Abstract
The immediate early (IE) 62 protein is the major varicella-zoster virus (VZV) regulatory factor. Analysis of the VZV genome revealed 40 predicted GC-rich boxes within 36 promoters. We examined effects of ectopic expression of Sp1-Sp4 on IE62-mediated transactivation of three viral promoters. Ectopic expression of Sp3 and Sp4 enhanced IE62 activation of ORF3 and gI promoters while Sp3 reduced IE62 activation of ORF28/29 promoter and VZV DNA replication. Sp2 reduced IE62 transactivation of gI while Sp1 had no significant influence on IE62 activation with any of these viral promoters. Electrophoretic mobility shift assays (EMSA) confirmed binding of Sp1 and Sp3 but not Sp2 and Sp4 to the gI promoter. Sp1–4 bound to IE62 and amino acids 238–258 of IE62 were important for the interaction with Sp3 and Sp4 as well as Sp1. This work shows that Sp family members have differential effects on IE62-mediated transactivation in a promoter-dependent manner.
Keywords: IE62, VZV, Sp1, Sp2, Sp3, Sp4, transcription, promoters
INTRODUCTION
Varicella-zoster virus (VZV) is an alphaherpesvirus that causes two diseases, varicella or chickenpox during primary infection and herpes zoster or shingles upon reactivation from latency. It has a 125 kb linear double-stranded DNA genome that encodes at least 71 genes (Arvin and Gilden, 2013). VZV gene expression during lytic infection is thought to occur in three kinetic stages, immediate early (IE), early and late. VZV utilizes the host cell RNA polymerase II (RNA Pol II) and the general transcription machinery of the cell for viral gene transcription as do the other herpesviruses. A few VZV regulatory proteins, including IE62, IE4, ORF61, IE63 and ORF10 are responsible for efficient viral gene expression (Arvin and Gilden, 2013).
IE62, a tegument protein, is the major VZV transactivator, regulates the expression from all of the VZV promoters tested to date (Kinchington et al., 2000; and Arvin and Gilden, 2013) and also activates a variety of cellular promoters (Perera, 2000). It is a 1310 amino acid protein with five distinct domains, based on comparisons with other herpesvirus IE transactivators (Cheung, 1989). Of these domains, II and IV are the most conserved at the amino acid level, while domain I, III and V show significant variability. Among the functions mapped to the various domains of IE62, domain I (aa 1–467) contains an acidic activation domain essential for the transcription activation function of IE62. Domain II (aa468–640) contains a highly conserved DNA binding and dimerization domain. These two domains have been shown to be a platform for the physical interaction of IE62 with several cellular transcription factors and VZV proteins. The VZV proteins ORF9 and IE4 bind to the acidic activation domain (Cilloniz et al., 2007 and Spengler et al., 2000) while IE63 interacts with the DNA binding and dimerization domain (Lynch et al., 2002). The cellular TATA box binding protein (TBP) also interacts with the DNA binding domain while the cellular transcription factors USF and Sp1 bind to the acidic activation domain (Peng et al., 2003; and Rahaus et al., 2003).
The Sp/KLF family contains at least twenty members identified thus far, with the prototypes being Sp1, Sp2, Sp3 and Sp4. Members of this family bind with varying affinities to sequences designated as ‘Sp1 sites’ (e.g., GC-boxes and CACCC-boxes). Sp family proteins are characterized by a highly conserved C-terminal DNA-binding domain containing three zinc fingers (Philipsen and Suske, 1999). Although conservation within their DNA binding domains means that Sp1-related proteins can interact with the same DNA sequences, the family members vary in their interaction with different sequences. For example, Sp1, Sp3, and Sp4 bind with higher affinity to GC-boxes than to CACCC-boxes (Hagen et al., 1992; Hagen et al., 1995; Kingsley and Winoto, 1992; and Thiesen and Bach, 1990), whereas Sp2 binds preferentially to CACCC-boxes over GC-boxes (Crossley et al., 1996; Matsumoto et al., 1998; and Shields and Yang, 1998). While Sp1, Sp2 and Sp3 are ubiquitously expressed in different tissues, Sp4 is expressed primarily in neurons and cells of neuronal origin.
Sp1 binds to several VZV promoters (Peng et al., 2003; and Ruyechan et al., 2003). The GC rich sequence that binds Sp1 has been found in the promoters of VZV ORF61, viral glycoproteins gI and gE, the VZV major single-strand binding protein and the VZV DNA polymerase catalytic subunit (Wang et al., 2009; Beraraducci et al., 2008; Peng et al., 2003; and Yang et al., 2004). Recently we reported that Sp3, in addition to Sp1, binds to the downstream region of VZV oriS and to the ORF3 promoter. Mutation of their binding sites reduced ORF62, ORF63 and ORF3 expression in reporter gene assays (Khalil et al., 2012; and Khalil et al., 2013). The contributions of Sp2 and Sp4 to VZV gene expression and replication remain unknown. The binding of Sp3 to the Sp1 binding sites in some VZV promoters, raised a specific question; is the involvement of these GC-rich sequences in IE62-mediated transactivation due to the binding of Sp3 rather than or in concert with Sp1?
In the work presented here, we extend our study of the role of Sp family members in VZV replication by examining the influence of the ectopic expression of four Sp family members, Sp1, 2, 3 and 4, on IE62 regulation of viral promoters that we showed previously to contain functional Sp1 sites. We demonstrate that Sp3 increases IE62-mediated transactivation of ORF3 and gI promoters as well as the model luciferase reporter pSp1-TA-Luc, while it reduces IE62 activation of the ORF28/29 promoter. In contrast, Sp2 reduced IE62-mediated transactivation of model promoters and the gI promoter, a finding consistent with the inhibitory effects typical of Sp2. Surprisingly, the ectopic expression of Sp1 had no effect on IE62-mediated transactivation of any of the VZV promoters or model promoters. Of particular interest, the ectopic expression of Sp3, which inhibited the dual ORF28/29 promoters, also inhibited oriS dependent DNA replication in DpnI assays. This work suggests that the Sp family members have significant differential effects on IE62-mediated activation of VZV genes and, contrary to previous hypotheses, Sp3 may play a much more important role than Sp1.
MATERIALS AND METHODS
Cells and viruses
MeWo cells, a human melanoma cell line was grown in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (Spengler et al., 2000). VZV strain MSP was propagated in MeWo cell monolayers as described by Lynch et al. (2002) and Peng et al. (2003).
Whole cell lysate and nuclear extract preparation
Whole cell lysates of mock transfected or overexpressing MeWo cells were prepared in lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1 mM EDTA, 0.1% Triton X-100 and protease inhibitor cocktail (Roche, Mannheim, GE, added per the manufacturer’s instructions) and analyzed for Sp1, Sp2, Sp3, and Sp4 expression by immunoblot as previously described (Khalil et al., 2008). Antibodies against Sp1, Sp2, Sp3, Sp4, and β-tubulin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Quantification of the relative amounts of these transcription factors normalized to β-tubulin in loading controls was performed using a Bio-Rad GS700 imagining densitometer (Bio-Rad, Hercules, CA). Statistical significance was determined by a one-way analysis of variance, followed by Tukey’s post hoc test.
Nuclear extracts of VZV infected MeWo cells were prepared as previously described (Lynch et al., 2002). Cells were incubated in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol) at 4 °C for 15 min to lyse the cells and release the cytoplasmic fraction. After centrifugation, the crude nuclear pellet was incubated on ice in buffer C (20 mM HEPES, pH 7.9, 25% (v/v) glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol). After centrifugation, the nuclear extract was dialyzed against buffer D (20mM HEPES, pH 7.9, 20% (v/v) glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol).
Plasmids
Luciferase reporter plasmids containing the ORF3, ORF28/29 and gI promoters were constructed as described (Khalil et al., 2013; Yang et al., 2004 and White et al., 2010). Wild type ORF3-Luc was constructed by inserting a 340 bp intergenic region between ORF3 and ORF4 into the pGL2 basic vector, flanked by firefly luciferase. Wild type R28/29F was constructed by inserting a 221 bp intergenic region between ORF28 and ORF29 into the basic pGL2 luciferase vector containing the Renilla and firefly luciferase reporters. Wild type gI-Luc was constructed by inserting the gI (VZV ORF67) promoter sequence into the pGL2 basic vector, flanked by firefly luciferase.
A chimeric pTA-Luc vector was constructed as described in Yang et al., (2006) based on the pGL-2 basic vector (Promega, Madison, WI). The TATA element and flanking sequences derived from the adenovirus major late promoter were inserted between the XhoI and HindIII sites of the pGL-2 vector to generate the pTA-Luc minimal reporter vector. The pSp1-TA-Luc plasmid was generated by insertion of the Sp1 consensus site (5-GGGGTGTGGGCGGGC-3) from the downstream region of VZV oriS, into the pTA-Luc vector, 25 bp upstream of the TATA box, using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The primers used for insertion of the Sp1 site were: 5’ GGGGAGGTACCAGCTGGGGTGTGGGCGGGCCT TACGCGTGCTAG-3’ and 5’-CTAGCACGCGTAAGAGCCCGCCCACACCCCGCTCG GTACCTCCCC-3’.
