Abstract
Here we investigated the binding of Dengue virus envelope glycoprotein domain III (DIII) by two broadly neutralizing antibodies (bNAbs), 4E11 and 4E5A. There are four serotypes of Dengue virus (DENV-1 to -4), whose DIII sequences vary by up to 49%. We used combinatorial alanine scanning mutagenesis, a phage display approach, to map functional epitopes (those residues that contribute most significantly to the energetics of antibody-antigen interaction) on these four serotypes. Our results showed that 4E11, which binds strongly to DENV-1, -2, and -3, and moderately to DENV-4, recognized a common conserved core functional epitope involving DIII residues K310, L/I387, L389, and W391. There were also unique recognition features for each serotype, suggesting that 4E11 has flexible recognition requirements. Similar scanning studies for the related bNAb 4E5A, which binds more tightly to DENV-4, identified broader functional epitopes on DENV-1. These results provide useful information for immunogen and therapeutic antibody design.
Keywords: Broadly neutralizing antibodies, Dengue virus, phage display, combinatorial scanning mutagenesis, Flavivirus
Graphical Abstract
INTRODUCTION
The specific recognition of an antigen by an antibody is the hallmark of humoral immunity; such interactions are essential for the function of numerous antibody or antibody-like molecules for use as therapeutic, diagnostic, or research reagents (French et al., 1989; Milstein and Rada, 1995; Neuberger, 2002). Significant effort has been expended toward understanding the physicochemical basis of high affinity antibody-antigen interactions (Da Silva et al., 2010; Fellouse et al., 2007; Fellouse et al., 2005; Fellouse et al., 2004; Persson et al., 2013), how these interactions develop through the process of affinity maturation in naturally-occurring antibodies, (Li et al., 2003; Wedemayer et al., 1997; Yin et al., 2003; Yin et al., 2001) and how the information from these studies can be exploited for functional design by combinatorial or computational protein engineering (Fellouse et al., 2004; Persson et al., 2013; Tharakaraman et al., 2013).
An added layer of complexity is that some antibody combining sites, natural or engineered, have “multispecific” properties in that they are able to bind in high affinity to multiple antigens with little or no non-specific activity against irrelevant targets (Bostrom et al., 2011; Bostrom et al., 2009; Cockburn et al., 2012a; Scheid et al., 2011; West et al., 2012; Wu et al., 2011). For engineered bispecific antibodies, this expanded recognition appears to require separate “functional paratopes” (i.e., those residues on the antibody combining site that contribute most to the energy of the binding reaction) for each of the binding targets (Bostrom et al., 2011; Bostrom et al., 2009). Prime examples of multispecific natural antibodies are broadly neutralizing virus antibodies (bNAbs), which are frequently derived from immunization or by mining antibody repertoires of vaccinees or survivors and can bind to glycoproteins with diverse primary sequences from different viral species or strains (Bostrom et al., 2011; Bostrom et al., 2009; Cockburn et al., 2012a; Scheid et al., 2011; Wu et al., 2011). In general, bNAbs target conserved and sometimes compact epitopes on viral glycoproteins (Ekiert et al., 2009; Wu et al., 2011), but, in some cases, larger structural paratopes (antigen contacting residues as determined from X-ray structures) engage structural epitopes that include positions that contain high antigenic diversity among viral strains or species (West et al., 2012). In several viral systems including Dengue, HIV-1, and influenza, a wealth of structural information on bNAbglycoprotein complexes provides some insight into the requisite features of the structural epitopes. However, less effort has been devoted toward understanding the functional features (i.e., epitope residues that contribute most significantly to the energy of the bNAb-glycoprotein interaction) by mutagenesis studies (Bedouelle et al., 2006; Lisova et al., 2014; Lisova et al., 2007).
Here, we obtained comprehensive insight into functional epitope requirements for recognition of Dengue virus (DENV) envelope glycoprotein E domain III (DIII) by two related broadly neutralizing antibodies, 4E11 and 4E5A, as a model system (Figure 1) (Bedouelle et al., 2006; Cockburn et al., 2012a; Lisova et al., 2014; Lisova et al., 2007; Tharakaraman et al., 2013). There are four co-circulating serotypes of Dengue virus (DENV-1, -2, -3, and -4), a mosquito-spread flavivirus that causes significant disease in endemic subtropical regions (Dengue Shock Syndrome, DSS, and Dengue Hemorrhagic Fever, DHF) (Gubler et al., 2007; Halstead and O’rourke, 1977; Sangkawibha et al., 1984). A particular challenge for DENV vaccine and immunotherapeutic development is antibody-dependent enhancement of infection (ADE), a phenomenon whereby serotype-specific antibodies resulting from a primary infection worsen the severity of secondary infections caused by other serotypes. In the current model for ADE, primary infection results in a febrile illness and subsequent lifelong immunity against the infecting serotype. However, antibodies from the primary infection are weakly cross-reactive and non-neutralizing against the other serotypes and lead to increased uptake of DENV by Fcγ-receptor positive cells and thus increased viremia and pathology upon secondary infection (Halstead and O’rourke, 1977; Sangkawibha et al., 1984). There is significant interest in identification and characterization of potent and broadly neutralizing antibodies, which would inhibit the infectious cycle of all serotypes and prevent ADE.
Figure 1.
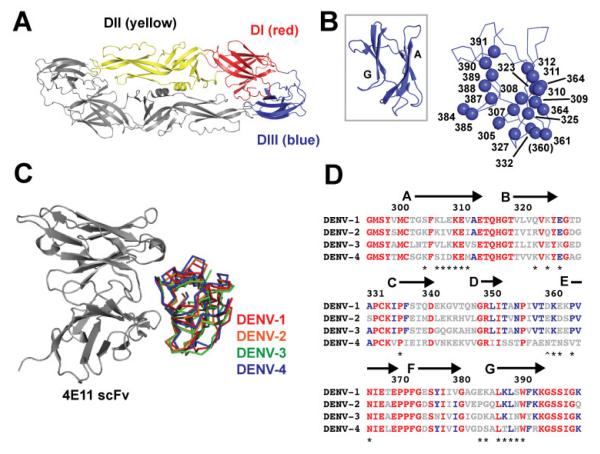
Structure of DENV DIII and library design. (A) Prefusion structure of homodimeric E, with the three domains on one of the subunits colored (DENV-2 shown as an example, from PDB code 1OK3, ref. 58). (B) View of the DENV-2 DIII as an example down the 4E11 structural epitope. The A- and G-strands are labeled in the inset, and the locations of the residues included in the combinatorial alanine scanning mutagenesis analysis are shown on the larger graphic, which has the same orientation as the inset. Position 360 was included in the analysis for DENV-2 only. (C) Overlay of the four DENV DIII domains bound to the 4E11 scFv (PDB codes 3UZQ, 3UZV, 3UZE, and 3UYP, ref. 18). (D) Amino acid alignment of the four DENV DIII domains. Residues that are absolutely conserved are colored red, those that are conserved in three of the serotypes are colored blue. The location of the β-strands is indicated with arrows; the residues subjected to combinatorial scanning mutagenesis are indicated with asterisks except for position 360 (^), which was analyzed in DENV-2 only.
A number of murine-derived DENV neutralizing antibodies, both serotype-specific as well as bNAbs, target DIII. However, potent bNAbs against other regions of the glycoprotein, such as the glycoprotein prefusion dimer interface, arise during the course of natural human infection (Austin et al., 2012; Beltramello et al., 2010; Brien et al., 2010; Dejnirattisai et al., 2015; Edeling et al., 2014; Gentry et al., 1982; Gromowski et al., 2010; Henchal et al., 1985; Kaufman et al., 1987; Li et al., 2012; Lok et al., 2008; Megret et al., 1992; Roehrig et al., 1998; Rouvinski et al., 2015; Shrestha et al., 2010; Sukupolvi-Petty et al., 2007; Wahala and de Silva, 2011; Williams et al., 2012). Glycoprotein E mediates entry into host cells and is located on the surface of the virus in an array of 90 homodimers parallel to the virus membrane (Figure 1A) (Kuhn et al., 2002). The E ectodomain is composed of three domains: domain I (DI) functions as a molecular hinge that links domains II and III to one another, domain II (DII) is the dimerization domain and contains the hydrophobic fusion loop necessary for virus fusion with host cell endosomal membranes, and domain III (DIII) has been implicated in receptor binding (Crill and Roehrig, 2001; Kuhn et al., 2002; Rey et al., 1995). DIII connects to a stem region and the transmembrane (TM) domain. After adsorption to host cells, DENV is taken up via receptor-mediated endocytosis and infects through pH-dependent virus membrane fusion in the late endosome (Harrison, 2008; van der Schaar et al., 2008; Zaitseva et al., 2010). Monoclonal antibody (mAb) 4E11 is a murine antibody that was isolated from a DENV-1 vaccinated mouse, binds to DIII and neutralizes all four serotypes in vitro albeit with less affinity and potency for DENV-4 (Cockburn et al., 2012a; Megret et al., 1992). MAb 4E11 was subsequently used as a template for computational redesign to develop a variant, 4E5A, with increased affinity for DENV-4 while maintaining affinity across the other three serotypes. The bNAb 4E5A has a 45-fold improved affinity and 75-fold improved neutralization potency for DENV-4 compared with 4E11 (Tharakaraman et al., 2013). In a recent report, additional computational optimization of 4E5A yielded Ab513, which exhibited further enhanced potency against DENV-4, was neutralizing across broad genotypes within the four serotypes, and was protective against clinical features of Dengue-related infections in humanized mice (Robinson et al., 2015). Thus, there is considerable interest in 4E11 and related mAbs as potential immunotherapeutic agents.
