Abstract
Vpx encoded by HIV-2 and SIVsm enhances retroviral reverse transcription in macrophages in vitro by mediating the degradation of the host SAMHD1 protein that hydrolyzes dNTPs and by elevating cellular dNTP levels. Here we employed RT-SHIV constructs (SIV encoding HIV-1 RT) to investigate the contribution of Vpx to the potency of NRTIs, which compete against dNTPs, in monocyte-derived macrophages (MDMs) and activated CD4+ T cells. Relative to HIV-1, both SIV and RT-SHIV exhibited reduced sensitivities to AZT, 3TC and TDF in MDMs but not in activated CD4+ T cells. However, when SIV and RT-SHIV constructs not coding for Vpx were utilized, we observed greater sensitivities to all NRTIs tested using activated CD4+ T cells relative to the Vpx-coding counterparts. This latter phenomenon was observed for AZT only when using MDMs. Our data suggest that Vpx in RT-SHIVs may underestimate the antiviral efficacy of NRTIs in a cell type dependent manner.
Keywords: Vpx, SAMHD1, Antiretrovirals, SIV, RT-SHIV, HIV-1
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection continues as a pandemic. The majority of those infected with HIV-1 will progress to AIDS unless strictly adhering to combined antiretroviral therapy (cART). All current optimal combined anti-HIV-1 treatment regimens include at least 2 nucleos(t)ide reverse transcriptase inhibitors (NRTIs) in combination with a third drug from one of the three following classes: (i) non-nucleoside reverse transcriptase inhibitors (NNRTIs), (ii) protease inhibitors (PIs) boosted with ritonavir, or (iii) an integrase/strand transfer inhibitor (INSTI) (Humphreys et al., 2010; Lee et al., 2014). Even with cART, however, the development of antiretroviral (ARV) resistance remains a formidable obstacle that often requires patients to switch to different drugs and may be considered the primary cause of reduced drug efficacy (Moniz et al., 2014; Stanojevic et al., 2014). Recent work, however, indicates that the host restriction factor, sterile alpha motif and histidine/aspartic acid containing domain protein 1 (SAMHD1), also affects NRTI efficacy against HIV-1 (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014). Understanding all facets of how currently available ARVs are affected at the molecular/cell level facilitates the conception of more efficacious drugs and facilitates progress toward a functional cure (Benard et al., 2011; Ekouevi et al., 2014; Ren et al., 2002; Witvrouw et al., 2004).
NRTIs exert their anti-HIV-1 effect by out-competing cellular dNTPs for incorporation into the growing proviral DNA during reverse transcription (Balzarini, 2000; Gao et al., 1993). The concentration of cellular dNTPs available for proviral DNA synthesis is affected by SAMHD1 phosphohydrolase activity and thought to impact non-dividing target cell types such as macrophages in particular. SAMHD1 phosphohydrolase activity leads to dNTPs degradation to deoxynucleosides (dNs) and triphosphates (Goldstone et al., 2011; Powell et al., 2011). SAMHD1 suppresses viral replication by depleting cellular dNTP concentrations, which, in turn, ultimately restricts the availability of dNTPs for proviral DNA synthesis in macrophages. Recently, studies have shown SAMHD1 does not degrade NRTIs containing chemically altered sugar moieties (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014). Thus, it is conceivable that SAMHD1, which delays HIV-1 reverses transcription by depleting dNTP substrates, further suppresses HIV-1 replication by enhancing NRTI potency against HIV-1. This favorable outcome, however, may not necessarily translate to the treatment of HIV-2 infections.
Human immunodeficiency virus type 2 (HIV-2) infection is an additional and important cause of HIV disease in West Africa and has limited spread to other regions of the world (da Silva et al., 2008; de Silva et al., 2013; Mansson et al., 2010). The 2013 World Health Organization guidelines recommend the combined use of either three NRTIs or two NRTIs plus one protease inhibitor (PI) as the initial cART regimen for HIV-2 infection (Benard et al., 2011; Gupta et al., 2014). However, the clinical, immunological, and virological outcomes for recommended HIV-2 cART are suboptimal (Benard et al., 2011; Ekouevi et al., 2014). Although there likely exist many factors that contribute to the suboptimal clinical outcome observed for the recommended treatment, there exists some in vitro evidence that may, at least, partially account for this observation. For example, it is well known that HIV-1 and HIV-2 NRTI-resistance mutation development differs in vivo (Smith et al., 2009). It has been suggested that structural differences in reverse transcriptase (RT) may account for the discrepancy between NRTI potency in HIV-1 and HIV-2 infections (Boyer et al., 2006) and recent work has highlighted the potential role of viral protein X (Vpx) in reducing NRTI efficacy in vitro (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014).
Vpx is encoded by HIV-2 and some lineages of simian immunodeficiency virus (SIV) but not HIV-1 (Fregoso et al., 2013). Vpx augments viral infectivity in non-dividing cells (Kappes et al., 1991) by the degradation of SAMDH1 (Hrecka et al., 2011; Laguette et al., 2011) and consequently increasing dNTP concentrations in primary human cells (Baldauf et al., 2012; Berger et al., 2011; Hrecka et al., 2011; Kim et al., 2012; Laguette et al., 2011; Lahouassa et al., 2012; St Gelais et al., 2012; White et al., 2012; Wu, 2012). In addition to hydrolyzing deoxynucleoside triphosphates, SAMHD1 has been reported to have ribonuclease activity (Choi et al., 2015; Ryoo et al., 2014), making its role in lentiviral infections more complex and not completely understood (Seamon et al., 2015; White et al., 2013). SAMHD1 is found in both cycling (activated CD4+ T cells) and non-dividing cells (macrophage, dendritic and resting CD4+ T cells) target cell types. However, SAMHD1 restriction of HIV has only been reported in non-dividing cells (Baldauf et al., 2012; Berger et al., 2011; Descours et al., 2012; Hrecka et al., 2011; Julg et al., 2010; Laguette et al., 2011; Riveira-Munoz et al., 2014). This lack of antiviral activity in cycling cells has been attributed to both reduction in SAMHD1 protein level and T592 phosphorylation of SAMHD1, which may regulate ribonuclease activity (Welbourn et al., 2013; White et al., 2013) and may influence dNTPase activity (Pauls et al., 2014a, 2014b; Ruiz et al., 2015; Yan et al., 2015). Therefore, SAMHD1 should have a minimal impact on HIV/SIV infection in activated CD4+ T cells. Vpx may have additional roles important for HIV-2/SIV biology, including modulation of the interferon response (Dragin et al., 2013; Goujon et al., 2013), assisting the preintegration complex transport into the nucleus (Cheng et al., 2008; Fujita et al., 2012; Pancio et al., 2000) or promoting cell cycle arrest (Bouzar et al., 2003).
