Summary
Aim
Drug addiction is characterized, in part, by deregulation of synaptic plasticity in circuits involved in reward, stress, cue learning, and memory. This study was designed to assess whether 185 variants in 32 genes central to synaptic plasticity and signal transduction contribute to vulnerability to develop heroin and/or cocaine addiction.
Methods
Analyses were conducted in a sample of 1860 subjects divided according to ancestry (African and European) and drug of abuse (heroin or cocaine).
Results
Eighteen SNPs in 11 genes (CDK5R1,EPHA4,EPHA6,FOSL2,MAPK3,MBP,MPDZ,NFKB1,NTRK2,NTSR1, and PRKCE) showed significant associations (P < 0.01), but the signals did not survive correction for multiple testing. SNP rs230530 in the NFKB1 gene, encoding the transcription regulator NF‐kappa‐B, was the only SNP indicated in both ancestry groups and both addictions. This SNP was previously identified in association with alcohol addiction. SNP rs3915568 in NTSR1, which encodes neurotensin receptor, and SNP rs1389752 in MPDZ, which encodes the multiple PDZ domain protein, were previously associated with heroin addiction or alcohol addiction, respectively.
Conclusions
The study supports the involvement of genetic variation in signal transduction pathways in heroin and cocaine addiction and provides preliminary evidence suggesting several new risk or protective loci that may be relevant for diagnosis and treatment success.
Keywords: African Americans, Cocaine addiction, Heroin addiction, Signal transduction, Synaptic plasticity
Introduction
Drug addiction is a brain disease characterized, in part, by uncontrolled use of drugs in spite of adverse consequences 1. With repeated drug use, addicts learn to associate the drug with cues in the environment and these cues can promote craving and relapse long after cessation of drug use. The repetitive drug use and withdrawal cause persistent changes to structure and function of key brain regions. Among the underlying mechanisms are deregulation of synaptic plasticity, gene expression, electrophysiological activity, and neural morphology 2. These changes are of particular importance in the cortico‐limbic–striatal circuits involved in reward, stress, cue learning, and habit formation including the major signaling pathways cAMP/PKA/CREB, DeltaFosB/Cdk5, and BDNF/ERK 2, 3, 4. This study focuses on signal transduction, which is a process of transmitting and amplifying signals from the cell surface to intracellular targets, and on synaptic plasticity that underlie memory formation.
Genetic variation is a critical determinant of individual differences in disease vulnerability and response to medical treatment. Twin and family studies documented a strong genetic influence on vulnerability for drug addiction (e.g., 5). Genetic factors may increase the effect of drugs of abuse on learning and memory and could be associated with specific addictive behaviors. Identifying variants that contribute to vulnerability to addictions can contribute to the existing knowledge of the neurobiological pathways that contribute to addiction and may help with treatment and prevention 6.
This study was designed to determine whether variations in selected 32 genes central to synaptic plasticity and signal transduction contribute to the susceptibility to heroin addiction (opiate dependence, OD) and/or cocaine dependence (CD) in populations of European (EA) and African (AA) ancestry. The genes include immediate‐early response genes, transcription factors and co‐factors, cell adhesion molecules that have a role in synaptic formation and maintenance, as well as neurotrophic factors and neuronal receptors. Previous association studies of polymorphisms in these genes with drug addiction reported the association of BDNF SNP rs6265 (Val66Met) with OD and methamphetamine dependence in Han Chinese 7 and European Americans 8, as well as of CREB1 SNP with OD in Indians 9. Alcohol dependence (AD) was associated with SNPs in BDNF, CREB1, MPDZ, NFKB1, NRXN1, NTRK2, and NTSR1 9, 10, 11, 12, 13, 14, 15, and nicotine dependence (ND) was associated with SNPs in NRXN1 and NRXN3 16, 17, 18.
The study extends our previous studies of heroin addiction in EA and AA 19, 20 with larger sample size and modified SNP content and enables a comparison between ancestries and drug‐specific addictions. This study also includes an AA cocaine group that was not previously analyzed for these genes. The samples analyzed in the current study were analyzed for genes in other systems (e.g., stress, dopaminergic), some of which could also be considered part of synaptic plasticity and signal transduction systems 21, 22, 23, 24.