Co-crystal structures of the 4E11 single chain variable fragment (scFv) in complex with DIII from each serotype revealed that the mAb binds in a similar location for all four serotypes (Figure 1C), and that a key aspect of recognition involves charge complementarity between the 4E11 paratope and DIII epitopes (Cockburn et al., 2012a). Limited scanning mutagenesis studies to dissect critical epitope-contributing residues on DIII for binding and neutralization to these and other related DIII mAbs have previously been described (Bedouelle et al., 2006; Lisova et al., 2007). However, we sought to leverage the high-throughput nature of combinatorial alanine scanning, a phage display technique, to gain deeper insight into similarities and differences among the recognition requirements for binding of 4E11 and 4E5A to varying serotypes (Bogan and Thorn, 1998; Clackson and Wells, 1995; Pál et al., 2005; Weiss et al., 2000). Furthermore, we sought to correlate binding requirements with neutralization breadth for both 4E11 and 4E5A to understand if and how the hotspot profile for DENV-4 differs for bNAb 4E5A compared with that of 4E11. The results reveal that recognition of DIIIs by 4E11 requires diverse functional epitopes, but that there is a common conserved core. Furthermore, 4E5A has extended functional epitopes on both DENV-1 and -4 DIII in comparison to 4E11, suggesting that its increased breadth requires important interactions with a broader set of residues at the epitope interface. These results provide information on the features that can be required for broad recognition by antibodies, and reveal functional details that can be used to help guide DENV DIII-based immunogen or therapeutic antibody design.
MATERIALS AND METHODS
Expression and purification of bNAbs 4E11 and 4E5A
The hybridoma cell line for 4E11 was provided by Dr. Fernando Arenzana-Seisdedos (Institute Pasteur, Paris). To purify 4E11 IgG, hybridoma cell supernatants were centrifuged at high speed to remove cell debris. 4E11 was then purified from the clarified supernatants by using protein A affinity agarose beads (Pierce, ThermoScientific, Rockford, IL) and the Gentle Antibody Elution System (Pierce, ThermoScientific, Rockford, IL) as per manufacturer’s protocol. 4E11 was desalted into 150 mM HEPES and 200 mM NaCl pH = 7.4 for use in experiments. The heavy and light chain variable domains of 4E5A were cloned into modified pMAZ-IgH and -IgL vectors (Mazor et al., 2007). These vectors were modified to express murine IgG2A constant domains 1, 2, and 3, and murine kappa light chain, respectively. 4E5A IgH and IgL were co-transfected into HEK 293F cells (Invitrogen, Grand Island, NY) using polyethylenimine (PEI) (Polysciences, Warrington, PA), and incubated for 5 days at 37°C with 8% CO2. IgGs were then purified and desalted as described above for the following experiments.
Phage display of DENV DIII
The HP153 vector was used to display the four DIII antigens on the surface of filamentous bacteriophage (Persson et al., 2013). The E. coli codon-optimized synthetic genes for DIII from each serotype (DENV-1: Guiana/FGA89/1989; DENV-2: Jamaica/1409/1983; DENV-3: Thailand/PaH881/1988; DENV-4: Burma/63632/1976) containing an N-terminal FLAG tag were obtained from Genewiz (South Plainfield, NJ). The DIII constructs were cloned into phagemid HP153 using NsiI and FseI restriction sites. This cloning strategy results in the bivalent display of DIII on the surface of phage as a fusion to the minor phage coat protein pIII with an N-terminal FLAG epitope for detection by the monoclonal antibody M2, all under the control of the pho A promoter. Phage particles expressing DIII were produced by separately electroporating each of the four phagemids (corresponding to DIII from DENV-1, -2, -3, and -4) into E. coli XL1-Blue cells and growing for 5 hours at 37°C in 2xYT media containing 10 μg/mL tetracycline and 50 μg/mL carbenicillin. This culture was coinfected with 1010 plaque forming units (PFU) of M13K07 helper phage for 1 hour at 37°C. 50 μg/mL of kanamycin was then added and the culture was grown overnight at 37°C. Phage were precipitated by addition of 4% (w/v) polyethylene glycol (PEG) 8000 and 3% (w/v) NaCl after removal of cells by centrifugation. Precipitated phage were centrifuged and subsequently resuspended in PBS containing 1% (w/v) BSA.
Functional display of DIII on the surface of phage was confirmed using a phage enzyme-linked immunosorbent assay (ELISA) in which 4E11 and 4E5A were coated on Costar EIA/RIA high-binding plates (Fisher Scientific, Nepean, ON, Canada) at 0.5 μg per well in PBS pH = 8.0 overnight at 4°C. All further incubations were completed at 37°C. Plates were blocked with 1% (w/v) BSA in PBS (pH = 7.4) for 2 hours. Wells were washed with PBS containing 0.05% (v/v) Tween-20 (PBS-T) and DIII-expressing phage were then added to the wells for 1 hour. Wells were washed 5 times with PBS-T. An anti-M13 HRP-conjugated antibody was then allowed to bind for 1 hour. The wells were washed with PBS-T and the anti-M13-HRP conjugate was detected using 3, 3′, 5, 5′-tetramethylbenzidine (TMB) (Sigma-Aldrich, St. Louis, MO).
Preparation of the DIII combinatorial scanning mutagenesis libraries
The structural epitope of each DIII serotype was split into two libraries in which the residues were allowed to vary between the wildtype (WT) identity and alanine (Ala) using degenerate codons based on the alanine scanning code (Weiss et al., 2000). This resulted in the production of 8 alanine-scanning libraries (two per serotype). For DENV-1 and -2, inactive templates, in which structural epitope residues were replaced with AGA or AGG rare arginine codons, were produced to serve as the templates for library synthesis. For DENV-3 and -4, the inactive template contained TAA stop codons in place of structural epitope residues. Kunkel mutagenesis was performed as previously described to replace inactive rare Arg or stop codons with library DNA using 5′-phosphorylated primers listed in the Supplemental Information (Sidhu and Weiss, 2004). Kunkel mutagenesis reactions contained 10 μg of uridine-enriched single stranded template DNA and a 3-fold excess of each library primer. Library DNA was subsequently purified and electroporated into high efficiency electrocompetent E. coli SS320 cells to yield from 1.4×107 to 5.6×1010 library clones. Each library synthesis resulted in the production of library sizes that exceeded the theoretical diversity by at least 100-fold; therefore, all sequences were adequately represented.
Library screening and analysis
Each alanine-scanning library (8 in total) was subjected to three rounds of selection against 4E11 or 4E5A (functional selection) and one to three rounds of selection against the anti-FLAG antibody, M2, to control for display bias (display selection). For the functional selections, each round consisted of coating seven wells with 0.5 μg of 4E11 or 4E5A and one well with PBS overnight at 4°C. Wells were blocked with 1% (w/v) BSA in PBS for 2 hours and library phage were then allowed to bind for 1 hour before washing 5 times with PBS-T. Bound phage were eluted by incubating with 100 mM Glycine pH = 2.0 for 5 minutes at room temperature (RT), and neutralizing with 2 M Tris pH = 7.5. M2 selections were performed similarly, except seven wells were coated with a 1:500 dilution of M2 in PBS pH = 8.0 overnight. After each round of selection, eluted phage were amplified by infecting E. coli XL1-Blue cells, coinfecting with M13K07 helper phage and precipitated as described above for further rounds of selection.
To analyze individual clones for binding to 4E11 and 4E5A individual clones were grown in 1 mL 2xYT media supplemented with 1010 PFU M13K07 helper phage, and 100 μg/mL carbenicillin in 96 deep-well plates overnight at 37°C. Cells were removed by centrifugation and the phage-containing supernatants were applied to plates containing 4E11, 4E5A or BSA. Wells were washed 5 times with PBS-T and were incubated with an anti-M13-HRP conjugate for 1 hour. Wells were washed with PBS-T and bound clones were detected using TMB substrate. Clones that demonstrated an ELISA-positive signal 10-fold higher for the 4E11 and 4E5A plates compared with the BSA control plates were sent to Genewiz for sequencing. The M2 display sorts were performed similarly, except the wells were coated with a 1:500 dilution of M2. The WT/Ala ratio was calculated at each position of diversity for each library and converted to a ΔΔG value using the equation: ΔΔG = RT ln (WT/Ala) for both the functional selection (4E11 or 4E5A) and the display selection (M2) as previously described (Pál et al., 2005; Weiss et al., 2000). ΔΔGAla-WT values were calculated as follows: ΔΔGAla-WT = ΔΔG (functional selection) – ΔΔG (display selection).
Single point mutations and competition ELISAs
Selected single point mutations for DENV-1 and DENV-2 DIII were prepared using Kunkel mutagenesis and the wildtype uridine-enriched single stranded DENV-1 or DENV-2 DIII phagemid DNA (dU-ssDNA). Mutation was confirmed by sequencing individual clones. Clones containing the correct mutations were electroporated into E. coli XL1-Blue cells as described above. Single point mutant phage clones were amplified and precipitated as described previously. Phage competition ELISAs were performed to determine the binding affinities to 4E11 of the mutant DENV-1 and DENV-2 DIII phage by competing with soluble WT DENV-1 or DENV-2 DIII, respectively. For competition ELISAs, 4E11 was coated at 0.5 μg per well overnight at 4°C. Plates were blocked with 1% (w/v) BSA in PBS for 2 hours and then washed 5 times with PBS-T. A 1:10 dilution of mutant or WT DENV-1 and DENV-2 DIII expressing phage was then incubated with varying concentrations of soluble WT DENV-1 (0 to 10 μM) and DENV-2 DIII (0 to 20 μM) for 1 hour. Plates were washed 5 times with PBS-T and incubated with an anti-M13-HRP conjugated antibody for 1 hour. Plates were washed 5 times with PBS-T and remaining bound phage were detected using TMB substrate. The data was graphed using GraphPad Prism (GraphPad Software, La Jolla, CA), IC50 values were obtained by fitting the competition ELISA data to a single phase exponential decay curve.
