Abstract
Background
To survive in a changing environment plants constantly monitor their surroundings. In response to several stresses and during photorespiration plants use reactive oxygen species as signaling molecules. The Arabidopsis thaliana catalase2 (cat2) mutant lacks a peroxisomal catalase and under photorespiratory conditions accumulates H2O2, which leads to activation of cell death.
Methods
A cat2 double mutant collection was generated through crossing and scored for cell death in different assays. Selected double mutants were further analyzed for photosynthetic performance and H2O2 accumulation.
Results
We used a targeted mutant analysis with more than 50 cat2 double mutants to investigate the role of stress hormones and other defense regulators in H2O2-mediated cell death. Several transcription factors (AS1, MYB30, MYC2, WRKY70), cell death regulators (RCD1, DND1) and hormone regulators (AXR1, ERA1, SID2, EDS1, SGT1b) were essential for execution of cell death in cat2. Genetic loci required for cell death in cat2 was compared with regulators of cell death in spontaneous lesion mimic mutants and led to the identification of a core set of plant cell death regulators. Analysis of gene expression data from cat2 and plants undergoing cell death revealed similar gene expression profiles, further supporting the existence of a common program for regulation of plant cell death.
Conclusions
Our results provide a genetic framework for further study on the role of H2O2 in regulation of cell death. The hormones salicylic acid, jasmonic acid and auxin, as well as their interaction, are crucial determinants of cell death regulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1964-8) contains supplementary material, which is available to authorized users.
Keywords: Programmed cell death, Reactive oxygen species, Salicylic acid, Jasmonic acid, Auxin, Lesion mimic mutants
Background
During their life cycle plants can experience multiple stress conditions – ranging from abiotic stresses (including fluctuating temperatures, water availability and light intensity) to biotic stresses (pathogen infection, insect attack). The early steps of plant defense against pathogens include recognition of conserved structures of the pathogen and activation of downstream responses [1]. In contrast, little information is available about the initial perception mechanisms during abiotic stresses. Down-stream from initial stress perception several signaling molecules have been identified, including changes in cytosolic Ca2+ concentration, production of reactive oxygen species (ROS) and activation of hormone signaling pathways including salicylic acid (SA), jasmonic acid (JA), ethylene and abscisic acid (ABA) [2]. Furthermore it is clear that defense signaling is not organized in linear parallel pathways, instead hormones have both synergistic and antagonistic interactions [2]. Consequently, the outcome of a stimulus is not determined by a single signaling molecule but a “web of interactions” involving multiple signaling pathways.
One of the early events after stress perception is the enhanced production of ROS, such as superoxide (O2.-) and hydrogen peroxide (H2O2) that are used as signaling molecules [3]. ROS signaling in turn alters the redox state, initiates Ca2+ signaling, activates protein kinases and cellular metabolic pathways [3]. Conversely, abiotic stress can alter the cell redox state which ultimately lead to increased ROS production [4]. Furthermore, ROS are produced in different cellular compartments which can activate separate signaling pathways [5]. This makes it important to distinguish the subcellular localization of ROS production and how these molecules integrate into other signaling pathways, including the regulation of programmed cell death (PCD) and changes in gene expression. Generally, ROS are removed by ROS scavenging enzymes including catalase and low molecular weight antioxidants including ascorbate and glutathione [4]. Catalases are localized in peroxisomes and detoxify H2O2 produced from glycolate oxidation [6]. Glycolate is formed during photorespiration when Rubisco uses O2 instead of CO2. It is estimated that a majority of H2O2 in the cell is produced during photorespiration and photosynthesis [7]. This makes photorespiration an important contributor to cellular redox state and this process may help protect cells and provide adaption to unfavorable conditions [8]. The importance of the interaction between ROS from different sources in defense signaling is reinforced by the study of plants deficient in both ASCORBATE PEROXIDASE and CATALASE, which are surprisingly more tolerant to different environmental conditions than plants deficient in only one scavenger [9]. Thus, ROS signaling is intimately connected to redox signaling as well as to the regulation of PCD [10].
PCD is an integral part of plant life and regulate stress, growth and developmental responses. Furthermore, in defense against pathogens, PCD at the site of infection known as the hypersensitive response (HR), is correlated with increased resistance to some pathogens [10]. Forward genetic screens in Arabidopsis thaliana have identified mutants with misregulated or spontaneous cell death. These mutants, called lesion mimic mutants (LMMs), have altered cell death phenotypes, which are usually accompanied by altered responses to pathogen infection [11, 12]. Since several LMMs exhibit large visible lesions on leaves and/or are dwarfs, they have been used in suppressor mutant screens or reverse genetics screens to identify additional regulators of PCD [11, 12]. SA has a central role in regulation of cell death. Many LMMs accumulate high amounts of SA and introduction of the bacterial enzyme salicylate hydroxylase (NahG) that degrades SA significantly reduces the extent of cell death in lsd1, acd5, acd6 and acd11 [12].
Arabidopsis cat2 mutants, deficient in CATALASE2, develop lesions that are day length and light intensity dependent [13–16]. PCD in cat2 can be prevented by introduction of a loss of function mutation in the main SA biosynthesis enzyme ISOCHORISMATE SYNTHASE1 (ICS1) (mutations in ICS1 are known as sid2 - salicylic acid induction deficient2) or by external supply of myoinositol, which repress ICS1 transcript accumulation [17, 18]. PCD in cat2 is long day-length dependent and is associated with changes in the glutathione redox status [13] but not in the ascorbate pool [16]. However, these responses are prevented when cat2 plants are grown at high CO2, which suppresses photorespiration and hence H2O2 production. In addition to lesion formation, the cat2 mutant also has an altered gene expression profile that includes genes related to SA and JA signaling [13, 18, 19]. Interestingly, altered expression of SA and JA marker genes is dependent on glutathione, the cat2 cad2 (cadmium sensitive2) double mutant with reduced glutathione biosynthesis, showed repression in SA and JA-associated gene expression [19, 20]. The target of glutathione in relation to SA signaling could be NPR1 (NONEXPRESSER OF PR GENES 1), but other as yet unidentified components may also be involved [19].
Lesion formation in cat2 is easily initiated through changes in growth conditions. In contrast to other LMMs, the exact source of initiating cell death in cat2 is also clear – increased H2O2 production in the peroxisome. Hence, cat2 can be used as a tool to understand the role of H2O2 in regulation of PCD. Here, we use cat2 to study the role of defense and hormone signaling in regulation of PCD employing a collection of 56 double and triple cat2 mutants. Through comparison of cat2 with other LMMs we are able to identify additional biological mechanisms involved in PCD regulation and establish a core set of PCD regulators.
Results and discussion
The cat2 mutant is a convenient tool to explore the role of H2O2 in regulation of PCD [15]. To systematically delineate the signal pathways downstream from ROS we used a genetic approach and crossed mutants related to plant hormones (ABA, JA, SA, ethylene and auxin) and defense signaling pathways into cat2 background (see Additional file 1: Table S1 for an overview of the genotypes used). Some of the cat2 double mutants used in this work have been independently generated and characterized in other reports [16, 18–20]. Most of these double mutants display the same phenotypes as in this study with some subtle differences. While cat2 sid2 [18] and cat2 apx1 (ascorbate peroxidase1) [21] had fully suppressed lesion formation in previous publications, in our growth conditions these double mutants had only partially suppressed lesion formation. Given that the cat2 phenotype is dependent on growth conditions, including photoperiod, light intensity, and likely also other factors such as humidity or soil nutrient content, some differences could be expected.
In vitro assay versus soil-grown plants
Two main assays were used to screen the mutant collection: (1) survival of in vitro grown plants on plates after restriction of gas exchange and altered day length, (2) lesion formation in 4 week old soil-grown plants. For restriction of gas exchange, the in vitro grown plants (14 days old) were transferred from short day (SD, 8/16 h day night) to photorespiration-inducing condition (long day; LD, 12/12 h) by sealing plates with parafilm to limit air influx. A similar assay but with different light conditions has been extensively used to study photorespiration and cat2 [22, 23]. The MS-agar media did not contain sugar, since in our growth conditions MS-media with sugar led to suppression of the cat2 phenotype, presumably due to reduced photosynthesis and repression of photorespiration (Additional file 2: Figure S1). The extent of cell death was evaluated during 8 days after changes of photoperiod (Table 1; Fig. 1, Additional file 3: Figure S2). Double mutants that led to increased survival included cat2 era1 (enhanced response to ABA1), cat2 axr1 (auxin resistant1), cat2 sgt1b, cat2 dnd1 (defense no death1) and cat2 as1 (asymmetric leaves1). Decreased survival was found in double mutants with defects in SA accumulation or SA signaling, cat2 sid2, cat2 eds1 (enhanced disease susceptibility1), cat2 ald1 (AGD2-like defense response protein1) and ascorbic acid biosynthesis cat2 vtc1 (vitamin c defective1) and cat2 vtc2 (Fig. 1).
Table 1.
