Abstract
Background
Pilocytic astrocytoma is the most common type of brain tumor in the pediatric population, with a generally favorable prognosis, although recurrences or leptomeningeal dissemination are sometimes also observed. For tumors originating in the supra-or infratentorial location, a different molecular background was suggested, but plausible correlations between the transcriptional profile and radiological features and/or clinical course are still undefined. The purpose of this study was to identify gene expression profiles related to the most frequent locations of this tumor, subtypes based on various radiological features, and the clinical pattern of the disease.
Methods
Eighty six children (55 males and 31 females) with histologically verified pilocytic astrocytoma were included in this study. Their age at the time of diagnosis ranged from fourteen months to seventeen years, with a mean age of seven years. There were 40 cerebellar, 23 optic tract/hypothalamic, 21 cerebral hemispheric, and two brainstem tumors. According to the radiological features presented on MRI, all cases were divided into four subtypes: cystic tumor with a non-enhancing cyst wall; cystic tumor with an enhancing cyst wall; solid tumor with central necrosis; and solid or mainly solid tumor. In 81 cases primary surgical resection was the only and curative treatment, and in five cases progression of the disease was observed. In 47 cases the analysis was done by using high density oligonucleotide microarrays (Affymetrix HG-U133 Plus 2.0) with subsequent bioinformatic analyses and confirmation of the results by independent RT-qPCR (on 39 samples).
Results
Bioinformatic analyses showed that the gene expression profile of pilocytic astrocytoma is highly dependent on the tumor location. The most prominent differences were noted for IRX2, PAX3, CXCL14, LHX2, SIX6, CNTN1 and SIX1 genes expression even within different compartments of the supratentorial region. Analysis of the genes potentially associated with radiological features showed much weaker transcriptome differences. Single genes showed association with the tendency to progression.
Conclusions
Here we have shown that pilocytic astrocytomas of three different locations can be precisely differentiated on the basis of their gene expression level, but their transcriptional profiles does not strongly reflect the radiological appearance of the tumor or the course of the disease.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1810-z) contains supplementary material, which is available to authorized users.
Keywords: Gene expression profiling, Location, Outcome, Pilocytic astrocytoma, Radiological appearance
Background
Pilocytic astrocytoma (PA) is the most common type of brain tumor in the pediatric population, comprising approximately 25 % of all primary tumors, with the most frequent occurrence taking place between 5–10 years of age. Fortunately this tumor has a generally good outcome, however recurrences or leptomeningeal dissemination are also sometimes observed. PAs can affect various anatomical structures, but there are three most common locations: cerebellum, optic tract with hypothalamus, and cerebral hemispheres. Those tumors are mainly sporadic, except for cases occurring in patients with neurofibromatosis type 1 and, less frequently, with Frasier and Noonan syndromes [1–3]. Molecularly, pilocytic astrocytoma is characterized by a relatively small number of chromosomal abnormalities with the most common alteration located at chromosome 7q34 comprising the BRAF oncogene [4, 5]. In the high-throughput analysis era, limited reports of this type of tumor were made using expression profiling. The presupposition concerning the molecular heterogeneity of pilocytic astrocytomas was defined previously by Wong et al. as the result of unsupervised hierarchical clustering. Without inference from a clinical outcome, they identified two subgroups of tumors. Such results could be a consequence of including two cases of subtotally resected tumors and, more importantly, the more aggressive variant of astrocytoma with pilomyxoid features [6]. Later, an assumption describing different expression profiles for PAs of various locations was given by Sharma et al., who showed the LHX2 gene expression to be connected with supratentorial location [7]. A following analysis made by Tchoghandjian et al. showed upregulation of LHX2 together with SIX6 in tumors originated from the hypothalamo-chiasmatic region [8]. At the same time, MATN2 and ALDH1L1 genes were assumed to be connected with plausible PAs progression despite total surgical resection [9, 10]. Children affected by this tumor usually have a good prognosis, although in some cases recurrence or leptomeningeal dissemination may be observed [11–14]. Thus there is an ongoing need to search for molecular markers influencing the clinical behavior of this tumor. On the basis of observations made to date we verified the hypothesis that the location of pilocytic astrocytomas is the major cause of their genomic differences, and tried to find genes connected with patient outcome and tumor appearance. The aim of this study was to identify gene expression profiles related to the most frequent locations, radiological features, and the clinical course of the disease in a representative group of Polish children with PAs.