Since NRTIs must outcompete dNTPs during reverse transcription (Gao et al., 1993), it is likely that an increase in dNTP concentrations may reduce NRTI efficacy in the context of HIV-2/SIV infections (Amie et al., 2013). Indeed, previous work from our group showed that pretreating activated CD4+ T cells and monocyte-derived macrophages (MDMs) with virus-like particles (VLPs) containing Vpx led to an increase in cellular dNTP concentrations that directly compete with NRTIs, consequently reducing their potency (Amie et al., 2013). More recently, Huber et al. showed similar results in which AZT, TDF, d4T and EFdA became less effective at inhibiting infection in cell lines not expressing SAMHD1, whereas ddI and 3TC sensitivity did not change (Huber et al., 2014). However, Ballana et al. (2014) reported that the presence of Vpx only reduced AZT and d4T potency out of a variety of NRTIs tested, suggesting that only thymidine analog NRTIs are affected in this context. These reported studies employed VSV-G-pseudotyped VLPs to introduce Vpx to target SAMHD1 in the cell types tested, which is a unnatural way to delivery Vpx, and did not directly evaluate the contribution of different RTs in these different viruses. Thus, here, we compared HIV-1, SIV (as a surrogate for HIV-2) and RT-SHIV to evaluate NRTI efficacy in the context of virion-associated Vpx and RT origin. We found that sensitivity to NRTIs was comparable for HIV-1, SIV and RT-SHIV in activated CD4+ T cells. Interestingly, SIVΔVpx and RT-SHIVΔVpx were more sensitive to AZT, 3TC and TDF as compared to parental control viruses in CD4+ T cells. When NRTI sensitivity was evaluated in MDMs, HIV-1, SIVΔVpx and RT-SHIVΔVpx infection were most sensitive to AZT, independent from the RT examined. The data reported here provide an additional explanation, that both cell type and virion-associated Vpx influences sensitivity to NRTIs.
Results
HIV-1, SIV and RT-SHIV show similar sensitivity to NRTI in activated CD4+ T cells using a single-cycle GFP expression infection assay
Previous studies tested the effect of the SAMHD1/Vpx interplay on HIV-1 infection (Baldauf et al., 2012; Bobadilla et al., 2012; Hrecka et al., 2011; Kim et al., 2012; Laguette et al., 2011; Lahouassa et al., 2012; Pauls et al., 2012; St Gelais et al., 2012; White et al., 2012, 2013). Fewer studies have examined the influence of SAMHD1 on NRTI efficacy, which employed Vpx-containing virus-like-particles (VLPs) to deliver the packaged Vpx protein to the target cells (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014). However, in natural infections, Vpx-encoding lentiviruses such as HIV-2 and SIVsm not only deliver their packaged Vpx protein to the target cells but also express Vpx from the integrated proviral DNAs in the infected cell. In addition, there exists evidence that the structural differences between the RTs encoded by HIV-1, HIV-2 and SIV to account for the observed differences in AZT resistance development (Herschhorn and Hizi, 2010; Ren et al., 2002). These structural differences appear to translate to the biochemical properties of RT such as enzyme/substrate fidelity and NRTI incorporation rates (Bakhanashvili and Hizi, 1992; Diamond et al., 2001; Lloyd et al., 2014; Skasko et al., 2009; Taube et al., 1997). Thus, it is conceivable that the differences between HIV-1 and SIV RTs may impact NRTI efficacy in addition to the SAMHD1-mediated dNTP metabolism alteration.
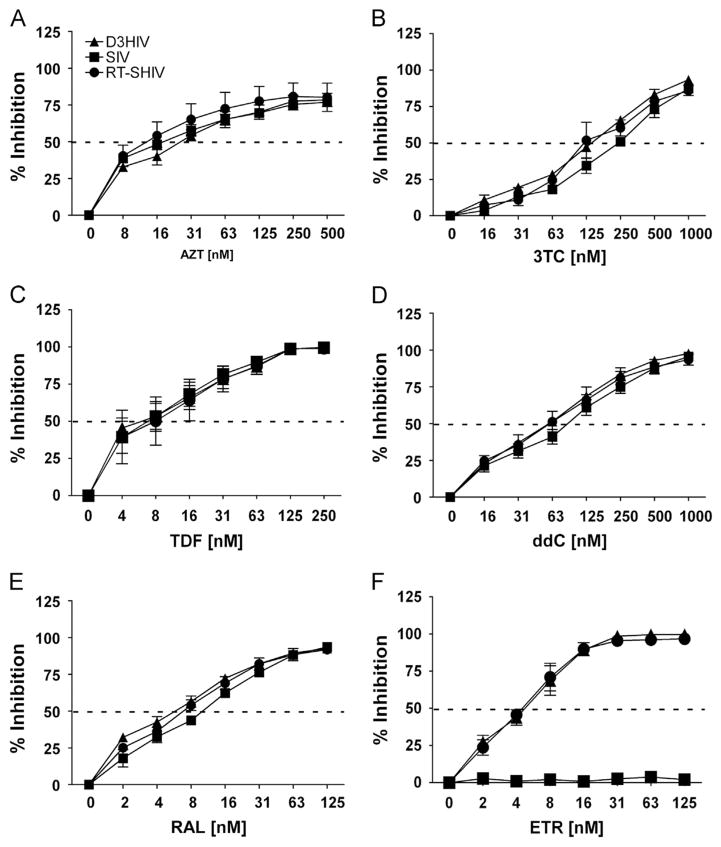
Activated CD4+ T cells are very permissive to HIV-1 infection as a result of high expression of the CD4 receptor on the cell surface, high cellular dNTP concentrations and modulation of SAMHD1 (Welbourn et al., 2013; White et al., 2013). First we evaluated the contribution of RT differences in the presence of Vpx to NRTI efficacy in activated CD4+ T cells. For this, we replaced SIV RT with HIV-1 RT to create a RT-SHIV vector. As shown in Fig. 1, we tested RT-SHIV (filled circle) in tandem with HIV-1 (filled triangle) and SIV (filled square) against a variety of NRTIs in our CD4+ T cell infection assay. We observed zidovudine (AZT; Fig. 1A), lamivudine (3TC; Fig. 1B), tenofovir disoproxil fumarate (TDF; Fig. 1C), and dideoxycytidine (ddC; Fig. 1D) showed little differences in sensitivity among these three viruses. As shown in Table 1, the ratio of IC50 for SIV and RT-SHIV to that of HIV-1, and IC50 of RT-SHIV to that of SIV, were less than two-fold for all drugs tested. As controls, we utilized the integrase inhibitor raltegravir (RAL; Fig. 1E), and the NNRTI etravirine (ETR; Fig. 1F). RAL inhibited infection of all three viral constructs within two-fold of each other (Table 1), whereas ETR only inhibited HIV-1 and RT-SHIV infections in CD4+ T cells, validating the RT differences between these three viruses.
Fig. 1.
