 were down-regulated in response to Ad5 infection. All significantly differentially expressed genes were further subjected to Gene Ontology analysis (GO) to identify functional categories of change in gene expression. As shown in Table 2A, Table 2B, transcripts in a number of critical function categories are over-represented. Most notably, genes involved in immune response (57 out of 64 genes (57/64)), defense response to virus (16/17), and antigen processing and presentation (27/31) were largely upregulated in the liver, although some were downregulated (Table 2A). Also largely upregulated were genes associated with regulating DNA-dependent transcription (82/119), signal transduction (76/109), cell adhesion (18/18), and cell communication (16/21) (Table 2A). In contrast, when the most down-regulated genes were investigated for GO function, there was over-representation of genes associated with oxidation–reduction process (214/249), metabolic process (82/101), carbohydrate metabolic process (45/69), DNA topological change (16/21), protein translation (15/18), and tricarboxylic acid cycle (7/7) (Table 2B). Differentially regulated genes are also overrepresented in the GO categories of peptidoglycan catabolic process and transmembrane transport. In the GO category of peptidoglycan catabolic process, 4 genes were upregulated and 4 genes were downregulated (p value=1.17e−9 ). In the GO category of transmembrane transport, 11 genes were upregulated and 12 genes were downregulated (p value=1.10e−08)
Table 2A.
Gene Ontology categories with genes largely up-regulated.
| GO Category | Number | # of Genes Up/Down regulated | Frac. | P value | Go term |
|---|---|---|---|---|---|
| GO:0006355 | 119 | 82/37 | 2.14 | 2.20E−16 | Regulation of transcription, DNA-dependent |
| GO:0007165 | 109 | 76/33 | 1.96 | 2.20E−16 | Signal transcduction |
| GO:0006955 | 64 | 57/7 | 1.15 | 2.20E−16 | Immune response |
| GO:0019882 | 31 | 27/4 | 0.56 | 2.20E−16 | Antigen processing and presentation |
| GO:0051607 | 17 | 16/1 | 0.31 | 4.58E−16 | Defense response to virus |
| GO:0006952 | 9 | 8/1 | 0.16 | 8.30E−10 | Defense response |
| GO:0007155 | 18 | 18/0 | 0.32 | 1.16E−09 | Cell adhesion |
| GO:0007154 | 21 | 16/5 | 0.38 | 1.91E−08 | Cell communication |
| GO:0016192 | 20 | 14/6 | 0.36 | 2.45E−08 | Vesicle-mediated transport |
Table 2B.
Gene Ontology categories with genes largely down-regulated.
| GO Category | Number | # of Genes Up/Down regulated | Frac. | P value | Go term |
|---|---|---|---|---|---|
| GO:0055114 | 249 | 35/214 | 4.47 | 2.20E−16 | Oxidation–reduction process |
| GO:0008152 | 101 | 19/82 | 1.81 | 2.20E−16 | Metabolic process |
| GO:0005975 | 69 | 24/45 | 1.24 | 2.20E−16 | Carbohydrate metabolic process |
| Go:0006265 | 21 | 5/16 | 0.38 | 2.20E−16 | DNA topological change |
| GO:0006418 | 18 | 3/15 | 0.32 | 3.36E−11 | tRNA aminoacylation for protein translation |
| GO:0046854 | 11 | 4/7 | 0.2 | 4.17E−09 | Phosphatidylinositol phosphorylation |
| GO:0048015 | 10 | 3/7 | 0.18 | 1.43E−08 | Phosphatidylinositol-mediated signaling |
| GO:0006099 | 7 | 0/7 | 0.13 | 1.14E−07 | Tricarboxylic acid cycle |
Table 3.
Regulation of genes involved in viral response.
| Locus ID | Gene | Fold change (mean±std. dev.) |
|---|---|---|
| locus_03211 | Toll-like receptor 7 | 10.3±0.197 |
| locus_05992 | Toll-like receptor 2 | 6.5±0.126 |
| locus_11815 | Mitochondrial antiviral-signaling protein-like | −1.5±0.056 |
| locus_06466 | 2′-5′-oligoadenylate synthase 2-like | 11.9±0.226 |
| locus_11667 | 2′-5′-oligoadenylate synthase 1A-like | 11.9±0.074 |
| locus_11853 | 59 kDa 2′-5′-oligoadenylate synthase-like protein-like | 4.1±0.067 |
| locus_16812 | 2′-5′-oligoadenylate synthetase 3-like | 22.6±0.146 |
| locus_29063 | 2′-5′-oligoadenylate synthetase 3 | 7.6±0.174 |
| locus_41602 | 2′-5′-oligoadenylate synthase 1-like | 23.3±0.017 |
| locus_41730 | 2′-5′-oligoadenylate synthase 3-like | 5.3±0.151 |
| locus_48794 | 2′-5′-oligoadenylate synthase 1-like | 1.5±0.033 |
| locus_32403 | Interferon-inducible protein 10 | 35.9±0.042 |
| locus_15775 | Interferon-activable protein 204 | 22.2±0.034 |
| locus_25036 | IL-6 | 6.1±0.104 |
| locus_06991 | Antigen-presenting glycoprotein CD1d1-like | 6.5±0.126 |
| locus_23284 | Proteoglycan 3-like | 6.1±0.094 |
| locus_11801 | Interferon-induced, double-stranded RNA-activated protein kinase | 26.5±0.093 |
Table 3 shows a more detailed depiction of differentially expressed genes in the GO category of anti-viral responses: 16 out of 17 genes were upregulated, including toll-like receptor 7 and 2, IL-6, and interferon-inducible proteins such as OAS family members and PKR.