Production of soluble DIII protein
Soluble DENV-2 DIII protein (construct “LDIIIH1CS”) was isolated as previously described (Liao and Kielian, 2005; Liao et al., 2010). A gene for expression of soluble DENV-1 DIII protein was produced by incorporating a His-8 tag and stop (TAA) codon into the DENV-1 DIII expressing phagemid at the end of the DIII sequence just prior to the phage pIII protein by Kunkel mutagenesis (Sidhu and Weiss, 2004). The mutation was confirmed by sequencing individual clones. Soluble wildtype DENV-1 DIII was produced by electroporating the His-8-stop DENV-1 DIII construct into E. coli BL21(DE3)STAR cells (Life Technologies, Carlsbad, CA). Cells were then grown in 25 mL 2xYT with 100 μg/mL Carbenicillin overnight at 37°C. Five mL of this overnight culture was then transferred to 100 mL low phosphate media (per liter: 3.57 g ammonium sulfate, 0.517 g sodium citrate monobasic anhydrous, 1.07 g potassium chloride, 5.36 g Hy-Case SF Casein acid hydrolysate from bovine milk, 5.36 g yeast extract, pH = 7.3 with KOH, after autoclaving add 7 mL 1 M MgSO4 and 14 mL 1 M Glucose) with 100 μg/mL Carbenicillin for 24 hours at 30°C. DENV-1 DIII was harvested by pelleting the cells, freezing the cell pellet, and then disrupting cell membranes by resuspending in 1x BugBuster (Novagen, Madison, WI) containing Deoxyribonuclease I (Life Technologies, Invitrogen, Carlsbad, CA) and a cOmplete mini EDTA-free protease inhibitor cocktail inhibitor (Lifesciences, Roche, Indianapolis, IN) in PBS pH = 7.4. Cell debris was pelleted by centrifugation at 16,000 rpm for 20 minutes. DIII-containing supernatant was then batch bound to Ni-NTA beads (Qiagen, Valencia, CA) for 2 hours at 4°C. Flow through was collected and the beads were washed and eluted in PBS pH = 7.4 with increasing concentrations of Imidazole (45 mM, 150 mM, 200 mM, and 1 M). Purified fractions of DIII were determined by SDS-PAGE, pooled, concentrated, and buffer exchanged into PBS pH = 7.4 using a 3,000 molecular weight cut off (MWCO) Amicon (Millipore, Merck KGaA, Darmstadt, Germany). DENV-1 DIII concentration was determined by Bradford assay.
RESULTS
Combinatorial alanine scanning library design, construction, and screening
DENV glycoprotein E DIII is a discretely folded β-sandwich domain comprised of seven antiparallel β-strands (A-G) (Figures 1B and 1C) (Modis et al., 2003; Rey et al., 1995). Overall, the DIII sequences among the serotypes are reasonably conserved, with pairwise amino acid identities ranging from 51-70% and similarities of 68-87% (Figure 1D and Supporting Information). The minimal DIII sequences from all four serotypes were expressed bivalently on M13 bacteriophage as fusions to the pIII coat protein under the control of an alkaline phosphatase promoter. Included in the display constructs were N-terminal FLAG epitopes, for detection of display. Monoclonal phage ELISAs indicated that all four DIII domains were expressed efficiently on phage, as detected by binding to immobilized anti-FLAG antibody M2, and functionally, as detected by binding to 4E11 (see Supporting Information).
The reported X-ray structures of the DIII domain from all four serotypes in complex with the single chain variable fragment (scFv) of 4E11 indicate that recognition of a lateral edge formed by the A and G β-strands is a significant component of 4E11-based recognition (Figures 1B and 1C) (Cockburn et al., 2012a). Several other bNAbs, and some serotype-specific mAbs also engage this epitope region, highlighting its potential as a neutralization target (Austin et al., 2012; Brien et al., 2010; Edeling et al., 2014; Gentry et al., 1982; Henchal et al., 1985; Kaufman et al., 1987; Li et al., 2012; Lok et al., 2008; Shrestha et al., 2010; Sukupolvi-Petty et al., 2007). Based on inspection of the 4E11 scFv-DIII complexes, 22 residues on DENV-1, -3 and -4 DIII and 23 residues on DENV-2 DIII were chosen for analysis via combinatorial alanine scanning (Figures 1B and 1D). Combinatorial alanine scanning is a high-throughput phage display method to determine energetic contributions of residues to intermolecular interactions (Weiss et al., 2000). In this approach, limited diversity phage libraries are produced in which the contact residues are allowed to vary between WT and Ala (in some cases, additional diversity is permitted due to the degeneracy of the genetic code). The “alanine scanning” libraries are subjected to selection against the target, here, 4E11 or 4E5A. The resulting population of ELISA-positive clones is analyzed for positional bias toward WT, indicative of a strong requirement for the WT side chain in binding. A parallel selection is performed to correct for display bias, here against anti-FLAG antibody M2 (a FLAG epitope is included at the N-terminus of the displayed DIIIs). The ratio of observed WT residues to Ala (WT/Ala) at any particular position can be determined and used to approximate the ratio of the association constants of WT and a WT→Ala mutant. Therefore, an energetic value can be computed and assigned for contribution of each residue to binding energy based on the sequencing data (ΔΔGAla-WT). This method allows for the rapid dissection of intermolecular interfaces; its high-throughput nature allows computation of ΔΔGAla-WT values for many more positions than is practical by traditional site-directed mutagenesis. Importantly, the results from combinatorial alanine scanning have been closely correlated with traditional mutagenesis studies in several systems (Da Silva et al., 2010; Pál et al., 2005; Vajdos et al., 2002; Weiss et al., 2000).
The residues chosen for analysis on DENV DIIIs included both direct contact residues (structural epitope), as defined by a 4.5 Å cutoff, but also residues that potentially played supporting conformational roles. About 29% of the mutagenized residues were conserved among all four serotypes. Limited diversity phage display libraries were constructed (two libraries/serotype) using the randomization scheme of Weiss et al., which allows variation among WT and Ala with additional diversity (3rd or 4th residues) at some positions due to the degeneracy of the genetic code (Weiss et al., 2000). All libraries were produced with sizes of ~108 clones or higher, exceeding the theoretical diversities by 100-fold or more thus ensuring adequate sampling of sequence space. Combinatorial alanine scanning libraries were each screened in parallel against both 4E11 (functional selection) and M2 (display selection). As described previously, the sequencing of ELISA-positive output populations from these was used to approximate energetic contributions to the intermolecular interface (ΔΔGAla-WT) (Pál et al., 2005; Weiss et al., 2000). The critical assumption in this analysis is that the ratio of WT to Ala (WT/Ala) in the selected sequence population is proportional to the ratio of the association constants of the WT protein and a WT→Ala point mutant (Weiss et al., 2000). Since display biases may be inherent for certain residues at particular positions or for some sequences, normalization for display via the M2 selection is a critical component of this analysis. The 4E11-selected populations contained a high percentage of clones with strong, positive and selective binding signals toward 4E11 (>50% of the clones had monoclonal phage ELISA signals greater than 5-fold over background wells containing BSA) . The sequence populations of the 4E11- and M2-selected clones were diverse in nature, though some siblings were apparent in the 4E11-selected populations. In the most abundant cases, a single sibling constituted ~5% of the overall population of selected clones.
Table 1 shows the numerical results of the sequence analysis from 60 – 157 clones per selection; these data are represented graphically, with the surface of the DIII structure color coded according to measured ΔΔGAla-WT values, in Figure 2. Screening of the DENV-4 structural epitope was incomplete because one of the two libraries, covering the “lower” portion of the epitope, was inefficient. Only one ELISA-positive sequence was identified from screening of approximately 200 clones from this library, and this clone maintained seven WT residues out of eleven varied structural epitope positions. This result suggests that mutations at these positions are not tolerated, likely because the binding of 4E11 to DENV-4 DIII is >500-fold weaker than for DENV-1-3 (reported KD of 4.1 μM compared to 0.08 – 8 nM) (Cockburn et al., 2012b). Thus, even minor perturbations in this C-terminal portion of the structural epitope cause a complete lack of binding and therefore very few members of the library contain functional activity. Among all positions for which data could be derived, the measured ΔΔGAla-WT values spanned a large range, with negative values at some positions, indicating a preference for Ala over WT. Numerous “hot spot” residues were identified on each serotype, defined here as ΔΔGAla-WT > 1.0 kcal/mol, and in several cases WT was absolutely conserved over Ala, allowing only a lower estimate of the value for ΔΔGAla-WT.
Table 1.