A pCMV62 plasmid expressing ORF62 under the control of the cytomegalovirus immediate-early (IE) promoter has been described previously (Perera et al., 1992 and 1993). The pCMV62-d20 was derived from pCMV62 by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) to delete 20 amino acids (238 to 258) in the ORF62 gene. The pCMV-Sp1, pCMV-Sp2, pCMV-Sp3 and pCMV-Sp4 expressing plasmids were purchased from Origene Technologies (Rockville, MD).
The p-Litmus R62/63F plasmid containing the wild type 1.5 kb region of DNA between ORF62 and ORF63 was constructed as described by Jones et al., (2006). It contains the complete sequence between the ORF62 and ORF63 genes including the VZV OriS structure cloned in an orientation between genes encoding the Renilla and firefly luciferases (Promega) so that the luciferase genes acted as reporters of ORF62 and ORF63 transcription respectively.
The pGEX-IE62 series was constructed as previously described by Peng et al., 2003 and Spengler et al., 2000, using the pGEX-4T-3 plasmid (Amersham Biosciences, Piscataway, NJ) to encode the N-terminal portions of the VZV ORF62 protein with an N-terminal GST tag. These were pGEX-IE62 (1–226) and pGEX-IE62 (1–299) and were generated by inserting each of the respective ORF62 coding sequences between the BamHI and SalI sites in the pGEX-4T-3 vector. The pGEX-IE62 (1–299d20) was derived from pGEX-IE62 (1–299) by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) to delete 20 amino acids (238 to 258) in the ORF62 gene.
Reporter gene assays
Luciferase reporter gene assays were performed in MeWo cells as previously described (Yang et al., 2004). Transfections were performed using 12-well plates. 2 × 105 MeWo cells were seeded in each well 24 h before transfection. Cells were transfected with one microgram of each reporter vector using Lipofectamine reagent (Invitrogen, Carlsbad, CA), along with 5 ng of pEF1a–RL plasmid (Promega, Madison, WI) for Sp1-TA-Luc, TA-Luc, ORF3 and gI promoter experiments or 0.4 µg of β-galactosidase (β-Gal)-expressing plasmid (Invitrogen, Carlsbad, CA) for ORF28/29 promoter experiments, as controls of transfection efficiency. The reporter plasmids were also co-transfected using 5 ng of wild type or d20 pCMV-ORF62 expressing plasmids and 0.6 µg of pCMV-Sp1, pCMV-Sp2, pCMV-Sp3 and pCMV-Sp4. Dual luciferase activities were normalized to Renilla luciferase activities or β-Gal. pcDNA was transfected along with the pCMV62 plasmids to equalize the amounts of both total DNA and the promoter constructs in each set of transfections.
Cells were lysed 48 h post transfection or super-infection in 250 µl of lysis buffer (50 mM HEPES, pH 7.4, 250 mM NaCl, 1% NP-40, 1 mM EDTA). Control experiments without transfection of pCMV-ORF62 were carried out for each plasmid to determine basal expression levels. Dual-luciferase assays were performed according to the manufacturer’s instructions. Transfection experiments were repeated at least three times.
EMSA and supershift analyses
40 bp oligonucleotide probes (IDT, Coralville, IA) containing the Sp1 site of the gI or ORF28/29 promoters were used in electrophoretic mobility shift assays (EMSAs). Probes were end-labeled with ATP [a-32P] using T4 kinase (Invitrogen, Carlsbad, CA). One hundred femtomoles of the labeled probes (~1 × 105 dpm) were incubated with 15 µg of VZV infected MeWo cell nuclear extract in a 10 µl reaction mixture in binding buffer: 40 mM HEPES, pH 7.9, 100 mM NaCl, 10 mM MgCl2, 200 µg/ml bovine serum albumin (BSA), 12% glycerol, 0.05% NP-40, 1 mM dithiothreitol, and 3 µg poly (dI.dC). In the supershift assays, rabbit polyclonal anti-Sp2, Sp3 and Sp4 antibodies obtained from Santa Cruz Biologicals (Santa Cruz, CA) or anti-Sp1 antibodies (Upstate, Temecula, CA) were added in 4 µg aliquots to reaction mixtures before the addition of the labeled probe to the nuclear extract-antibody mixture. The samples were analyzed by electrophoresis on a 5% polyacrylamide (37.5:1 acrylamide/bisacrylamide) gel followed by autoradiography.
DpnI replication assays
MeWo cells were transfected with Lipofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Four µl of Lipofectamine reagent was used per µg of transfected DNA in each transfection. Transfections were performed in 100-mm-diameter dishes, with 2.1 × 106 MeWo cells per dish seeded in 12 ml of complete growth medium. The medium was replaced three hours before transfection and cells were 80% confluent at the time of transfection. Origin-dependent DNA replication experiments were performed as described previously (Khalil et al., 2008). In the ectopic expression experiments 6 µg of pCMV-Sp1, pCMV-Sp2, pCMV-Sp3 or pCMV-Sp4-expressing plasmids were transfected in the presence of 5 µg of wild type pLitmus R62/63F plasmid. At 6 h post-transfection, cells were superinfected with VZV strain MSP by adding a ratio of 0.4 infected cells per 1 uninfected cell to each monolayer. 6 µg of the empty cloning vector, pcDNA, were transfected along with the plasmids ectopically expressing the cellular transcription factors to equalize the amount of both total DNA and CMV promoter in each set of transfections. Total cellular DNA was prepared 48 h after superinfection, and the DNA was isolated by phenol-chloroform extraction followed by ethanol precipitation. The DNA was digested with DpnI and EcoRI (Stow and Davison, 1986), and analyzed by Southern blot hybridization. The blots were probed with a 476-bp PCR product prepared from pLitmus R62/63F by using primers 5′-TA GGCCACCACTTCAAGAACTCTGT-3′ and 5′-AGCAAAAGGCCAGCAAAAGGCCA GG-3′; the probe was end labeled with [α-32P] ATP by using T4 kinase (Invitrogen, Carlsbad, CA).
The resulting bands were quantified by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) analysis. The ratio of replicated plasmid to input plasmid represents the replication efficiency of the test plasmid. The data from representative experiments are presented as means of results from triplicate DpnI replication assays. Statistical significance was determined by a one-way ANOVA followed by Tukey’s post hoc test.
GST-tag protein affinity pull-down assays
Following induction with IPTG, crude lysates of E. coli expressing GST and GST-IE62 fusions were prepared and clarified as previously described (Peng et al., 2003). Two hundred microliter aliquots of the bacterial lysates were added to 50 µl glutathione-Sepharose beads and incubated for 1 h at 4°C. Then 250 µg of VZV infected MeWo cells nuclear extracts in 250 µl buffer D was added to the protein-coupled beads. Incubation was performed for 3 h at 4 °C and followed by repeated washings with 0.1% Triton X-100 in buffer D; bound protein was eluted with 80 µl 2x SDS-PAGE sample buffer by boiling for 5 min and then analyzed by SDS-PAGE and immunoblotting.
RESULTS
Predicted Sp1 binding sites within putative VZV promoters
The number of predicted Sp1 binding sites in putative VZV promoters was determined using the two Sp1 binding sites previously identified in the VZV ORF3 and gI promoters as the template for a bioinformatics search. These were 5′-CCCGCCC-3′ in the ORF3 promoter and 5′-CACGCCCC-3′ in the gI promoter (Peng et al., 2003; Khalil et al., 2013). The search was done with NCBI Blastn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and a conservative promoter size of 200 to 300 bp upstream from the translational start sites of the VZV genes was chosen. Forty Sp1 sites were identified within putative promoter regions (Table 1). Of the 36 promoters identified, we chose three that we had studied previously, (ORF3, ORF28/29 and gI), to assess how over-expression of Sp family members affected VZV genes transcription.
Table 1.