Validation of microarray data by qRT-PCR
To validate the differential gene expression profiles obtained by microarray analysis, qRT-PCR was performed. First, we confirmed the suitability of four housekeeping genes, namely PPI, RPS6KB1, HBP2, and GAPDH. These genes range from low, medium–low, medium, and high regarding the abundance of their expression, and their expression was unaffected by Ad5 infection. Therefore, these genes were appropriate to serve as endogenous controls. We then examined the relative expression of a few selected target genes which are differentially expressed as found in our microarray analysis. The selected target genes for validation covered a full expression abundance spectrum, ranging from low (prostaglandin G/H synthase 1 precursor, PTGS; 2′-5′-oligoadenylate synthase 3, OAS3); medium–low (eukaryotic translation initiation factor 2 alpha kinase2, PKR); medium (interferon-inducible protein 10, ICP10; prostaglandin reductase 1, PTGR1), and high (interferon-induced protein with tetratricopeptide repeat3-like, IFIT3; MHC-II β chain, MHCIIβ). These selected targets also represent genes involved in the inflamatory response (PTGR1, PTGS1), the immune response (MHC II), interferon-inducible genes (IFP10, IFIT3), and interferon-inducible antiviral effector genes (PKR, OAS3).
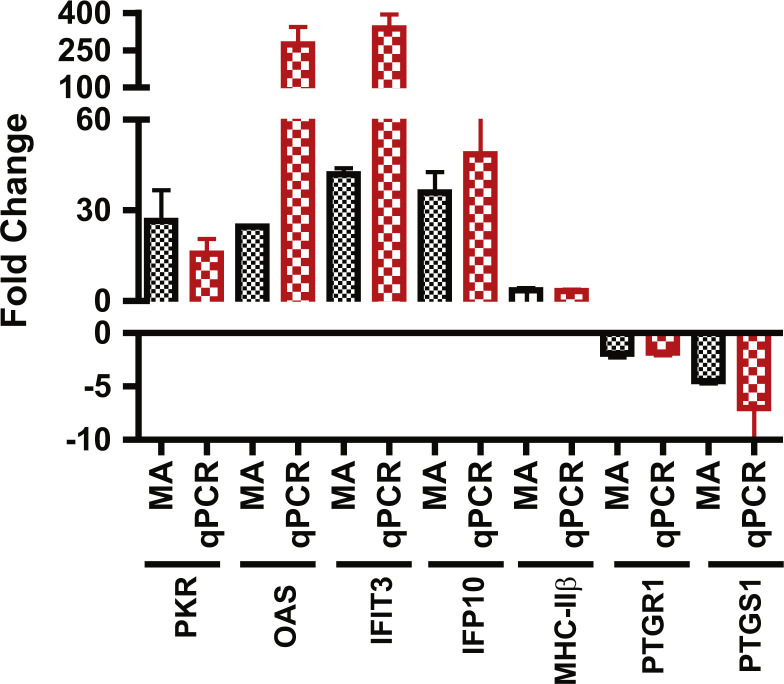
The overall results of qRT-PCR were in agreement with the microarray analysis ( Fig. 1); less than two-fold differences were obtained between the microarray and qRT-PCR data in five out of seven genes tested. Two genes (OAS3, IFIT3) showed 8–9 fold differences between these two types of assays, which probably reflects the intrinsic sensitivity differences between these two techniques.
Fig. 1.
Validation of Microarray (MA) data of selected genes by qRT-PCR. The mRNA expression levels of various genes were determined by qRT-PCR to validate the expression data of the microarray analysis. The fold change represents the mRNA expression level in Ad5-infected hamsters over that of untreated hamsters. For qRT-PCR (red bar), the value represents mean±SD of three biological replicates without pooling RNA. For microarray (MA, black bar), the value represents the average of two biological replicates.
The number of Class II Histocompatibility Antigen (MHC II) positive cells is increased in the liver of Ad5-infected hamsters
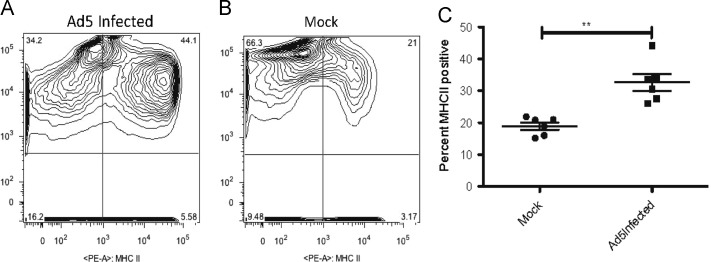
To assess whether the increase for the mRNA levels seen for MHC II translated into increases at the protein levels, we assayed by flow cytometry single-cell suspensions of livers collected from mock-infected and Ad5-infected animals. We found that there was a significant elevation in the number of MHC II positive cells in the liver of Ad5-infected hamsters compared to mock-infected animals ( Fig. 2). The mean fluorescence intensity of the MHC II positive cells also increased approximately 3-fold. These data indicate that the elevation in the MHC II mRNA levels detected by the microarray and qRT-PCR were a good indication of the actual protein levels of MHC II in the sample.
Fig. 2.
The number of MHC II positive cells is elevated in the liver of Ad5-infected hamsters. Combined results from two independent experiments are shown. A, B: Flow cytometry data from two individual animals. C: The symbols represent data from individual animals, horizontal bars symbolizes the mean, and the whiskers signify the standard deviation. ** p<0.01.
Discussion
Global transcriptome representing whole Syrian golden hamster transcripts
In this report, we extend previously described Syrian hamster transcriptome data (see Introduction) and we provide novel information on the pathogenesis of Ad5 in a permissive animal model. We present the sequence of the hamster transcriptome, the construction of a unique hamster microarray, and the characterization of the global transcriptional response in the liver of hamsters to i.v. infection with Ad5. Our transcriptome covers hamster transcripts derived from ten pooled tissues. Our study has increased the number of non-redundant transcripts for the Syrian hamster from 11,648 to 42,707, representing 34,191 unique genes, resulting in 4× increased gene discovery in our data set. The larger tissue sampling combined with LPS treatment and deeper sequencing all contributed to the increased gene discovery.