Dose–response of NRTIs in CD4+ Tcells challenged with VSV-G-pseudotyped HIV-1, SIV and RT-SHIV. Percent inhibition (y-axis) of CD4+ Tcells treated with increasing concentrations (x-axis) of AZT (A), 3TC (B), TDF (C) ddC (D), RAL (E), and ETR (F), and then challenged with VSV-G-pseudotyped HIV-1 (filled diamonds), SIV (filled squares) and RT-SHIV (filled circles). Dashed line transects curve to show where 50% inhibition occurs. Data points depict the means and standard error of the means from three to four independent experiments.
Table 1.
IC50 vales for CD4 + T cells.
| IC50 ± SD [nM]
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | n | HIV-1
|
SIV
|
RT-SHIV
|
Fold change
|
|||||||
| Mean | SD | Mean | SD | Mean | SD | SIV/HIV-1 | RT-SHIV/HIV-1 | RT-SHIV/SIV | ||||
| AZT | 3 | 14.8 | 4.3 | 9 | 1.5 | 8.7 | 2.6 | 0.6 | 0.6 | 1 | ||
| 3TC | 3 | 193 | 44 | 289 | 30 | 154 | 40 | 1.5 | 0.8 | 0.5 | ||
| TDF | 4 | 6 | 2.5 | 8.6 | 5.7 | 6.4 | 2.7 | 1.4 | 1.1 | 0.7 | ||
| ddC | 3 | 66.8 | 35.3 | 96.8 | 35.3 | 57.2 | 8.7 | 1.5 | 0.9 | 0.6 | ||
| ETR | 3 | 4.9 | 1.7 | – | – | 4.4 | 1.4 | – | 0.9 | – | ||
| RAL | 3 | 5 | 1.4 | 9.7 | 3 | 6.3 | 1 | 1.9 | 1.3 | 0.6 | ||
| SIV
|
SIVΔVpx
|
RT-SHIV
|
RT-SHIVΔVpx
|
|||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | SIV/SIVΔVpx | RT-SHIV/RT-SHIVΔVpx | |||
|
| ||||||||||||
| AZT | 3 | 120 | 102 | 9 | 4.2 | 84.4 | 66.2 | 8 | 3.9 | 13.3 | 10.6 | |
| 3TC | 3 | 347 | 83.2 | 114 | 29 | 206 | 61.6 | 67 | 21.7 | 3 | 3.1 | |
| TDF | 3 | 12.7 | 9.8 | 2.1 | 1.4 | 10.9 | 8.4 | 1.9 | 1.2 | 6 | 5.7 | |
| RAL | 3 | 9.7 | 3 | 13.9 | 9.2 | 6.3 | 1.1 | 6 | 1.1 | 0.7 | 1.1 | |
| ETR | 4 | – | – | – | – | 2.9 | 1.6 | 2.3 | 1.1 | – | 1.3 | |
Vpx contributes to NTRI sensitivity in activated CD4+ T cells
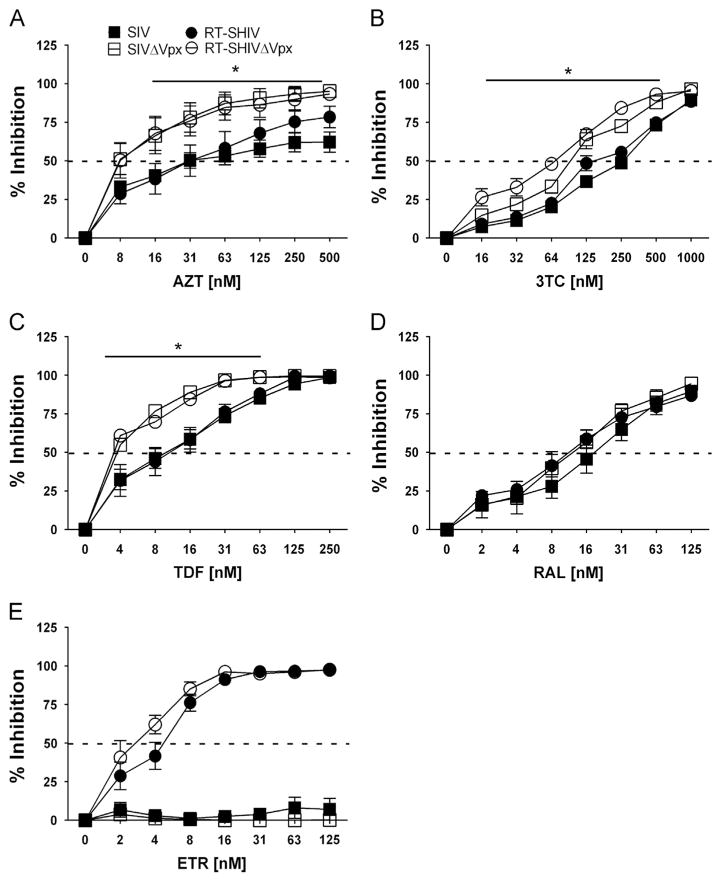
To evaluate the contribution of Vpx to NRTI efficacy, SIVΔVpx and RT-SHIVΔVpx vectors were generated and tested in tandem with SIV and RT-SHIV against a limited number of NRTIs. We found that SIVΔVpx (open square) versus SIV (filled square) and RT-SHIVΔVpx (open circle) versus RT-SHIV (filled circle) exhibited significantly (*) lower IC50s to AZT, 3TC and TDF (Fig. 2A, B and C, respectively). For the controls, all viruses were similarly sensitive to RAL (Fig. 2D), whereas only RT-SHIV and RT-SHIVΔVpx viruses were sensitive to ETR (Fig. 2E; 1.3-fold) as expected. Fold change comparisons for the viruses are displayed in Table 1.
Fig. 2.
Dose–response of NRTIs in activated CD4+ T cells challenged with molecular clones that code, or do not code, for Vpx. Percent inhibition (y-axis) of CD4+ T cells treated with increasing concentrations (x-axis) of AZT (A), 3TC (B), TDF (C), RAL (D) and ETR (E) before challenge with SIV (filled squares), SIVΔVpx (open squares), RT-SHIV (filled circles) and RT-SHIVΔVpx (open circles). Dashed line transects curve to show where 50% inhibition occurs. Significant differences in dose–response curves are indicated with an asterisk (*). Data points depict the means and standard error of the means from three to four independent experiments.
Since activated CD4+ T cells already have high cellular dNTP levels well above the Km for reverse transcriptase (Kennedy et al., 2010; Lenzi et al., 2014), we did not expect Vpx to affect viral sensitivity to any NRTIs. Our observation implies that the virion-associated Vpx may have additional roles in regulating viral sensitivity to NRTIs in activated CD4+ T cells, in addition to regulating SAMHD1 antiviral activities in macrophages (Kim et al., 2012; St Gelais and Wu, 2011).