Combinatorial alanine scanning results against 4E11A.
| DENV-1 DIIIB |
DENV-2 DIIIB |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Res. | WT/Ala (4E11) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
Res. | WT/Ala (4E11) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
| S305 | 1.0 | 1.4 | −0.18 | - | - | K305 | 5.7 | 2.3 | 0.54 | 0.63 (E) | 0.54 (T) |
| K307 | 4.6 | 3.9 | 0.10 | 0.40 (E) | 0.0 (T) | K307 | 35 | 2.2 | 1.7 | 1.9 (E) | 0.67 (T) |
| L308 | 4.6 | 3.6 | 0.15 | 0.14 (P) | −0.03 (V) | I308 | 15 | 4.7 | 0.70 | 0.65 (T) | 0.45 (V) |
| E309 | 1.5 | 1.4 | 0.04 | - | - | V309 | 1.7 | 2.4 | −0.21 | - | - |
| K310 | 24 | 3.2 | 1.2 | 1.7 (E) | 0.47 (T) | K310 | 11 | 1.9 | 1.0 | 1.6 (E) | 0.90 (T) |
| E311 | 1.5 | 2.8 | −0.36 | - | - | E311 | 2.3 | 2.0 | 0.07 | - | - |
| V312 | 2.7 | 1.6 | 0.29 | - | - | I312 | 20 | 1.8 | 1.4 | 1.5 (T) | 0.46 (V) |
| Q323 | 2.8 | 1.5 | 0.39 | 0.50 (E) | 0.24 (P) | R323 | 0.64 | 0.94 | −0.23 | −0.79 (G) | −0.86 (P) |
| K325 | 3.9 | 2.6 | 0.23 | 1.3 (E) | −1.0 (T) | Q325 | 1.9 | 1.7 | 0.07 | −0.5 (E) | > 2.6 (P)C |
| E327 | 2.3 | 3.0 | −0.16 | - | - | E327 | 6.0 | 1.5 | 0.84 | - | - |
| P332 | 40 | 5.1 | 1.2 | - | - | P332 | 52 | 0.90 | 2.4 | - | - |
| E360 | 2.1 | 1.5 | 0.22 | - | - | ||||||
| K361 | 2.2 | 5.5 | −0.55 | −0.08 (E) | 0.03 (T) | K361 | 1.5 | 0.94 | 0.27 | 0.91 (E) | 0.39 (T) |
| E362 | 1.5 | 1.8 | −0.12 | - | - | D362 | 1.9 | 1.7 | 0.05 | - | - |
| P364 | 0.77 | 0.99 | −0.15 | - | - | P364 | 1.1 | 0.75 | 0.24 | - | - |
| N366 | 0.95 | 1.9 | −0.41 | −0.42 (D) | 0.07 (T) | N366 | 2.3 | 2.5 | −0.05 | 0.25 (D) | 0.24 (T) |
| E384 | 2.4 | 2.3 | 0.01 | - | - | P384 | 7.0 | 0.60 | 1.5 | - | - |
| K385 | 2.9 | 2.2 | 0.16 | 0.36 (E) | 0.13 (T) | G385 | 1.4 | 2.4 | −0.31 | - | - |
| L387 | 18 | 2.6 | 1.1 | 1.2 (P) | 0.15 (V) | L387 | 12 | 0.88 | 1.6 | > 2.7 (P)C | 0.50 (V) |
| K388 | 2.6 | 1.3 | 0.41 | 0.80 (E) | 0.36 (T) | K388 | 0.95 | 1.0 | −0.03 | −0.53 (E) | 0.02 (T) |
| L389 | 8.9 | 2.0 | 0.89 | 2.0 (P) | 0.23 (V) | L389 | 14 | 1.0 | 1.6 | > 2.5 (P)C | 0.39 (V) |
| S390 | 1.5 | 1.1 | 0.18 | - | - | N390 | 2.2 | 1.5 | 0.25 | 1.4 (D) | 0.14 (T) |
| W391 | 5.2 | 1.6 | 0.69 | 0.67 (G) | 0.34 (S) | W391 | 77 | 1.2 | 2.5 | 3.4 (G) | 2.4 (S) |
|
| |||||||||||
| DENV-3 DIIIB |
DENV-4 DIIIB |
||||||||||
| WT/Ala (4E11) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
WT/Ala (4E11) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
||
|
| |||||||||||
| T305 | 0.83 | 0.77 | 0.04 | - | - | K305 | 4.7 | 2.1 | 0.49 | 0.92 (E) | 0.09 (T) |
| V307 | 4.2 | 2.7 | 0.25 | - | - | S307 | 78 | 6.9 | 1.5 | - | - |
| L308 | 32 | 5.9 | 1.0 | > 1.6 (P)C | 1.3 (V) | I308 | >121 | 18 | > 1.2C | > 1.7 (T)C | 0.39 (V) |
| K309 | 0.39 | 1.3 | −0.70 | −0.22 (E) | −0.35 (T) | D309 | 0.01 | 0.58 | −2.3 | - | - |
| K310 | 6.4 | 2.8 | 0.51 | 1.2 (E) | 0.21 (T) | K310 | 157 | 4.8 | 2.1 | > 3.5 (E)C | > 28 (T)C |
| E311 | 3.0 | 1.6 | 0.38 | - | - | E311 | 5.1 | 3.0 | 0.31 | - | - |
| V312 | 16 | 5.3 | 0.67 | - | - | M312 | 10 | 5.0 | 0.43 | > 1.7 (T)C | −0.17 (V) |
| K323 | 8.5 | 3.1 | 0.60 | −0.50 (E) | −0.64 (T) | K323 | NDD | ||||
| E325 | 22 | 7.8 | 0.62 | - | - | K325 | NDD | ||||
| K327 | 52 | 2.5 | 1.8 | > 2.8 (E)C | 1.2 (T) | E327 | NDD | ||||
| P332 | >114 | 1.1 | > 2.8C | - | - | P332 | NDD | ||||
| K361 | 1.4 | 1.2 | 0.08 | 0.59 (E) | −0.25 (T) | T361 | 3.6 | 2.3 | 0.28 | - | - |
| E362 | 2.0 | 2.4 | −0.11 | - | - | N362 | 3.6 | 3.4 | 0.03 | −0.29 (D) | 0.10 (T) |
| P364 | 0.95 | 0.68 | 0.20 | - | - | V364 | 6.5 | 4.4 | 0.23 | - | - |
| N366 | 1.1 | 1.7 | −0.27 | −0.16 (D) | −0.35 (T) | N366 | 1.1 | 0.60 | 0.37 | −0.42 (D) | −0.19 (T) |
| D384 | 1.2 | 1.7 | −0.21 | - | - | D384 | NDD | ||||
| K385 | 5.8 | 3.1 | 0.38 | 0.70 (E) | 0.89 (T) | S385 | NDD | ||||
| L387 | >110 | 1.5 | > 2.6C | > 2.4 (P)C | 2.0 (V) | L387 | NDD | ||||
| K388 | 33 | 1.1 | 2.0 | > 2.8 (E)C | 1.3 (T) | T388 | NDD | ||||
| I389 | > 93 | 1.8 | > 2.4C | > 2.1 (T)C | 0.77 (V) | L389 | NDD | ||||
| N390 | 1.7 | 1.2 | 0.22 | 1.3 (D) | 1.6 (T) | H390 | NDD | ||||
| W391 | 16 | 1.2 | 1.6 | 2.0 (G) | 1.5 (S) | W391 | NDD | ||||
Calculated WT/Ala ratios for 4E11 and M2 and ΔΔGAla-WT are listed. In cases where additional residues were permitted, the ΔΔGm2-WT and ΔΔGm3-WT values are also provided. Mutations that were largely deleterious (≥ 1 kcal/mol) are shown in bold.
Sample sizes were greater than 100 in most cases (see Supporting Information).
In some cases, a single sequence containing a mutation was not isolated in the selected pool, therefore only a lower estimate could be provided.
Not determined. Values for positions located in the “lower” segment of the DENV-4 DIII epitope could not be obtained due to inefficient library screening.
Figure 2.
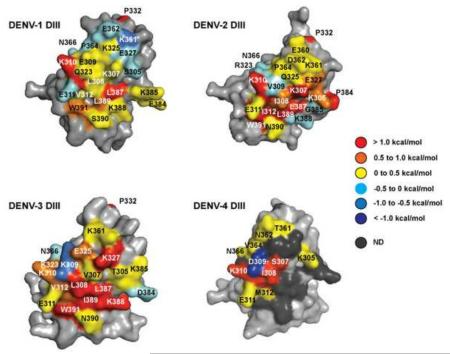
Heat maps of functional epitopes on DENV-1 through -4 for 4E11 binding color-coded based on observed ΔΔGAla-WT values. Residues in DENV-4 that could not be mapped are shown as dark grey. A conserved, core functional epitope is comprised of K310, L/I387, L389, and W391. In addition, recognition features unique to each serotype were also observed.
Comparison of functional epitopes across DENV serotypes for recognition by 4E11
Figure 2 shows that the topological landscape of the functional epitopes varies among the four serotypes. For example, only two residues on DENV-1 DIII were “hot spot” positions, K310 on the A-strand and L387 on the G-strand. Residues L389 and W391 on the G-strand contributed less significantly (0.5 – 1 kcal/mol) but together with K310 and L387 form a nearly contiguous and mostly hydrophobic core epitope. By contrast, the functional epitopes on DENV-2 and DENV-3 are more extended in nature with additional contributing residues from the A and G strands. In addition to the conserved K310 and L387 as observed with DENV-1, the hotspot residues in DENV-2 include K307, I312, L389, and W391 with significant but smaller contributions from K305 and E327. In DENV-3, hot spot residues include L308, K327, L387, K388, I389, and W391; residue K310 contributed more moderately as did V312, K323, and E325. In addition, P322, which was not a direct contact residue but appeared to possibly play a supporting structural role, was strongly preferred in DENV-1, -2, and -3. The lower half of the structural epitope on DENV-4 could not be scanned, including the G strand, but among those assayed, A strand residues S307, I308, and K310 were all hot spot residues.