Methods
Study Population
The study included 1860 subjects (38% females) that were divided into five groups according to their predominant ancestry and drugs of abuse (heroin or cocaine): (1) EA OD ± CD, (2) AA OD ± CD, (3) AA CD without OD, (4) EA control, and (5) AA control (Table 1). The subjects in the “OD ± CD” groups (1 and 2) were former heroin addicts in methadone maintenance treatment that had a history of at least 1 year of multiple daily uses of heroin. About half of them also had a history of cocaine addiction. The “CD without OD” group (3) included subjects with a history of cocaine addiction that had no history of heroin addiction. A third of them had history of alcohol addiction (AD), but AD was not a factor in the analysis. This study is a major expansion of our previous studies of OD 19, 20 for which we added 465 EA subjects and 481 AA subjects and included an AA “CD without OD” group.
Table 1.
Groups description
| Ancestry | Heroin addiction | Cocaine addiction | Controls | Total |
|---|---|---|---|---|
| (OD ± CD) | (CD) | |||
| EA | 827 (1) | – | 232 (4) | 1059 |
| AA | 315 (2) | 279 (3) | 207 (5) | 801 |
| Total | 1142 | 279 | 439 | 1860 |
The EA samples included subjects with >70% European, Middle‐Eastern (ME), or combined ancestry contributions based on Structure analysis (see below) from the United States (n = 744) and Israel (n = 315). A more homogenous subsample that included only samples with European contributions of >50% was also used to assess a potential effect of population substructure (EA OD ± CD; n = 636, EA control; n = 189).
The AA sample included subjects with >50% African ancestry contribution. Self‐identified Hispanics and AA subjects with >25% contribution of any major ancestry other than European/ME were not included.
Ascertainment of cases and controls was made by personal interview using several instruments: the Addiction Severity Index 25, KMSK 26, and the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM‐IV). Diagnosis was based on life‐time DSM‐IV criteria. Subjects were recruited at the Rockefeller University Hospital, the Manhattan campus of the VA NY Harbor Health Care System, and the Dr. Miriam and Sheldon G. Adelson Clinics for Drug Abuse Treatment and Research in Las Vegas and Israel.
The exclusion criteria from the control sample were as follows: (1) drinking to intoxication and/or using illicit drugs in the last month or more than twice a week for more than six consecutive months, and (2) cannabis use for more than 12 days in the last month or more than twice a week for >4 years.
The study was approved by the Institutional Review Boards of the Rockefeller University Hospital, the VA New York Harbor Healthcare System, and the Tel Aviv Sourasky Medical Center (Helsinki Committee). All subjects signed informed consent for genetic studies.
SNP Selection and Genotyping
A total of 32 genes related to synaptic plasticity and signal transduction were selected based on the “addiction array” 27 with some modifications (Tables 2 and S1). The “addiction array” included tagging SNPs in 23 genes of these systems that aimed to capture the maximum haplotype information. The modified Illumina GoldenGate custom panel (GS0013101‐OPA) used in the current study contained 185 SNPs in these genes, including the “addiction array” SNPs, except for 39 SNPs that were excluded due to failure or low frequency in the relevant populations, and 32 SNPs which were added based on functionality or reported association with related phenotypes (Table S1). SNPs were genotyped at the Rockefeller University Genomics Resource Center and analyzed with BeadStudio software v2.3.43 as described 19.
Table 2.
Gene list
| Symbol | Description | No. of SNPsa |
|---|---|---|
| ADCY7 | Adenylate cyclase 7 | 9 |
| BDNF | Brain‐derived neurotrophic factor | 13 (4) |
| CDK5R1 | Cyclin‐dependent kinase 5, regulatory subunit 1 (p35) | 2 |
| CREB1 | cAMP responsive element binding protein 1 | 8 |
| EFNA1 | ephrin‐A1 | 3 (1) |
| EPHA4 b | EPH receptor A4 (ephrin receptor) | (2) |
| EPHA6 b | EPH receptor A6 (ephrin receptor) | (1) |
| EPHB1 | EPH receptor B1 (ephrin receptor) | 5 (3) |
| FEV | FEV (ETS Oncogene Family) | 3 |
| FOS | v‐fos FBJ murine osteosarcoma viral oncogene homolog | 2 |
| FOSB b | FBJ murine osteosarcoma viral oncogene homolog B | (4) |
| FOSL1 | FOS‐like antigen 1 (FRA‐1) | 3 |
| FOSL2 | FOS‐like antigen 2 (FRA‐2) | 3 |
| GSK3B | Glycogen synthase kinase 3 beta | 13 |
| JUN | Jun proto‐oncogene | 3 |
| MAPK1 | Mitogen‐activated protein kinase 1 (ERK2) | 15 |
| MAPK14 | Mitogen‐activated protein kinase 14 | 7 |
| MAPK3 | Mitogen‐activated protein kinase 3 | 2 |
| MBP b | Myelin basic protein | (2) |
| MPDZ | Multiple PDZ domain protein | 11 |
| NCDN | Neurochondrin (norbin) | 2 (1) |
| NEGR1 | Neuronal growth regulator 1 | 2 (1) |
| NFKB1 b | Nuclear factor kappa‐light‐chain‐enhancer of activated B cells | (6) |
| NGFB | Nerve growth factor, beta polypeptide | 9 |
| NRGN b | Neurogranin | (1) |
| NRXN1 b | Neurexin 1 | (4) |
| NRXN3 b | Neurexin 3 | (1) |
| NTRK1 b | Neurotrophic tyrosine kinase, receptor, type 1 (TRK‐A, NGFB receptor) | (2) |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 (TRK‐B, BDNF receptor) | 15 |
| NTSR1 | Neurotensin receptor 1 (high affinity) | 10 |
| NTSR2 | Neurotensin receptor 2 | 3 |
| PRKCE | Protein kinase C, epsilon | 19 |
Number in parenthesis refers to SNPs that were not included in the original “addiction array” 27.