List of the VZV promoters that contain predicted Sp1 binding site(s).
| VZV gene | No. of Sp1 sites | Sp1 site sequence |
|---|---|---|
| ORF3 | 1 | CCCGCCC |
| ORF4 | 1 | CCGCC |
| ORF6 | 2 | GGAGCCGCCC CCGCC |
| ORF10 | 1 | CGGCGGG |
| ORF13 | 1 | GGCGG |
| ORF18 | 1 | GGAGGCGGG |
| ORF23 | 1 | GCACCGCCCC |
| ORF25 | 1 | CGGCGG |
| ORF28&29 | 1 | CCCCACGCCC |
| ORF32 | 1 | CCGCACCCG |
| ORF33 | 4 | GGCGGCG CGGCGGC CGGCGG GCCGCCC |
| ORF35 | 1 | CCGCCGCCG |
| ORF37 | 2 | CCCGCC GCCGCC |
| ORF38&39 | 1 | GGCGGGG |
| ORF40 | 2 | CCGCCCCCG CCCGCAC |
| ORF41 | 1 | CCACGCCC |
| ORF44 | 1 | CCCGGGCGGC |
| ORF45 | 1 | CCGCC |
| ORF46 | 1 | CCGCC |
| ORF50 | 1 | CGCCGCCG |
| ORF51 | 2 | GAGGCGGC CCGCACCC |
| ORF52 | 1 | CGGGGCGG |
| ORF56 | 1 | CGGCGG |
| ORF61 | 1 | GGCGG |
| ORF62 & ORF71 | 1 | CGAGGCGGG |
| ORF63 & ORF70 | 1 | CTCCCGCCCCGG |
| ORF64 | 1 | CCGCCCCCCC |
| ORF65&66 | 1 | CTCCGCCCTC |
| ORF67 | 1 | CACGCCCC |
| ORF68 | 1 | GGGCGGGG |
| ORF69 | 1 | GGGGGGGCGG |
Ectopic expression of Sp family members influences IE62-mediated transactivation differentially in a promoter dependent manner
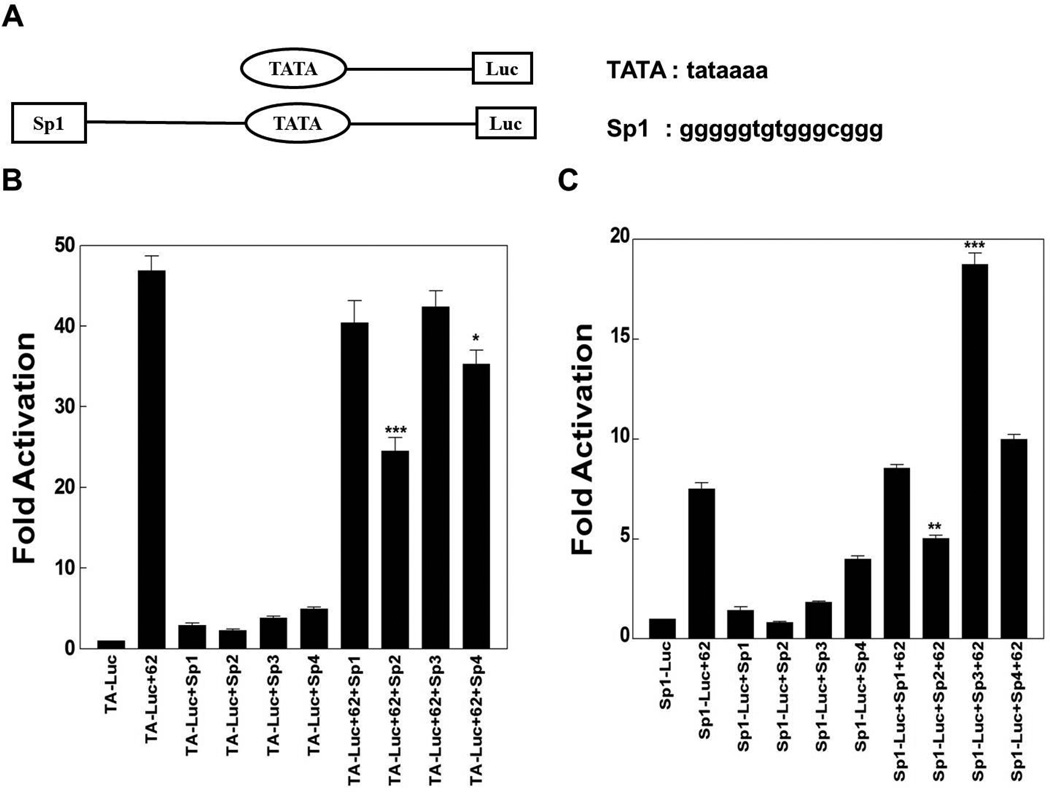
In the first set of experiments, reporter gene assays were done in the context of pCMV-62 transfection in the presence and absence of plasmids expressing Sp1, Sp2, Sp3 and Sp4, using two reporters containing model promoters. One model reporter, pTA-Luc, is a minimal promoter including only a TATA element. The second model reporter contains the Sp1 site identified in the downstream region of VZV oriS (Khalil et al., 2008) at a distance of 25 nucleotides upstream of the same TATA element (Fig. 1A). Luciferase activities obtained from each reporter plasmid in the absence of ORF62, but with ectopic expression of Sp family members, represented basal activity levels and were normalized to 1. Reporter gene activities in the presence of ORF62 transfection and/or plasmids over-expressing Sp1, Sp2, Sp3 and Sp4 were displayed as induction (n-fold) of luciferase activities relative to the basal activity.
Fig. 1.
The influence of ectopic expression of Sp1, Sp2, Sp3 and Sp4 on IE62 mediated transactivation of model promoters. A) Schematic of the model promoters and sequences of the consensus TATA element and the Sp1 site in the downstream region of VZV oriS. Results of triplicate transient transfection assays determining the effect of Sp family members’ overexpression on IE62 mediated transactivation are shown for B) the TA-Luc model promoter and C) the Sp1-TA-Luc model promoter. Luciferase activities in the absence of IE62 and ectopic expression of Sp family members were normalized to one. Promoter activities in the presence of IE62 and ectopic expression of Sp1, Sp2, Sp3, Sp4 or both are reported as induction (n-fold) of the luciferase activity over the basal activity. Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
As shown in Fig. 1B and C, IE62 activated the two model reporters, as did the Sp family members Sp1, Sp3 and Sp4, but to a lesser extent. Ectopic expression of Sp2 reduced IE62 activation of the two reporters significantly while Sp1 had no statistically significant influence. In contrast, Sp3 activated the IE62-mediated transactivation of pSp1-TA-Luc reporter significantly but had no significant effect on IE62 activation of the pTA-Luc reporter. Sp4 overexpression reduced IE62 activation of the pTA-Luc reporter significantly but had no statistically significant influence on IE62-mediated transactivation of the pSp1-TA-Luc reporter.