Quantification of cat2 and cat2 double mutant survival and lesion formation
| Functional group | Genotypes | Survival (%) | p | Lesion (%) | ±SE | p | Reference to single mutant |
|---|---|---|---|---|---|---|---|
| Col-0 | 100.0 | * | 0 | 0 | * | ||
| cat2 | 30.0 | 5.8 | 0.3 | [16] | |||
| Double mutants with cat2 | |||||||
| Abscisic acid | era1 | 96.7 | * | 0 | 0 | * | [56] |
| abcg25 | 32.9 | 2.4 | 1.2 | * | [99] | ||
| abcg40 | 6.7 | 3.0 | 1.6 | * | [100] | ||
| abi1-1 | 70.0 | * | 2.7 | 2.2 | * | [101] | |
| Auxin/Jasmonic acid | axr1 | 88.9 | * | 0 | 0 | * | [51] |
| sgt1b | 83.3 | * | 0.9 | 0.3 | * | [53] | |
| Cell death regulation | rcd1 | 48.8 | 0 | 0 | * | [34] | |
| dnd1 | 100.0 | * | 0 | 0 | * | [30] | |
| bag4 | 93.3 | * | 7.4 | 3.2 | [102] | ||
| Ethylene | ein2 | 32.5 | 6.8 | 2.6 | [103] | ||
| etr1-3 | 46.7 | 6.4 | 1.7 | [104] | |||
| G-protein | agb1 | 53.9 | 2.7 | 0.5 | * | [105] | |
| gpa1 | 73.3 | * | 2.4 | 0.7 | * | [105] | |
| agb1 gpa1 | 58.5 | 0.9 | 0.5 | * | |||
| Jasmonic acid | jar1 | 96.7 | * | 7.9 | 1.4 | * | [106] |
| aos | 13.3 | 5.1 | 2.2 | [107] | |||
| MAP kinase signaling | mpk3 | 73.3 | * | 3.9 | 1.5 | [108] | |
| mpk6 | 66.7 | * | 3.5 | 1.4 | * | [108] | |
| mkk1 | 36.7 | 7.3 | 1.4 | [108] | |||
| mkk2 | 44.4 | 5.5 | 2.7 | [108] | |||
| mkk3 | 58.9 | 2.5 | 0.7 | * | [108] | ||
| ibr5 | 40.0 | 3.6 | 1.1 | * | [109] | ||
| ROS biosynthesis/scavenger | rbohD | 10.0 | 3.1 | 1.6 | * | [25] | |
| rbohF | 38.3 | * | 8.0 | 1.3 | * | [25] | |
| vtc1-1 | 13.3 | * | 0 | 0 | * | [110] | |
| vtc2-1 | 0 | * | 3.1 | 2.4 | [110] | ||
| apx1 | 53.3 | 2.2 | 1.1 | * | [111] | ||
| oxi1 | 16.7 | 4.5 | 0.9 | [112] | |||
| Salicylic acid/Defense | sid2 | 0 | * | 0.4 | 0.3 | * | [113] |
| npr1 | 66.7 | * | 1.4 | 0.6 | * | [114] | |
| eds1 | 0 | * | 2.0 | 1.9 | * | [115] | |
| pad4 | 53.3 | 3.1 | 1.2 | * | [116] | ||
| ald1 | 0 | * | 4.5 | 2.1 | [38] | ||
| fmo1 | 36.7 | 3.6 | 1.7 | [37] | |||
| cbp60g | 23.3 | 5.4 | 4.4 | [117] | |||
| mos3 | 23.3 | 3.2 | 1.7 | * | [40] | ||
| mos6 | 10.0 | 3.3 | 1.9 | * | [40] | ||
| snc1-11 | 0 | * | 4.2 | 1.3 | [41] | ||
| rar1-21 | 40.0 | 1.5 | 1.1 | * | [118] | ||
| Transcription factors | wrky25 | 10.0 | * | 8.6 | 1.9 | * | [65] |
| wrky33 | 70.0 | * | 3.0 | 1.5 | * | [119] | |
| wrky70 | 86.7 | * | 2.1 | 1.3 | * | [62] | |
| myb30 | 85.0 | * | 2.1 | 0.9 | * | [67] | |
| myc2 | 100.0 | * | 2.9 | 1.2 | * | [120] | |
| abi4-1 | 56.7 | * | 3.1 | 1.2 | * | [121] | |
| wrky50 | 83.3 | * | 2.3 | 1.6 | * | [122] | |
| wrky51 | 40.0 | 3.5 | 1.8 | [122] | |||
| wrky50 wrky51 | 28.9 | 4.2 | 1.2 | ||||
| as1 | 100.0 | * | 0 | 0 | * | [64] | |
| Histone methylases | atx1 | 51.7 | 2.5 | 0.9 | * | [68] | |
| atx2 | 0 | * | 2.4 | 0.6 | * | [68] | |
| atxr7 | 70.0 | * | 3.1 | 1.3 | * | [123] | |
| Others | wakl10 | 76.7 | * | 2.5 | 1.5 | * | [124] |
| PLD | 42.2 | 3.9 | 1.8 | [125] | |||
| bak1-4 | 0 | * | 7.7 | 1.1 | * | [35] | |
| β-cas | 3.3 | 8.4 | 4.1 | [126] | |||
Survival was scored in a plate assay for a period of 8 days after transfer to LD and restricted gas exchange which promotes photorespiration. The survival score was estimated 8 days after transfer, for a time course of survival see Additional file 3: Figure S2. Asterisks indicate statistically significant difference of survival between cat2 and double mutant during LD treatment (p <0.05, n = 30, General Linear model for testing interactions). Lesions area (%; mean ± SE) were measured from 4 week old LD soil-grown plants.
Significantly different results from cat2 single mutant are denoted by asterisks (p <0.05; n = 30, Fisher LSD)
Fig. 1.
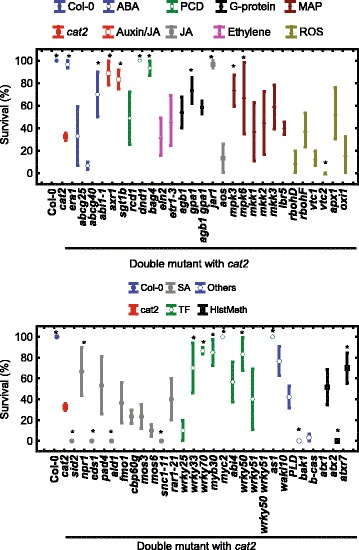
Survival of cat2 and cat2 double mutants in the in vitro assay. Fourteen-day old seedlings were transferred from SD (8 h/16 h) to LD (12 h/12 h) and gas exchange was restricted by sealing of plates with parafilm. Survival was scored after 8 days. Asterisks indicate significant difference from cat2 single mutant at the end of LD treatment (n = 30; p <0.05, Fisher LSD). For a version of this figure with a time course of survival, see Additional file 2: Figure S1
The restricted gas exchange assay used 2 week old in vitro grown seedlings. To assess the cell death response in soil-grown plants, lesion size was measured and cell death was visualized with trypan blue stain in 4 week old plants (Table 1, Fig. 2, Additional files 4, 5 and 6: Figures S3–S5). The two assays, restricted gas exchange versus lesion formation in soil grown plants, gave mostly similar results with some exceptions: cat2 jar1 (jasmonate resistant1) and cat2 bag4 (BCL-2-associated athanogene 4) plants had high survival rate in restricted gas exchange, but displayed enhanced cell death in soil-grown plants compared to cat2. Contrary, cat2 vtc1, cat2 atx2 (trithorax protein like2) and mutants with decreased SA accumulation cat2 ald1, cat2 eds1 and cat2 sid2, died 8 days after transfer to restricted gas exchange although mature plants had no or few visual lesions, respectively (Table 1, Fig. 2, Additional files 4, 5, 6 and 7: Figures S3–S6).
Fig. 2.
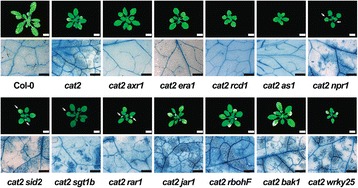
Formation of lesions in soil-grown cat2 and cat2 double mutants. Selected double mutants without lesions (cat2 axr1, cat2 era1, cat2 rcd1, cat2 as1), with few lesions (cat2 npr1, cat2 sid2, cat2 sgt1b, cat2 rar1) and with more lesions than cat2 (cat2 jar1, cat2 bak1, cat2 wrky25). Cell death is indicated by trypan blue stain of 4 weeks old plants. White scale bar shows 1 cm and black scale bar 600 μm. All mutants used in this study are in Additional files 3, 4 and 5: Figures S2–S4
In conclusion, both assays gave consistent results, with a few important exceptions. Ascorbic acid and SA double mutants were sensitive in restricted gas exchange but tolerant in soil-grown plants, conversely the cat2 jar1 and cat2 bag4 exhibited better performance in vitro. One additional factor that distinguishes the restricted gas exchange plate assay from the soil grown plants is temperature. Although the plates were incubated in a temperature controlled growth cabinet, the sealing of the plates led to an increased temperature and humidity inside the plates (2–3 °C and air humidity 98 %, measured with sensor for temperature and humidity DS1923, Digi-Key Corporation, Canada). In Arabidopsis high temperature alters the balance of defense signaling between SA and JA/ethylene, and in some LMMs the lesion formation is suppressed at higher growth temperature [24]. We speculate that some of the difference between restricted gas exchange and soil grown plants could be related to temperature and humidity, especially in SA mutants, and further study of cat2 double mutants in different temperatures could give insight into cross talk between temperature and H2O2 signaling.
In the following sections we will highlight some of the double mutant data in relation to the functional category of the double mutants.
The role of ROS production and ROS scavenging
A major source of extracellular ROS in plants is the NADPH oxidase, which are encoded by RBOH (RESPIRATORY BURST OXIDASE HOMOLOGUE) genes [25]. RBOHD and RBOHF are the main RBOH genes expressed in leaves and have been extensively characterized for their role in stress responses [26]. In avirulent pathogen infection RBOHF contributes more to cell death regulation than RBOHD [25]. The function of RBOH proteins may also be regulated by hetero-trimeric G-proteins [27]. In soil-grown plants cat2 rbohF but not cat2 rbohD had more lesions than cat2 (Table 1, Figs. 1 and 2) consistent with a previous report [28]. Furthermore, reduced lesion formation was observed in alpha and beta subunits of the heterotrimeric G-protein cat2 agb1, cat2 gpa1 and cat2 agb1 gpa1 (Table 1). Hence, a ROS signal from RBOHF may be required to restrict the spread of cell death.