Systemic adenovirus infection causes a massive change in regulation of the hamster liver transcriptome
Investigations into interactions among Ad and specific cellular proteins and pathways in cell culture have provided insights about Ad and host responses. Genome-wide gene expression analyses have been performed by several laboratories using different cell lines, cell culture conditions, cells (non-cancerous versus cancer; proliferating or quiescent), wild-type Ad, various replication-competent and replication-defective vectors, and different conditions such as time post-infection and multiplicity of infection. Several microarray studies were conducted with Ad5 or the closely related Ad2 in non-cancerous human cells, namely in IMR-90 cells (a normal diploid cell derived from lungs of a 16 week female fetus) (Granberg et al., 2006, Stilwell and Samulski, 2004, Zhao et al., 2007), WI-38 cells (diploid cell line derived from normal embryonic lung culture) (Rao et al., 2006), synchronized human foreskin fibroblasts (Miller et al., 2007), or normal human bronchial epithelial cells (Dorer et al., 2011). Then, more recently, Ad2-induced transcriptome changes were studied in detail in IMR-90 cells using a sensitive cDNA deep sequencing strategy (Zhao et al., 2012). In general, these studies depict a struggle between the virus and the host cell in which the virus assumes control of the cell and the cell fights back with anti-viral defenses. The response begins within minutes of the virus interacting with the cell receptors and continues throughout the periods of virus early gene expression, viral DNA replication and early-late gene expression, and very late gene expression and virus assembly. Three of the microarray studies identified 988 (Zhao et al., 2007), about 2000 (Miller et al., 2007), or 943 (424 up, 519 down) (Dorer et al., 2011) cellular genes that were differentially up- or down-regulated by a factor of 2, and the RNA sequencing approach found 3791 genes that were differentially regulated (Zhao et al., 2012). Of these genes, it’s not clear whether the changes are caused by the action of the virus directly, e.g. through the activity of the E1A transcriptional co-regulator, or by the cell “sensing” that it has been infected. Altogether, these studies revealed that Ad infection leads to induction of genes involved in cell cycle regulation (activation), cell proliferation, chromatin reorganization, DNA replication, RNA transcription and processing, nucleotide metabolism, and the Wnt signaling pathway. Some but not all genes whose promoters contain binding sites for the E2F transcription factor were conspicuously up-regulated (Miller et al., 2007, Zhao et al., 2012). When quiescent human foreskin fibroblasts were infected, there was a reversal of the quiescent state similar to that seen in the cellular response to serum (Miller et al., 2007). In contrast to the up-regulated genes, there is repression of genes involved in cell differentiation, various signaling pathways (TGFβ, Rho, G-protein, Map kinase, STAT, NFκB), cell death regulation, and metabolism (Dorer et al., 2011, Zhao et al., 2012). Transcriptional activity associated with the p53 transcription factor was suppressed (Miller et al., 2007).
Another study in IMR-90 cells employed chromatin immunoprecipitation combined with microarray (ChIP–chip) technology to analyze global changes in gene expression at early (6 h), intermediate (12 h), and late times p.i. with h5dl1500, an Ad5 mutant that expresses only the 243 amino acid (243R) version of E1A protein (Ferrari et al., 2008), see review by Ferrari et al. (2009). Interaction of the conserved region 1 (CR1) and conserved region 2 (CR2) domains of the E1A 243R protein with RB family proteins leads to the release of free E2F family members to activate (or, with some E2F members, to repress) transcription of target genes. Also, interaction of the 243R N-terminus with histone acetyltransferase proteins (p300/CBP) regulates chromatin structure by altering the global pattern of histone modification, in particular H2K18ac, an epigenetic marker for actively transcribed genes (Ferrari et al., 2012, Horwitz et al., 2008). Ferrari et al. (2008) found that transcription of ~70% of cellular genes was altered in cells infected by H5dl1500 and that these changes were linked to E1A binding to RB members and p300/CBP. In brief, at early times p.i., E1A caused induction of genes that function in cell cycle regulation, and at late times (24 h p.i.) E1A activity led to repression of genes that function in development, differentiation, and cell signaling. Cell genes involved in defense against virus infection were induced at 6 h but repressed by 24 h p.i. The results of Ferrari et al. (2008) are generally consistent with the gene profiling data discussed earlier and they appear to make sense: cell cycle and DNA replication genes needed by the virus are induced and antiviral defense genes and cell differentiation genes that are harmful or not needed by the virus are repressed.
In addition to non-cancerous cell lines, microarray gene profiling studies have been conducted in several cancer cell lines infected by wild type Ad or Ad vectors, namely HeLa (Granberg et al., 2005, Zhao et al., 2003), Huh7 (Martina et al., 2007), and melanoma (Dorer et al., 2011, Volk et al., 2005) cells. In general, in these cells Ad infection affected genes involved in cell growth arrest, cell metabolism, and antiviral defense. Interestingly, fewer genes were dysregulated in cancer cells than in normal cells, consistent with the idea that infection of normal cells by Ad induces a cancer cell-like state (Ferrari et al., 2009, Ferrari et al., 2008, Horwitz et al., 2008).
A very recent study used microarray technology to examine cellular transcription in response to induction of the Ad2-coded E4orf3 protein in cells stably transfected with the gene for E4orf3 (Vink et al., 2015). E4orf3 functions in Ad replication by causing cellular proteins to re-localize into filamentous nuclear inclusions termed “tracks”. Multiple cellular proteins that are re-localized into these tracks function in the DNA damage response, the interferon response, and cellular transcription. Upon induction of E4orf3 in the transfected cells, more than 400 cellular genes were differentially regulated; 70% of the genes were upregulated and 30% were downregulated. With at least 8 of these genes, dysregulation was linked to nuclear track formation, i.e. physical relocation of cellular proteins into the tracks (Vink et al., 2015). Many of the genes with altered expression influence cellular growth properties and a variety of the proteins coded by these genes localize to the extracellular space or plasma membrane. Some genes affected by E4orf3 are also involved in antiviral activity.