HIV-1, SIV and RT-SHIV demonstrate different sensitivities towards NRTIs in MDMs
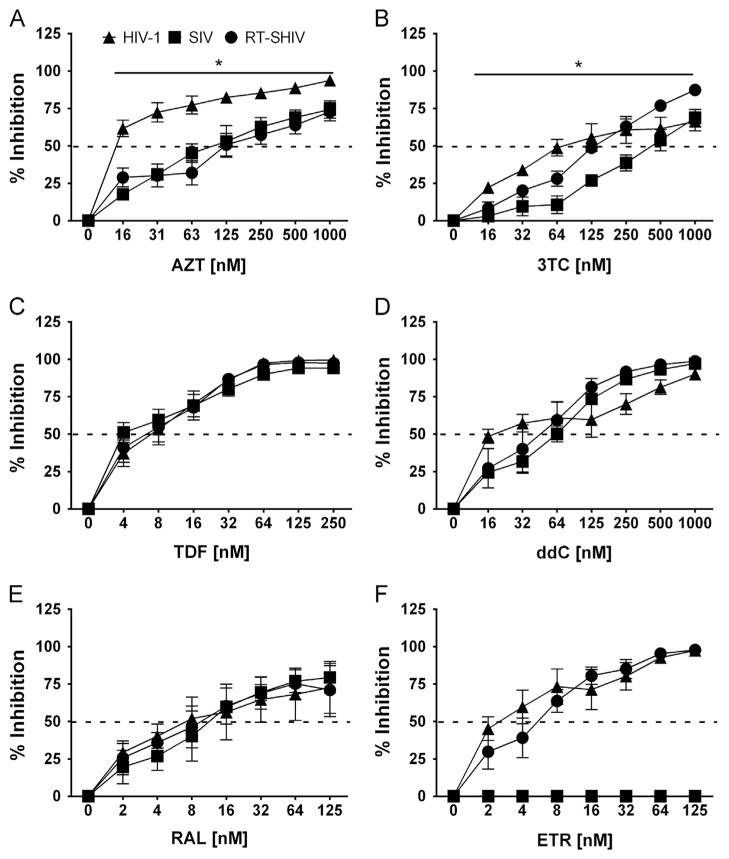
Previous studies reported VLPs reduce anti-HIV-1 activity of various NRTIs primarily in macrophages, supporting that SAMHD1 affects NRTI sensitivity of HIV-1 by modulating cellular dNTP levels (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014). Using a single-cycle GFP expression infection assay for MDMs, we observed significantly greater HIV-1 sensitivity to AZT (Fig. 3A) relative to SIV and RT-SHIV (Table 2). 3TC (Fig. 3B), HIV-1 was 2.6-fold more sensitive than SIV, whereas RT-SHIV was comparable to HIV having 1.3-fold difference with their IC50 values. All three viruses showed comparable trends in sensitivity for TDF (Fig. 3C) and ddC (Fig. 3D), with IC50 values within two-fold of each other (Table 2). Moreover, RAL (Fig. 3E) inhibited infection of all three viral constructs comparably, whereas ETR (Fig. 3F) only inhibited HIV-1 and RT-SHIV infection, confirming the RT differences in these three viral constructs.
Fig. 3.
Dose–response of ARVs in MDMs challenged with VSV-G-pseudotyped HIV-1, SIV and RT-SHIV expressing GFP in vitro. Percent inhibition (y-axis) of MDMs treated with increasing concentrations (x-axis) of AZT (A), 3TC (B), TDF (C) ddC (D), RAL (E), and ETR (F), and then challenged with VSV-G-pseudotyped HIV-1 (filled diamonds), SIV (filled squares) and RT-SHIV (filled circles). Dashed line transects curve to show where 50% inhibition occurs. Significant differences in dose–response curves are indicated with an asterisk (*). Data points depict the means and standard error of the means from four to five independent experiments.
Table 2.
IC50 vales for MDMs.
| IC50 ± SD [nM]
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | n | HIV-1
|
SIV
|
RT-SHIV
|
Fold change
|
|||||||
| Mean | SD | Mean | SD | Mean | SD | SIV/HIV-1 | RT-SHIV/HIV-1 | RT-SHIV/SIV | ||||
| AZT | 3 | 10.1 | 4.6 | 164 | 144 | 217 | 186 | 16.2 | 21.4 | 1.3 | ||
| 3TC | 3 | 131 | 71 | 338 | 20 | 166 | 66 | 2.6 | 1.3 | 0.5 | ||
| TDF | 4 | 8.1 | 5.5 | 7.3 | 5.4 | 4.3 | 2.4 | 0.9 | 0.5 | 0.6 | ||
| ddC | 3 | 33.7 | 45.1 | 62 | 22 | 54.7 | 45.2 | 1.8 | 1.6 | 0.9 | ||
| ETR | 3 | 4.8 | 4.3 | – | – | 6.1 | 4.2 | – | 1.3 | – | ||
| RAL | 3 | 19.6 | 1.3 | 12.2 | 3.5 | 12.4 | 5.4 | 0.6 | 0.6 | 1 | ||
| SIV
|
SIVΔVpx
|
RT-SHIV
|
RT-SHIVΔVpx
|
|||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | SIV/SIVΔVpx | RT-SHIV/RT-SHIVΔVpx | |||
|
| ||||||||||||
| AZT | 5 | 105 | 196 | 16.4 | 2.6 | 143 | 201 | 11.3 | 6 | 6.4 | 12.7 | |
| 3TC | 5 | 123 | 237 | 103 | 94 | 119 | 35.4 | 68.5 | 71.1 | 1.2 | 1.7 | |
| TDF | 5 | 4.2 | 3.2 | 11.4 | 11.6 | 10.4 | 6.6 | 9.7 | 6.4 | 0.4 | 1.1 | |
| RAL | 4 | 26 | 20 | 16.1 | 14.5 | 10.1 | 9.1 | 4.1 | 2.9 | 1.6 | 2.5 | |
| ETR | 4 | – | – | – | – | 12 | 4 | 5 | 1.4 | – | 2.4 | |
Recently, Huber et al. and Ballana et al. showed Vpx+VLP treatment decreased sensitivity towards AZT using both cell lines and human primary MDMs and CD4+ T cells, while observing very little change in sensitivities towards other NRTIs (Ballana et al., 2014; Huber et al., 2014). Our observation that the IC50 values of SIV and RT-SHIV to most NRTIs tested are relatively similar to these reports (Table 2), suggesting that the RT difference in SIV and RT-SHIV does not significantly contribute to their sensitivity to NRTIs tested in MDMs.