These genes were not represented in the original “addiction array” and are not covered with tag SNPs.
Structure Analysis
A total of 155 ancestry informative markers (AIMs) were genotyped with the modified array (GS0013101‐OPA). Assessment of ancestry contribution using AIMs was performed by Structure 2.2 with seven clusters (K) using data from 155 AIMs. Each subject was anchored against genotypes of 1051 samples from 51 worldwide populations represented in the Human Genome Diversity Cell Line Panel, as described 28. The EA sample included subjects with >70% contribution in the European or the ME clusters or a combined total from the two clusters. The European and ME clusters show relative low population differentiation 29, 30.
Statistical Analysis
Pairwise linkage disequilibrium (LD) (D′ and r 2) was estimated using Haploview 4.2. LD blocks were identified using the D′ confidence interval bound of 0.7–0.98 31. Exact tests for deviation from Hardy–Weinberg equilibrium (HWE) were performed with the PLINK program. Association analyses were conducted using PLINK for each SNP separately by logistic regression, under dominant or recessive model assumptions. Four association analyses were performed independently for EA OD ± CD (1), AA OD ± CD (2), AA CD without OD (3), and groups 2 and 3 combined. To assess the effect of including subjects with high ME contribution in the EA sample, an additional analysis of EA OD ± CD subsample that included only samples with European contributions of >50% was conducted as described above. Correction for multiple testing was performed by permutation test (n = 100,000) for each model of inheritance, using PLINK.
Results
The study included 1860 subjects that were divided into five groups according to their predominant ancestry and main drugs of abuse (heroin or cocaine): (1) EA OD ± CD, (2) AA OD ± CD, (3) AA CD without OD, (4) EA control, and (5) AA control (Table 1). The ancestry of all subjects was verified using Structure analysis of 155 AIMs and was used to define the groups (see Methods). There was no evidence for substructure among the case/control subgroups for each ancestry group. The three AA groups (cases and control) had an average range of 80% (SD = 0.1) African ancestry and 10% (SD = 0.08) European ancestry, as described 22.
A total of 185 SNPs from 32 genes related to synaptic plasticity and signal transduction (Tables 2 and S1) were analyzed in four independent analyses under two models of inheritance (dominant or recessive): EA OD ± CD (1 vs. 4), AA OD ± CD (2 vs. 5), AA CD without OD (3 vs. 5), as well as AA OD ± CD and CD without OD (2 + 3 vs. 5) (Table 1). An EA OD ± CD subgroup that included only subjects with European contributions of >50% was subsequently analyzed to rule out an effect of population substructure on the results. Thirty SNPs were excluded from the EA analysis, and 11 SNPs were excluded from the AA analyses, based on low frequency (minor allele frequency [MAF] < 0.05), including four SNPs that were excluded from all analyses (Table S1). Five SNPs showed significant deviation from HWE (P < 0.01) in the AA control sample and were excluded from AA analyses (Table S1). The minor allele of 53 SNPs in EA was the major allele in AA (Table S1).