The cat2 mutation impairs H2O2 scavenging, thus we expected that further removal of the important ROS scavenger ascorbic acid would lead to enhanced damage. This was indeed the case for cat2 vtc1 and cat2 vtc2 when assayed with restricted gas exchange (Table 1). In contrast, soil-grown cat2 vtc1 had abolished visible lesion formation (Table 1, Additional file 7: Figure S6). The vtc1 mutant is defective at an early step of ascorbic acid biosynthesis and in addition to lower ascorbic acid content also lacks biosynthesis of intermediates in cell-wall polysaccharide synthesis. Furthermore, vtc1 has a pleiotropic phenotype including spontaneous cell death (Additional file 7: Figure S6; [29]). Thus, we suggest that in adult plants the vtc1 mutation leads to activation of plant defenses that protect against damage from cat2 induced H2O2 production.
The role of cell death regulators
The defense no death1 (dnd1) mutant is a conditional LMM, where dnd1 exhibit spontaneous cell death in some growth conditions (Additional file 7: Figure S6) [30, 31]. In cat2 dnd1 there was a reduced amount of lesions and increased survival compared to cat2 (Table 1, Additional file 7: Figure S6). The exact step at which dnd1 regulates cell death is currently unknown. However, the protein encoded by DND1, CYCLIC NUCLEOTIDE GATED CHANNEL2 (CNGC2), regulates Ca2+ transport and activation of nitric oxide signaling pathways [32]. Since Ca2+ is required for execution of cell death in catalase silenced tobacco [33], it is possible that DND1/CNGC2 mediates a Ca2+ signal required for initiation of cell death.
The radical-induced cell death1 (rcd1) mutant is sensitive to apoplastic ROS, but tolerant to chloroplastic ROS induced damage caused by treatment with methyl viologen [34]. Likewise, intracellular ROS from cat2 did not cause lesions in cat2 rcd1 (Fig. 2). Thus, plants are able to distinguish the source and location of ROS production (apoplast versus chloroplast and peroxisome) and RCD1 acts as a central regulator to determine the switch from life to death [34].
BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE) is a co-receptor of multiple receptor like kinases (RLK) and a regulator of cell death [35]. In cat2 bak1-4 there was increased sensitivity to restricted gas exchange and increased lesion formation is soil-grown plants (Table 1, Figs. 1 and 2). BAK1 may regulate cell death responses through its interaction with BIR1 and regulation of ROS production [35, 36]. Thus, signaling via BAK1 could be required to restrict PCD in cat2 through regulation of a ROS signal.
The role of salicylic acid and TIR-NB-LRRs
Increased ROS production leads to increased biosynthesis of all major defense-related hormones – JA, SA, ethylene and ABA. SA is a crucial regulator of multiple plant defenses, and at high concentrations promotes cell death [12]. The amount of cell death in cat2 is reduced when crossed the SA biosynthesis mutant sid2 (Fig. 2). Several proteins are required for systemic accumulation of SA and function upstream and/or in parallel to SA and include ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), PHYTOALEXIN DEFICIENT 4 (PAD4), AGD2-LIKE DEFENSE RESPONSE PROTEIN 1 (ALD1) and FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) [37, 38]. These regulators were also found as mutants that suppress cell death [39]. Lesion formation was reduced in cat2 eds1 and cat2 pad4 (Table 1), but given the multitude of signal pathways where EDS1 and PAD4 are involved, it is not straightforward to pinpoint exactly at which point these proteins regulate cell death.
Downstream from SA, NONEXPRESSOR OF PR GENES1 (NPR1) is required for execution of many SA responses. Loss of function in npr1 attenuated cell death in cat2 (Table 1; Fig. 2; [19]). Several other regulators of SA and defense signaling have been identified through mutant screens for altered defense responses and include the dominant mutant suppressor of npr1-1, constitutive 1 (snc1) that has a mutation in a TIR-NB-LRR (Toll Interleukin1 receptor-nucleotide binding-Leu- rich repeat) and mos3 and mos6 (modifiers of snc1) that encode components of the nucleocytoplasmic trafficking machinery [40]. A loss of function allele in snc1-11 restore normal growth to several LMMs; mkp1 (map kinase phosphatase1), srfr1 (suppressor of RPS4-RLD1) and nudt6 nudt7 (nudix hydrolase homolog6 nudix hydrolase homolog7) directly showing that an appropriate level of signaling through NB-LRR proteins regulates cell death [41–43]. Indeed, several defense responses is regulated by the stability of NB-LRRs through RAR1 (REQUIRED FOR MLA12 RESISTANCE 1) [44]. Of the different mutations that have an impact on defense signaling down-stream or in parallel to SA, the cat2 mos3 and cat2 mos6 had fewer lesions than cat2 indicating that execution of cell death requires nuclear localization of a yet to be identified cell death regulator (Table 1). Furthermore, cat2 rar1 also had fewer lesions supporting a role for NB-LRR signaling in cell death execution [40–44]. Overall, using both a SA biosynthesis mutant and multiple mutants for regulators of SA accumulation, SA signaling and defense signaling, give support for the crucial role of SA as a regulator of cell death.
The role of ethylene, jasmonic acid and auxin
Like SA, the role for JA and ethylene in regulation of cell death is complex. In different LMMs JA and ethylene can act as either positive or negative PCD regulators [12, 31]. Impaired ethylene signaling did not modulate defense reactions in cat2, since cat2 ein2 and cat2 etr1-3 survival rate and lesion development did not differ from cat2.
In response to biotic and abiotic stress several oxylipins are produced, where jasmonic acid (JA) is the best characterized. However, some of the pre-cursors of JA have biological activities of their own and for example 12-oxo-phytodienoic acid (OPDA) is perceived through its own receptor CYCLOPHILIN 20–3 to regulate cysteine biosynthesis and the cellular redox status [45]. The aos (allene oxide synthase) mutant is deficient in both OPDA and JA biosynthesis and in jar1 (jasmonate resistant1) the formation of the biologically active JA-Ile is deficient [46]. However, jar1 is not completely blocked in JA signaling; while aos is male sterile, jar1 is fully fertile suggesting redundant pathways in the final step of JA-Ile biosynthesis [46]. Cell death in cat2 aos was similar to cat2, indicating a limited role for JA in ROS induced cell death (Table 1). However, cat2 jar1 had increased amount of cell death in soil grown plants (Table 1, Fig. 2). Since the main difference between these mutants is that aos also lacks OPDA, the contrasting results for cat2 aos and cat2 jar1 could indicate distinct roles for OPDA and JA in H2O2 induced cell death. A differential role between JA and OPDA has also been observed in the flu mutant where cell death is initiated through singlet oxygen production [47]. In flu, cell death is reduced when JA biosynthesis is abolished, but when both OPDA and JA biosynthesis were removed through aos no impact on cell death was observed [47]. Thus, both in H2O2 and singlet oxygen induced cell death, OPDA and JA appear to have distinct roles. Alternatively, the specific timing of biosynthesis of biologically active JA-Ile could be important for execution of PCD. Further investigation of other JA mutants, e.g. the JA receptor coi1 could help clarify the role of JA in PCD regulation.
Auxin is a central regulator of plant development. Recently auxin has also been identified as an important regulator of stress responses, including PCD regulation in cat2 [23] and mitochondria retrograde signaling [48, 49]. The auxin insensitive mutant axr1 (auxin resistant1) is also JA insensitive [50]. AXR1 encodes a protein involved in regulation of the activity of Skp1-Cullin-F-box (SCF) complexes, which in turn regulate protein degradation [51]. Both the JA receptor COI1 and the auxin receptor TIR1 are regulated through this mechanism, explaining the auxin and JA insensitivity of axr1. SGT1b is also a protein involved in regulation of SCF complexes, and sgt1b has multiple phenotypes including auxin insensitivity, altered pathogen responses and defense signaling, and regulation of cell death [52, 53]. SGT1b is also part of a complex with RAR1 and HSP90 (HEAT SHOCK PROTEIN 90) to ensure proper function of TIR-NB-LRRs [54]. The phenotypes of cat2 axr1, cat2 rar1 and cat2 sgt1b were similar, high survival in restricted gas exchange assay and few lesions in soil-grown plants (Table 1, Figs. 1 and 2). Given the importance of protein degradation in all aspects of plant signaling and the multiple phenotypes of axr1, sgt1b and rar1 it is difficult to pinpoint exactly where AXR1, RAR1 and SGT1b might regulate ROS induced PCD, however, auxin or JA might be involved.
The role of abscisic acid
ABA is an important hormone in abiotic stress responses (cold, salt and drought), in biotic stress responses through interaction with SA, and as a regulator in high light signaling [55]. ABA signaling is blocked in abi1-1 (ABA insensitive1). In cat2 abi1 there was increased survival as well as fewer lesions in soil-grown plants (Table 1 and Fig. 1). A mutant in the beta subunit of farnesyl transferase, era1 (enhanced response to ABA1), has multiple phenotypes including increased sensitivity to ABA, altered developmental, stress and defense responses [56, 57]. Protein farnesylation is modification to proteins to change their subcellular localization . The cat2 era1 double mutant was resistant to restricted gas exchange and did not develop lesions in soil-grown plants (Table 1, Figs. 1 and 2), thus a regulator of PCD might be farnesylated for proper function.
The role of transcription factors and chromatin environment
ROS induced cell death caused by ozone treatment requires changes in gene expression since the extent of cell death can be decreased with a transcriptional inhibitor [58]. When bZIP10, a positive regulator of cell death [59], was knocked out in cat2 bzip10, a reduced amount of cell death was observed [18]. The cat2 lesion phenotype was also suppressed by cpsf30 (cleavage and polyadenylation specificity factor30), which regulates mRNA polyadenylation [60], thus both transcription and proper mRNA processing are regulatory steps in the execution of cell death. Analysis of gene expression signatures in response to several ROS treatments suggests the involvement of several TFs in regulation of ROS responses [61], hence TFs other than bZIP10 are likely involved in the regulation of cell death in cat2.