Despite these differences, there appear to be some similarities among the contributing functional epitopes. Residues K310 (A strand) and residues L387, L389 (I389 in DENV-3), and W391 from the G strand form a “core functional epitope” that contributes significantly to binding in DENV-1-3. Additionally, K310 on DENV-4 was shown to be important for recognition, from the limited scanning results. The core functional epitope residues are conserved across the four serotypes, except for a relatively conservative L → I substitution at position 389 in DENV-3, and likely function as a critical recognition motif for 4E11. The quantified ΔΔGAla-WT for core functional epitope residues is higher than 1 kcal/mol in most cases. Values for L389 and W391 were below this threshold in DENV-1 (0.89 and 0.69 kcal/mol, respectively) as was K310 in DENV-3 (0.51 kcal/mol) but these side chains nonetheless contributed significantly to the interaction; notably, L387 and I389 were completely intolerant to Ala substitution in DENV-3. At all four core functional epitope positions, the combinatorial alanine scanning randomization scheme allowed for third and fourth side chain variants, many of which were also poorly tolerated. For example, a K310E mutation was disfavored in all four serotypes, with ΔΔGm2-WT values greater than 1 kcal/mol in all cases; accommodation of a K310T mutation varied across serotypes. Mutations L387P and L389P were strongly destabilizing, with Pro not observed in any selected clones at either position in DENV-2 and DENV-3. The more conservative L387V and L389V mutations were accommodated to various degrees in DENV-1, -2, and -3 with DENV-3 being the most stringent. A W391G mutation was poorly tolerated in DENV-1, -2, and -3 with ΔΔGm2-WT higher than 1 kcal/mol in the latter two; a W391S mutation was somewhat destabilizing in DENV-1 but significantly destabilizing in DENV-2 and DENV-3.
In addition to the core functional epitope, each serotype also contains unique hot spots that are only partially conserved across the serotypes and lie outside of a central hydrophobic patch made up of the A and G strands (Cockburn et al., 2012a). K307 (S307 in DENV-4) is a hotspot only in DENV-2 and -4, while residue I312 is unique to DENV-3 as are residues K327 and K388 for DENV-3. L308 in DENV-3 and the corresponding I308 in DENV-4 were hotspot residues, the corresponding positions were less important in DENV-2 (I308) and DENV-1 (L308). These results suggest that 4E11 binding involves recognition of the core functional epitope, but that 4E11 is ultimately flexible in its binding requirements to allow broad and tight recognition across DENV-1 to -3. Certain aspects of this recognition appear to be important for recognition by DENV-4.
Relation to previously reported site-directed mutagenesis studies
The novelty of the current study relative to previous work lies in the high-throughput nature of the combinatorial alanine scanning method, which allowed comprehensive analysis of thermodynamic contributions for nearly all 4E11 structural epitope residues in the four DENV serotypes. Previously reported site-directed mutagenesis studies on this interaction focused only on a single serotype (DENV-1) and a subset of residues (Bedouelle et al., 2006; Lisova et al., 2014; Lisova et al., 2007). In addition, an important aspect of combinatorial alanine scanning is benchmarking to previous site-directed mutagenesis studies when available. For example, the ΔΔGAla-WT values from combinatorial alanine scanning and traditional site-directed mutagenesis studies correlate closely in the case of hGH-hGHR, and to lesser extents in other systems (Clackson and Wells, 1995; Da Silva et al., 2010; Pál et al., 2005; Weiss et al., 2000). We therefore compared our alanine scanning results with previously published mutagenesis data by Bedouelle and coworkers for DENV-1 DIII in which individual DIII alanine mutants were purified and tested for their binding affinities for 4E11 (Bedouelle et al., 2006; Lisova et al., 2014; Lisova et al., 2007). The Bedouelle study encompassed 14 total A/G strand residues, 11 of which overlapped with our subset, and three of which are not included in our analysis. Overall, the range of positive ΔΔGAla-WT values obtained by combinatorial scanning mutagenesis was smaller than those previously reported by site-directed mutagenesis studies. Bedouelle and coworkers defined “hot spot” positions as those with ΔΔGAla-WT > 1.5 kcal/mol whereas we, and others who have utilized combinatorial alanine scanning, have defined hot spot positions as > 1.0 kcal/mol.
Residues K307, L308, K310, and L389 were identified as hot spot residues with ΔΔGAla-WT values ranging from 1.8 to 5.1 kcal/mol by Bedouelle and coworkers (Lisova et al., 2007). Residues K310 and L389 were also important in combinatorial alanine scanning (ΔΔGAla-WT of 1.2 and 0.9 kcal/mol, with L389 falling just below the hot spot cutoff) but K307 and L308 were less important (ΔΔGAla-WT of 0.1 and 0.2 kcal/mol). A possible explanation for the discrepancy of ΔΔGAla-WT values at K307 and L308 reported here and in the Bedouelle studies is that WT→Ala mutations are deleterious to folding; we observed elevated WT/Ala ratios in both the functional (4E11) and display (M2) selections. Display efficiency of a protein is thought to be linked to folding stability and thus high WT/Ala ratios in the display selection may be indicative of defects in folding (Weiss et al., 2000). However, we note that the range of elevated WT/Ala ratios we observed, 3.6 - 4.6, were much more muted than the preference that would result in ΔΔGAla-WT values observed in the Bedouelle site-directed mutagenesis study (2.2 and 1.1 kcal/mol, respectively, which corresponds to 7- and 50-fold preferences in KD).
Although below their hot spot cutoff, E309A and V312A were also destabilizing (1.1 and 1.3 kcal/mol) in the Bedouelle studies. However, from our analysis, ΔΔGAla-WT values at these positions by combinatorial alanine scanning were lower (0.04 and 0.29 kcal/mol). To further explore the contribution of these two residues, we prepared individual site-directed E309A and V312A point mutants of phage-expressed DIII. A competition ELISA was performed, in which the capacity of phage-expressed DENV-1 (WT and mutants) to bind immobilized 4E11 in the presence of increasing amounts of soluble WT DENV-1 DIII was examined (Figure 3A). It is expected that, if a particular point mutant has drastically decreased binding affinity for 4E11 relative to WT, then those phage would be much more effectively competed for binding by soluble WT DENV-1 DIII protein than the WT DENV-1 DIII expressing phage. The IC50 values from this analysis indicated only minor differences between WT and E309A DENV-1 DIII phage (3.1 μM vs. 1.4 μM), and a stronger but still moderate effect for the V312A mutant (0.3 μM). These results are internally consistent with the combinatorial alanine scanning results that suggest E309A has no perturbing effect on binding, and V312A has an intermediate effect. The reasons for the discrepancy in our results with those of Bedouelle and coworkers are not immediately obvious. However, the overall comparisons among combinatorial scanning data and previously published data suggest that strong hot spots, that do not affect display efficiency, can be identified with strong correlation by combinatorial scanning mutagenesis.
Figure 3.
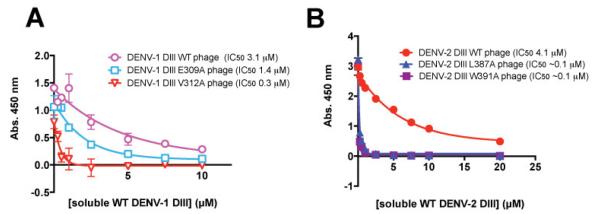
Validation of combinatorial alanine scanning results for DENV-1 (A) and DENV-2 (B) point mutants by competition phage ELISA. (A) Competition of phage expressing WT, E309A, and V312A DENV-1 DIII for binding to immobilized 4E11 by soluble WT DENV-1 DIII protein. (B) Similar analysis for WT, L387A, and W391A DENV-2 DIII expressing phage for competition against soluble WT DENV-2 DIII protein.
On DENV-1, L387A and W391A mutations were found deleterious for binding in site-directed mutagenesis studies, in agreement with our studies, but the ΔΔGAla-WT values varied slightly in the previous report (0.7 and 1.2 kcal/mol, respectively) than those observed here (1.1 and 0.7 kcal/mol, respectively). Notably, these two positions form part of the “core functional epitope” and were much more strongly preferred in DENV-2 and -3 (ΔΔGAla-WT > 1 kcal/mol). To further explore the role of these mutations, and to further validate the combinatorial scanning mutagenesis data in DENV-2, where previous site directed mutagenesis data are not available, we prepared individual WT→Ala variants at these positions in DENV-2 DIII on phage and performed competitive phage ELISA with soluble, WT DENV-2 DIII protein (Figure 3B). Soluble WT DENV-2 DIII protein competed with binding of WT DENV-2 DIII phage, as expected, with an IC50 of 4.1 μM. However, both L387A and W391A DIII phage were competed much more effectively by soluble WT DENV-2 DIII protein, with IC50 values ~40-fold lower. The higher capacity of WT DENV-2 DIII protein to compete the DENV-2 DIII L387A and W391A phage relative to WT DENV-2 DIII phage indicate that the affinity for these two point mutants is much lower than WT DENV-2 DIII for binding to 4E11 and support conclusions by combinatorial scanning mutagenesis that these DENV-2 DIII residues are critical for binding 4E11.
Our analysis further identified residue P332 as a strong hot spot residue in DENV-1 (ΔΔGAla-WT of 1.2 kcal/mol). A P332A mutation was not previously assayed by site-directed mutagenesis studies for 4E11, as it lies in the turn region between the B and C strands and does not make direct contact with 4E11. However, P332A mutation was assayed previously by site-directed mutagenesis studies against a similar antibody, 1A1D-2, and found to be important for binding in that interaction (Gromowski et al., 2010). Furthermore, P332 appears to play a critical supporting role for DIII recognition by 4E11 as it is both conserved and had a high ΔΔGAla-WT value for DENV-1, -2, and -3 by our analysis (this residue could not be scanned in DENV-4).