A total of 45 SNPs in 21 genes showed nominally significant associations (P < 0.05) with OD and/or CD in either EA or AA (Table S2). The 14 SNPs (in eight genes) with the most significant associations (P < 0.01) are listed in Table 3, including two CDK5R1 SNPs in complete LD and three SNP pairs in moderate to strong LD. None of the signals survived correction for multiple testing. Associations with OD in EA (P < 0.01) were indicated for nine SNPs in CDK5R, NFKB1, NTRK2, NTSR1, and PRKCE. Associations with OD ± CD, CD, or both, in AA (P < 0.01), were indicated for SNPs in EPHA6, FOSL2, MPDZ, NFKB1, and PRKCE. The only genes that were indicated in both EA and AA were NFKB1 and PRKCE. NFKB1 SNP rs230530 was the only SNP that showed associations in both ancestry groups. All the associations that survived the P = 0.01 cutoff were of SNPs in non‐coding regions, including two SNPs in the CDK5R1 upstream region and one SNP in MPDZ 3′ UTR. The associations that survived the P = 0.05 cutoff also included two nonsynonymous SNPs (EFNA1 rs4745, MBP rs470797) and one synonymous SNP (NTRK1 rs6337) (Table S2).
Table 3.
The most significant association results (P < 0.01)
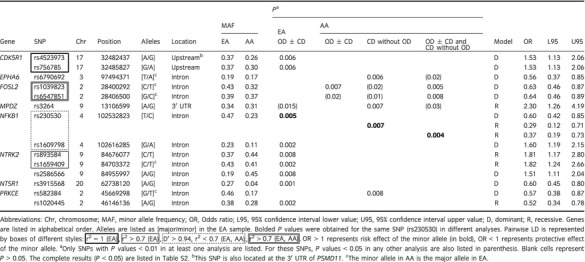
The analysis of the more homogeneous EA subsample that includes only samples with >50% European contribution revealed results similar to those of the analysis of the whole EA sample. The main exception was the 3ʹ UTR MAPK3 SNP rs7698 that did not reach significance in the original analysis (P = 0.07) but showed strong association with a protective effect under the dominant model in this analysis (P < 0.007, OR = 0.57, 95% CI = 0.38–0.86). In addition, four SNPs, including the synonymous MBP SNP rs470797 (Tyr96=), and the intronic EPHA4 rs2288629 that did not reach the significance threshold (P < 0.01) in the original analysis, reach this threshold in this analysis (Table S2).
Discussion
The study identified nominally significant associations of heroin and/or cocaine addiction with SNPs in a number of genes related to signal transduction and synaptic plasticity in subjects with predominantly European and/or African ancestry. These findings support the hypothesis of genetic contributions to drug addictions in these systems. The results may also be relevant to treatment effectiveness in general and for memory manipulation treatment in particular 32. There are only a few reports of association of variants in these genes with heroin or cocaine addiction (see Introduction) so the majority of the results (on the gene and/or SNP level) may be considered novel. However, associations of several variants or other variants in these genes were indicated in other studies of drug addictions or related phenotypes and may indicate non‐specific susceptibility. As the associations did not survive correction for multiple testing, they should be considered tentative until further corroboration. Nevertheless, a hypothesis‐driven study of genes with known or potential addiction‐related functionality may not require as stringent a threshold for significance as a hypothesis‐free study.
Additional support for the findings of this study comes from previous association studies of alcohol addiction (see Introduction) that identified NFKB1 SNPs rs230530 and rs1609798 as well as MPDZ SNP rs1389752 10, 12, 13. The current study extends our previous studies of heroin addiction in samples of predominantly European and African ancestry 19, 20. NTSR1 SNP rs3915568 indicated in the current study was associated with OD in EA in our previous study that included approximately half of the current sample, but did not survive the original cutoff of P < 0.01 19. Although the current analysis is not a replication, it corroborates this finding.
One intriguing result is the associations of NFKB1 SNP rs230530 with OD ± CD in EA, CD without OD in AA, and OD + CD in AA. This is the only SNP that was indicated with strong associations in both ancestries in this study. A protective effect of the NFKB1 SNP rs230530 minor C allele was found in EA and AA, under different models of inheritance. NFKB1 SNP rs230530 is also in LD with NFKB1 SNPs rs230529 and rs4699030 that were associated with treatment‐refractory schizophrenia in Han Chinese 33. The second NFKB1 SNP (rs1609798) identified in association with OD ± CD in EA showed a risk effect of the minor allele. NFKB1 (nuclear factor kappa‐light‐chain‐enhancer of activated B cells 1) encodes the transcription factor NF‐kappa B (NF‐kB) that is activated by synaptic activity as well as mu opioid receptor agonists and may play important roles in the process of learning and memory 34. Pharmacological and genetic manipulations of NF‐kB signaling are being developed for treatment of several disorders including cancer, Alzheimer's disease and schizophrenia 35.