Microarray based gene expression analysis of Ad-induced transcriptome changes have also been documented in a mouse cell culture system (mouse embryo fibroblasts, MEFs) as well as in non-permissive C57BL/6 J mice following systemic infection with a replication-defective E1- and E3-minus Ad5-based vector (Hartman et al., 2007, Hartman et al., 2008). While these studies revealed differences in the expression of genes belonging to particular function groups, these studies only analyzed the host transcriptome response in a non-permissive host for Ad infection, and did not address the role of viral replication and viral late gene expression in modulating host responses.
As discussed earlier, the liver is the major site of Ad5 replication in Syrian hamsters following i.v. administration of Ad5 (Lichtenstein et al., 2009, Thomas et al., 2008, Toth et al., 2008, Ying et al., 2009). Accordingly, we developed a hamster expression microarray to investigate the transcriptional response in the liver to systemic Ad5 infection. Our analysis revealed that approximately 16% of total unique transcripts were differentially expressed, indicating an extensive change in liver transcription. Functional analysis using GO categories revealed that genes involved in the immune response, defense response to virus, and antigen processing and presentation were largely upregulated (Table 2A). The same is true with genes associated with regulating DNA-dependent transcription, signal transduction, cell adhesion, and cell communication (Table 2A). Cell adhesion and cell-cell communication are critical in immune response networks where various lymphocytes, antigen-presenting cells, and antigen interact actively. Up-regulation of genes in these categories is further indication of an activated immune response to virus infection. The most down-regulated genes were associated with oxidation–reduction process, metabolic process, carbohydrate metabolic process, DNA topological change, protein translation, and tricarboxylic acid cycle (Table 2A, Table 3). Our results are in good agreement with the microarray analysis of gene expression in the liver of C57BL/6 J mice infected i.v. with a replication-defective Ad5 vector (Hartman et al., 2007). The latter study found that up-regulated genes are over-represented in categories involving the defense response, immune response, antigen presentation, signal transduction, and RNA processing, whereas down-regulated genes are over-represented in categories associated with metabolism, oxidoreduction, chromatin maintenance, and remodeling.
Robust up-regulation of genes involved in antiviral response
Taking a closer examination of differentially expressed genes involved in anti-viral responses, receptors engaging in the inflammatory immune response such as toll-like receptor 2 and 7 were upregulated (Table 3). Other genes associated with the inflammatory response such proteoglycan 3 and cytokine IL-6 were also upregulated. Most notably, a cluster of genes including PKR and oligoadenylate synthase family members OAS1-3 were upregulated. PKR and OAS are two major IFN-induced effectors demonstrated to possess antiviral activities against various viruses through different mechanisms of action. Following activation by viral double-stranded RNA (dsRNA), OAS synthesizes 2′,5′-oligoadenylates that in turn activate RNAse L, which in turn triggers cellular and viral RNA cleavage. IFN-induced PKR, following binding of dsRNA, inhibits cellular and viral protein translation by phosphorylation of eukaryotic translation initiation factor 2α (Randall and Goodbourn, 2008, Sadler and Williams, 2008).
Our data imply that innate immune responses mediated by activation of toll-like receptors, induction of proinflammatory cytokines and IFN pathways provide a first line of host defense against systemic Ad5 in the liver at the early encounter of infection, and that innate immune responses play a crucial role in controlling virus infection, likely mediated through the action of Kupffer cells. We have shown that the number of MHC II positive cells increased in the liver of Ad5 infected hamsters, and that these cells up-regulate the surface expression of MHC II, indicating up-regulation of the antigen presentation process. These flow cytometry data corroborate data obtained using microarray and qRT-PCR. This elevation in the number of MHC II positive cells in the liver is somewhat unexpected, inasmuch as Ad5 is known to preferentially infect and kill Kupffer cells, the major MHC II positive cell population in the liver, within minutes of intravenous injection (Smith et al., 2008). Further, the Ad E1 A protein down-regulates the expression of MHC II RNA (Kretsovali et al., 1998). Clearly, further investigation is needed to discover the nature of the MHC II positive cells in the liver of Ad5-infected hamsters.
In summary, our current study significantly expanded the number of non-redundant transcripts of the Syrian hamster transcriptome to 42,707, representing 34,191 unique genes, and generated a microarray platform for global transcription analysis. By using these new platforms, we describe the first global transcriptional analysis of gene expression in the liver of Ad5-infected Syrian hamsters. We anticipate that the novel platforms developed in this study will facilitate further development of new tools for this important animal model, and will be beneficial to the whole hamster research community.
Materials and methods
Animals
Female Syrian hamsters (Mesocricetus auratus) were obtained from Harlan Laboratories, Indianapolis, IN. All studies were approved by the Institutional Animal Care and Use Committee of Saint Louis University and were conducted according to federal and institutional regulations.
RNA extraction
To extract RNA for transcriptome sequencing, two female Syrian hamsters (5–6 weeks old), one untreated and one treated intraperitoneally with lipopolysaccharide (LPS) for 18 h, were anesthetized with CO2 and then sacrificed. Whole blood was collected by cardiac puncture, and bone marrow was flushed from the femur. Both blood and bone marrow samples were placed into RNA Protect Animal Blood tubes (Qiagen, Valencia, CA). Eight different organs were collected and frozen immediately in liquid nitrogen (Table 1). Total RNAs from blood and bone marrow were extracted using RNeasy Protect Animal Blood kit (Qiagen). RNA was extracted from all organs except liver by homogenizing the samples in Trizol reagent and then extracting the RNA from the homogenate. To extract liver RNA the sample was homogenized in RNALater buffer and the homogenate was processed using the RNeasy mini kit (Qiagen). All RNA samples were treated with RNase-free DNase followed by RNA cleanup to eliminate DNA contamination. RNA integrity was determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), while the RNA yield was determined on a NanoDrop-2000 spectrophotometer. To obtain a diverse representation of the whole transcriptome, total RNAs from all the organs were pooled from an untreated and a LPS-treated hamster (Table 1). The pooled RNA quality, as assessed by Bioanalyzer (Agilent), had a RNA integrity number (RIN) of 8.5.