Many TF mutations reduced cell death in cat2 (abi4-1, as1, myb30, myc2, wrky33 and wrky70) or led to enhanced cell death (wrky25) (Table 1, Figs. 1 and 2). Since several of these TFs either regulate JA responses (AS1, MYC2, WRKY70; [62, 63], SA responses (WRKY70; [62]), pathogen responses (AS1, WRKY25, WRKY33, WRKY70; [64–66]) or cell death (MYB30; [67]), this reinforces the importance of SA and JA in regulation of gene expression required for execution of PCD in cat2. To regulate gene expression TFs needs access to DNA, which is affected by how tightly DNA is packed in histones and chromatin. Histones can be post-translationally modified, including methylation by ARABIDOPSIS TRITHORAX1 (ATX1) [68]. Lesion development was decreased in cat2 atx1, cat2 atx2 and cat2 atxr7 indicating a regulatory role for histone methylation and chromatin packaging in cell death regulation (Table 1).
At a first glance it might appear counter-intuitive that such a large number of TFs would influence cell death regulation. However, high resolution TF-promoter interaction assay for genes encoding components related to the cell wall identified on average five TFs bound to each promoter [69]. Assuming that genes encoding proteins involved in PCD would have a similar promoter-TF relationship, we could expect that there are even more TFs than the ones identified here to be part of PCD regulation.
The role of MAP kinase signaling
Mitogen-activated protein kinase (MPK) cascades regulate many aspects of development and stress responses in plants [70]. MAP kinase kinase kinases (MPKKKs) activate MAPK kinase kinases (MPKKs) that in turn activate MAP kinase (MPKs) that phosphorylate and activate target proteins, including TFs like WRKY2, WRKY25, WRKY33, WRKY34 and ERF6 [71, 72]. The cat2 mpk3 and cat2 mpk6 double mutants had increased survival in restricted gas exchange and cat2 mpk6 exhibited fewer lesions in soil-grown plants (Table 1, Fig. 1, Additional file 4: Figure S3). MKK3 is one of MPKKs activating MPK6 [73], and cat2 mkk3 also had a reduced amount of lesions. MKK3-MPK6 can phosphorylate MYC2 and regulate plant responses to blue light [74]; the similar phenotypes of cat2 mkk3, cat2 mpk6 and cat2 myc2 indicates that a similar signal pathway may take place during cat2 lesion formation.
Maximum photosynthetic efficiency
Regulation of lesion size takes place at separate steps – lesion initiation, spreading and containment [75]. Since the restricted gas exchange and lesion development assays used rather long time scales, a separate method that follow early signaling in cat2 could give information whether the increased tolerance of selected cat2 double mutants were related to the initiation or the spreading phase. A sensitive assay to detect early stress symptoms is to measure the maximum photosynthetic efficiency of PSII (Fv/Fm) in dark adapted plants [23, 76]. In Arabidopsis photorespiration mutants, transferred from elevated CO2 to ambient CO2 conditions or from low to high light, a rapid onset of photoinhibition was accompanied by a decline in PSII maximum efficiency [77]. A subset of cat2 double mutants with either strongly enhanced or reduced cell death were grown on plates for 14 days and assayed for photosynthetic efficiency over a period of 7 days after shifting to continuous light conditions (Fig. 3. NB, these growth conditions were different from the ones in Fig. 1 and Table 1). The PSII maximum efficiency declined dramatically in cat2 compared to wild type, at the end of the experiment average values were 0.45 (cat2) and 0.75 (Col-0). For mutants with enhanced cell death (cat2 bak1, cat2 jar1, cat2 rbohF, cat2 wrky25); their Fv/Fm decreased to 0.3, however, this did not differ significantly from cat2. Double mutants without lesions (cat2 axr1, cat2 era1 and cat2 rcd1; Table 1, Fig. 2) were able to stabilize their photosynthetic efficiency and their Fv/Fm were significantly higher than cat2 1 week after the shift to photorespiratory conditions (Fig. 3; for a version of Fig. 3 with error bars see Additional file 8: Figure S7). However, of these double mutants only cat2 axr1 had stable photosynthetic efficiency at early time points during photorespiratory conditions and could indicate a role for AXR1 already in the early process of PCD regulation.
Fig. 3.
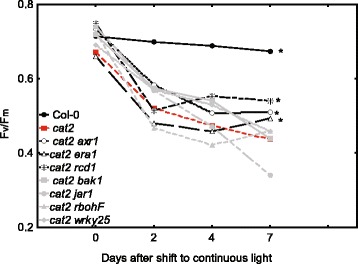
Maximum photosynthetic efficiency (Fv/Fm) in dark adapted plants after transfer to continuous light and restricted gas-exchange. Fourteen-days old seedlings were transferred from 70 μmol m−2 s−1 light intensity and 8 h day/16 h night to 120 μmol m−2 s−1 continuous light. Fv/Fm was measured on shift day, 2nd, 4th, and 7th. The experiment was repeated three times using 20 plants per repeat (n = 60). Asterisks indicate significantly different Fv/Fm values of double mutant in time scale and compared to cat2 (p <0.05, General Linear Model)
H2O2 accumulation
H2O2 is a major byproduct of photorespiration and H2O2 is a regulator of PCD. Since cat2 is deficient in H2O2 scavenging, we measured H2O2 concentration in 4 weeks old plants at 12:00 hours to further explore the mechanism underlying absent or increased lesion development in a subset of cat2 double mutants. Our initial hypothesis was that double mutants with fewer lesions would have less H2O2 and vice versa for double mutants with increased cell death.
Most of the double mutants with decreased lesion formation also had less H2O2 (cat2 era1, cat2 as1, cat2 rcd1 and cat2 sid2; Fig. 4). However, there were two exceptions, cat2 axr1 and cat2 dnd1, that have no lesions but similar H2O2 content as the single cat2 mutant. For double mutants with increased lesion formation (cat2 jar1, cat2 rbohF and cat2 wrky25) the H2O2 concentrations were the same as in cat2. Interestingly, cat2 bak1 had reduced H2O2 production, even though cat2 bak1 had more lesions than cat2. Overall this suggests that there is no strict correlation between H2O2 accumulation and cell death, instead as long as the initial steps towards PCD have been executed in cat2, further H2O2 accumulation appears to not influence the PCD process.
Fig. 4.
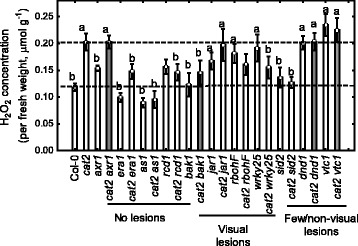
H2O2 concentration in cat2 and selected double mutants in 4 weeks old plants measured with Amplex Red. Letter “a” indicates significant difference from Col-0 and letter “b” significant difference from cat2 (mean ± SE, n = 6)
Similarities between cat2 and other mutants that develop spontaneous lesions
Arabidopsis mutants that spontaneously develop lesions, the LMMs, have long been used to study regulation of cell death and defense signaling [11, 12]. For three of these mutants extensive collections of double or triple mutants have been generated to identify regulators of cell death: accelerated cell death6 (acd6) [78], dnd1 [31] and syp121 syp122 [39]. All three mutants acd6, dnd1 and syp121 syp122 have constitutively activated defense pathway and increased SA accumulation. We compared the regulators of PCD in cat2 (Table 1) with the corresponding information from acd6, dnd1 and syp121 syp122 to determine whether cell death in cat2 is unique or if there is a common genetic program for execution of cell death. In all four mutants SA plays a major role, mutations in sid2, npr1, pad4 and eds1 reduce the extent of cell death (Table 1; [31, 78, 39]. In cat2, dnd1 and syp121 syp122 the rar1 mutation also led to less lesion formation (this mutant was not tested in acd6). In contrast ald1 and fmo1 mutations protected acd6, dnd1 and syp121 syp122, but not cat2. Overall this emphasizes the role of SA in cell death execution and that PAD4, EDS1, RAR1 and SGT1B are regulators of cell death downstream from activation of cell death by different mutations in acd6, dnd1, syp121 syp122 and cat2. In our growth conditions the sid2 and eds1 mutations only led to partial reduction of the lesion formation in cat2, similarly acd6, dnd1 and syp121 syp122 were only partially rescued by these mutations. In contrast introduction of both sid2 and eds1 mutations led to increased plant health in acd6, dnd1 and syp121 syp122 compared to introduction of only one mutation. This prompted us to generate the cat2 sid2 eds1, cat2 sid2 fmo1 and cat2 sid2 ald1 triple mutants (Fig. 5). Although, cat2 sid2 fmo1 and cat2 sid2 ald1 had less lesions compared to cat2 fmo1 and cat2 ald1 the cell death area did not differ from cat2 sid2. Of the different triple mutants cat2 sid2 eds1 completely lacked lesions, indicating the importance of SA and EDS1 acting together as regulators in execution of cell death. Indeed, inactivation of both SA (via the sid2 mutation) and eds1 leads to a compromised HR [79]. The cat2 dnd1 double mutant represented a case where both of the single mutants are LMMs (Additional file 7: Figure S6). The amount of cell death was reduced in cat2 dnd1 compared to cat2 (Table 1). Since sid2 suppress the extent of cell death in both cat2 and dnd1, the triple mutant cat2 sid2 dnd1 was tested (Fig. 5). Visible lesions were absent, but there were small patches of dead cells visualized with trypan blue stain in a pattern different compared to cat2 dnd1 (Fig. 5). In future studies there will be a need to move to assays that more emphasize the behavior of specific cells rather than the average of whole leaves. We conclude that cell death in cat2 is largely executed through a similar genetic program as in other lesion mimic mutants, including the dependence on SA and EDS1.
Fig. 5.