Three intronic NTRK2 SNPs with unknown function showed association with OD ± CD in EA in the current study. In addition, several SNPs in BDNF, NGFB, and NTRK1 showed nominally significant associations (P < 0.05) in at least one analysis. NTRK2 encodes the tyrosine kinase TrkB receptor. This receptor autophosphorylation is dependent upon association with brain‐derived neurotrophic factor (BDNF) that is involved in synaptic plasticity and mediates memory consolidation 36. Many drugs of abuse lead to changes in BDNF expression in neural circuits relevant for addiction 34. Association studies of the functional BDNF SNP rs6265 (Val66Met) with OD gave inconsistent results. A meta‐analysis of 20 studies revealed association with methamphetamine dependence in South Asians and OD in Chinese subjects 7. This SNP was not associated with OD ± CD or CD without OD in this study.
NTSR1 SNP rs3915568 indicated in the current study corroborate our previous study of OD in EA 19. NTSR1 encodes one of the neurotensin receptors. Neurotensin modulates dopamine and other neurotransmitter systems involved in addiction and reward pathways and its effect depends on the location of the receptors 37. Different NTSR1 SNPs were associated with alcohol dependence and working memory in Han Chinese 15, 38, but there are no LD data for these SNPs and the SNP identified in the current study.
Two SNPs in complete LD located upstream of CDK5R1 were associated with OD ± CD in EA. There is no information on the functionality of these SNPs, although they may be involved in gene expression regulation. Interestingly, one of the SNPs is also located at the 3′ UTR of the adjacent PSMD11 (proteasome 26S subunit, non‐ATPase, 11) that is involved in ATP‐dependent degradation of ubiquitinated proteins. CDK5R1 (CDK5 regulatory subunit 1) encodes p35 that is an activator of CDK5 (serine/threonine kinase cyclin‐dependent kinase 5) 39. Rodent studies showed that Cdk5/p35 acts as downstream regulators of the prolonged activation of dopamine signaling after chronic exposure to cocaine 40, 41 and are involved in synaptic plasticity by affecting dendritic spine formation, ion channel conductance, and transcription 42. Rat study showed that CDK5 negatively regulates postsynaptic signaling of dopamine in the striatum and is also a key player in the regulation of the mu and delta opioid receptors 43. Decreased levels of CDK5/p35 were found in the postmortem prefrontal cortex of opioid addicts compared with healthy controls 44.
One intronic EPHA6 SNP was indicated in the study in association with CD without OD in AA, and several SNPs in genes encoding other members of this family (EFNA1, EPHA4, and EPHB1) were also indicated under the less stringent cutoff (P < 0.05), including a non‐synonymous EFNA1 SNP. The ephrin receptor tyrosine kinase (EPH) and its ligand, ephrins, control multiple cellular responses including neural plasticity 45. The functional EFNA1 3′ UTR SNP rs12904 that overlaps a miR‐200c binding site and was associated with cancer 46 did not show association signal in the current study.
Two PRKCE SNPs were associated with OD ± CD in EA or with CD without OD in AA. Protein kinase C (PKC) is a family of serine/threonine kinases that are involved in diverse cellular signaling pathways. PRKCE encodes protein kinase C epsilon (PKCε) that was shown in rodent studies to have a role in behavioral responses to ethanol and nicotine 47. A different PRKCE SNP was reported to be associated with suicide attempts in meta‐analysis of mood disorder patients, but there is no LD information that may connect it to the SNPs identified in this study 48.
Two intronic FOSL2 SNPs in strong LD showed association with OD ± CD in AA. Fos‐related antigen 2 (FRA‐2/FOSL2) belongs to the transcription factor complex AP‐1 which includes the various isoforms of Fos and Jun and upregulates transcription of many genes, including genes involved in synaptic plasticity and long‐term memory. Exposure to drugs of abuse induces all Fos family transcription factors in several brain regions with persistent accumulation of DeltaFosB 49. To the best of our knowledge, this is the first report on association of SNPs in the FOS family genes with drug addiction. Two FOS SNPs were recently associated with schizophrenia in Armenians 50. Of those two SNPs, SNP rs7101 was genotyped in the current study but was excluded due to technical problem.