Methods
Patient samples
Eighty-six children with pilocytic astrocytoma who were operated on at the Department of Neurosurgery, Polish Mother’s Memorial Hospital, Research Institute in Lodz were included in this study. The group was comprised of 55 males and 31 females. The median age of patients at the time of diagnosis was 7 years (ranging from 14 months to 17 years). There were 40 cerebellar, 23 optic tract and hypothalamic, 21 cerebral hemispheric, and 2 brainstem tumors (Fig. 1). All specimens were diagnosed at the Department of Molecular Pathology and Neuropathology, Medical University of Lodz, according to the WHO criteria [1].
Fig. 1.
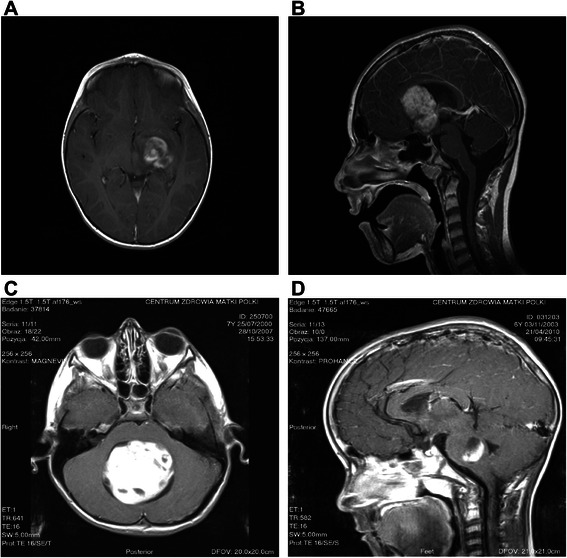
Location of pilocytic astrocytoma. a cerebral hemispheric tumor. b optic tract and hypothalamic tumor. c cerebellar tumor. d brainstem tumor. MRI scans after contrast administration
In all patients, preoperative MRI scans with and without contrast administration were obtained. For assessing the radiological features of tumors, we adopted the classification of radiological subtypes of PA proposed by Pencalet et al., commonly used in analyses of this tumor [15, 16]. According to the radiological features presented on MRI, all tumors were divided into four subtypes: cystic with a non-enhancing cyst wall, cystic with an enhancing cyst wall, solid with central necrosis, and solid or mainly solid tumors (Fig. 2).
Fig. 2.

Radiological type of pilocytic astrocytoma. a cystic tumor with an enhancing cyst wall, R1. b cystic tumor with a non-enhancing cyst wall, R2. c solid tumor with central necrosis, R3. d solid or mainly solid tumor, R4. MRI scans after contrast administration
In 81 cases primary surgical resection was the only and curative treatment, while in five cases progression of the disease, requiring additional treatment, was noted. In two cases clinical manifestation of neurofibromatosis type 1 (NF1) was observed. The clinical data of patients included in this study are presented in Table 1. All samples were collected using the protocols approved by the Bioethics Medical University Committee (Approval No. RNN/154/06/KE).
Table 1.
Clinicopathologic features of pilocytic astrocytoma patients
| Variable | Number | Percent |
|---|---|---|
| Gender | ||
| Male | 55 | 64 % |
| Female | 31 | 36 % |
| Age | ||
| 0-9 years | 53 | 62 % |
| 10-17 years | 33 | 38 % |
| Histopathology | ||
| Pilocytic astrocytoma | 86 | 100 % |
| Location | ||
| Cerebellum | 40 | 47 % |
| Optic tracts/Hypothalamus | 23 | 27 % |
| Hemisphere | 21 | 24 % |
| Brainstem | 2 | 2 % |
| Radiological appearance | ||
| Solid or mainly solid | 43 | 50 % |
| Cystic/Enhanced | 24 | 28 % |
| Cystic/Non enhanced | 10 | 12 % |
| Largely necrotic | 9 | 10 % |
| Extent of resection | ||
| Gross total | 63 | 73 % |
| Partial | 23 | 27 % |
| Clinical course and current patient status | ||
| Cured | 81 | 94 % |
| Alive | 80 | 99 % |
| Dead | 1 | 1 % |
| Progressive | 5 | 6 % |
| Alive | 4 | 99 % |
| Dead | 1 | 1 % |
| Genetic conditions | ||
| NF1 excluded | 84 | 98 % |
| NF1 confirmed | 2 | 2 % |
| Total | 86 | 100 % |
Written informed parental consent was obtained from all patients under 16 (75 patients). In eleven older patients the participants gave their own consent according to the Polish law. All data were processed and stored in compliance with the Helsinki Declaration.