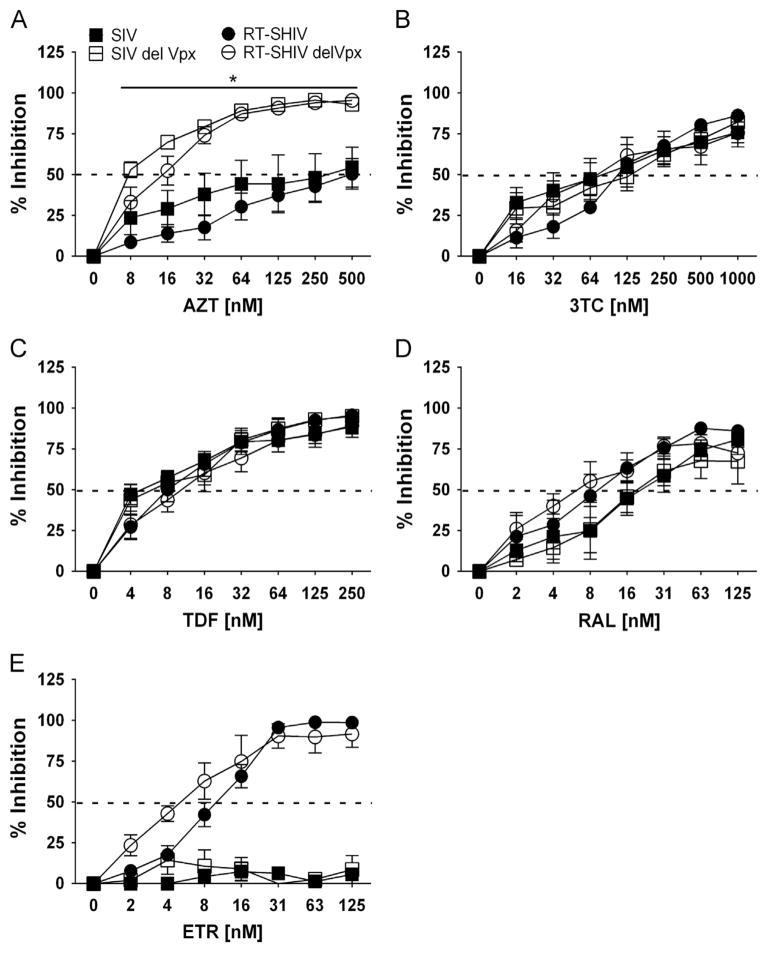
Depletion of Vpx results in an increase sensitivity towards AZT in MDMs
Since previous studies used VLPs containing Vpx to counteract SAMHD1, we directly tested the role of virion-associated Vpx in viral sensitivity to NRTIs in MDMs. The SIVΔVpx (open squares) and RT-SHIVΔVpx (open circles) vectors (Fig. 4A) were more sensitive to AZT as compared to their parental vectors, SIV (filled squares) and RT-SHIV (filled circles). As displayed in Table 2, the IC50 values for SIV/SIVΔVpx and RT-SHIV/RT-SHIVΔVpx were 6.4-and 12.7-fold, respectively. The sensitivity of these viruses to 3TC (Fig. 4B) and TDF (Fig. 4C) were comparable and IC50 values were within 2-fold range (Table 2). The RAL (Fig. 4D) and ETR (Fig. 4E) controls were more effective in RT-SHIVΔVpx than RT-SHIV vector, give fold changes of 2.5- and 2.4-fold, respectively. Overall, these data indicate that the ΔVpx viruses were more sensitive to AZT treatment in MDMs, whereas Vpx does not significantly influence viral sensitivity towards other NRTIs.
Fig. 4.
Dose–response of NRTIs in MDMs challenged with molecular clones that code, or do not code, for Vpx. Percent inhibition (y-axis) of MDMs treated with increasing concentrations (x-axis) of AZT (A), 3TC (B), TDF (C), RAL (D) and ETR (E) before challenge with SIV (filled squares), SIVΔVpx (open squares), RT-SHIV (filled circles) and RT-SHIVΔVpx (open circles). Dashed line transects curve to show where 50% inhibition occurs. Significant differences in dose–response curves are indicated with an asterisk (*). Data points depict the means and standard error of the means from four to five independent experiments.
Discussion
HIV-1 does not code for Vpx. However, non-human primate models used to evaluate anti-HIV-1 RT inhibitor efficacy often employ SIV encoding HIV-1 RT in lieu of SIV RT; HIV-1 RT is substituted in place of SIV RT given the differences between these enzymes and NNRTI efficacy (Ambrose et al., 2004, 2014; Balzarini et al., 1995; North et al., 2014; Wang et al., 2014; Witvrouw et al., 2004). Although the importance of substituting HIV-1 RT in lieu of SIV RT in this context is recognized, the relationship between Vpx and evaluation of NRTI efficacy in the context of RT-SHIV constructs, to our knowledge, has not been addressed prior to this report.
Vpx-mediated degradation of SAMHD1 increases dNTP concentrations in MDMs (Lahouassa et al., 2012). This would conceivably increase dNTP/NRTI-TP competition as substrates for RT (Amie et al., 2013; Ballana et al., 2014; Huber et al., 2014). Recent work has suggested that RT efficiencies are different among SIV and HIV-1 according to dNTP concentrations (Lenzi et al., 2014; Skasko et al., 2009) and postulates that HIV-1 RT evolved to be more efficient at low dNTP concentrations. Thus, the observation that NRTIs have similar lower activities against SIV and RT-SHIV in macrophages relative to HIV-1 and ΔVpx counterpart constructs is expected since the presence of Vpx increases dNTP concentrations in the cytoplasm where reverse transcription takes place; higher dNTP concentrations is likely directly proportional to NRTI/dNTP competition.
In this study we evaluated the contributions of virion-associated Vpx, and also RT-SHIV to allow us to test the role of RTs in NRTI sensitivity with respect to Vpx. For MDMs, our studies showed that SIV and RT-SHIV were less sensitive towards AZT, ddC and 3TC as compared to HIV-1. The sensitivity of HIV-1 to AZT, 3TC and ddC in MDMs were consistent with recent reports (Ballana et al., 2014; Huber et al., 2014). SIVΔVpx and RT-SHIVΔVpx were more sensitive to AZT as compared to SIV and RT-SHIV, however sensitivity was not altered for 3TC and TDF in MDMs (Fig. 4B and C, and Table 2). For activated CD4+ T cells, the sensitivity towards NRTIs did not significantly change for HIV-1, SIV and RT-SHIV (Fig. 1 and Table 1). However, deletion of Vpx influenced the viral sensitivity to AZT, 3TC, and TDF in activated CD4+ T cells (Fig. 2A C), but not for RAL and ETR, which do not require phosphorylation to become active.
Treatment of activated CD4+ T cells and MDMs with NRTIs led to different cellular concentrations of NRTI-TP (Gavegnano et al., 2013). Huber et al. (2014) hypothesized that kinases associated with AZT and d4T phosphorylation may be the rate-limiting step in their activation, and influence the levels of dNTPs and NRTI-TPs. Since SAMHD1 hydrolyzes canonical dNTP, but not NRTI-TPs, the Vpx effect on NRTI sensitivity reduction could be primarily due to 1) increase of dNTP biosynthesis, 2) decrease of dNTP degradation, and 3) decrease of NRTI-TP synthesis. While it is not clear that SAMHD1 is directly involved in all of these three possibilities, it is plausible that Vpx can influence the NRTI sensitivity using all three possibilities in different cell types.
Importantly, this work suggests that instead of structural and enzymatic differences between RTs, Vpx influences NRTI sensitivity in different cell types. Therefore, special consideration may be needed when evaluating the sensitivity profiles of NRTIs when using RT-SHIVs coding for Vpx, since an underestimate of the antiviral efficacy of NRTIs may be occurring.