The MPDZ 3′ UTR SNP rs3264 showed association with CD without OD in AA (P < 0.01) and also with OD ± CD in EA and OD ± CD in AA (P < 0.05). Two other MPDZ intronic SNPs were associated with OD ± CD in EA (rs1389752) or CD without OD in AA (rs1999395) (P < 0.05). Association of MPDZ variants with AD was reported 12, 13 including SNP rs1389752 that was indicated in the current study. MPDZ encodes for the scaffolding multiple‐PDZ‐domain protein (MPDZ/MUPP1) that impacts signaling 51.
The analysis of the more homogenous EA OD ± CD subsample that was conducted to assess the effect population substructure revealed a limited effect. Three of the SNPs indicated in this sub analysis are in genes that were not indicated by the main analysis (P < 0.01), including the 3ʹ UTR MAPK3 SNP rs7698, the synonymous MBP SNP rs470797 (Tyr96=), and the intronic EPHA4 rs2288629, although the last two SNPs were indicated in the less stringent analysis (P < 0.05). Although there are limited data in the public databases on the allele frequencies of these SNPs in ME populations, these findings suggest that they differ in MAF from European populations, as was shown in our previous study of NGFB 52.
The mitogen‐activated protein kinases (MAPK) are part of a signaling pathway that is involved in diverse processes including stress response and drug addiction 53, 54. Specific MAPK inhibitors were indicated as potential treatment of drug addiction 55. In addition to MAPK3 SNP rs7698, several SNPs in genes encoding other members of this family (MAPK1, and MAPK14) were indicated under the less stringent cutoff (P < 0.05, Table S2). There is no information about the functionality of these SNPs and there is only one study showing association of SNPs in these genes with a drug addiction‐related phenotype (MAPK1 SNP with alcohol consumption) 13. Comparison of the associations with OD ± CD in EA and OD ± CD and/or CD without OD in AA reveals two genes (NFKB1 and PRKCE) and only one SNP (NFKB1 rs230530) in common. Six additional genes showed association signals in both ancestry groups under the P < 0.05 cutoff. The results suggest shared and population‐specific genetic risk factors for addiction, but may also be a consequence of limited power in specific analyses, effect of population admixture, distinct allele frequencies, and/or different LD patterns. Identifying SNPs associated with addiction in different ancestry groups is relevant for population‐specific diagnosis and treatment.
In summary, the study suggests numerous potential susceptibility loci in synaptic plasticity and signal transduction pathways for heroin and cocaine addiction in subjects of African and European ancestry. Future studies are necessary to corroborate the results and to evaluate the potential contribution of the findings for diagnosis and treatment.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Table S1 SNP list.
Table S2 Association results (P < 0.05).
Acknowledgments
This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Clinical and Translational Science Award UL1RR024143 from the National Center for Advancing Translational Sciences of the NIH (B. Coller), and NSFC grant 31470070 from the Chinese Government (J. Ott).
We thank all the clinical staff including S. Linzy, E. Ducat, and B. Ray. We are grateful to P.‐H. Shen and D. Goldman for Structure analysis. We thank C. Zhao and B. Zhang for their excellent assistance in genotyping.