To extract RNA for microarray analysis, four female hamsters were divided into two groups; two hamsters in each group were administered i.v. with either phosphate buffered saline (PBS) or 1010 plaque forming units (pfu) of Ad5 in a total volume of 200 µl. Hamsters were sacrificed 18 h later and liver RNA from each hamster was extracted using the RNeasy mini kit.
To extract RNA for microarray validation, two groups of three hamsters each were treated with PBS or Ad5 i.v. and then liver RNA was extracted as described as above.
Library construction and sequencing
Approximately 450 ng (quantified by Qubit; Life Technologies) of pooled RNA was used as input for library construction. For sequencing, a library was generated using Illumina TruSeq RNA preparation kit version 2 following the manufacturer׳s directions. The library was sequenced on Illumina HiSeq to obtain 100 base pair (bp) paired-end reads.
Sequence assembly and annotation
Raw Illumina RNAseq data were adapter trimmed, validated, and filtered based on quality. Quality-passed data were assembled via the de novo RNA-seq assembler Trinity (Grabherr et al., 2011, Haas et al., 2013). Trinity assembled contigs were validated via ESTscan and custom scripts, resulting in a set of 49,176 high-confidence transcripts. These high-confidence transcripts denote the assembled transcriptome. Each protein sequence from the assembled transcriptome was annotated using BLASTP versus the NCBI/Genbank NR protein database, and using the protein domain/motif prediction program InterProScan. BLASTP matching against protein database typically allows for better annotations because wobble positions and sequencing errors are eliminated in the alignment. The sequencing and summary statistics are shown in Table 1.
Microarray design and analysis
While RNA-Seq is likely to replace microarrays eventually, microarrays are still used because they are highly time and cost-effective and because methods of normalization and analysis of data are more robust. In our hands two-color microarrays perform as well, if not better than RNA-Seq. Therefore, we designed microarrays based on the hamster transcriptome. The 60-mer probes were designed using the high-confidence transcripts. Briefly, each transcript was divided into non-overlapping (contiguous) 60 nucleotide oligonucleotide (oligos). We calculated four parameters for each oligo: uniqueness, sequence complexity, self-annealing and melting temperature (T M). The uniqueness parameter is the binding energy (Allawi and SantaLucia, 1998) calculated using the sequence of the oligo to the reverse-complementary sequence and to its closest match identified by BLASTN against the high-confidence transcripts. Sequence complexity is calculated using the Ziv and Lempel algorithm (Ziv and Lempel, 1977). The T M is calculated using equation and parameters as previously described (Bolton and Mc Carthy, 1962). The designed probes were submitted to Agilent for printing of the arrays in the 8×60K format, where each slide has eight identical arrays with 61,657 60-mer-oligonucleotide probes.
Quantitative reverse transcriptase RT-PCR (qRT-PCR)
For qRT-PCR, 2 µg of each RNA and 50 pmol of oligo (dT) primer were used for in vitro reverse transcription (RT) using the High Capacity cDNA Reverse Transcription kit (ABI, Forster city, CA). SYBR Green based qPCR was used to specifically detect target gene mRNA. The primers were designed using Primer 3 software and synthesized by Integrated DNA Technologies (Coralville, IA). The sequence of the primers is shown in Supplementary Table S1. The PCR was set up in a 20 μl volume containing 1x SYBR® select master mix (ABI), 250 nM forward and reverse primers, and 2 μl of the diluted RT template. Quantification was done in triplicate for each sample using an ABI model 7500 genetic analyzer with the following cycling parameters: 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Dissociation curve analysis was performed following PCR to verify the specificity of the amplified product.
The data were analyzed using the Δ∆Ct method. Four housekeeping genes were used as endogenous controls for normalization, namely peptidyl-prolyl cis–trans isomerase-like 1 (PPI), ribosomal protein S6 kinase polypeptide 1 (RPS6KB1), histone-binding protein RBBP7 isoform2 (HBP2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Briefly, the Ct of each target gene in a treated hamster was first normalized to the Ct of the endogenous control (ΔCt) and then compared to the same normalized gene in a mock-treated (calibrator) hamster to determine the ∆∆Ct. The final value is displayed as the relative fold change between the Ad5-infected and mock-treated hamsters.
Flow cytometry
Syrian hamsters were mock-infected or infected i.v. with 1.2×1010 pfu of Ad5 (n=3 for both groups; the experiment was repeated once). At 18 h post injection, the animals were sacrificed, and the liver was perfused with PBS. The liver was harvested, and the left lateral lobe was homogenized by forcing it through a 100 μm cell strainer (Fisher Scientific, Waltham, MA). The cell suspensions were washed using PBS. The cells were incubated with Live/Dead Fixable Aqua cell stain (Life Technologies, Grand Island, NY), washed twice with FACS buffer (PBS containing 2% fetal bovine serum), and then re-suspended in FACS buffer containing a 1:2000 dilution of PE-conjugated anti-mouse/rat MHC class II (I-Ek; clone 14-4-4 S). After 1 h incubation, the cells were washed twice with PBS, fixed in 2% paraformaldehyde solution for 5 min, washed in PBS, re-suspended in FACS buffer, and analyzed on a BD FACS LSRII analyzer.
Acknowledgment
This research was supported by Grant R01 CA118022 from the National Institutes of Health.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2015.07.024.