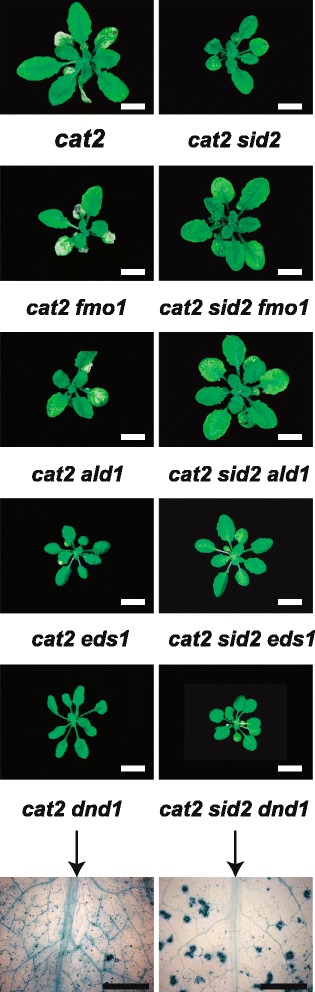
Lesion development in soil-grown cat2 double and triple mutants. Cell death is indicated by trypan blue stain for weeks old plants. White scale bar shows 1 cm and black scale bar 600 μm
Gene expression analysis of cat2 in comparison with cell death mutants
If cell death is executed through a common genetic program we would also expect to see a similar gene expression profile of cat2 in comparison with other LMMs. We obtained array data from three studies with cat2 performed on the Affymetrix ATH1 chip. Vanderauwera et al. [80] used plants silenced for catalase in a high light experiment with two time points, 3 and 8 h. Queval et al. [81] grew cat2 in high CO2 followed by transfer to ambient CO2 in either short day or long days for 2 and 4 days. Sewelam et al. [82] grew cat2 and transgenic GO5 lines in high CO2 followed by transfer to ambient CO2 (GO5 plants express GLYCOLATE OXIDASE in the chloroplast, these plants produce H2O2 in the chloroplast during photorespiratory conditions). These experimental designs allow the identification of H2O2 responsive transcripts after the shift to photorespiratory conditions. The array raw data was normalized and clustered with Bayesian hierchical clustering together with array data from LMMs and various other treatments that induce ROS production or cell death (see Methods for full list of experiments). The gene list used in the cluster analysis was the gene ontology category cell death (488 genes, Additional file 9: Table S2).
Four major clusters were obtained (Fig. 6). The Queval [81] experiment gave weaker changes in gene expression compared to the other experiments, possibly due to the use of a later time point (2 days versus 3 to 8 h for the other experiments). Cluster 1 contained genes with strongly induced gene expression in LMMs; these genes also had high expression in cat2, GO5, by ROS treatment (ozone), and during pathogen infection and senescence. The genes in this cluster (Additional file 9: Table S2) included genes that in our mutant analysis were important in lesion formation, including SID2, PAD4, WRKY25 and WRKY33. Cluster 2 was more heterogeneous in terms of expression pattern, but had genes with increased expression in both cell death mutants, cat2 and GO5 plants from the Sewelem [82] experiment. Cluster 3 contained genes with decreased expression and cluster 4 genes with minor altered expression by most treatments. From the hormone perspective, SA and BTH (Benzothiadiazole, a SA analog) gave a similar expression profile as cat2 and other cell death mutants, reinforcing the crucial role of SA in regulation of cell death.
Fig. 6.
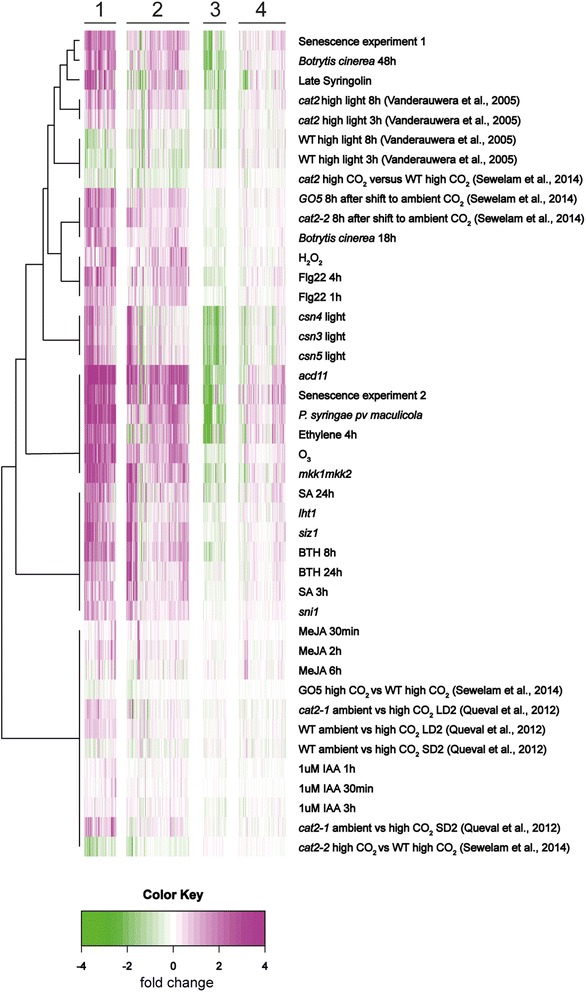
Cluster analysis of cell death related genes in cat2 and other LMMs. Experiments performed on the Affymetrix ATH1 chip were obtained from cat2, LMMs, constitutive defense mutants, hormone treatments, senescence and biotic and abiotic stresses (see Methods for a full list of experiments). The gene ontology category cell death (488 genes) was used for Bayesian hierarchal clustering. Values are mean of log2 ratio of the treatment and control expressions. Magenta and green indicate increased and decreased expression compared with untreated or wild type plants, respectively
We conclude that the shift to photorespiratory conditions in cat2 and promotion of H2O2 production leads to a gene expression profile that is also seen in plants undergoing cell death activated by either mutation or through treatments. Thus, both double mutant analysis in cat2 and gene expression profiles, suggest that plants have a core program that includes SA and EDS1 for execution of PCD and that H2O2 may be one of the key signals of this program.
Localized gene expression in cat2
Microarray analysis performed with cat2 was done with either whole rosettes or whole leaves [80–82]. This sampling strategy is likely to average out any cell specific responses across the whole leaf. In particular, it would be informative if genes related to PCD are expressed only at the lesions in cat2, or if altered expression is spreading outwards from the lesions. Here we crossed cat2 with several promoter-uidA fusions, which allows GUS staining and visualization of gene expression activity at the site of lesion formation in cat2 and surrounding tissues in 4 week old plants (Table 2, Fig. 7).
Table 2.
Summary of promoter GUS line expression in Col-0 and cat2 background
Fig. 7.

Local gene expression in cat2. Histochemical analyses of uidA expression in Col-0 and cat2 in 4 week old soil-grown plants. Promoter-uidA lines included PR1, PDF1.1, DR5 and FMO1 (Table 2)
Expression of the auxin reporter DR5-uidA in cat2 decreased in the whole leaf (Fig. 7), consistent with a previous report [83]. The lack of localized DR5 expression at the site of lesions would suggest that auxin is not directly involved in the PCD process and could instead play a role in reprogramming of leaf growth in response to elevated ROS [83, 84]. A screen for chemicals that rescue the photoinhibition in cat2 revealed that both auxin and synthetic auxin-like molecules can rescue cat2 phenotypes [23]. Thus, mutations in auxin signaling components axr1, sgt1b and as1 (Table 1) as well as application of auxin rescue cat2 phenotypes, whereas DR5 expression suggest decreased auxin signaling in cat2. Given the central role for auxin as a growth hormone it is possible that different branches of auxin signaling could be activated during development versus PCD or higher H2O2 levels. This has been observed in mitochondria retrograde signaling, where two specific AUXIN RESPONSE FACTORS (ARF7 and ARF19) act as negative regulators [49]. Introduction of other auxin reporter constructs in cat2, including DII-Venus [85] could help to unravel the role of auxin in the PCD process.
One of the most commonly used marker genes to study SA signaling is PR1 (PATHOGENESIS-RELATED GENE 1). Expression of PR1 was very high around lesions (Fig. 7), this could indicate that SA biosynthesis is initiated at the cells where PCD is taking place. Expression of PDF1.1 (PLANT DEFENSIN 1.1) is high at sites of B. cinerea infection and by paraquat treatment [86]. In cat2 PDF1.1 expression was high in leaves with lesions, but interestingly the signal was strongest some distance away from lesions, suggesting that expression of this gene is a systemic response. Expression of FMO1 is induced in response to pathogens in an EDS1/PAD4 dependent manner [37], by ROS treatment and during cell death [87]. Expression of FMO1 was more localized in cat2 than either PR1 or PDF1.1 (Fig. 7).
Overall the results using GUS lines clearly show that there is a spatial component to gene expression induced during lesion formation in cat2. Future studies in the context of gene expression should aim towards more cell specific assays, with the goal to find the genes directly related to lesion formation versus genes responsible for initiating defenses further away in the leaf and in systemic tissues.
Conclusion
Based on the data presented here and in previous research on cat2 [15, 18, 21, 23, 28]; we suggest a model for signaling events leading to cell death in cat2 (Fig. 8). After a certain threshold concentration of H2O2 has built up in cat2 the lesion process is initiated. Subsequently two main pathways are activated: SA and its associated signaling partners including EDS1; and auxin/JA. These two pathways are likely to exhibit extensive crosstalk. Transcriptional regulation is essential for cell death control and includes several TFs (including AS1, WRKY33, WRKY70, MYB30 and MYC2) and proteins that interact with TFs (RCD1). The exact molecular function for other cell death regulators remain to be identified, as an example BAK1, a co-receptor of many plasma membrane receptors, is part of a larger complex that regulate ROS production via RBOHs [88]. The enhanced cell death of cat2 bak1 and cat2 rbohF (Table 1), could hint towards a role for regulated apoplastic ROS production as a signal to inhibit the spread of cell death. Using assays that directly target protein-DNA interaction (chromatin immunoprecipitation) could help unravel which targets genes of AS1, MYB30 and WRKY70 may be involved in execution of cell death.