References
- 1. Kreek MJ, Levran O, Reed B, Schlussman SD, Zhou Y, Butelman ER. Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J Clin Invest 2012;122:3387–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology 2008;33:3–17. [DOI] [PubMed] [Google Scholar]
- 3. Torregrossa MM, Corlett PR, Taylor JR. Aberrant learning and memory in addiction. Neurobiol Learn Mem 2011;96:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward‐related learning and memory. Annu Rev Neurosci 2006;29:565–598. [DOI] [PubMed] [Google Scholar]
- 5. Tsuang MT, Lyons MJ, Meyer JM, et al. Co‐occurrence of abuse of different drugs in men: the role of drug‐specific and shared vulnerabilities. Arch Gen Psychiatry 1998;55:967–972. [DOI] [PubMed] [Google Scholar]
- 6. Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci 2005;8:1450–1457. [DOI] [PubMed] [Google Scholar]
- 7. Haerian BS. BDNF rs6265 polymorphism and drug addiction: a systematic review and meta‐analysis. Pharmacogenomics 2013;14:2055–2065. [DOI] [PubMed] [Google Scholar]
- 8. Greenwald MK, Steinmiller CL, Sliwerska E, Lundahl L, Burmeister M. BDNF Val(66)Met genotype is associated with drug‐seeking phenotypes in heroin‐dependent individuals: a pilot study. Addict Biol 2013;18:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pal A, Chakraborty J, Das S. Association of CREB1 gene polymorphism with drug seeking behaviour in eastern Indian addicts. Neurosci Lett 2014;570:53–57. [DOI] [PubMed] [Google Scholar]
- 10. Edenberg HJ, Xuei X, Wetherill LF, et al. Association of NFKB1, which encodes a subunit of the transcription factor NF‐kappaB, with alcohol dependence. Hum Mol Genet 2008;17:963–970. [DOI] [PubMed] [Google Scholar]
- 11. Xu K, Anderson TR, Neyer KM, et al. Nucleotide sequence variation within the human tyrosine kinase B neurotrophin receptor gene: association with antisocial alcohol dependence. Pharmacogenomics J 2007;7:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karpyak VM, Kim JH, Biernacka JM, et al. Sequence variations of the human MPDZ gene and association with alcoholism in subjects with European ancestry. Alcohol Clin Exp Res 2009;33:712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tabakoff B, Saba L, Printz M, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol 2009;7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hishimoto A, Liu QR, Drgon T, et al. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum Mol Genet 2007;16:2880–2891. [DOI] [PubMed] [Google Scholar]
- 15. Ma H, Huang Y, Zhang B, et al. Association between neurotensin receptor 1 gene polymorphisms and alcohol dependence in a male Han Chinese population. J Mol Neurosci 2013;51:408–415. [DOI] [PubMed] [Google Scholar]
- 16. Nussbaum J, Xu Q, Payne TJ, et al. Significant association of the neurexin‐1 gene (NRXN1) with nicotine dependence in European‐ and African‐American smokers. Hum Mol Genet 2008;17:1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high‐density genome wide association study for nicotine dependence. Hum Mol Genet 2007;16:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Docampo E, Ribases M, Gratacos M, et al. Association of neurexin 3 polymorphisms with smoking behavior. Genes Brain Behav 2012;11:704–711. [DOI] [PubMed] [Google Scholar]
- 19. Levran O, Londono D, O'Hara K, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav 2008;7:720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levran O, Londono D, O'Hara K, et al. Heroin addiction in African Americans: a hypothesis‐driven association study. Genes Brain Behav 2009;8:531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levran O, Randesi M, da Rosa JC, et al. Overlapping dopaminergic pathway genetic susceptibility to heroin and cocaine addictions in African Americans. Ann Hum Genet 2015;79:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levran O, Randesi M, Li Y, et al. Drug addiction and stress‐response genetic variability: association study in African Americans. Ann Hum Genet 2014;78:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levran O, Peles E, Randesi M, et al. Stress‐related genes and heroin addiction: a role for a functional FKBP5 haplotype. Psychoneuroendocrinology 2014;45:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levran O, Peles E, Randesi M, et al. Dopaminergic pathway polymorphisms and heroin addiction: further support for association of CSNK1E variants. Pharmacogenomics 2014;15:2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McLellan AT, Kushner H, Metzger D, et al. The fifth edition of the addiction severity index. J Subst Abuse Treat 1992;9:199–213. [DOI] [PubMed] [Google Scholar]
- 26. Kellogg SH, McHugh PF, Bell K, et al. The Kreek–McHugh–Schluger–Kellogg scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend 2003;69:137–150. [DOI] [PubMed] [Google Scholar]
- 27. Hodgkinson CA, Yuan Q, Xu K, et al. Addictions biology: haplotype‐based analysis for 130 candidate genes on a single array. Alcohol Alcohol 2008;43:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ducci F, Roy A, Shen PH, et al. Association of substance use disorders with childhood trauma but not African genetic heritage in an African American cohort. Am J Psychiatry 2009;166:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science 2002;298:2381–2385. [DOI] [PubMed] [Google Scholar]