Materials and methods
Cells and cell culture
Monocytes were isolated from whole blood (New York Blood Service, Long Island New York) by using MACS® CD14+ beads as described previously (Hollenbaugh et al., 2013) and cultured in the presence of 5 ng/ml human GM-CSF (Miltenyi Biotec). The resulting MDMs were utilized on day 7 for all experiments. From monocyte-depleted PBMCs, MACS CD4 beads were used to enrich for CD4+ T cells as previously described (Hollenbaugh et al., 2011). Cells were cryopreserved (10 million cells/ml, −80 °C) until reanimated for experimentation. Upon reanimation in DMEM (5% FBS/30 million cells), 60–120 million CD4+ T cells were activated using 5 μg/ml of PHA (Sigma) and 10 ng/ml IL-2 (NIH AIDS Reagent Program). On day 3 post-reanimation, fresh medium and 10 ng/ml IL-2 were added to cells and cells were permitted to expand for a further 48 h after which cells were utilized in experiments.
Drugs
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Zidovudine (AZT), Etravirine (ETR) TMC125 (Cat#11609) from Tibotec, Inc., Raltegravir (RAL) (Cat#11680) from Merck & Company, Inc., and tenofovir disoproxil fumarate (TDF; Cat#10198). 3TC and ddC were purchased from Sigma-Aldrich.
Molecular clones and cloning
DHIV3-GFP plasmid encodes the HIV-1 NL4-3 genome with the enhanced GFP gene in place of the HIV-1 nef gene and has a deleted envelope gene (Andersen et al., 2006). pSIVmac239-GFP was a kind gift from Dr. Diaz-Griffero (Albert Einstein College of Medicine, NYC). 5′-pSHIV, which contained the HIV-1 HXB2 RT gene, was a kind gift from Dr. North (Emory University). We sub-cloned the HIV-1 RT from p5′–SHIV into pSIVmac239-GFP by digesting with NheI and PacI followed by site-directed mutagenesis (Quikchange, Agilent) of the primer-binding site to make it complementary to tRNALys sequence (Das et al., 1997). The resulting construct is the pSIVmac239-GFP with the HXB2 RT gene instead of the SIV RT gene. We called the construct ‘pRT-SHIVmac239-GFP′. Next, pSIVmac239-GFP vector, pRT-SHIVmac239-GFP and pSIVmac239 SpX ΔVpx, (the latter construct kindly provided by NIH AIDS Reagent Program), were digested with NheI and ClaI. The ΔVpx fragment was then subcloned into pSIVmac239-GFP and pRT-SHIVmac239-GFP to generate ‘pSIVmac239-ΔVpx-GFP′ and ‘pRT-SHIV-mac239-ΔVpx GFP′, respectively.
Generation of viruses
293FT cells (Invitrogen) were transfected with 20 μg of pVSV-G and 40 μg of pDHIV3 (HIV-1), pSIVmac239-GFP (SIV), pRT-SHIVmac239-GFP (RT-SHIV), pSIVmac239-ΔVpx-GFP (SIVΔVpx), or pRT-SHIVmac239-ΔVpx-GFP (RT-SHIVΔVpx) via polyethylenimine (PEI; 1 mg/mL) in a ratio of 1 μg of DNA to 3 μl PEI. Twenty-four hours post-transfection, the culture medium was removed and replaced with fresh DMEM containing 5% FBS with penicillin and streptomycin. Cell supernatants were harvested 48 h and 72 h post-transfection, centrifuged at 2500 rpm for 10 min to remove cellular debris. Virus was concentrated from cell supernatants via ultracentrifugation (22,000 rpm for 1.5 h in a SW32Ti rotor) and resuspended in cell culture medium after a short high-speed centrifugation to remove any excess debris (13,000 rpm for 1 min) and stored at −80 °C until use. VSV-G-pseudotyped viruses were assessed five days post-transduction in MDMs and 2 days post-transduction in CD4+ T cells for infectivity/GFP expression using a MACSQuant Analyzer. Data files were analyzed using the FlowJo software (TreeStar). Data was graphed using Prism software.
Evaluation of drug efficacy
Activated CD4+ T cells were seeded into a 96-well plate at 100,000 cells/well. Drugs were diluted from stock concentrations to 1000 nM and 125 nM for NRTIs and non-NRTIs, respectively. Drugs were then serially diluted to achieve 7 total dilutions (in order to obtain a dose–response curve) and then added to cells immediately. Cells were permitted to incubate with drugs for 2 h before the addition of virus and drug was present throughout the experiment. Seventy-two hours post-infection, cells were treated with DAPI (0.001 μg/μl), to account for dead cells during FACS collection (MACSQuant FACS instrument). Data files were analyzed using the FlowJo software.
Drug concentrations were prepared for experiments utilizing MDMs (250,000 cells/well, 24-well plate) as described for CD4+ T cells above. As with CD4+ T cells, MDMs were also permitted to incubate with drugs for 2 h prior to infections. Culture medium was changed 48 h post-infections with fresh medium containing drug (at concentrations used at experiment initiation). At day 5, MDMs were harvested by removing medium and adding 250 μl of 2.5% Trypsin to each experimental well and allowing incubating for 5 min at 37 °C, after which time the cells were gently scraped and transferred to the eppendorf tube and pelleted for 60 s at 6000 rpm. Pellets were suspended in 50 μl medium contain DAPI (0.001 μg/μl), to exclude for dead cells, and evaluated for GFP expression via flow cytometry (MACSQuant FACS machine).
Determination of IC50 values, graphing of dose–response curves and statistical analysis
Prism software was used for plotting the data. All the data sets were compared for significant differences (*) using Two-way ANOVA analysis and then Tukey multiple comparison test. To determine IC50 values, the mean values for each independent donor were combined and evaluated using log[inhibitor] vs. normalized response – variable slope. Graphs are plotted as the means and standard error of the means (SEM) for the donors.
Supplementary Material
Acknowledgments
This study was supported by NIH AI049781 (B.K.), GM104198 (B.K.), 5P30-AI-50409 Emory Centers for AIDS Research (R.F.S.), MH100999 (R.F.S), and the Department of Veterans Affairs (R.F.S.).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2015.08.006.