Appendix A. Supplementary materials
Supplementary data
References
- Allawi H.T., SantaLucia J., Jr. Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- Berk A.J. Adenoviridae. In: Knipe D.M., Howley P.M., editors. Lippincott williams & Wilkins; Philadelphia, PA: 2013. pp. 1704–1731. [Google Scholar]
- Bhathena J., Kulamarva A., Martoni C., Urbanska A.M., Malhotra M., Paul A., Prakash S. Diet-induced metabolic hamster model of nonalcoholic fatty liver disease. Diabetes, Metab. Syndr. Obes.: Targets Ther. 2011;4:195–203. doi: 10.2147/DMSO.S18435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E.T., Mc Carthy B.J. A general method for the isolation of RNA complementary to DNA. Proc. Natl. Acad. Sci. USA. 1962;48:1390–1397. doi: 10.1073/pnas.48.8.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolanza S., Alzuguren P., Bunuales M., Qian C., Prieto J., Hernandez-Alcoceba R. Human adenovirus replicates in immunocompetent models of pancreatic cancer in Syrian hamsters. Hum. Gene Ther. 2007;18:681–690. doi: 10.1089/hum.2007.017. [DOI] [PubMed] [Google Scholar]
- Cerullo V., Koski A., Vaha-Koskela M., Hemminki A. Chapter eight--Oncolytic adenoviruses for cancer immunotherapy: data from mice, hamsters, and humans. Adv. Cancer Res. 2012;115:265–318. doi: 10.1016/B978-0-12-398342-8.00008-2. [DOI] [PubMed] [Google Scholar]
- Churchill G.A. Using ANOVA to analyze microarray data. Biotechniques. 2004;37(173–175):177. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- Dhar D., Spencer J.F., Toth K., Wold W.S. Effect of preexisting immunity on oncolytic adenovirus vector INGN 007 antitumor efficacy in immunocompetent and immunosuppressed Syrian hamsters. J. Virol. 2009;83:2130–2139. doi: 10.1128/JVI.02127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D., Spencer J.F., Toth K., Wold W.S.M. Pre-existing immunity and passive immunity to Adenovirus 5 prevents toxicity caused by an oncolytic adenovirus vector in the Syrian hamster model. Mol. Ther. 2009;17:1724–1732. doi: 10.1038/mt.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D., Toth K., Wold W.S. Syrian hamster tumor model to study oncolytic Ad5-based vectors. Methods Mol. Biol. 2012;797:53–63. doi: 10.1007/978-1-61779-340-0_4. [DOI] [PubMed] [Google Scholar]
- Dhar D., Toth K., Wold W.S. Cycles of transient high-dose cyclophosphamide administration and intratumoral oncolytic adenovirus vector injection for long-term tumor suppression in Syrian hamsters. Cancer Gene Ther. 2014;21:171–178. doi: 10.1038/cgt.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard A., Matthan N.R., Lichtenstein A.H. Use of hamster as a model to study diet-induced atherosclerosis. Nutr. Metab. 2010;7:89. doi: 10.1186/1743-7075-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer D.E., Holtrup F., Fellenberg K., Kaufmann J.K., Engelhardt S., Hoheisel J.D., Nettelbeck D.M. Replication and virus-induced transcriptome of HAdV-5 in normal host cells versus cancer cells--differences of relevance for adenoviral oncolysis. PLoS One. 2011;6:e27934. doi: 10.1371/journal.pone.0027934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H., Zivcec M., Gardner D., Falzarano D., LaCasse R., Rosenke R., Long D., Haddock E., Fischer E., Kawaoka Y., Feldmann H. Vol. 207. 2013. A Syrian golden hamster model recapitulating ebola hemorrhagic fever; pp. 306–318. (J. Infect. Dis.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria M. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 2008;21:704–715. doi: 10.1128/CMR.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espitia C.M., Zhao W., Saldarriaga O., Osorio Y., Harrison L.M., Cappello M., Travi B.L., Melby P.C. Duplex real-time reverse transcriptase PCR to determine cytokine mRNA expression in a hamster model of New World cutaneous leishmaniasis. BMC Immunol. 2010;11:31. doi: 10.1186/1471-2172-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Berk A.J., Kurdistani S.K. Viral manipulation of the host epigenome for oncogenic transformation. Nat. Rev. Genet. 2009;10:290–294. doi: 10.1038/nrg2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Su T., Li B., Bonora G., Oberai A., Chan Y., Sasidharan R., Berk A.J., Pellegrini M., Kurdistani S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen B.B., Holbrook M.R. Animal models of highly pathogenic RNA viral infections: hemorrhagic fever viruses. Antivir. Res. 2008;78:79–90. doi: 10.1016/j.antiviral.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granberg F., Svensson C., Pettersson U., Zhao H. Modulation of host cell gene expression during onset of the late phase of an adenovirus infection is focused on growth inhibition and cell architecture. Virology. 2005;343:236–245. doi: 10.1016/j.virol.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Granberg F., Svensson C., Pettersson U., Zhao H. Adenovirus-induced alterations in host cell gene expression prior to the onset of viral gene expression. Virology. 2006;353:1–5. doi: 10.1016/j.virol.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., Macmanes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., Leduc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Z.C., Appledorn D.M., Amalfitano A. Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications. Virus Res. 2008;132:1–14. doi: 10.1016/j.virusres.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman Z.C., Kiang A., Everett R.S., Serra D., Yang X.Y., Clay T.M., Amalfitano A. Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J. Virol. 2007;81:1796–1812. doi: 10.1128/JVI.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook M.R., Gowen B.B. Animal models of highly pathogenic RNA viral infections: encephalitis viruses. Antivir. Res. 2008;78:69–78. doi: 10.1016/j.antiviral.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Horwitz G.A., Zhang K., McBrian M.A., Grunstein M., Kurdistani S.K., Berk A.J. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison M.G. Adenovirus infections in transplant recipients. Clin. Infect. Dis. 2006;43:331–339. doi: 10.1086/505498. [DOI] [PubMed] [Google Scholar]