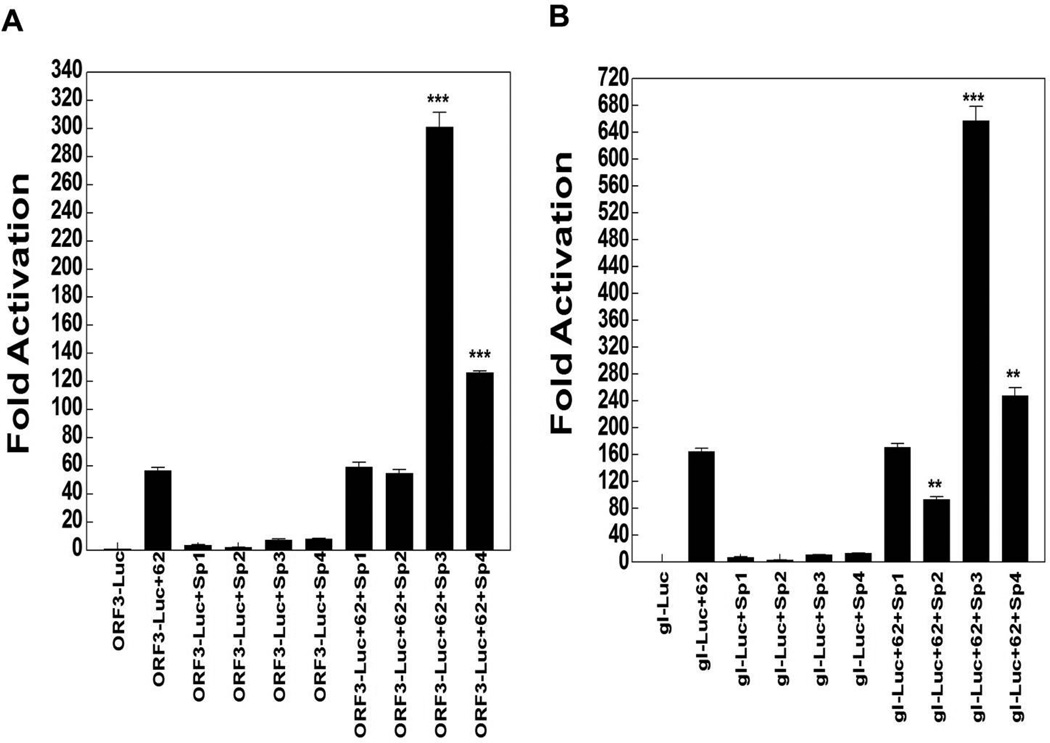
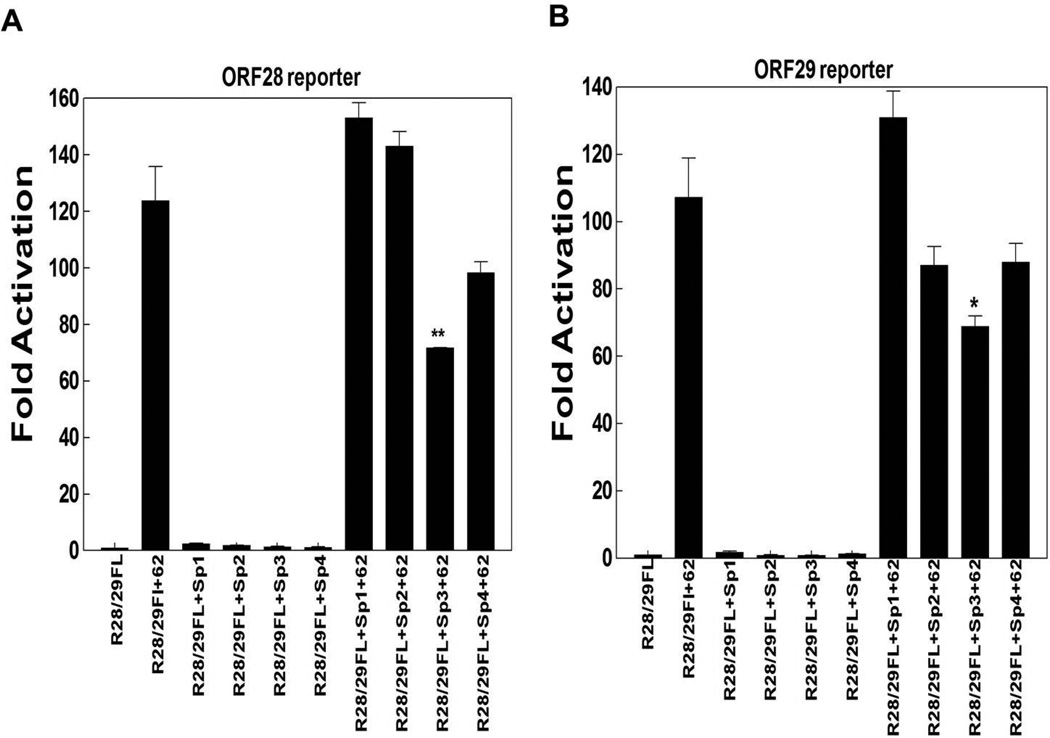
In the next series of experiments, we tested the ORF3, gI and ORF28/29 promoters in reporter gene assays. IE62 activated all of these reporters, while Sp1, Sp3 and Sp4 activated significantly ORF3 and gI promoters, as shown in Fig. 2. Sp1 overexpression activated ORF3 and gI promoter 4- and 8-fold respectively while Sp3 overexpression activated the two promoters 7- and 11-fold respectively. Also, Sp4 ectopic expression activated ORF3 and gI promoters 8- and 13-fold respectively. Ectopic expression of both Sp3 and Sp4 enhanced IE62-mediated transactivation of ORF3 and gI promoters by 6 fold and 2 fold respectively (Fig. 2A and B). However, Sp2 overexpression reduced IE62 activation of the gI promoter by about 50% but not the ORF3 promoter, while Sp1 had no statistically significant effect on IE62 activation of either of these two promoters. Interestingly, only Sp3 reduced significantly IE62-mediated transactivation of the ORF28/29 promoter by about 50% (Fig. 3A and B). In contrast, the inhibition of the IE62 activation of the ORF28/29 promoter by Sp2 and Sp4 overexpression was statistically insignificant.
Fig. 2.
The influence of ectopic expression of Sp1, Sp2, Sp3 and Sp4 on IE62 mediated transactivation of selected VZV promoters. Results from triplicate transient transfection assays showing the effect of the Sp family members on IE62 mediated transactivation are presented with A) ORF3 promoter, B) gI promoter. Luciferase activities in the absence of IE62 and ectopic expression of Sp family members were normalized to one. Promoter activities resulting from the presence of IE62 and the ectopic expression of Sp1, Sp2, Sp3, Sp4 or both are reported as induction (n-fold) of the luciferase activity over the basal activity. Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
Fig. 3.
The influence of ectopic expression of Sp1, Sp2, Sp3 and Sp4 on IE62 mediated transactivation of selected VZV promoters. Results from triplicate transient transfection assays showing the effect of the Sp family members on IE62 mediated transactivation are presented with A) ORF28 promoter, B) ORF29 promoter. Luciferase activities in the absence of IE62 and ectopic expression of Sp family members were normalized to one. Promoter activities resulting from the presence of IE62 and the ectopic expression of Sp1, Sp2, Sp3, Sp4 or both are reported as induction (n-fold) of the luciferase activity over the basal activity. Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
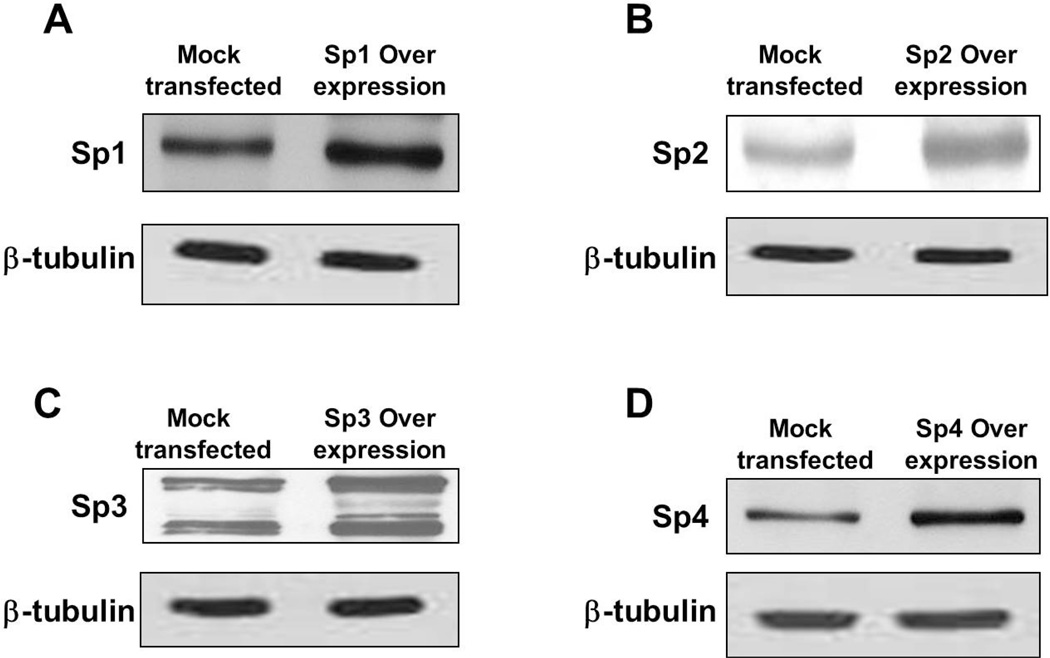
The increase in the expression levels of Sp family members in the overexpression experiments was determined using western blot analysis and was shown to be ~2-fold (Fig. 4).
Fig. 4.
Western blot analysis examining the levels of (A) Sp1, (B) Sp2, (C) Sp3 and (D) Sp4 in mock transfected and overexpression whole cell lysates of MeWo cells. Equal amounts of protein were added in each lane. β-tubulin was used as a loading control.