References
- Ambrose Z, Boltz V, Palmer S, Coffin JM, Hughes SH, Kewalramani VN. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J Virol. 2004;78:13553–13561. doi: 10.1128/JVI.78.24.13553-13561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose Z, Kline C, Polacino P, Hu SL. Dysregulation of multiple inflammatory molecules in lymph node and ileum of macaques during RT-SHIV infection with or without antiretroviral therapy. J Med Primatol. 2014;43:298–309. doi: 10.1111/jmp.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amie SM, Daly MB, Noble E, Schinazi RF, Bambara RA, Kim B. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J Biol Chem. 2013;288:20683–20691. doi: 10.1074/jbc.M113.472159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, DeHart JL, Zimmerman ES, Ardon O, Kim B, Jacquot G, Benichou S, Planelles V. HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS Pathog. 2006;2:e127. doi: 10.1371/journal.ppat.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhanashvili M, Hizi A. Fidelity of the RNA-dependent DNA synthesis exhibited by the reverse transcriptases of human immunodeficiency virus types 1 and 2 and of murine leukemia virus: mispair extension frequencies. Biochemistry. 1992;31:9393–9398. doi: 10.1021/bi00154a010. [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1689. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballana E, Badia R, Terradas G, Torres-Torronteras J, Ruiz A, Pauls E, Riveira-Munoz E, Clotet B, Marti R, Este JA. SAMHD1 specifically affects the antiviral potency of thymidine analog hiv reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2014;58:4804–4813. doi: 10.1128/AAC.03145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini J. Effect of antimetabolite drugs of nucleotide metabolism on the anti-human immunodeficiency virus activity of nucleoside reverse transcriptase inhibitors. Pharmacol Ther. 2000;87:175–187. doi: 10.1016/s0163-7258(00)00050-4. [DOI] [PubMed] [Google Scholar]
- Balzarini J, Weeger M, Camarasa MJ, De Clercq E, Uberla K. Sensitivity/resistance profile of a simian immunodeficiency virus containing the reverse transcriptase gene of human immunodeficiency virus type 1 (HIV-1) toward the HIV-1-specific non-nucleoside reverse transcriptase inhibitors. Biochem Biophys Res Commun. 1995;211:850–856. doi: 10.1006/bbrc.1995.1890. [DOI] [PubMed] [Google Scholar]
- Benard A, van Sighem A, Taieb A, Valadas E, Ruelle J, Soriano V, Calmy A, Balotta C, Damond F, Brun-Vezinet F, Chene G, Matheron S, Group AECS. Immunovirological response to triple nucleotide reverse-transcriptase inhibitors and ritonavir-boosted protease inhibitors in treatment-naive HIV-2-infected patients: the ACHIEV2E collaboration study group. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2011;52:1257–1266. doi: 10.1093/cid/cir123. [DOI] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobadilla S, Sunseri N, Landau NR. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 2012;20:514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzar AB, Guiguen F, Morin T, Villet S, Fornazero C, Garnier C, Gallay K, Gounel F, Favier C, Durand J, Balleydier S, Mornex JF, Narayan O, Chebloune Y. Specific G2 arrest of caprine cells infected with a caprine arthritis encephalitis virus expressing vpr and vpx genes from simian immunodeficiency virus. Virology. 2003;309:41–52. doi: 10.1016/s0042-6822(03)00014-x. [DOI] [PubMed] [Google Scholar]
- Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2006;2:e10. doi: 10.1371/journal.ppat.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Belshan M, Ratner L. Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 Vpx-mediated preintegration complex. J Virol. 2008;82:1229–1237. doi: 10.1128/JVI.00540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ryoo J, Oh C, Hwang S, Ahn K. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology. 2015;12:46. doi: 10.1186/s12977-015-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva ZJ, Oliveira I, Andersen A, Dias F, Rodrigues A, Holmgren B, Andersson S, Aaby P. Changes in prevalence and incidence of HIV-1, HIV-2 and dual infections in urban areas of Bissau, Guinea-Bissau: is HIV-2 disappearing? AIDS. 2008;22:1195–1202. doi: 10.1097/QAD.0b013e328300a33d. [DOI] [PubMed] [Google Scholar]
- Das AT, Klaver B, Berkhout B. Sequence variation of the human immunodeficiency virus primer-binding site suggests the use of an alternative tRNA (Lys) molecule in reverse transcription. J Gen Virol. 1997;78:837–840. doi: 10.1099/0022-1317-78-4-837. [DOI] [PubMed] [Google Scholar]
- de Silva TI, van Tienen C, Onyango C, Jabang A, Vincent T, Loeff MF, Coutinho RA, Jaye A, Rowland-Jones S, Whittle H, Cotten M, Hue S. Population dynamics of HIV-2 in rural West Africa: comparison with HIV-1 and ongoing transmission at the heart of the epidemic. AIDS. 2013;27:125–134. doi: 10.1097/QAD.0b013e32835ab12c. [DOI] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Kimata J, Kim B. Identification of a simian immunodeficiency virus reverse transcriptase variant with enhanced replicational fidelity in the late stage of viral infection. J Biol Chem. 2001;276:23624–23631. doi: 10.1074/jbc.M102496200. [DOI] [PubMed] [Google Scholar]
- Dragin L, Nguyen LA, Lahouassa H, Sourisce A, Kim B, Ramirez BC, Margottin-Goguet F. Interferon block to HIV-1 transduction in macrophages despite SAMHD1 degradation and high deoxynucleoside triphosphates supply. Retrovirology. 2013;10:30. doi: 10.1186/1742-4690-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekouevi DK, Tchounga BK, Coffie PA, Tegbe J, Anderson AM, Gottlieb GS, Vitoria M, Dabis F, Eholie SP. Antiretroviral therapy response among HIV-2 infected patients: a systematic review. BMC Infect Dis. 2014;14:461. doi: 10.1186/1471-2334-14-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9:e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Nomaguchi M, Adachi A, Otsuka M. SAMHD1-dependent and -independent functions of HIV-2/SIV Vpx protein. Front Microbiol. 2012;3:297. doi: 10.3389/fmicb.2012.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao WY, Cara A, Gallo RC, Lori F. Low levels of deoxynucleotides in peripheral blood lymphocytes: a strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA. 1993;90:8925–8928. doi: 10.1073/pnas.90.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavegnano C, Detorio MA, Bassit L, Hurwitz SJ, North TW, Schinazi RF. Cellular pharmacology and potency of HIV-1 nucleoside analogs in primary human macrophages. Antimicrob Agents Chemother. 2013;57:1262–1269. doi: 10.1128/AAC.02012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Goujon C, Schaller T, Galao RP, Amie SM, Kim B, Olivieri K, Neil SJ, Malim MH. Evidence for IFNalpha-induced, SAMHD1-independent inhibitors of early HIV-1 infection. Retrovirology. 2013;10:23. doi: 10.1186/1742-4690-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Williams B, Montaner J. Realizing the Potential of Treatment as Prevention: Global ART Policy and Treatment Coverage. Current HIV/AIDS reports. 2014 doi: 10.1007/s11904-014-0230-z. [DOI] [PubMed] [Google Scholar]
- Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell Mol Life Sci: CMLS. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Gee P, Baker J, Daly MB, Amie SM, Tate J, Kasai N, Kanemura Y, Kim DH, Ward BM, Koyanagi Y, Kim B. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 2013;9:e1003481. doi: 10.1371/journal.ppat.1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Munger J, Kim B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology. 2011;415:153–159. doi: 10.1016/j.virol.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AD, Michailidis E, Schultz ML, Ong YT, Bloch N, Puray-Chavez MN, Leslie MD, Ji J, Lucas AD, Kirby KA, Landau NR, Sarafianos SG. SAMHD1 has differential impact on the efficacies of HIV nucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2014;58:4915–4919. doi: 10.1128/AAC.02745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys EH, Chang LW, Harris J. Antiretroviral regimens for patients with HIV who fail first-line antiretroviral therapy. The Cochrane database of systematic reviews. 2010:CD006517. doi: 10.1002/14651858.CD006517.pub2. [DOI] [PubMed] [Google Scholar]
- Julg B, Pereyra F, Buzon MJ, Piechocka-Trocha A, Clark MJ, Baker BM, Lian J, Miura T, Martinez-Picado J, Addo MM, Walker BD. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2010;51:233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes JC, Conway JA, Lee SW, Shaw GM, Hahn BH. Human immunodeficiency virus type 2 vpx protein augments viral infectivity. Virology. 1991;184:197–209. doi: 10.1016/0042-6822(91)90836-z. [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Gavegnano C, Nguyen L, Slater R, Lucas A, Fromentin E, Schinazi RF, Kim B. rNTPs as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem. 2010;285:39380–39391. doi: 10.1074/jbc.M110.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem. 2012;287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FJ, Amin J, Carr A. Efficacy of initial antiretroviral therapy for HIV-1 infection in adults: a systematic review and meta-analysis of 114 studies with up to 144 weeks’ follow-up. PloS One. 2014;9:e97482. doi: 10.1371/journal.pone.0097482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi GM, Domaoal RA, Kim DH, Schinazi RF, Kim B. Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses. Retrovirology. 2014;11:111. doi: 10.1186/s12977-014-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SB, Kent SJ, Winnall WR. The high cost of fidelity. AIDS Res Hum Retroviruses. 2014;30:8–16. doi: 10.1089/aid.2013.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson F, Camara C, Biai A, Monteiro M, da Silva ZJ, Dias F, Alves A, Andersson S, Fenyo EM, Norrgren H, Unemo M. High prevalence of HIV-1, HIV-2 and other sexually transmitted infections among women attending two sexual health clinics in Bissau, Guinea-Bissau, West Africa. Int J STD AID. 2010;21:631–635. doi: 10.1258/ijsa.2010.009584. [DOI] [PubMed] [Google Scholar]
- Moniz P, Alcada F, Peres S, Borges F, Baptista T, Miranda AC, Antunes I, Aldir I, Ventura F, Nina J, Mansinho K. Durability of first antiretroviral treatment in HIV chronically infected patients: why change and what are the outcomes? J Int AIDS Soc. 2014;17:19797. doi: 10.7448/IAS.17.4.19797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TW, Villalobos A, Hurwitz SJ, Deere JD, Higgins J, Chatterjee P, Tao S, Kauffman RC, Luciw PA, Kohler JJ, Schinazi RF. Enhanced antire-troviral therapy in rhesus macaques improves RT-SHIV viral decay kinetics. Antimicrob Agents Chemother. 2014;58:3927–3933. doi: 10.1128/AAC.02522-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancio HA, Vander Heyden N, Ratner L. The C-terminal proline-rich tail of human immunodeficiency virus type 2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J Virol. 2000;74:6162–6167. doi: 10.1128/jvi.74.13.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E, Badia R, Torres-Torronteras J, Ruiz A, Permanyer M, Riveira-Munoz E, Clotet B, Marti R, Ballana E, Este JA. Palbociclib, a selective inhibitor of cyclin-dependent kinase4/6, blocks HIV-1 reverse transcription through the control of sterile alpha motif and HD domain-containing protein-1 (SAMHD1) activity. AIDS. 2014a;28:2213–2222. doi: 10.1097/QAD.0000000000000399. [DOI] [PubMed] [Google Scholar]
- Pauls E, Ballana E, Este JA. Nucleotide embargo by SAMHD1: a strategy to block retroviral infection. Antivir Res. 2012 doi: 10.1016/j.antiviral.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol. 2014b;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW. The Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Bird LE, Chamberlain PP, Stewart-Jones GB, Stuart DI, Stammers DK. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc Natl Acad Sci USA. 2002;99:14410–14415. doi: 10.1073/pnas.222366699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveira-Munoz E, Ruiz A, Pauls E, Permanyer M, Badia R, Mothe B, Crespo M, Clotet B, Brander C, Ballana E, Este JA. Increased expression of SAMHD1 in a subset of HIV-1 elite controllers. J Antimicrob Chemother. 2014;69:3057–3060. doi: 10.1093/jac/dku276. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Pauls E, Badia R, Torres-Torronteras J, Riveira-Munoz E, Clotet B, Marti R, Ballana E, Este JA. Cyclin D3-dependent control of the dNTP pool and HIV-1 replication in human macrophages. Cell Cycle. 2015;14:1657–1665. doi: 10.1080/15384101.2015.1030558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon KJ, Sun Z, Shlyakhtenko LS, Lyubchenko YL, Stivers JT. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skasko M, Diamond TL, Kim B. Mechanistic variations among reverse transcriptases of simian immunodeficiency virus variants isolated from African green monkeys. Biochemistry. 2009;48:5389–5395. doi: 10.1021/bi900346m. [DOI] [PubMed] [Google Scholar]
- Smith RA, Anderson DJ, Pyrak CL, Preston BD, Gottlieb GS. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J Infect Dis. 2009;199:1323–1326. doi: 10.1086/597802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, Wu L. SAMHD1: a new insight into HIV-1 restriction in myeloid cells. Retrovirology. 2011;8:55. doi: 10.1186/1742-4690-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic M, Siljic M, Salemovic D, Pesic-Pavlovic I, Zerjav S, Nikolic V, Ranin J, Jevtovic D. Ten years survey of primary HIV-1 resistance in Serbia: the occurrence of multiclass resistance. AIDS Res Hum Retroviruses. 2014;30:634–641. doi: 10.1089/aid.2013.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube R, Avidan O, Hizi A. The fidelity of misinsertion and mispair extension throughout DNA synthesis exhibited by mutants of the reverse transcriptase of human immunodeficiency virus type 2 resistant to nucleoside analogs. Eur J Biochem. 1997;250:106–114. doi: 10.1111/j.1432-1033.1997.00106.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Yao N, Cong Z, Jiang H, Qin C, Wei Q. Prophylactic and therapeutic effect of AZT/3TC in RT-SHIV infected Chinese-origin rhesus macaques. AIDS Res Ther. 2014;11:12. doi: 10.1186/1742-6405-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol. 2013;87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Carlos Valle-Casuso J, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology. 2012;436:81–90. doi: 10.1016/j.virol.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther. 2004;9:57–65. [PubMed] [Google Scholar]
- Wu L. SAMHD1: a new contributor to HIV-1 restriction in resting CD4+ T-cells. Retrovirology. 2012;9:88. doi: 10.1186/1742-4690-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J. CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J Biol Chem. 2015;290:13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.