- Johnson K.C., Yongky A., Vishwanathan N., Jacob N.M., Jayapal K.P., Goudar C.T., Karypis G., Hu W.S. Exploring the transcriptome space of a recombinant BHK cell line through next generation sequencing. Biotechnol. Bioeng. 2014;111:770–781. doi: 10.1002/bit.25135. [DOI] [PubMed] [Google Scholar]
- Jove M., Ayala V., Ramirez-Nunez O., Serrano J.C., Cassanye A., Arola L., Caimari A., Del Bas J.M., Crescenti A., Pamplona R., Portero-Otin M. Lipidomic and metabolomic analyses reveal potential plasma biomarkers of early atheromatous plaque formation in hamsters. Cardiovasc. Res. 2013;97:642–652. doi: 10.1093/cvr/cvs368. [DOI] [PubMed] [Google Scholar]
- Julander J.G., Ennis J., Turner J., Morrey J.D. Treatment of yellow fever virus with an adenovirus-vectored interferon, DEF201, in a hamster model. Antimicrob. Agents Chemother. 2011;55:2067–2073. doi: 10.1128/AAC.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander J.G., Furuta Y., Shafer K., Sidwell R.W. Activity of T-1106 in a hamster model of yellow Fever virus infection. Antimicrob. Agents Chemother. 2007;51:1962–1966. doi: 10.1128/AAC.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesel J., Miller C., Abu-Amer Y., Aurora R. Systems level analysis of osteoclastogenesis reveals intrinsic and extrinsic regulatory interactions. Dev. Dyn. 2007;236:2181–2197. doi: 10.1002/dvdy.21206. [DOI] [PubMed] [Google Scholar]
- Kretsovali A., Agalioti T., Spilianakis C., Tzortzakaki E., Merika M., Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein D.L., Spencer J.F., Doronin K., Patra D., Meyer J., Shashkova E.V., Kuppuswamy M., Dhar D., Thomas M.A., Tollefson A.E., Zumstein L.A., Wold W.S.M., Toth K. An acute toxicology study with INGN 007, an oncolytic adenovirus vector, in mice and permissive Syrian hamsters; comparisons with wild-type Ad5 and a replication-defective adenovirus vector. Cancer Gene Ther. 2009;16:644–654. doi: 10.1038/cgt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014;27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina Y., Avitabile D., Piersanti S., Cherubini G., Saggio I. Different modulation of cellular transcription by adenovirus 5, DeltaE1/E3 adenovirus and helper-dependent vectors. Virus Res. 2007;130:71–84. doi: 10.1016/j.virusres.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Matthes-Martin S., Boztug H., Lion T. Diagnosis and treatment of adenovirus infection in immunocompromised patients. Expert Rev. Anti-Infect. Ther. 2013;11:1017–1028. doi: 10.1586/14787210.2013.836964. [DOI] [PubMed] [Google Scholar]
- Matthes-Martin S., Feuchtinger T., Shaw P.J., Engelhard D., Hirsch H.H., Cordonnier C., Ljungman P. European guidelines for diagnosis and treatment of adenovirus infection in leukemia and stem cell transplantation: summary of ECIL-4 (2011) Transpl. infect. Dis. 2012;14:555–563. doi: 10.1111/tid.12022. [DOI] [PubMed] [Google Scholar]
- Miller D.L., Myers C.L., Rickards B., Coller H.A., Flint S.J. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 2007;8:R58. doi: 10.1186/gb-2007-8-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath T.P., Lee C.K., Julander J.G., Brown A., Beasley D.W., Watts D.M., Hayman E., Guertin P., Makowiecki J., Crowell J., Levesque P., Bowick G.C., Morin M., Fowler E., Trent D.W. Inactivated yellow fever 17D vaccine: development and nonclinical safety, immunogenicity and protective activity. Vaccine. 2010;28:3827–3840. doi: 10.1016/j.vaccine.2010.03.023. [DOI] [PubMed] [Google Scholar]
- Morrey J.D., Day C.W., Julander J.G., Olsen A.L., Sidwell R.W., Cheney C.D., Blatt L.M. Modeling hamsters for evaluating West Nile virus therapies. Antivir. Res. 2004;63:41–50. doi: 10.1016/j.antiviral.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Morrey J.D., Taro B.S., Siddharthan V., Wang H., Smee D.F., Christensen A.J., Furuta Y. Efficacy of orally administered T-705 pyrazine analog on lethal West Nile virus infection in rodents. Antivir. Res. 2008;80:377–379. doi: 10.1016/j.antiviral.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama E., Saijo M. Animal models for Ebola and Marburg virus infections. Front. Microbiol. 2013;4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rao X.M., Zheng X., Waigel S., Zacharias W., McMasters K.M., Zhou H.S. Gene expression profiles of normal human lung cells affected by adenoviral E1B. Virology. 2006;350:418–428. doi: 10.1016/j.virol.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Roberts A., Vogel L., Guarner J., Hayes N., Murphy B., Zaki S., Subbarao K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79:503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immun. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkovsky U., Vargas L., Florescu D.F. Adenovirus: current epidemiology and emerging approaches to prevention and treatment. Curr. Infect. Dis. Rep. 2014;16:416. doi: 10.1007/s11908-014-0416-y. [DOI] [PubMed] [Google Scholar]
- Schmucki R., Berrera M., Kung E., Lee S., Thasler W.E., Gruner S., Ebeling M., Certa U. High throughput transcriptome analysis of lipid metabolism in Syrian hamster liver in absence of an annotated genome. BMC Genom. 2013;14:237. doi: 10.1186/1471-2164-14-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.S., Xu Z., Tian J., Stevenson S.C., Byrnes A.P. Interaction of systemically delivered adenovirus vectors with Kupffer cells in mouse liver. Hum. Gene Ther. 2008;19:547–554. doi: 10.1089/hum.2008.004. [DOI] [PubMed] [Google Scholar]