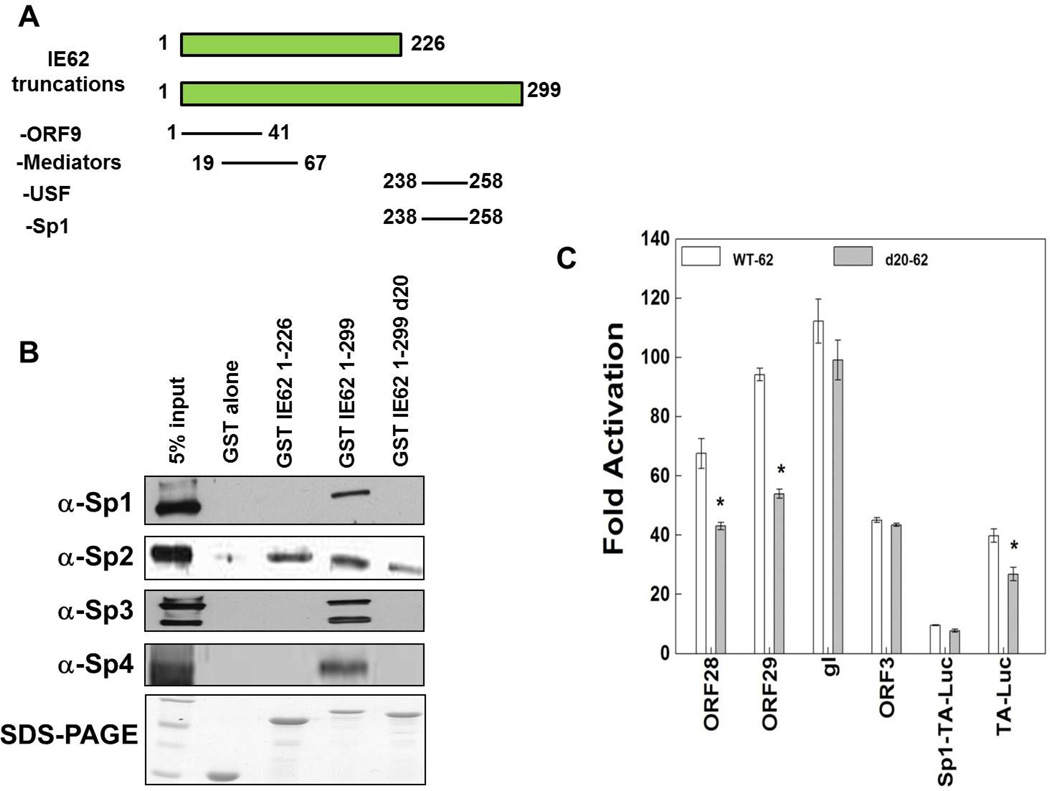
Sp family members interact physically with the IE62 acidic activation domain, and deletion of IE62 aa 238–258 affects the interaction, as well as IE62 mediated transactivation
A physical interaction has been demonstrated between Sp1 and IE62, and has been suggested to be part of the mechanism of joint Sp1-IE62 activation of VZV promoters (Peng et al., 2003). Based on our findings above, we next determined if there were physical interactions between IE62 and the other Sp family members Sp2, Sp3 and Sp4. Prior work had shown that Sp1 is capable of binding to IE62 in the absence of other viral proteins (Peng et al., 2003), and amino acids 226–299 of IE62 were necessary for the interaction with Sp1 in protein pull-down experiments.
GST-IE62 fusion proteins containing N-terminal fragments of IE62 used to show this interaction (Fig. 5A), including GST-IE62 (1–226), GST-IE62 (1–299), and GST-IE62 (1–299)d20 [in which amino acids 238–258 are deleted] were tested in screens utilizing VZV-infected MeWo cell nuclear extracts. These experiments showed that Sp3 and Sp4 bound to IE62 (1–299) and not IE62 (1–226). In addition, we narrowed down the binding region for all three factors to IE62 (238–258) (Fig. 5B). In contrast, Sp2 bound to IE62 (1–226).
Fig. 5.
The role of IE62 amino acids 238–258 in interaction with Sp family members and IE62-mediated transactivation. A) A schematic diagram shows the IE62 truncations used in the GST pull-down experiment and viral and cellular protein interactions-with these truncations. B) GST pull-down analyses of the region of IE62 that interacts with the cellular transcription factors, Sp1, Sp2, Sp3 and Sp4. Truncated GST-tagged IE62 proteins were coupled to glutathione-Sepharose beads and incubated with nuclear extracts of VZV infected MeWo cells. GST alone was used as a control. The binding of Sp1, Sp2, Sp3 and Sp4 to the GST-IE62 fusions was examined by Western blotting (upper panels). The lowest panel is a Coomassie blue stain showing the levels of the GST-tagged IE62 fusion proteins eluted from the glutathione beads. C) Results of the triplicate transient transfection assays determining the effect of the deletion of amino acids 238–258 of IE62 on IE62 mediated activation of the model and VZV promoters studied above.
Next, we examined the influence of the deletion of amino acids 238–258 on IE62 transcriptional activation. In this set of experiments, reporter gene assays were done in the context of transfection of either wild type pCMV-62 or pCMV-62 with a deletion of amino acids 238–258. We used the model reporters, pTA-Luc and pSp1-TA-Luc, as well as the ORF3-Luc, gI-Luc and R28/29F reporters. Luciferase activities obtained from each reporter plasmid in the absence of ORF62 transfection represented the basal level for the plasmid and were normalized to 1. Reporter gene activities in the presence of ORF62 transfection were reported as induction (n-fold) of luciferase activities relative to the basal activity. The deletion of amino acids 238–258 reduced IE62-mediated transactivation of ORF28/29 and pTA-Luc promoters significantly but had no influence on ORF3-Luc, gI-Luc and pSp1-TA-Luc, as shown in Fig. 5C.
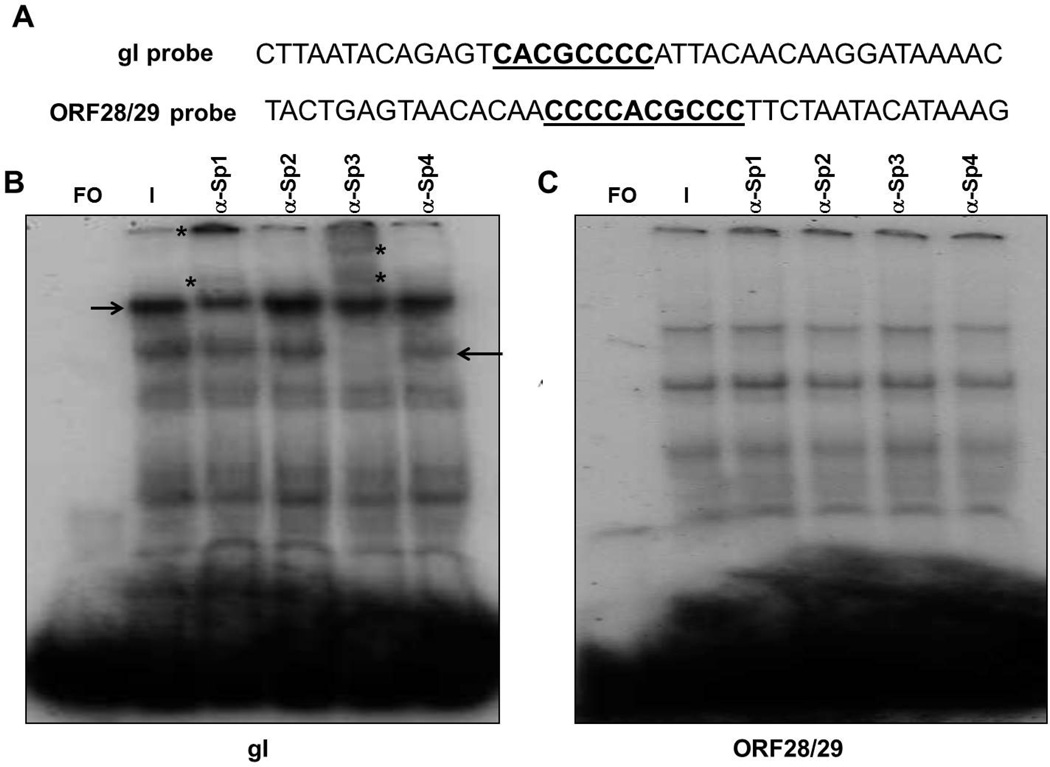
Sp1 and Sp3 bind to the predicted Sp1 binding site within the gI promoter but not to the ORF28/29 promoter
In these experiments, we used EMSA and supershift assays to establish the binding of the four Sp family members to predicted binding sites within the gI and ORF28/29 promoters. Experiments were done using 40 bp duplex oligonucleotides containing wild type Sp1 sites within these promoters and nuclear extracts from VZV infected MeWo cells (Fig. 6).
Fig. 6.
EMSA and supershift assays using 40 bp oligonucleotides containing the wild-type Sp1 site of the gI promoter or the ORF28/29 promoter. A) Sequences of the two oligonucleotides used in the assays. The Sp1 sites appear in bold underlined. B) EMSA and supershift assays using a 40 bp gI probe and C) EMSA and supershift assays using a 40 bp ORF28/29 probe. Supershift assays were done in the presence of VZV-infected cell nuclear extracts using polyclonal antibodies against Sp1, Sp2, Sp3 and Sp4. Lanes: FO, free oligonucleotide; I, VZV-infected nuclear extract. The positions of the original complexes are indicated by arrows and the positions of the supershifted bands are indicated by asterisks.
EMSA and supershift assays using oligonucleotides containing the predicted Sp1 binding site from the gI promoter revealed multiple complexes including two major faster-migrating complexes and two minor slower-migrating complexes (Fig. 6B). Antibody supershift assays were then performed to assess the binding of the Sp family members to this site. The anti-Sp1 antibody supershifted one of the major complexes while anti-Sp3 antibody supershifted the other major complex (Fig. 6B).