Fig. 8.
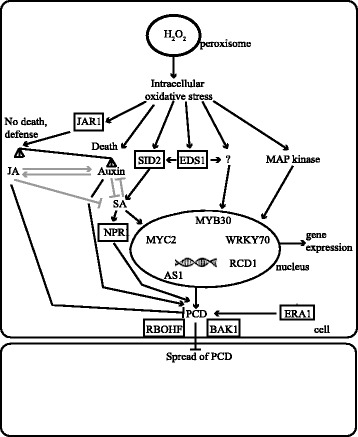
Model for ROS in cell death regulation. Intracellular H2O2 production caused by the cat2 mutation lead to increased concentration of hormones (SA and JA, through SID2 and JAR1) and activation of multiple signaling pathways. The balance and timing of biosynthesis of auxin, SA and JA is one important determinant of cell death. In parallel, MPK signaling and other unknown signaling pathways may directly target TFs and regulate their activity and interaction with DNA with a subsequent change in gene expression. Other mechanisms in the nucleus also provide a suitable environment for accurate gene expression, including RCD1 that interacts with TFs. A yet to be identified regulator of cell death is farnesylated for proper function by ERA1. Extensive cross-talk between hormone signaling pathways allow fine tuning of the signal. Controlled ROS production via BAK1 and RBOHs may provide a signal to neighboring cells leading to propagation of cell death
Methods
Plant material and growth conditions
All the mutants were in Columbia-0 (Col-0) background and Col-0 was used as control plant for all experiments. Arabidopsis mutants were obtained from NASC [130] or were gifts, see Additional file 1: Table S1 for more details [89, 90]. Double and triple mutants were constructed using cat2 (SALK_076998) as the pollen acceptor. Double and triple mutants were screened for visible cat2 mutant phenotypes (dwarf, bleached leaves, visual cell death) and subsequently genotyped using PCR based CAPS, dCAPS [91] and T-DNA markers (Additional file 1: Table S1). After screening of double/triple homozygotes in the F2 generation, all mutants were confirmed in F3 or F4 generations.
Sterilized seeds were placed on Murashige and Skoog (MS) medium (1/2 MS salts, and 0.7 % agar), stratified for 3 days and transferred to growth chamber. One week old plants were transplanted into pots with 1:1 peat:vermiculate mixture with one Col-0, one cat2, one of the single mutants and the corresponding cat2 double mutant (4 plants per pot). Plants were subsequently grown on soil for 3 weeks in a controlled chambers (MCA1600, Snijders Scientific, Netherlands), at 21 °C/19 °C under a 12 h day/12 h night regime, light intensity 120 μmol m−2 s−1, and 70 % relative humidity.
For the in vitro plate assays, plants were grown on plates in 8 h day/16 h night during 14 days, subsequently sealed with parafilm M and transferred to 12 h/12 h regime for 8 days (survival estimation). Chamber temperature was kept 23 °C throughout day (Sanyo chambers, Sanyo Electric Co, Japan).
Estimation of lesions and cell death
Four week old plants were photographed and visual lesion size was calculated from photos using Adobe Photoshop Professional CS5.1. The mutant collection was screened 6 times with 5 plants per repeat for lesion size. Fully expanded leaves were used for trypan blue staining to confirm cell death [92].
Survival during restricted gas exchange was calculated from photos scoring dead plants per mutant. The mutant screen was repeated three times with 10 plants per repeat.
Photosynthetic performance
Plants were grown on plates at 70 μmol m−2 s−1 for 2 weeks in SD (8 h/16 h), subsequently sealed with parafilm (no air influx) and transferred to continuous light (120 μmol m−2 s−1 irradiance) to promote photorespiration. Chlorophyll fluorescence was measured during 7 days (on shift day, 2nd, 4th and 7th day). Using Imaging PAM-2000 (Heinz Walz GmbH, Germany), the photosystem II (PSII) maximum efficiency (Fv/Fm) was determined in which Fv denotes variable fluorescence (ability of PSII to perform photochemistry) and Fm maximal fluorescence (PSII centers closed).
Histochemical analyses and H2O2 quantification
GUS staining was performed according to [93]. For H2O2 quantification 100 mg of leaf material was collected from 4 weeks old plants and frozen in liquid nitrogen at 12:00 when CATALASE2 expression and photorespiration are presumably at their highest level [6, 8]. Frozen samples were milled using Silamat S6 (Ivoclar Vivadent, NY, USA) and extracted with 500 μl of 50 mMK2HPO4, pH 7.2. The slurry was cleared by centrifugation at 16,000 g for 15 min at 4 °C, and 50 μl of supernatant was incubated with 0.1 mM Amplex Red (Thermo Fisher Scientific Inc., MA, USA) and 0.2 mM Horseradish peroxidase at room temperature for 30 min [94]. Absorbance was detected at 570 nm wavelength (Multiskan FC, Thermo Scientific Inc., MA, USA). Calibration curves were calculated in 0, 5, 10, 25, 50 and 75 μM H2O2 range.
Analysis of gene expression array data
Affymetrix raw data were processed with robust multiarray average normalization using Bioconductor limma and affy packages in R [95, 96]. Raw data from the Affymetrix ATH1-121501 platform was obtained from several data sources: NASC Arrays [131] (BTH, NASCARRAYS-392; Senescence experiment 1, NASCARRAYS-52; Senescence experiment 2, NASCARRAYS-150; SA, NASCARRAYS-192). ArrayExpress [132] (MeJA, E-ATMX-13; Syringolin, E-MEXP-739; catalase deficient Arabidopsis and high light, E-MEXP-449) Gene Expression Omnibus [133] (H2O2, GSE5530; csn3, csn4 and csn5, GSE9728; lht1, GSE19109; mkk1mkk2, GSE10646; sni1, GSE6827; siz1, GSE6583; SA 24 h, GSE14961; Ethylene, GSE14247; Flg22, GSE5615; Botrytis cinerea, GSE5684; Pseudomonas syringae ES4326, GSE18978; ozone, GSE5722; auxin, GSE39384; cat2 and GO5, GSE54534; cat2 and daylength, GSE27985). Raw data for acd11 were obtained from John Mundy. Gene expression for each experiment was computed by log2 fold changes between treatment and control, or between wild type and mutant. The processed data was discretized and clustered using Bayesian Hierarchical Clustering [97]. Bootstrap analysis was done as previously described in [98].
Statistical analyses
One-way ANOVA was used to test differences in survival, PCD and H2O2 between mutants (Statistica 7.1, Stat Soft Inc., OK, USA). After confirmation of statistically significant F-value, the pair-wise comparison between cat2 and double mutants was performed with Fisher LSD test. Analysis of variance (GLM procedure) was used for multiple comparisons between mean values of factor levels to find significant interactions, thus, the effect was tested for categorical predictor variables (time from shift, cat2 versus double mutant in binary system) with single dependent variable (survival or Fv/Fm).
Acknowledgements
Tuomas Puukko and Taras Kazantsev provided superb technical assistance. We thank Fuqiang Cui, Melanie Carmody, Luis Morales, Kirk Overmyer, Cezary Waszczak and Michael Wrzaczek for comments on the manuscript. This work was supported by grants from European Social Fund (Mobilitas Top Researchers grant MTT9 and Mobilitas Post-doc grant MJD115), by the Academy of Finland (grants: 135751, 140981 and 273132) and the Academy of Finland Center of Excellence in Primary Producers 2014–2019.
Abbreviations
- ABA
Abscisic acid
- Fv/Fm
Photosystem II maximum photosynthetic efficiency
- HR
Hypersensitive response
- JA
Jasmonic acid
- H2O2
Hydrogen peroxide
- LD
Long day
- LMM
Lesion mimic mutant
- MPK
Mitogen-activated protein kinases
- MS
Murashige and Skoog medium
- OPDA
12-oxo-phytodienoic acid
- PCD
Programmed cell death
- RBOHs
Respiratory burst oxidase homologs
- ROS
Reactive oxygen species
- SA
Salicylic acid
- SD
Short day
- TF
Transcription factor
Additional files
Primers and restriction enzymes used for mutant genotyping. Additional information about the mutants used. (XLSX 14 kb)
The cat2 phenotype depends on sucrose concentration in the agar media. Ten days old seedlings in different sucrose concentrations (0, 0.5, 1 and 2 %). At 0 % sucrose the seedlings were bleached, with increased sucrose concentration the seedlings were green. (TIFF 2004 kb)
Survival of cat2 and cat2 double mutants in the in vitro plate assay. Fourteen-days old seedlings were transferred from SD (8 h/16 h) to LD (12 h/12 h) and survival was scored on the 1st, 5th, 8th days after shift to LD. The experiment was repeated three times using 10 plants (mean±SE, n=30). (EPS 14277 kb)
Lesion formation in cat2 and cat2 double mutants in 4 week old soil-grown plants in LD (12 h/12 h). Cell death is indicated by trypan blue stain. White scale bar shows 1 cm and black scale bar 600 μm. The experiment was repeated six times using 5 plants per repeat. (EPS 36576 kb)
Lesion formation in cat2 and cat2 double mutants in 4 week old soil-grown plants in LD (12 h/12 h). Cell death is indicated by trypan blue stain. White scale bar shows 1 cm and black scale bar 600 μm. The experiment was repeated six times using 5 plants per repeat. (EPS 36989 kb)
Lesion formation in cat2 and cat2 double mutants in 4 week old soil-grown plants in LD (12 h/12 h). Cell death is indicated by trypan blue stain. White scale bar shows 1 cm and black scale bar 600 μm. The experiment was repeated six times using 5 plants per repeat. (EPS 30832 kb)
Microscopic cell death in dnd1, vtc1 and corresponding double mutants with cat2. Cell death is indicated with trypan blue stain. White scale bar shows 1 cm and black scale bar 600 μm. (EPS 23350 kb)
Maximum photosynthetic efficiency (Fv/Fm; mean±SE) in dark adapted plants after transfer to continuous light and restricted gas-exchange. Fourteen-days old seedlings were transferred from 70 μmol m−2 s−1 light intensity and 8h day/ 16h night to 120 μmol m−2 s−1 continuous light. Fv/Fm was measured on shift day, 2nd, 4th, and 7th. The experiment was repeated three times using 20 plants per repeat (n=60). Asterisks indicate significantly different Fv/Fm values of double mutant in time scale and compared to cat2 (p<0.05, General Linear Model). (EPS 815 kb)
A list of the genes used in the cat2 gene expression cluster analysis. (XLSX 15 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MB provided experimental design and plant material. EK performed experiments and statistical analyses. EX analyzed array data. EK and MB wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Eve Kaurilind, Email: eve.kaurilind@ut.ee.