- Spencer J.F., Sagartz J.E., Wold W.S.M., Toth K. New pancreatic carcinoma model for studying oncolytic adenoviruses in the permissive Syrian hamster. Cancer Gene Ther. 2009;16:912–922. doi: 10.1038/cgt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffend N.A., Meisel R.L. Aggressive experience increases dendritic spine density within the nucleus accumbens core in female Syrian hamsters. Neuroscience. 2012;227:163–169. doi: 10.1016/j.neuroscience.2012.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stercz B., Nagy K., Ongradi J. Adenovirus infections in immunocompromised patients. Orvosi Hetil. 2012;153:1896–1904. doi: 10.1556/OH.2012.29496. [DOI] [PubMed] [Google Scholar]
- Stilwell J.L., Samulski R.J. Role of viral vectors and virion shells in cellular gene expression. Mol. Ther.: J. Am. Soc. Gene Ther. 2004;9:337–346. doi: 10.1016/j.ymthe.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Tchitchek N., Safronetz D., Rasmussen A.L., Martens C., Virtaneva K., Porcella S.F., Feldmann H., Ebihara H., Katze M.G. Sequencing, annotation and analysis of the Syrian hamster (Mesocricetus auratus) transcriptome. PLoS One. 2014;9:e112617. doi: 10.1371/journal.pone.0112617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.A., Spencer J.F., La Regina M.C., Dhar D., Tollefson A.E., Toth K., Wold W.S. Syrian hamster as a permissive immunocompetent animal model for the study of oncolytic adenovirus vectors. Cancer Res. 2006;66:1270–1276. doi: 10.1158/0008-5472.CAN-05-3497. [DOI] [PubMed] [Google Scholar]
- Thomas M.A., Spencer J.F., Toth K., Sagartz J.E., Phillips N., Wold W.S.M. Immunosuppression enhances oncolytic adenovirus replication and anti tumor efficacy in the Syrian hamster model. Mol. Ther. 2008;16:1665–1673. doi: 10.1038/mt.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.A., Spencer J.F., Wold W.S.M. Use of the Syrian hamster as an animal model for oncolytic adenovirus vectors. Methods Mol. Med. 2007;130:169–183. doi: 10.1385/1-59745-166-5:169. [DOI] [PubMed] [Google Scholar]
- Toth K., Spencer J.F., Dhar D., Sagartz J.E., Buller R.M., Painter G.R., Wold W.S.M. Hexadecyloxypropyl-cidofovir, CMX001, prevents adenovirus-induced mortality in a permissive, immunosuppressed animal model. Proc. Natl. Acad. Sci. USA. 2008;105:7293–7297. doi: 10.1073/pnas.0800200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth K., Spencer J.F., Wold W.S.M. Immunocompetent, semi-permissive cotton rat tumor model for the evaluation of oncolytic adenoviruses. In: Tollefson A.E., Wold W.S.M., editors. Adenovirus Methods and Protocols. Humana Press; 2007. pp. 157–168. [DOI] [PubMed] [Google Scholar]
- Vink E.I., Zheng Y., Yeasmin R., Stamminger T., Krug L.T., Hearing P. Impact of adenovirus E4-ORF3 oligomerization and protein localization on cellular gene expression. Viruses. 2015;7:2428–2449. doi: 10.3390/v7052428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan N., Yongky A., Johnson K.C., Fu H.Y., Jacob N.M., Le H., Yusufi F.N., Lee D.Y., Hu W.S. Global insights into the Chinese hamster and CHO cell transcriptomes. Biotechnol. Bioeng. 2015;112:965–976. doi: 10.1002/bit.25513. [DOI] [PubMed] [Google Scholar]
- Volk A.L., Rivera A.A., Page G.P., Salazar-Gonzalez J.F., Nettelbeck D.M., Matthews Q.L., Curiel D.T. Employment of microarray analysis to characterize biologic differences associated with tropism-modified adenoviral vectors: utilization of non-native cellular entry pathways. Cancer Gene Ther. 2005;12:162–174. doi: 10.1038/sj.cgt.7700776. [DOI] [PubMed] [Google Scholar]
- Wold W.S., Toth K. Syrian hamster as an animal model to study oncolytic adenoviruses and to evaluate the efficacy of antiviral compounds. Adv. Cancer Res. 2012;115:69–92. doi: 10.1016/B978-0-12-398342-8.00003-3. [DOI] [PubMed] [Google Scholar]
- Wold W.S.M., Ison M.G. Adenoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2013. p. 1732. [Google Scholar]
- Ying B., Toth K., Spencer J.F., Meyer J., Tollefson A.E., Patra D., Dhar D., Shashkova E.V., Kuppuswamy M., Doronin K., Thomas M.A., Zumstein L.A., Wold W.S., Lichtenstein D.L. INGN 007, an oncolytic adenovirus vector, replicates in Syrian hamsters but not mice: comparison of biodistribution studies. Cancer Gene Ther. 2009;16:625–637. doi: 10.1038/cgt.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.A., Spencer J.F., Ying B., Tollefson A.E., Toth K., Wold W.S. The role of cyclophosphamide in enhancing antitumor efficacy of an adenovirus oncolytic vector in subcutaneous Syrian hamster tumors. Cancer Gene Ther. 2013;20:521–530. doi: 10.1038/cgt.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.A., Spencer J.F., Ying B., Toth K., Wold W.S. The effects of radiation on antitumor efficacy of an oncolytic adenovirus vector in the Syrian hamster model. Cancer Gene Ther. 2013;20:531–537. doi: 10.1038/cgt.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Dahlo M., Isaksson A., Syvanen A.C., Pettersson U. The transcriptome of the adenovirus infected cell. Virology. 2012;424:115–128. doi: 10.1016/j.virol.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Zhao H., Granberg F., Elfineh L., Pettersson U., Svensson C. Strategic attack on host cell gene expression during adenovirus infection. J. Virol. 2003;77:11006–11015. doi: 10.1128/JVI.77.20.11006-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Granberg F., Pettersson U. How adenovirus strives to control cellular gene expression. Virology. 2007;363:357–375. doi: 10.1016/j.virol.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Ziv J., Lempel A. A universal algorithm for sequential data compression. IEEE Trans. Inf. Theory. 1977;23:337–343. [Google Scholar]
- Zivcec M., Safronetz D., Haddock E., Feldmann H., Ebihara H. Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus) J. Immunol. Methods. 2011;368:24–35. doi: 10.1016/j.jim.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data