Next, a similar series of experiments was done to detect the binding of Sp family members to the ORF28/29 promoter. Again, numerous complexes were formed using the Sp1 site containing oligonucleotides from the ORF28/29 promoter (Fig. 6C). Antibody supershift assays were then performed to determine the binding of the Sp1, 2, 3 and 4 cellular transcription factors to these sequences. No supershifted band was detected in the presence of any of the four antibodies.
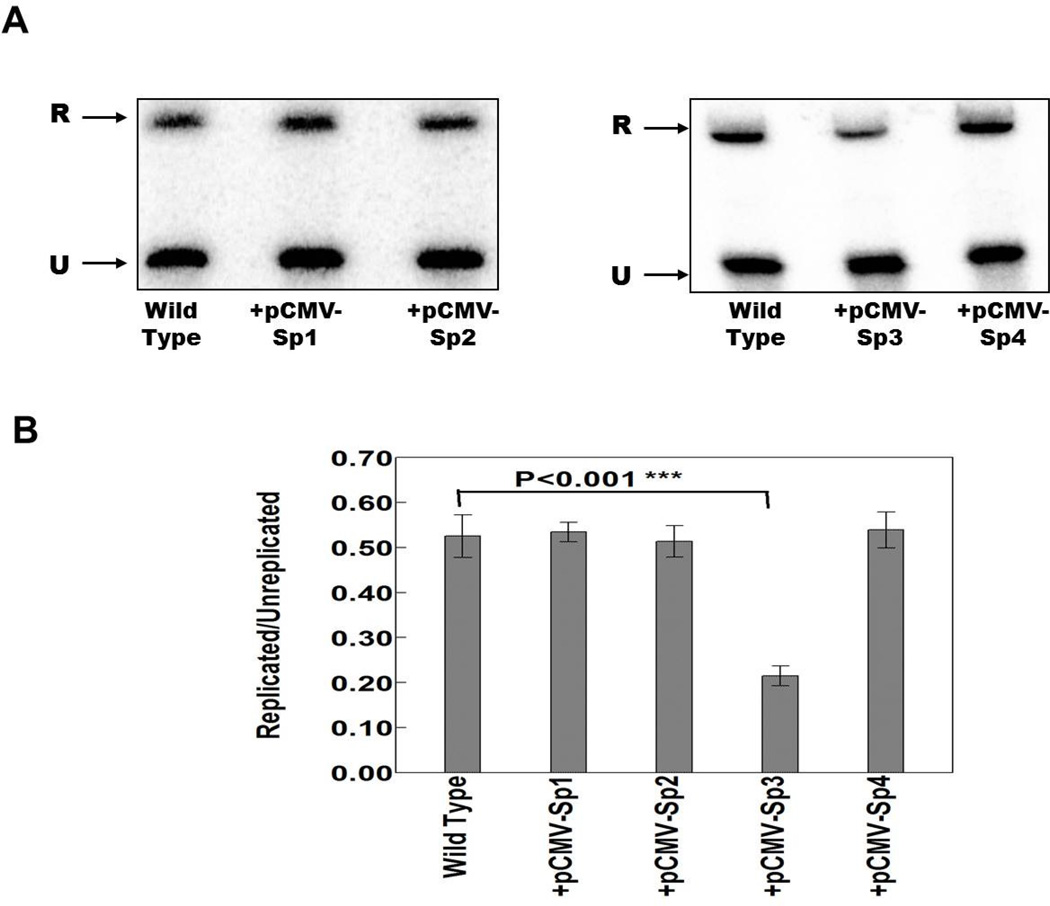
Ectopically expressed Sp3 decreases the efficiency of VZV origin-dependent DNA replication
In our previous work we established that Sp1 and Sp3 bound to the downstream region of the VZV oriS (Khalil et al., 2008). Mutation of their binding sites not only inhibited binding but also suppressed VZV origin-dependent DNA replication in DpnI assays (Khalil et al., 2012). We performed DpnI replication assays in the presence of ectopically expressed Sp family members in order to further examine the influence of these cellular transcription factors on VZV DNA replication. These experiments involved co-transfection of the pLitmus R62/63F plasmid (Khalil et al., 2011 and Khalil et al., 2012) containing the VZV oriS sequence, with individual plasmids expressing each of the Sp factors into MeWo cells, followed by VZV superinfection at a ratio of 0.4/1 infected to uninfected cells.
In the first series of experiments, triplicate DpnI assays were done using wild type pLitmus R62/63F plasmid with and without plasmids expressing Sp1, Sp2, Sp3 and Sp4. As shown in Fig. 7, only Sp3 was associated with a highly significant decrease in DNA replication. Sp1, Sp2 and Sp4 had no significant effects. The decrease observed with Sp3 was consistent with this factor acting as a repressor of ORF28 and ORF29 expression in reporter gene assays.
Fig. 7.
The effect of the ectopic expression of the cellular transcription factors Sp1, Sp2, Sp3 and Sp4 on origin dependent DNA replication from the wild type pLitmus R62/63F plasmid containing the VZV oriS. A) Typical southern blot analysis of the effects of the ectopic expression of Sp family members on oriS-dependent DNA replication. The upper band (R) indicates the position of DpnI resistant DNA resulting from replication within the MeWo cells. The lower band (U) indicates the position of unreplicated input plasmid. B) Histogram summarizing the data from three independent DpnI replication assays. Statistical significance was determined by a one-way ANOVA analysis of variance followed by Tukey’s post hoc test. Error bars indicate standard error.
DISCUSSION
All of the VZV promoters that have been examined to date are activated by the major virus transactivator IE62, either with or without a functional TATA box. There are at least two VZV promoters (for ORF3 and ORF10) that are known to have no TATA element (Khalil et al., 2013 and Che et al., 2007). In contrast, all known VZV promoters contain predicted or defined binding sites for one or more cellular transcription factors including ATF, USF, YY1 and Sp1. The binding of these factors to the viral promoters influences gene expression both in vitro and in vivo (Berarducci et al., 2007; Che et al., 2007; Ito et al., 2003; Meier et al., 1994; Narayanan et al., 2005; Peng et al., 2003; Rahaus and Wolff, 2003; Rahaus et al., 2003; Wang et al., 2009; Yang et al., 2004; Khalil et al., 2013 and Khalil et al., 2014).
Nine Sp family members, Sp1-Sp9, belong to the Sp/KLF family of transcription factors. These proteins bind with high affinity to GC-rich sequences (Suske, 1999). About half of the known or putative VZV promoters contain GC boxes that could function as Sp1 binding site, as shown in Table 1. Sp1 was the first Sp family member found to bind GC boxes in authentic VZV gene promoters, including those for gI, ORF28/29, gE and ORF61 and was assumed to be the major player in the Sp family for VZV.
Sp family members not only play an important role in VZV replication but also in replication of other herpesviruses. In the case of HSV-1, Sp1 and Sp3 are involved in the DNA replication process (Nguyen-Huynh and Schaffer, 1998). Mutations of the Sp1/Sp3 binding site in the HSV oriS decreased DNA replication efficiency by approximately 60%. Also, Sp1 binds to HSV promoters, including those for ICP-4, thymidine kinase, UL19, ICP34.5 and UL50, and is required for promoter activation (Jones et al., 1985; Jones and Tjian, 1985; Huang and Wagner, 1994; Chung et al., 1995; and Pande et al., 1998). In the case of HCMV, Sp1 binds to the DNA polymerase (UL54) promoter and is required for the activation of this promoter by HCMV immediate early proteins IE72 and IE86 (Luu and Flores, 1997). Moreover, there are two Sp1/Sp3 binding sites identified within the major immediate early (MIE) promoter of HCMV, and mutation of these sites inhibits transcription from this promoter (Isomura et al., 2005). In EBV, Sp1 and Sp3 interacts with the viral DNA polymerase and its processivity factor to stimulate DNA replication (Baumann et al., 1999; and Gruffat et al., 1995). Sp1 and Sp3 have been shown to bind to BRLF1 and ED-L1E promoters and binding site mutations inhibited activation of these promoters (Ragoczy and Miller, 2001; and Tsai et al., 1999).
In this study, we expanded our previous investigation of the role of Sp family members in VZV IE62-mediated transactivation, by determining the influence of ectopic overexpression of Sp1–4 on transcriptional activation of IE62 with model and VZV promoters containing GC-rich boxes. First, we identified the binding of Sp3 to the GC box of the gI promoter, which had been shown previously to be an Sp1-binding site. No binding of Sp2 and Sp4 to this sequence was observed. These results are similar to those described earlier with the ORF3 promoter and the downstream region of the VZV oriS. They also imply the need to retest all of the previously studied GC boxes in VZV promoters for the binding of other Sp family members.