Enjun Xu, Email: enjun.xu@helsinki.fi.
Mikael Brosché, Phone: +358294159436, Email: mikael.brosche@helsinki.fi.
References
- 1.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 2.Kohli A, Sreenivasulu N, Lakshmanan P, Kumar PP. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013;32:945–957. doi: 10.1007/s00299-013-1461-y. [DOI] [PubMed] [Google Scholar]
- 3.Wrzaczek M, Brosché M, Kangasjärvi J. ROS signaling loops — production, perception, regulation. Curr Opin Plant Biol. 2013;16:575–582. doi: 10.1016/j.pbi.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Foyer CH, Bloom AJ, Queval G, Noctor G. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol. 2009;60:455–484. doi: 10.1146/annurev.arplant.043008.091948. [DOI] [PubMed] [Google Scholar]
- 5.Vaahtera L, Brosché M, Wrzaczek M, Kangasjärvi J. Specificity in ROS signaling and transcript signatures. Antioxid Redox Signal. 2014;21:1422–1441. doi: 10.1089/ars.2013.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterhansel C, Maurino VG. Photorespiration redesigned. Plant Physiol. 2011;155:49–55. doi: 10.1104/pp.110.165019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. doi: 10.1034/j.1399-3054.2003.00223.x. [DOI] [Google Scholar]
- 8.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal. 2008;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 9.Rizhsky L, Hallak-Herr E, Van Breusegem F, Rachmilevitch S, Barr JE, Rodermel S, Inzé D, Mittler R. Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J. 2002;32:329–342. doi: 10.1046/j.1365-313X.2002.01427.x. [DOI] [PubMed] [Google Scholar]
- 10.Dickman MB, Fluhr R. Centrality of host cell death in plant-microbe interactions. Annu Rev Phytopathol. 2013;51:543–570. doi: 10.1146/annurev-phyto-081211-173027. [DOI] [PubMed] [Google Scholar]
- 11.Bruggeman Q, Raynaud C, Benhamed M, Delarue M. To die or not to die? Lessons from lesion mimic mutants. Plant Physiol. 2015;6:24. doi: 10.3389/fpls.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorrain S, Vailleau F, Balagué C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 13.Mhamdi A, Hager J, Chaouch S, Queval G, Han Y, Taconnat L, Saindrenan P, Gouia H, Issakidis-Bourguet E, Renou J-P, Noctor G. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010;153:1144–1160. doi: 10.1104/pp.110.153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Breusegem FV, Noctor G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- 15.Mhamdi A, Noctor G, Baker A. Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys. 2012;525:181–194. doi: 10.1016/j.abb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakière B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007;52:640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- 17.Chaouch S, Noctor G. Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 2010;188:711–718. doi: 10.1111/j.1469-8137.2010.03453.x. [DOI] [PubMed] [Google Scholar]
- 18.Chaouch S, Queval G, Vanderauwera S, Mhamdi A, Vandorpe M, Langlois-Meurinne M, Breusegem FV, Saindrenan P, Noctor G. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 2010;153:1692–1705. doi: 10.1104/pp.110.153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G. Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H(2)O(2) to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal. 2013;18:2106–2121. doi: 10.1089/ars.2012.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Mhamdi A, Chaouch S, Noctor G. Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 2013;36:1135–1146. doi: 10.1111/pce.12048. [DOI] [PubMed] [Google Scholar]
- 21.Vanderauwera S, Suzuki N, Miller G, van de Cotte B, Morsa S, Ravanat J-L, Hegie A, Triantaphylidès C, Shulaev V, Montagu MCEV, Breusegem FV, Mittler R. Extranuclear protection of chromosomal DNA from oxidative stress. Proc Natl Acad Sci. 2011;108:1711–1716. doi: 10.1073/pnas.1018359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanderauwera S, Vandenbroucke K, Inzé A, van de Cotte B, Mühlenbock P, Rycke RD, Naouar N, Gaever TV, Montagu MCEV, Breusegem FV. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc Natl Acad Sci. 2012;109:20113–20118. doi: 10.1073/pnas.1217516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerchev P, Mühlenbock P, Denecker J, Morreel K, Hoeberichts FA, Van Der Kelen K, Vandorpe M, Nguyen L, Audenaert D, Van Breusegem F. Activation of auxin signalling counteracts photorespiratory H2O2-dependent cell death. Plant Cell Environ. 2015;38:253–265. doi: 10.1111/pce.12250. [DOI] [PubMed] [Google Scholar]
- 24.Alcázar R, Parker JE. The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 2011;16:666–675. doi: 10.1016/j.tplants.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Torres MA, Dangl JL, Jones JDG. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Torres MA, Morales J, Sánchez-Rodríguez C, Molina A, Dangl JL. Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol Plant Microbe Interact. 2013;26:686–694. doi: 10.1094/MPMI-10-12-0236-R. [DOI] [PubMed] [Google Scholar]
- 28.Chaouch S, Queval G, Noctor G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J Cell Mol Biol. 2012;69:613–627. doi: 10.1111/j.1365-313X.2011.04816.x. [DOI] [PubMed] [Google Scholar]
- 29.Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 2005;139:1291–1303. doi: 10.1104/pp.105.067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Bent AF. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu E, Brosché M. Salicylic acid signaling inhibits apoplastic reactive oxygen species signaling. BMC Plant Biol. 2014;14:155. doi: 10.1186/1471-2229-14-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell. 2007;19:1081–1095. doi: 10.1105/tpc.106.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjärvi J, Inzé D, Van Breusegem F. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 2003;33:621–632. doi: 10.1046/j.1365-313X.2003.01655.x. [DOI] [PubMed] [Google Scholar]
- 34.Ahlfors R, Lång S, Overmyer K, Jaspers P, Brosché M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M, Inzé D, Palva ET, Kangasjärvi J. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein–protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 36.Kim BH, Kim SY, Nam KH. Assessing the diverse functions of BAK1 and its homologs in arabidopsis, beyond BR signaling and PTI responses. Mol Cells. 2013;35:7–16. doi: 10.1007/s10059-013-2255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Lenk A, Andersson MX, Gjetting T, Pedersen C, Nielsen ME, Newman M-A, Hou B-H, Somerville SC, Thordal-Christensen H. A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol Plant. 2008;1:510–527. doi: 10.1093/mp/ssn011. [DOI] [PubMed] [Google Scholar]
- 40.Monaghan J, Germain H, Weihmann T, Li X. Dissecting plant defence signal transduction: modifiers of snc1 in Arabidopsis. Can J Plant Pathol. 2010;32:35–42. doi: 10.1080/07060661003621001. [DOI] [Google Scholar]
- 41.Bartels S, Anderson JC, Besteiro MAG, Carreri A, Hirt H, Buchala A, Métraux J-P, Peck SC, Ulm R. MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Lu Y, Liu P, Wen W, Zhang J, Ge X, Xia Y. The ammonium/nitrate ratio is an input signal in the temperature-modulated, SNC1-mediated and EDS1-dependent autoimmunity of nudt6-2 nudt7. Plant J. 2013;73:262–275. doi: 10.1111/tpj.12032. [DOI] [PubMed] [Google Scholar]
- 44.Hubert DA, He Y, McNulty BC, Tornero P, Dangl JL. Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB-LRR disease resistance protein regulation. Proc Natl Acad Sci. 2009;106:9556–9563. doi: 10.1073/pnas.0904877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park S-W, Li W, Viehhauser A, He B, Kim S, Nilsson AK, Andersson MX, Kittle JD, Ambavaram MMR, Luan S, Esker AR, Tholl D, Cimini D, Ellerström M, Coaker G, Mitchell TK, Pereira A, Dietz K-J, Lawrence CB. Cyclophilin 20–3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci. 2013;110:9559–9564. doi: 10.1073/pnas.1218872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danon A, Miersch O, Felix G, op den Camp RGL, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005;41:68–80. doi: 10.1111/j.1365-313X.2004.02276.x. [DOI] [PubMed] [Google Scholar]
- 48.Ivanova A, Law SR, Narsai R, Duncan O, Lee J-H, Zhang B, Aken OV, Radomiljac JD, van der Merwe M, Yi K, Whelan J. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiol. 2014;165:1233–1254. doi: 10.1104/pp.114.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F. Mitochondrial perturbation negatively affects auxin signaling. Mol Plant. 2014;7:1138–1150. doi: 10.1093/mp/ssu071. [DOI] [PubMed] [Google Scholar]
- 50.Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002;130:887–894. doi: 10.1104/pp.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 2007;52:114–123. doi: 10.1111/j.1365-313X.2007.03211.x. [DOI] [PubMed] [Google Scholar]