Comparison of functional epitopes on DENV-1 and DENV-4 for recognition by 4E5A
Antibody 4E5A is a computationally designed variant of 4E11 that has higher binding and neutralization activity against DENV-4 while retaining strong activity against DENV-1, -2, and -3 (Tharakaraman et al., 2013). The sequence of 4E5A differs from 4E11 at one heavy chain position and four light chain positions (Tharakaraman et al., 2013). The difference in activity between 4E11, which binds and effectively neutralizes DENV-1, -2, and -3 but has more modest activity against DENV-4, and 4E5A, which is potent against all four serotypes, provides an opportunity to explore whether functional epitopes vary among similar bNAbs that vary in the breadth of their activity. The combinatorial alanine scanning analysis was performed for DENV-1 and DENV-4 DIII against 4E5A (Figure 4). In contrast to 4E11, the selection of DENV-4 DIII alanine scanning libraries proceeded efficiently and therefore the functional epitope could be mapped to completion for both DENV-1 and DENV-4. It is possible that complete scanning for DENV-4 against 4E5A was efficient because it has 45-fold higher affinity than 4E11, thus individual WT→Ala mutations were tolerated enough to allow analysis in this case. The results from this analysis are presented numerically in Table 2 and graphically in Figure 4.
Figure 4.
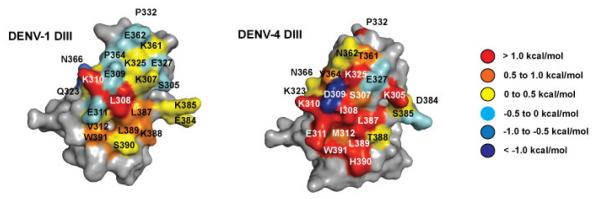
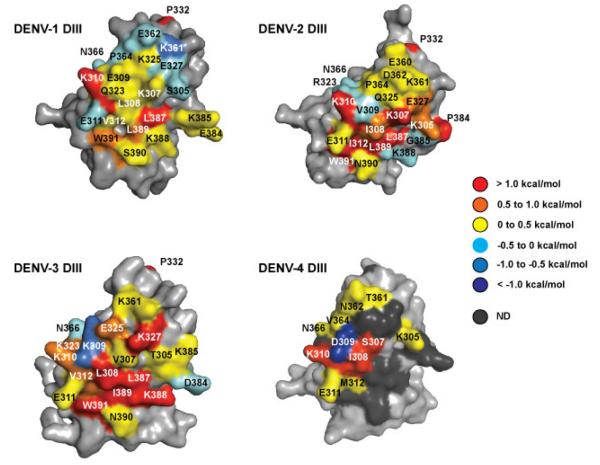
Heat maps of functional epitopes on DENV-1 and -4 for 4E5A binding color-coded based on observed ΔΔGAla-WT values. For both serotypes, an extended functional epitope was observed in comparison to 4E11.
Table 2.
Combinatorial alanine scanning results against 4E5A.
| DENV-1 DIII |
DENV-4 DIII |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Res. | WT/Ala (4E5A) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
Res. | WT/Ala (4E5A) |
WT/Ala (M2) |
ΔΔGAla-WT (kcal/mol) |
ΔΔGm2-WT (kcal/mol) |
ΔΔGm3-WT (kcal/mol) |
| S305 | 0.92 | 1.4 | −0.26 | - | - | K305 | 13 | 2.1 | 1.1 | 1.4 (E) | 1.2 (T) |
| K307 | 6.0 | 3.9 | 0.27 | 0.53 (E) | 0.34 (T) | S307 | 20 | 6.9 | 0.63 | - | - |
| L308 | 35 | 3.6 | 1.4 | 0.73 (P) | 0.23 (V) | I308 | > 78 | 18 | > 0.89A | 1.0 (T) | 0.41 (V) |
| E309 | 1.0 | 1.4 | −0.20 | - | - | D309 | 0.05 | 0.58 | −1.5 | - | - |
| K310 | 44 | 3.2 | 1.6 | 2.0 (E) | 1.0 (T) | K310 | 100 | 4.8 | 1.8 | > 3.2 (E) | 2.1 (T) |
| E311 | 2.0 | 2.8 | −0.20 | - | - | E311 | 16 | 3.0 | 1.0 | - | - |
| V312 | 2.0 | 1.6 | 0.12 | - | - | M312 | 19 | 5.0 | 0.79 | 1.6 (T) | 0.28 (V) |
| Q323 | 2.2 | 1.5 | 0.23 | 0.67 (E) | 0.00 (P) | K323 | 5.8 | 3.2 | 0.36 | - | - |
| K325 | 5.5 | 2.6 | 0.45 | 0.06 (E) | −0.19 (T) | K325 | 61 | 3.9 | 1.6 | 1.7 (E) | 0.73 (T) |
| E327 | 2.9 | 3.0 | −0.02 | - | - | E327 | 1.5 | 2.2 | −0.23 | - | - |
| P332 | 4.7 | 5.1 | −0.05 | - | - | P332 | > 75 | 1.0 | > 1.5 | - | - |
| K361 | 5.5 | 5.5 | 0.00 | −0.28 (E) | 0.02 (T) | T361 | 5.4 | 2.3 | 0.52 | - | - |
| E362 | 1.3 | 1.8 | −0.21 | - | - | N362 | 5.9 | 3.4 | 0.32 | −0.02 (D) | 0.51 (T) |
| P364 | 0.66 | 0.99 | −0.25 | - | - | V364 | 14 | 4.4 | 0.68 | - | - |
| N366 | 0.79 | 1.9 | −0.52 | −0.05 (D) | 0.11 (T) | N366 | 0.82 | 0.60 | 0.19 | −0.31 (D) | −0.39 (T) |
| E384 | 2.6 | 2.3 | 0.08 | - | - | D384 | 2.6 | 3.0 | −0.10 | - | - |
| K385 | 2.9 | 2.2 | 0.15 | 0.67 (E) | 0.04 (T) | S385 | 3.4 | 2.5 | 0.19 | - | - |
| L387 | 8.8 | 2.6 | 0.72 | 0.93 (P) | 0.38 (V) | L387 | 17 | 0.98 | 1.7 | > 1.6 (P) | 0.82 (V) |
| K388 | 4.3 | 1.3 | 0.72 | 0.75 (E) | 1.0 (T) | T388 | 2.4 | 1.4 | 0.30 | - | - |
| L389 | 7.0 | 2.0 | 0.75 | 0.67 (P) | 0.15 (V) | L389 | > 52 | 2.7 | > 1.8 | > 1.9 (P) | 0.38 (V) |
| S390 | 1.4 | 1.1 | 0.13 | - | - | H390 | 21 | 1.8 | 1.5 | 2.4 (D) | 0.62 (P) |
| W391 | 5.5 | 1.6 | 0.72 | 0.45 (G) | 0.35 (S) | W391 | 14 | 2.4 | 1.1 | 2.5 (G) | 0.88 (S) |
A lower estimate only for ΔΔGAla-WT could be obtained for I308, because no Ala mutations were observed in the selected pool. Although the calculated lower estimate falls below the “hot spot” threshold, this residue was classified nonetheless as a hot spot due to the clear preference for WT over Ala in the 4E5A binding selection.
Comparison of DENV-1 functional epitopes for 4E11 and 4E5A indicates that the 4E5A functional epitope is larger and more extended for at least DENV-1, with L308 and K388 playing stronger roles in the 4E5A interaction. The 4E5A epitope contains fewer residues that meet the criteria of a hot spot (ΔΔGAla-WT > 1 kcal/mol) than 4E11, but several additional residues had contributions > 0.5 kcal/mol. The analysis also revealed a total of 10 hot spot residues for DENV-4 DIII recognition by 4E5A, four of which were also part of the partially mapped functional epitope of 4E11. In line with the higher affinity of 4E5A for DENV-4 DIII, we found that 4E5A requires extended interactions involving many residues on both the A and G strands. All four residues from the core functional epitope of 4E11 were also required for recognition of both DENV-1 and DENV-4 by 4E5A. Together, these results suggest that both bNAbs recognize similar features across the different serotypes, but that key mutations within 4E5A increase favorable interactions with DENV-4 DIII while maintaining affinity across the other three serotypes.
DISCUSSION
Co-crystal structures of DIII from all four serotypes in complex with the 4E11 scFv suggested that the strictly conserved, charged residues K310 and E311 are important for antibody-antigen recognition across the four serotypes (Cockburn et al., 2012a). In partial agreement with these conclusions, our mutagenesis data revealed that K310 is critically important for the binding energetics and recognition of 4E11 across the four serotypes as it was identified as a hotspot with ΔΔG > 1 kcal/mol for three of the serotypes (DENV-1, -2, -4). However, the requirement for E311 was not evident from our analysis, as this position did not have high ΔΔGAla-WT values for any of the serotypes. From the crystallographic studies, it was postulated that bNAb 4E11 recognizes DIII from all four serotypes by flexibly binding to DIII in order to partially conserve charge complementarity between the paratope and epitope. The charge complementarity is maintained by adjacent patches of positive and negative charge that arise from CDR-H2 residue ArgH94 and CDR-L3 residue GluL55 in the 4E11 paratope (Cockburn et al., 2012a). Supporting evidence for this hypothesis is evident in the co-crystal structure of DENV-3, in which the overall positioning of 4E11 in relation to DIII is shifted, so that the charged side chains of ArgH94 and GluL55 interact with the B-strand residues E325 and K327 on DENV-3 DIII to conserve charge complementarity across the paratope-epitope. Interestingly, our mutagenesis data correlates with this, as E325 and K327 were both significant contributors to binding of 4E11 to DENV-3 DIII, a unique feature that was not observed in DENV-1 and -2 (Figure 2).