Yang et al., (2004) demonstrated binding of Sp1 to the 221 bp intergenic region between ORF28 and 29, which includes the promoter elements of both genes, using magnetic bead recruitment assays. However, no other Sp family member was tested in these experiments. Mutation of the GC box (the same one that we tested in our EMSA and supershift assays for the binding of Sp family members) in the magnetic bead recruitment assay inhibited the interaction of Sp1 with this 221 bp sequence. Interestingly, none of the Sp family members tested bound to the GC box of the ORF28/29 promoter using EMSA and supershift assays under our experimental conditions. These results do not exclude completely the possibility of the binding of Sp1 (and probably other Sp family members) to the ORF28/29 promoter as shown in magnetic bead recruitment assays. The size of the duplex oligonucleotides used in the EMSA and supershift assays is 40 bp, which is short compared to the size of the probe (221 bp) used in the magnetic bead recruitment assays. This difference may affect the binding that might result from cooperative interaction of the Sp family members and the promoter, as shown by Yang et al., (2004).
Most surprisingly, ectopic overexpression of Sp1 had no effect on IE62 transactivation of any of the model or VZV promoters that we studied. In contrast, the overexpression of Sp3 enhanced both IE62-mediated transactivation of the model promoter containing the Sp1 site from the downstream region of VZV oriS, as well as VZV promoters that contained functional GC boxes (e.g. ORF3 and gI). Also, the ectopic expression of Sp3 reduced IE62 activation of the ORF28/29 promoters that do not contain a functional GC box. This all suggests that the positive influence of Sp3 on IE62 transcriptional activation is due to the binding of Sp3 to the promoter. These results also indicate that although Sp1 binds to all of the VZV promoters that contain GC boxes and physically interacts with IE62, it has no obvious role in IE62-mediated transactivation, at least with the promoters we have studied. This implies that the effects seen on activation of promoters following mutation of the GC rich element (e.g. ORF61, gE, Berarducci et al., 2007 and, Wang et al., 2009) are probably due to the binding of Sp3 rather than Sp1.
Sp1 and Sp3 are ubiquitously expressed in cell types and tissues that support VZV lytic replication but are present at low levels in neuronal cells and tissues where the virus is able to establish latency. About half of the 71 VZV promoters contain putative Sp family binding sites and Sp3 overexpression clearly shows substantial IE62 activation of the VZV promoters that contain functional GC boxes. This might explain why VZV gene expression is upregulated during lytic infection in tissues expressing Sp3 (and Sp1) at a high level, while it is down-regulated during latency in neuronal tissues that express both at only low levels.
The ectopic expression of Sp4 enhanced IE62 activation of gI and ORF3 promoters but the level was less than with Sp3. Sp4 is expressed in several cell lines at a level less than that of Sp1, Sp2 and Sp3, but is expressed more abundantly than other Sp family member in cells of neuronal origin (Lerner et al., 2005). Thus, it might play an important role in the reactivation of VZV from latency in these tissues by enhancing IE62-activation of VZV promoters. The overexpression of Sp2 displayed a negative influence over IE62-mediated transactivation of the model promoters and the gI promoter, which reflects the general transcriptional inhibitory influence of Sp2.
The inhibition by Sp3 of oriS-dependent DNA replication in the DpnI assays seems likely to be an indirect effect of Sp3 on IE62-mediated transactivation of the ORF28/29 promoter. We know from our prior work that mutation of the Sp1/Sp3 site in the downstream region of VZV oriS inhibits oriS-dependent DNA replication in MeWo cells (Khalil et al., 2012).
Sp2, Sp3 and Sp4 bind to domain I of IE62, as does Sp1. The amino acids 238–258 of IE62 are important for the physical interaction of Sp3 and Sp4 with IE62, but Sp2 binds to a different region of IE62 (amino acids 1–226) which includes the N-terminal acidic activation domain. The deletion of amino acids 238–258 of IE62 reduced significantly the activation by IE62 of the promoters that lack functional GC boxes (like the TA-Luc and ORF28/29 promoters) but had no effect on the promoters with a functional GC-rich element, such as the ORF3 and gI promoters and Sp1-TA-Luc. This latter effect may be due to the ability of Sp1 and Sp3 to bind to these promoters and to their ability to aid in the stabilization of the transcription complex in the absence of a physical interaction between IE62 and Sp1 and/or Sp3. On the other hand, with the TA-Luc and ORF28/29 promoters there is no binding of Sp1 or Sp3 and thus no physical interaction between IE62 and these two proteins, which could also influence the stabilization of the transcription complex. All the data on the Sp family members’ interactions with IE62 and VZV promoters, as well as their influence on IE62-mediated transactivation are summarized in Table 2.
Table 2.
Summary of the Sp family members’ activities and interactions.
| Sp1 | Sp2 | Sp3 | Sp4 | |
|---|---|---|---|---|
| IE62 interaction | + IE62 (238–258) |
+ IE62 (1–226) |
+ IE62 (238–258) |
+ IE62 (238–258) |
| DNA replication | ns | ns | −− | ns |
| ORF28/29 promoter | ns | ns | −− | ns |
| ORF3 promoter | ns | ns | +++ | + |
| gI promoter | ns | −− | +++ | + |
| TA-Luc | ns | −− | ns | − |
| Sp1-TA-Luc | ns | −− | ++ | ns |
| gI promoter binding | + | − | + | − |
(+) means increasing or interacting, (−) means inhibiting, (ns) means non significant influence.
Sp family members, Sp1–4, have been shown to play important roles during differentiation and development of human cells, as well as in cell cycle regulation. Sp1, Sp3 and Sp4 interact with several proteins involved in cell cycle regulation, such as E2F family members and retinoblastoma-like proteins (Rb and P130), as well as interacting with each other (e.g, Sp1 binds to both Sp3 and Sp4) (Karlseder et al., 1996; Rotheneder et al., 1999; Chang et al., 2001; and Zhang et al., 2003). Thus the possibility exists that part of the ability of these factors (specifically Sp3 and Sp4) to enhance IE62-mediated transactivation may be due to their ability to recruit additional cellular factors involved in transcriptional activation.
The results of this study suggest that, at least in MeWo cells, Sp3, rather than Sp1, may act as a major player in IE62-mediated transactivation of the VZV promoters containing a functional GC-rich sequence. Applying this idea to other VZV promoters that have been shown to contain functional GC-rich sequences (e.g. ORF61 and gE) will help in understanding this possible new role for Sp3 in VZV replication. The same may be true for other herpesviruses; for example, looking at binding of Sp1 and Sp3 to HSV oriS and EBV oriLyt, may open the door to investigating which of them (or both) influences herpesvirus replication in general.
The further study of cellular transcription factors affecting VZV replication is essential for a better understanding of how the lytic and latent stages of virus replication are controlled. In this study, we have established the importance of the Sp family members, in particular Sp3, in mediating VZV IE62 transcriptional activation, and in controlling viral replication.
-
-
Sp family members bind to IE62 and influence IE62-mediated transactivation.
-
-
Sp1 and Sp3 bind to many VZV promoters but only the over expression of Sp3 enhances IE62 transactivation.
-
-
Sp3 and Sp4 activation of IE62-mediated transactivation depends on the presence of functional GC box within the promoter.