- 30. Atzmon G, Hao L, Pe'er I, et al. Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet 2010;86:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 32. Xue YX, Luo YX, Wu P, et al. A memory retrieval‐extinction procedure to prevent drug craving and relapse. Science 2012;336:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liou YJ, Wang HH, Lee MT, et al. Genome‐wide association study of treatment refractory schizophrenia in Han Chinese. PLoS ONE 2012;7:e33598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russo SJ, Mazei‐Robison MS, Ables JL, Nestler EJ. Neurotrophic factors and structural plasticity in addiction. Neuropharmacology 2009;56(Suppl 1):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattson MP, Culmsee C, Yu Z, Camandola S. Roles of nuclear factor kappaB in neuronal survival and plasticity. J Neurochem 2000;74:443–456. [DOI] [PubMed] [Google Scholar]
- 36. Barker JM, Taylor JR, De Vries TJ, Peters J. Brain‐derived neurotrophic factor and addiction: pathological versus therapeutic effects on drug seeking. Brain Res 2014;doi: 10.1016/j.brainres.2014.10.058. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boules MM, Fredrickson P, Muehlmann AM, Richelson E. Elucidating the role of neurotensin in the pathophysiology and management of major mental disorders. Behav Sci (Basel) 2014;4:125–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J, Chen C, Chen C, et al. Neurotensin receptor 1 gene (NTSR1) polymorphism is associated with working memory. PLoS ONE 2011;6:e17365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arif A. Extraneuronal activities and regulatory mechanisms of the atypical cyclin‐dependent kinase Cdk5. Biochem Pharmacol 2012;84:985–993. [DOI] [PubMed] [Google Scholar]
- 40. Takahashi S, Ohshima T, Cho A, et al. Increased activity of cyclin‐dependent kinase 5 leads to attenuation of cocaine‐mediated dopamine signaling. Proc Natl Acad Sci U S A 2005;102:1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chergui K, Svenningsson P, Greengard P. Cyclin‐dependent kinase 5 regulates dopaminergic and glutamatergic transmission in the striatum. Proc Natl Acad Sci U S A 2004;101:2191–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai KO, Ip NY. Recent advances in understanding the roles of Cdk5 in synaptic plasticity. Biochim Biophys Acta 2009;1792:741–745. [DOI] [PubMed] [Google Scholar]
- 43. Beaudry H, Mercier‐Blais AA, Delaygue C, et al. Regulation of mu and delta opioid receptor functions: involvement of cyclin‐dependent kinase 5. Br J Pharmacol 2015;172:2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferrer‐Alcon M, La Harpe R, Guimon J, Garcia‐Sevilla JA. Downregulation of neuronal cdk5/p35 in opioid addicts and opiate‐treated rats: relation to neurofilament phosphorylation. Neuropsychopharmacology 2003;28:947–955. [DOI] [PubMed] [Google Scholar]
- 45. Lai KO, Ip NY. Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Curr Opin Neurobiol 2009;19:275–283. [DOI] [PubMed] [Google Scholar]
- 46. Li Y, Nie Y, Cao J, et al. G‐A variant in miR‐200c binding site of EFNA1 alters susceptibility to gastric cancer. Mol Carcinog 2014;53:219–229. [DOI] [PubMed] [Google Scholar]
- 47. Newton PM, Kim JA, McGeehan AJ, et al. Increased response to morphine in mice lacking protein kinase C epsilon. Genes Brain Behav 2007;6:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perlis RH, Huang J, Purcell S, et al. Genome‐wide association study of suicide attempts in mood disorder patients. Am J Psychiatry 2010;167:1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nestler EJ. Review. Transcriptional mechanisms of addiction: role of DeltaFosB. Philos Trans R Soc Lond B Biol Sci 2008;363:3245–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boyajyan A, Zakharyan R, Atshemyan S, Chavushyan A, Mkrtchyan G. Schizophrenia‐associated risk and protective variants of c‐Fos encoding gene. Recent Adv DNA Gene Seq 2015. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 51. Romero G, von Zastrow M, Friedman PA. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Adv Pharmacol 2011;62:279–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levran O, Peles E, Hamon S, et al. Nerve growth factor beta polypeptide (NGFB) genetic variability: association with the methadone dose required for effective maintenance treatment. Pharmacogenomics J 2012;12:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valjent E, Pages C, Herve D, Girault JA, Caboche J. Addictive and non‐addictive drugs induce distinct and specific patterns of ERK activation in mouse brain. Eur J Neurosci 2004;19:1826–1836. [DOI] [PubMed] [Google Scholar]
- 54. Girault JA, Valjent E, Caboche J, Herve D. ERK2: a logical AND gate critical for drug‐induced plasticity? Curr Opin Pharmacol 2007;7:77–85. [DOI] [PubMed] [Google Scholar]
- 55. Bruchas MR, Schindler AG, Shankar H, et al. Selective p38alpha MAPK deletion in serotonergic neurons produces stress resilience in models of depression and addiction. Neuron 2011;71:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 SNP list.
Table S2 Association results (P < 0.05).