Our results suggest that broad recognition for at least DENV-1, -2, and -3 requires recognition of a common conserved functional epitope core consisting of K310, L387, L/I389, and W391. Requirements for DENV-2 and -3 appear more stringent than DENV-1, as indicated by the broader contiguous residues with strong ΔΔGAla-WT values. The ΔΔGAla-WT values obtained by combinatorial scanning mutagenesis reliably identified strongly contributing residues and were internally consistent with competitive ELISA experiments with specific single point mutations in DENV-1 and DENV-2 DIII. A limitation combinatorial scanning mutagenesis is the fact that it does not allow for error estimates within a given dataset, but this is mitigated by the rapidity with which functional data can be obtained. The analysis provided here, which involved assays for 89 total residues across four different DIII variants, and against two different antibodies, underscores the utility of the approach for obtaining large datasets. Individual purification and characterization of single point mutants to obtain this level of insight is laborious and not practical for most laboratories. The analysis reported here also augments previous studies employing less comprehensive analysis of epitope residues on DENV-1 DIII for 4E11 or DENV-2 DIII for the related antibody 1AID-2 (Gromowski et al., 2010; Lisova et al., 2007). The novel conclusions from our analysis are that the core functional epitope exists for DENV-1 through -3, but that broad recognition also relies on subtle differences among the serotypes. For example, DENV-2 and DENV-3 functional epitopes for recognition by 4E11 are larger than that of DENV-1, encompassing additional A/G strand residues that lie outside of the core functional epitope. Furthermore, our analysis revealed differences in recognition requirements for DENV-1 and DENV-4 for binding to 4E5A or 4E11.
Antibody 4E5A was reported to have a 45-fold higher binding affinity for DENV-4, improved affinity for DENV-2 and DENV-3, and a slightly lower affinity for DENV-1 (Tharakaraman et al., 2013). The improved affinity for DENV-4 was evident in our combinatorial scanning data, as we were able to fully map the functional epitope of DENV-4 DIII for 4E5A. Of the 11 fully scanned residues for 4E11 binding, only three were found to be hotspots of binding energy, whereas for 4E5A binding, one additional hotspot residue was identified among these 11 residues and six residues were identified as hotspot residues in the lower half of the epitope.
For DENV-1, Tharakaraman and coworkers reported a KD value of 0.5 nM for binding to 4E11 and a KD value of 1.8 nM for binding to 4E5A (Tharakaraman et al., 2013). While this difference in affinity for DENV-1 is not substantial, it is interesting that our combinatorial scanning mutagenesis analysis revealed differences in the DENV-1 functional epitopes, with a broader functional epitope required for 4E5A. Antibodies 4E5A and 4E11 differ by five residues, and the mutations in 4E5A relative to 4E11 are thought to directly contact S305, K310, E311, Q323, E327, and T329 on DENV-1. All of these residues except T329 were included in our scanning analysis, but interestingly, substantial differences in contribution to the antibody-antigen interface were not detected. Instead, our results imply that the mutations in 4E5A relative to 4E11 serve to expand contributing residues on both the A and G strands (increased ΔΔGAla-WT values for L308, V312, and K388 in 4E5A relative to 4E11). Thus, in this case, the designed mutations to endow 4E5A with broader specificity also increased the contributing surface area to DENV-1 in the central portion of the structural epitope. This information could not be inferred from previous structural or biochemical experiments on 4E11 and 4E5A. Given recent interest in therapeutic antibodies targeting DENV DIII (Robinson et al., 2015), the features of recognition highlighted by our results could be used for further optimization of DIII-targeting mAbs or of immunogens designed to elicit similar responses.
Murine-derived neutralizing antibodies with a range of specificity profiles target DIII, with the A and G strands constituting a critical neutralization epitope (Austin et al., 2012; Brien et al., 2010; Cockburn et al., 2012a; Edeling et al., 2014; Gentry et al., 1982; Henchal et al., 1985; Kaufman et al., 1987; Megret et al., 1992; Midgley et al., 2012; Roehrig et al., 1998; Shrestha et al., 2010; Sukupolvi-Petty et al., 2007). An interesting comparison arises when the previously mapped functional epitope of murine DENV-2 mAb 1A1D-2 (Gromowski et al., 2010) is compared to our results with 4E11. MAb 1A1D-2 is highly related to 4E11 (>85% identity in heavy and light chain Fvs), but was isolated following immunization with DENV-2 rather than DENV-1. The structures of 1A1D-2 and 4E11 in complex with DENV-2 DIII are highly similar, engaging similar structural epitopes. Gromowski et al. mapped the functional epitope on 1A1D-2 by site directed mutagenesis and ELISA, and identified K307, K310, I312, and W391 as strongly contributing residues (≥ 10-fold reduction in binding upon mutation to alanine) and E311 and L389 among those that contributed substantially (4- to 10-fold, respectively). In addition, K307Q and K307T escape mutants were obtained from passaging of the virus in the presence of 1A1D-2. Many of these features are maintained in the 4E11 functional epitope for DENV-2 mapped here, with the inclusion of L387 and L389 among the most important contributing residues. These results illustrate that similar, but not identical, functional epitopes may be engaged by neutralizing antibodies that target the same structural epitope.
Human neutralizing antibodies that arise from natural DENV infections generally target other regions of the E glycoprotein, although some DIII-specific human mAbs have been described (Beltramello et al., 2010; de Alwis et al., 2011; Wahala and de Silva, 2011; Williams et al., 2012). Nonetheless the potency of the murine DIII mAbs has led to significant interest in DIII as a target for immunotherapeutic development as well as immunogen design (Bernardo et al., 2008; Block et al., 2010; Izquierdo et al., 2008). Due to the potential for ADE, it is beneficial to focus on bNAbs and immunogens that elicit bNAbs. Our results suggest that requirements for broad recognition of DIII include the core functional epitope of K310, L387, L/I389, and W391, at least for A/G strand-targeting antibodies. DIII vaccination studies have suggested that recombinant DIII alone, from a single serotype, elicits mostly a serotype-specific and non-neutralizing immune response. In one study, the majority of elicited antibodies in response to DIII immunization were localized to the AB loop and were poorly or non-neutralizing (Li et al., 2013). Our analysis suggests that targeting the immune response or therapeutic antibody design toward the critical functional core of DIII could improve efficiency of bNAb development or design of immunogens that elicit bNAbs.
Antibody-protein antigen interactions are prime examples of specific protein-protein interactions that are essential to biology. The functional requirements for most antibody-protein interactions are presumably similar to those of most other protein-protein interactions and involve burial of a few critical hydrophobic hot spot residues. However, the relationship between functional requirements for multiple antigens in multispecific antibodies (e.g., bNAbs) has not been as clearly delineated. Our results indicate that broad recognition of DIII by the two different but related bNAbs 4E11 and 4E5A have distinct functional epitopes despite the overall similarity of binding. Therefore, plasticity in recognition by the combining site is a key factor for broad recognition. Whether or not a set of general set of rules can be established for bNAb recognition remains to be determined, but the results presented herein provide a template for at least one case.
Supplementary Material
-
➢
DENV E DIII is a target of broadly neutralizing antibodies.
-
➢
There are four serotypes of DENV, the DIII sequences diverge by up to 49%.
-
➢
The recognition requirements for two bNAbs for binding to DIII was determined by phage display.
-
➢
This information will be useful for immunogen or therapeutic antibody design.
ACKNOWLEDGMENT
The authors thank Dillon Ade for his help in screening DIII alanine-scanning libraries for DENV-1 and DENV-2, Aihua Zheng for providing soluble purified DENV-2 DIII, and Youqing Xiang and Susan Buhl for assistance with production of 4E11.
FUNDING STATMENT
This work was supported by the NIH (R01-AI090249 to J. R. L., and R01-AI075647 to M. K.). J.C.F. was supported in part by NIH Cellular and Molecular Biology and Genetics Training Program T32-GM007491.
ABBREVIATIONS
- DENV
Dengue virus
- DHF
Dengue hemorrhagic fever
- DSS
Dengue shock syndrome
- ADE
antibody dependent enhancement of infection
- bNAb
broadly neutralizing antibody
- DIII
domain III
- DI
domain I
- DII
domain II
- mAb
monoclonal antibody
- IgH
immunoglobulin heavy chain
- IgL
immunoglobulin light chain
- HEK
human embryonic kidney cells
- PEI
polyethylenimine
- PFU
plaque forming units
- PEG
polyethylene glycol
- PBS
phosphate buffered saline
- PBS-T
phosphate buffered saline with tween 20
- BSA
bovine serum albumin
- HRP
horse radish peroxidase
- TMB
3,3′,5,5′-Tetramethylbenzidine
- WT
wildtype
- Ala
alanine
- RT
room temperature
- ELISA
enzyme-linked immunosorbent assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPORTING INFORMATION AVAILABLE. ELISA of DIII phage binding to 4E11 and M2 (anti-FLAG); sequence identity among DENV DIIIs; primers used for library construction; number of sequences used for analysis; full results from combinatorial alanine scanning mutagenesis.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing financial interest(s).