ACKNOWLEDGEMENTS
This work was supported by grants AI018449, AI053846 and AI020459 from the National Institutes of Health, and grants from the John R. Oishei Foundation and the National Shingles Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Arvin AM, Gilden D. In: Varicella-Zoster Virus. Fields Virology. Howley PM, Knipe DM, editors. Vol. 2. Philadelphia: Lippincott Williams and Wilkins Press; 2013. pp. 2015–2184. [Google Scholar]
- 2.Baumann M, Feederle R, Kremmer E, Hammerschmidt W. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 1999;18:6095–6105. doi: 10.1093/emboj/18.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berarducci B, Sommer M, Zerboni L, Rajamani J, Arvin A. Cellular and viral factors regulate the varicella-zoster virus gE promoter during viral replication. J. Virol. 2007;81:10258–10267. doi: 10.1128/JVI.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang YC, Illenye S, Heintz NH. Cooperation of E2F–P130 and Sp1-pRb complexes in repression of the Chinese hamster dhfr gene. Mol. Cell Biol. 2001;21:1121–1131. doi: 10.1128/MCB.21.4.1121-1131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Che X, Berarducci B, Sommer M, Ruyechan WT, Arvin AM. The ubiquitous cellular transcriptional factor USF targets the varicella-zoster virus open reading frame 10 promoter and determines virulence in human skin xenografts in SCIDhu mice in vivo. J. Virol. 2007;81:3229–3239. doi: 10.1128/JVI.02537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung AK. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic acids Res. 1989;17:4637–4646. doi: 10.1093/nar/17.12.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung IK, Soisson SM, Muller MT. Clustering of Sp1 sites near the promoter region of ICP34.5 in herpes simplex virus type 1. J. Biol. Chem. 1995;117:19–22. doi: 10.1093/oxfordjournals.jbchem.a124708. [DOI] [PubMed] [Google Scholar]
- 8.Cilloniz C, Jackson W, Grose C, Czechowski D, Hay J, Ruyechan WT. The varicella-zoster virus (VZV) ORF9 protein interacts with the IE62 major VZV transactivator. J. Virol. 2007;81:761–774. doi: 10.1128/JVI.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruffat H, Renner O, Pich D, Hammerschmidt W. Cellular proteins bind to the downstream component of the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 1995;69:1878–1886. doi: 10.1128/jvi.69.3.1878-1886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J. Biol. Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- 13.Huang CJ, Wagner EK. The herpes simplex virus type 1 major capsid protein (VP5-UL19) promoter contains two cis-acting elements influencing late expression. J. Virol. 1994;68:5738–5747. doi: 10.1128/jvi.68.9.5738-5747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isomura H, Stinski MF, Kudoh A, Daikoku T, Shirata N, Tsurumi T. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 2005;79:9597–9607. doi: 10.1128/JVI.79.15.9597-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito H, Sommer MH, Zerboni L, He H, Boucaud D, Hay J, Ruyechan W, Arvin AM. Promoter sequences of varicella-zoster virus glycoprotein I targeted by cellular transactivating factors Sp1 and USF determine virulence in skin and T cells in SCIDhu mice in vivo. J. Virol. 2003;77:489–498. doi: 10.1128/JVI.77.1.489-498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones KA, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- 17.Jones KA, Yamamoto KR, Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 18.Jones JO, Sommer MH, Stamatis S, Arvin AM. Mutational analysis of the varicella-zoster virus ORF62/63 intergenic region. J. Virol. 2006;80:3116–3121. doi: 10.1128/JVI.80.6.3116-3121.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil MI, Hay J, Ruyechan WT. The cellular transcription factors Sp1 and Sp3 suppress varicella zoster virus origin-dependent DNA replication. J. Virol. 2008;82:11723–11733. doi: 10.1128/JVI.01322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil MI, Arvin A, Jones J, Ruyechan WT. A sequence within the varicella-zoster virus (VZV) OriS is a negative regulator of DNA replication and is bound by a protein complex containing the VZV ORF29 protein. J. Virol. 2011;85:12188–12200. doi: 10.1128/JVI.05501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalil MI, Robinson M, Sommer M, Arvin A, Hay J, Ruyechan WT. An Sp1/Sp3 site in the downstream region of varicella zoster virus oriS influences origin-dependent DNA replication and flanking gene transcription and is important for VZV replication in vitro and in human skin. J. Virol. 2012;86:13070–13080. doi: 10.1128/JVI.01538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalil MI, Sommer M, Arvin A, Hay J, Ruyechan WT. Regulation of the Varicella-zoster virus ORF3 promoter by cellular and viral factors. Virology. 2013;440:171–181. doi: 10.1016/j.virol.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil MI, Sommer M, Arvin A, Hay J, Ruyechan WT. Cellular transcription factor YY1 mediates the varicella-zoster virus (VZV) IE62 transcriptional activation. Virology. 2014;449:244–253. doi: 10.1016/j.virol.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinchington P, Fite K, Turse SE. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J. Virol. 2000;74:2265–2277. doi: 10.1128/jvi.74.5.2265-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingsley C, Winoto A. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J. Biol. Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- 28.Luu P, Flores O. Binding of Sp1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J. Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch JM, Kenyon TK, Grose C, Hay J, Ruyechan WT. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology. 2002;302:71–82. doi: 10.1006/viro.2002.1555. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto N, Laub F, Aldabe R, Zhang W, Ramirez F, Yoshida T, Terada M. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J. Biol. Chem. 1998;273:28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 31.Meier JL, Luo X, Sawadogo M, Straus SE. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol. Cell Biol. 1994;14:6896–6906. doi: 10.1128/mcb.14.10.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 2005;280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen-Huynh AT, Schaffer PA. Cellular transcription factors enhance herpes simplex virus type 1 oriS-dependent DNA replication. J. Virol. 1998;72:3635–3645. doi: 10.1128/jvi.72.5.3635-3645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pande NT, Petroski MD, Wagner EK. Functional modules important for activated expression of early genes of herpes simplex virus type 1 are clustered upstream of the TATA box. Virology. 1998;246:145–157. doi: 10.1006/viro.1998.9189. [DOI] [PubMed] [Google Scholar]
- 35.Peng H, He H, Hay J, Ruyechan WT. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J. Biol. Chem. 2003;278:38068–38075. doi: 10.1074/jbc.M302259200. [DOI] [PubMed] [Google Scholar]
- 36.Perera LP, Mosca JD, Sadeghi-Zadeh M, Ruyechan WT, Hay J. The varicella-zoster virus immediate early protein, IE62, can positively regulate its cognate promoter. Virology. 1992;191:346–354. doi: 10.1016/0042-6822(92)90197-w. [DOI] [PubMed] [Google Scholar]
- 37.Perera LP, Mosca JD, Ruyechan WT, Hayward GS, Straus SE, Hay J. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 1993;67:4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perera LP. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J. Biol. Chem. 2000;275:487–496. doi: 10.1074/jbc.275.1.487. [DOI] [PubMed] [Google Scholar]
- 39.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragoczy T, Miller G. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 2001;75:5240–5251. doi: 10.1128/JVI.75.11.5240-5251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahaus M, Desloges N, Yang M, Ruyechan WT, Wolff MH. Transcription factor USF, expressed during the entire phase of varicella-zoster virus infection, interacts physically with the major viral transactivator IE62 and plays a significant role in virus replication. J. Gen. Virol. 2003;84:2957–2967. doi: 10.1099/vir.0.19335-0. [DOI] [PubMed] [Google Scholar]
- 42.Rahaus M, Wolff MH. Reciprocal effects of varicella-zoster virus (VZV) and AP1: activation of jun, fos and ATF-2 after VZV infection and their importance for the regulation of viral genes. Virus Res. 2003;92:9–21. doi: 10.1016/s0168-1702(02)00310-6. [DOI] [PubMed] [Google Scholar]
- 43.Rotheneder H, Geymayer S, Haidweger E. Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J. Mol. Biol. 1999;293:1005–1015. doi: 10.1006/jmbi.1999.3213. [DOI] [PubMed] [Google Scholar]
- 44.Ruyechan WT, Peng H, Yang M, Hay J. Cellular factors and IE62 activation of VZV promoters. J. Med. Virol. 2003;70:S90–S94. doi: 10.1002/jmv.10328. [DOI] [PubMed] [Google Scholar]
- 45.Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spengler ML, Ruyechan WT, Hay J. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology. 2000;272:375–381. doi: 10.1006/viro.2000.0389. [DOI] [PubMed] [Google Scholar]
- 47.Stow ND, Davison AJ. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J. Gen. Virol. 1986;67:1613–1623. doi: 10.1099/0022-1317-67-8-1613. [DOI] [PubMed] [Google Scholar]
- 48.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 49.Thiesen HJ, Bach C. Target Detection Assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on Sp1 protein. Nucleic acids Res. 1990;18:3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai CN, Lee CM, Chien CK, Kuo SC, Chang YS. Additive effect of Sp1 and Sp3 in regulation of the ED-L1E promoter of the EBV LMP 1 gene in human epithelial cells. Virology. 1999;261:288–294. doi: 10.1006/viro.1999.9851. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Sommer M, Rajamani J, Arvin AM. Regulation of the ORF61 promoter and ORF61 functions in varicella-zoster virus replication and pathogenesis. J. Virol. 2009;83:7560–7572. doi: 10.1128/JVI.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White K, Peng H, Hay J, Ruyechan WT. Role of the IE62 consensus binding site in transactivation by the varicella-zoster virus IE62 protein. J. Virol. 2010;84:3767–3779. doi: 10.1128/JVI.02522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M, Hay J, Ruyechan WT. The DNA element controlling expression of the varicella-zoster virus open reading frame 28 and 29 genes consists of two divergent unidirectional promoters which have a common USF site. J. Virol. 2004;78:10939–10952. doi: 10.1128/JVI.78.20.10939-10952.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang M, Peng H, Hay J, Ruyechan WT. Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF. J. Virol. 2006;80:7339–7353. doi: 10.1128/JVI.00309-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Li Y, Dai C, Yang J, Mundel P, Liu Y. Sp1 and Sp3 transcription factors synergistically regulate HGF receptor gene expression in kidney. Am. J. Physiol. Renal Physiol. 2003;284:F82–F94. doi: 10.1152/ajprenal.00200.2002. [DOI] [PubMed] [Google Scholar]