- 52.Gray WM, Muskett PR, Chuang H, Parker JE. Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Türk F, Can C, Dangl JL, Holub EB. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 55.Galvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, Mullineaux PM. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell. 2009;21:2143–2162. doi: 10.1105/tpc.108.061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 57.Goritschnig S, Weihmann T, Zhang Y, Fobert P, McCourt P, Li X. A novel role for protein farnesylation in plant innate immunity. Plant Physiol. 2008;148:348–357. doi: 10.1104/pp.108.117663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overmyer K, Brosché M, Pellinen R, Kuittinen T, Tuominen H, Ahlfors R, Keinänen M, Saarma M, Scheel D, Kangasjärvi J. Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol. 2005;137:1092–1104. doi: 10.1104/pp.104.055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaminaka H, Näke C, Epple P, Dittgen J, Schütze K, Chaban C, Holt BF, Merkle T, Schäfer E, Harter K, Dangl JL. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25:4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruggeman Q, Garmier M, de Bont L, Soubigou-Taconnat L, Mazubert C, Benhamed M, Raynaud C, Bergounioux C, Delarue M. The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 2014;165:732–746. doi: 10.1104/pp.114.236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Breusegem FV. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/pp.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 64.Nurmberg PL, Knox KA, Yun B-W, Morris PC, Shafiei R, Hudson A, Loake GJ. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci. 2007;104:18795–18800. doi: 10.1073/pnas.0705586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007;7:2. doi: 10.1186/1471-2229-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Birkenbihl RP, Diezel C, Somssich IE. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses towards botrytis cinerea infection. Plant Physiol. 2012;159(1):266–85. doi: 10.1104/pp.111.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raffaele S, Rivas S, Roby D. An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 2006;580:3498–3504. doi: 10.1016/j.febslet.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 68.Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, Al-Abdallat A, Xia Y, Lu G, Ladunga I, Avramova Z. The highly similar arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 2008;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda-Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–575. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–266. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- 71.Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu J-L, Micheelsen P, Rocher A, Petersen M, Newman M-A, Bjørn Nielsen H, Hirt H, Somssich I, Mattsson O, Mundy J. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meng X, Xu J, He Y, Yang K-Y, Mordorski B, Liu Y, Zhang S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. The mitogen-activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sethi V, Raghuram B, Sinha AK, Chattopadhyay S. A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell. 2014;26:3343–3357. doi: 10.1105/tpc.114.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Overmyer K, Brosché M, Kangasjärvi J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003;8:335–342. doi: 10.1016/S1360-1385(03)00135-3. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi S, Bauwe H, Badger M. Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol. 2007;144:487–494. doi: 10.1104/pp.107.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Badger MR, Fallahi H, Kaines S, Takahashi S. Chlorophyll fluorescence screening of Arabidopsis thaliana for CO2 sensitive photorespiration and photoinhibition mutants. Funct Plant Biol. 2009;36:867–873. doi: 10.1071/FP09199. [DOI] [PubMed] [Google Scholar]
- 78.Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, Lu H. Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics. 2011;189:851–859. doi: 10.1534/genetics.111.132332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venugopal SC, Jeong R-D, Mandal MK, Zhu S, Chandra-Shekara AC, Xia Y, Hersh M, Stromberg AJ, Navarre D, Kachroo A, Kachroo P. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Queval G, Neukermans J, Vanderauwera S, Van Breusegem F, Noctor G. Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ. 2012;35:374–387. doi: 10.1111/j.1365-3040.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 82.Sewelam N, Jaspert N, Kelen KVD, Tognetti VB, Schmitz J, Frerigmann H, Stahl E, Zeier J, Breusegem FV, Maurino VG. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol Plant. 2014;7:1191–1210. doi: 10.1093/mp/ssu070. [DOI] [PubMed] [Google Scholar]
- 83.Gao X, Yuan H-M, Hu Y-Q, Li J, Lu Y-T. Mutation of Arabidopsis CATALASE2 results in hyponastic leaves by changes of auxin levels. Plant Cell Environ. 2014;37:175–188. doi: 10.1111/pce.12144. [DOI] [PubMed] [Google Scholar]
- 84.Tognetti VB, Mühlenbock P, Van Breusegem F. Stress homeostasis – the redox and auxin perspective. Plant Cell Environ. 2012;35:321–333. doi: 10.1111/j.1365-3040.2011.02324.x. [DOI] [PubMed] [Google Scholar]
- 85.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 86.De Coninck BMA, Sels J, Venmans E, Thys W, Goderis IJWM, Carron D, Delauré SL, Cammue BPA, De Bolle MFC, Mathys J. Arabidopsis thaliana plant defensin AtPDF1.1 is involved in the plant response to biotic stress. New Phytol. 2010;187:1075–1088. doi: 10.1111/j.1469-8137.2010.03326.x. [DOI] [PubMed] [Google Scholar]
- 87.Brosché M, Blomster T, Salojärvi J, Cui F, Sipari N, Leppälä J, Lamminmäki A, Tomai G, Narayanasamy S, Reddy RA, Keinänen M, Overmyer K, Kangasjärvi J. Transcriptomics and functional genomics of ROS-induced cell death regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, Zipfel C. Direct regulation of the NADPH oxidase RBOHD by the PRR-Associated Kinase BIK1 during plant immunity. Mol Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 89.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 90.Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313X.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 92.Cui F, Brosché M, Sipari N, Tang S, Overmyer K. Regulation of ABA dependent wound induced spreading cell death by MYB108. New Phytol. 2013;200:634–640. doi: 10.1111/nph.12456. [DOI] [PubMed] [Google Scholar]
- 93.Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. CSHL Press; 2002.
- 94.Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, Brearley C, Hell R, Marin E, Pogson BJ. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 96.Smyth GK. Limma: Linear Models fo Microarray data. In: Bioinformatics and Computational Biology Solution Using R and Bioconductor. Springer; 2005. p. 397–420.
- 97.Savage RS, Heller K, Xu Y, Ghahramani Z, Truman WM, Grant M, Denby KJ, Wild DL. R/BHC: fast Bayesian hierarchical clustering for microarray data. BMC Bioinformatics. 2009;10:242. doi: 10.1186/1471-2105-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. Transcriptional regulation of the CRK/DUF26 group of Receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010;10:95. doi: 10.1186/1471-2229-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Merilo E, Jalakas P, Kollist H, Brosché M. The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity, elevated CO2, and exogenous ABA. Mol Plant. 2015;8:657–659. doi: 10.1016/j.molp.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 100.Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci. 2010;107:2361–2366. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng W-H, Chiang M-H, Hwang S-G, Lin P-C. Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol. 2009;71:61–80. doi: 10.1007/s11103-009-9509-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doukhanina EV, Chen S, van der Zalm E, Godzik A, Reed J, Dickman MB. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J Biol Chem. 2006;281:18793–18801. doi: 10.1074/jbc.M511794200. [DOI] [PubMed] [Google Scholar]
- 103.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 104.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 105.Jones AM, Ecker JR, Chen J-G. A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 2003;131:1623–1627. doi: 10.1104/pp.102.017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Kangasjärvi J. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002;31:1–12. doi: 10.1046/j.1365-313X.2002.01328.x. [DOI] [PubMed] [Google Scholar]
- 108.Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 109.Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Conklin PL, Saracco SA, Norris SR, Last RL. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics. 2000;154:847–856. doi: 10.1093/genetics/154.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peleg-Grossman S, Melamed-Book N, Cohen G, Levine A. Cytoplasmic H2O2 prevents translocation of NPR1 to the nucleus and inhibits the induction of PR genes in Arabidopsis. Plant Signal Behav. 2010;5:1401–1406. doi: 10.4161/psb.5.11.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Forzani C, Carreri A, de la Fuente van Bentem S, Lecourieux D, Lecourieux F, Hirt H. The Arabidopsis protein kinase Pto-interacting 1–4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. FEBS J. 2011;278:1126–1136. doi: 10.1111/j.1742-4658.2011.08033.x. [DOI] [PubMed] [Google Scholar]
- 113.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 114.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/S0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 115.Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against pseudomonas syringae. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 119.Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 120.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao Q-M, Venugopal S, Navarre D, Kachroo A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011;155:464–476. doi: 10.1104/pp.110.166876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yun H, Hyun Y, Kang M-J, Noh Y-S, Noh B, Choi Y. Identification of regulators required for the reactivation of FLOWERING LOCUS C during Arabidopsis reproduction. Planta. 2011;234:1237–1250. doi: 10.1007/s00425-011-1484-y. [DOI] [PubMed] [Google Scholar]
- 124.Meier S, Ruzvidzo O, Morse M, Donaldson L, Kwezi L, Gehring C. The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell. 2012;24:2200–2212. doi: 10.1105/tpc.111.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Machingura M, Sidibe A, Wood AJ, Ebbs SD. The β-cyanoalanine pathway is involved in the response to water deficit in Arabidopsis thaliana. Plant Physiol Biochem. 2013;63:159–169. doi: 10.1016/j.plaphy.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 127.Shapiro AD, Zhang C. The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 2001;127:1089–1101. doi: 10.1104/pp.010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Olszak B, Malinovsky FG, Brodersen P, Grell M, Giese H, Petersen M, Mundy J. A putative flavin-containing mono-oxygenase as a marker for certain defense and cell death pathways. Plant Sci. 2006;170:614–623. doi: 10.1016/j.plantsci.2005.10.016. [DOI] [Google Scholar]
- 130.The European Arabidopsis Stock Centre. http://arabidopsis.info/. Accessed 1 May 2015.
- 131.NASC`s International Affymetric Service. http://affymetrix.arabidopsis.info/link_to_iplant.shtml. Accessed 1 May 2015.
- 132.ArrayExpress. http://www.ebi.ac.uk/arrayexpress/. Accessed 1 May 2015.
- 133.NCBI Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/. Accessed 1 May 2015.