REFERENCES
- Austin SK, Dowd KA, Shrestha B, Nelson CA, Edeling MA, Johnson S, Pierson TC, Diamond MS, Fremont DH. Structural basis of differential neutralization of DENV-1 genotypes by an antibody that recognizes a cryptic epitope. PLoS pathogens. 2012;8:e1002930. doi: 10.1371/journal.ppat.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle H, Belkadi L, England P, Guijarro JI, Lisova O, Urvoas A, Delepierre M, Thullier P. Diversity and junction residues as hotspots of binding energy in an antibody neutralizing the dengue virus. FEBS Journal. 2006;273:34–46. doi: 10.1111/j.1742-4658.2005.05045.x. [DOI] [PubMed] [Google Scholar]
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NTH, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, De Silva AM. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell host & microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo L, Hermida L, Martin J, Alvarez M, Prado I, López C, Martínez R, Rodríguez-Roche R, Zulueta A, Lazo L. Anamnestic antibody response after viral challenge in monkeys immunized with dengue 2 recombinant fusion proteins. Archives of virology. 2008;153:849–854. doi: 10.1007/s00705-008-0050-9. [DOI] [PubMed] [Google Scholar]
- Block OK, Rodrigo WSI, Quinn M, Jin X, Rose RC, Schlesinger JJ. A tetravalent recombinant dengue domain III protein vaccine stimulates neutralizing and enhancing antibodies in mice. Vaccine. 2010;28:8085–8094. doi: 10.1016/j.vaccine.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of molecular biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Bostrom J, Haber L, Koenig P, Kelley RF, Fuh G. High affinity antigen recognition of the dual specific variants of herceptin is entropy-driven in spite of structural plasticity. PLoS One. 2011;6:e17887. doi: 10.1371/journal.pone.0017887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom J, Yu S-F, Kan D, Appleton BA, Lee CV, Billeci K, Man W, Peale F, Ross S, Wiesmann C. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–1614. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. Journal of virology. 2010;84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012a;20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Cockburn JJ, Sanchez MEN, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Seisdedos FA, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012b;20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. Journal of virology. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva GF, Harrison JS, Lai JR. Contribution of light chain residues to high affinity binding in an HIV-1 antibody explored by combinatorial scanning mutagenesis. Biochemistry. 2010;49:5464–5472. doi: 10.1021/bi100293q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brian J, Tsai W-Y. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS neglected tropical diseases. 2011;5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nature immunology. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Austin SK, Shrestha B, Dowd KA, Mukherjee S, Nelson CA, Johnson S, Mabila MN, Christian EA, Rucker J. Potent Dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS pathogens. 2014;10:e1004072. doi: 10.1371/journal.ppat.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger M-A, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. Journal of molecular biology. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. Journal of molecular biology. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DL, Laskov R, Scharff MD. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- Gentry M, Henchal E, McCown J, Brandt W, Dalrymple J. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. The American journal of tropical medicine and hygiene. 1982;31:548–555. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- Gromowski GD, Roehrig JT, Diamond MS, Lee JC, Pitcher TJ, Barrett AD. Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology. 2010;407:237–246. doi: 10.1016/j.virol.2010.06.044. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff L. Flaviviruses. Fields virology. 2007;1:1153–1253. [Google Scholar]
- Halstead S, O’rourke E. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. The Journal of experimental medicine. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nature structural & molecular biology. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal E, McCown J, Burke D, Seguin M, Brandt W. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. The American journal of tropical medicine and hygiene. 1985;34:162–169. doi: 10.4269/ajtmh.1985.34.162. [DOI] [PubMed] [Google Scholar]
- Izquierdo A, Bernardo L, Martin J, Santana E, Hermida L, Guillén G, Guzmán MG. Serotype-specificity of recombinant fusion proteins containing domain III of dengue virus. Virus research. 2008;138:135–138. doi: 10.1016/j.virusres.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Kaufman B, Summers P, Dubois D, Eckels K. Monoclonal antibodies against dengue 2 virus E-glycoprotein protect mice against lethal dengue infection. The American journal of tropical medicine and hygiene. 1987;36:427–434. doi: 10.4269/ajtmh.1987.36.427. [DOI] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P-C, Liao M-Y, Cheng P-C, Liang J-J, Liu I-J, Chiu C-Y, Lin Y-L, Chang G-JJ, Wu H-C. Development of a humanized antibody with high therapeutic potential against dengue virus type 2. PLoS neglected tropical diseases. 2012;6:e1636. doi: 10.1371/journal.pntd.0001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Q, Qiu L-W, Chen Y, Wen K, Cai J-P, Chen J, Pan Y-X, Li J, Hu D-M, Huang Y-F. Dengue virus envelope domain III immunization elicits predominantly cross-reactive, poorly neutralizing antibodies localized to the AB loop: implications for dengue vaccine design. Journal of General Virology. 2013;94:2191–2201. doi: 10.1099/vir.0.055178-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nature Structural & Molecular Biology. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. The Journal of cell biology. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Sánchez-San Martín C, Zheng A, Kielian M. In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. Journal of virology. 2010;84:5730–5740. doi: 10.1128/JVI.00170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisova O, Belkadi L, Bedouelle H. Direct and indirect interactions in the recognition between a cross-neutralizing antibody and the four serotypes of dengue virus. Journal of Molecular Recognition. 2014;27:205–214. doi: 10.1002/jmr.2352. [DOI] [PubMed] [Google Scholar]
- Lisova O, Hardy F, Petit V, Bedouelle H. Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus. Journal of General Virology. 2007;88:2387–2397. doi: 10.1099/vir.0.83028-0. [DOI] [PubMed] [Google Scholar]
- Lok S-M, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nature structural & molecular biology. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Mazor Y, Barnea I, Keydar I, Benhar I. Antibody internalization studied using a novel IgG binding toxin fusion. Journal of immunological methods. 2007;321:41–59. doi: 10.1016/j.jim.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Megret F, Hugnot J, Falconar A, Gentry M, Morens D, Murray J, Schlesinger J, Wright P, Young P, Van Regenmortel M. Use of recombinant fusion proteins and monoclonal antibodies to define linear and discontinuous antigenic sites on the dengue virus envelope glycoprotein. Virology. 1992;187:480–491. doi: 10.1016/0042-6822(92)90450-4. [DOI] [PubMed] [Google Scholar]
- Midgley CM, Flanagan A, Tran HB, Dejnirattisai W, Chawansuntati K, Jumnainsong A, Wongwiwat W, Duangchinda T, Mongkolsapaya J, Grimes JM. Structural analysis of a dengue cross-reactive antibody complexed with envelope domain III reveals the molecular basis of cross-reactivity. The Journal of Immunology. 2012;188:4971–4979. doi: 10.4049/jimmunol.1200227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein C, Rada C. The maturation of the antibody response. Immunoglobulin Genes. 1995;57:81. [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. Novartis Medal Lecture. Antibodies: a paradigm for the evolution of molecular recognition. Biochemical Society Transactions. 2002;30:341–350. doi: 10.1042/bst0300341. [DOI] [PubMed] [Google Scholar]
- Pál G, Fong SY, Kossiakoff AA, Sidhu SS. Alternative views of functional protein binding epitopes obtained by combinatorial shotgun scanning mutagenesis. Protein science. 2005;14:2405–2413. doi: 10.1110/ps.051519805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Ye W, Wernimont A, Adams JJ, Koide A, Koide S, Lam R, Sidhu SS. CDR-H3 diversity is not required for antigen recognition by synthetic antibodies. Journal of molecular biology. 2013;425:803–811. doi: 10.1016/j.jmb.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. 1995. [DOI] [PubMed]
- Robinson LN, Tharakaraman K, Rowley KJ, Costa VV, Chan KR, Wong YH, Ong LC, Tan HC, Koch T, Cain D. Structure-Guided Design of an Anti-dengue Antibody Directed to a Non-immunodominant Epitope. Cell. 2015 doi: 10.1016/j.cell.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246:317–328. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney M-C, Kikuti CM, Sanchez MEN, Dejnirattisai W, Wongwiwat W, Haouz A. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015 doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand I. The 1980 outbreak. American Journal of Epidemiology. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O’Brien KM, Nelson CA, Johnson S, Fremont DH. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS pathogens. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu SS, Weiss GA. Constructing phage display libraries by oligonucleotide-directed mutagenesis. Phage Display: A practical approach. 2004;2:27–41. [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH. Type-and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. Journal of virology. 2007;81:12816–12826. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Robinson LN, Hatas A, Chen Y-L, Siyue L, Raguram S, Sasisekharan V, Wogan GN, Sasisekharan R. Redesign of a cross-reactive antibody to dengue virus with broad-spectrum activity and increased in vivo potency. Proceedings of the National Academy of Sciences. 2013;110:E1555–E1564. doi: 10.1073/pnas.1303645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajdos FF, Adams CW, Breece TN, Presta LG, de Vos AM, Sidhu SS. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. Journal of molecular biology. 2002;320:415–428. doi: 10.1016/S0022-2836(02)00264-4. [DOI] [PubMed] [Google Scholar]
- van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS pathogens. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, de Silva AM. The human antibody response to dengue virus infection. Viruses. 2011;3:2374–2395. doi: 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proceedings of the National Academy of Sciences. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proceedings of the National Academy of Sciences. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KL, Wahala WM, Orozco S, de Silva AM, Harris E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology. 2012;429:12–20. doi: 10.1016/j.virol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Beuscher AE, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. Journal of molecular biology. 2003;330:651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- Yin J, Mundorff EC, Yang PL, Wendt KU, Hanway D, Stevens RC, Schultz PG. A comparative analysis of the immunological evolution of antibody 28B4. Biochemistry. 2001;40:10764–10773. doi: 10.1021/bi010536c. [DOI] [PubMed] [Google Scholar]
- Zaitseva E, Yang S-T, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS pathogens. 2010;6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.