RNA isolation
Total RNA was extracted from the snap-frozen tumor tissues stored at–80 °C after excision, using the acid phenol-guanidinum extraction method, purified using commercially available sets (RNeasy Mini Kit, Qiagen) and treated with DNAase (Qiagen) [17]. In order to obtain a high amount of RNA, macrodissection was used in all cases. Specimens were visually assessed by the pathologist to confirm that at least 50 % of tumor cells within the sample and areas with highest content of neoplastic tissue were used for direct RNA extraction. The quantity of RNA was measured using the NanoDrop 1000 (Thermo Scientific). RNA samples’ quality was analysed using 2000 Bioanalyzer (Agilent Technologies), and after capillary electrophoresis the RNA integrity number (RIN) was generated by the software for each specimen.
cRNA synthesis and hybridization
250 ng RNA of each sample selected for array analysis (50 cases) was used for cDNA and subsequent cRNA synthesis (GeneChip® 3′ IVT Expression Kit, Affymetrix). The amplified and biotinylated complementary RNA (cRNA) was purified and fragmented using heat and Mg2+, and then underwent hybridization (45 °C, 16 hours) with GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix), followed by array staining (Streptavidin, Alexa Fluor 610-R-phycoerythrin conjugate, Molecular Probes). All procedures were performed according to the manufacturer’s instructions (Affymetrix). Arrays were scanned using the GeneChip Scanner 3000 (Affymetrix).
Microarray analysis
Quality control of microarray data was carried out according to standard protocols, based on R/Bioconductor packages (ver. 2.3.5). Data were pre-processed using the GC-Robust Multi-array Average (GC-RMA) procedure, Normalized Unscaled Standard Error (NUSE) and Relative Log Expression (RLE) measures were calculated to verify the technical homogeneity of the dataset. On the basis of quality control, 47 out of 50 microarrays were then classified to the bioinformatic analyses.
Transcripts showing minimal variation of expression across the set of arrays were excluded from the analysis. Genes with expression differed by at least 1.5 times from the median in at least 10 % of the arrays, with variance significantly larger than the median variance (p ≤ 0.01) retained. For the selection of genes’ differentiating subgroups, the Welch t-test with false discovery rate (FDR) estimation was used. A global test was applied to test whether the expression profiles differed between the classes by permuting the labels of which arrays corresponded to which classes. Biological relevance and contribution in cellular processes of obtained sets was analyzed by Gene Ontology classification on the basis of the Gene Ontology Consortium database (http://www.geneontology.org). For selected genes the gene set enrichment analysis, with curated and motif gene set collections, was performed to analyze the signaling pathways (Molecular Signatures Database v. 3.0, http://www.broadinstitute.org/gsea/msigdb/index.jsp). These analyses were performed using Kolmogorov-Smirnov, the Least Squares test, and the Gene Set Analysis method (p ≤ 0.001). Statistical analysis was carried out by BRB-Array Tools (ver. 4.1.0, http://linus.nci.nih.gov/BRB-ArrayTools.html, developed by Dr R. Simon and BRB-Array Tools Development Team) and R/Bioconductor packages (http://www.bioconductor.org).
Validation of the microarray data
Correlation analysis of RT-qPCR and microarray expression values were carried out for 39 independent samples equally diversified according to the three tumor locations: cerebral hemispheric tumors (M1), optic tract and hypothalamic tumors (M2), cerebellar tumors (M3). TaqMan® Gene Expression Assays by TaqMan® real time PCR with TaqMan® Universal PCR Master Mix (Applied Biosystems, UK) was used following the manufacturer’s instructions on a Rotor Gene 6000 instrument (Qiagene-Corbett Life Science, Sydney, Australia) for selected genes (Additional file 1: Table S1). The PCR reactions for each assay were run in triplicate and the results were averaged. The normalized relative expression level of the genes of interest was calculated according to the method described by Pfaffl and Vandesompele et al., with GAPDH used as a reference gene [18, 19]. Statistical comparison of three subgroups was made on the basis of the Kruskal-Wallis nonparametric test, with post hoc pairwise comparisons using the Dwass-Steel-Critchlow-Fligner test. Statistical significance was assumed for p ≤ 0.05.
Results
We performed bioinformatic analysis of the global gene expression of 47 childhood pilocytic astrocytoma with respect to the selected clinical features. After pre-processing of the data 21,910 probesets showed significant variance and were further analysed. For the purposes of bioinformatic analysis, all analyzed samples were divided, on the basis of pivotal clinical data, into eight subgroups: cerebral hemispheric tumors (M1); optic tract and hypothalamic tumors (M2); cystic cerebellar tumors with an non-enhanced cyst (M3R1); cystic cerebellar tumors with an enhanced cyst (M3R2); solid cerebellar tumors with central necrosis (M3R3); solid or mainly solid cerebellar tumors (M3R4); tumors linked to the neurofibromatosis type 1 (NF1); and progressive tumors (P2).
During the comparison of these eight subgroups using the parametric Welch t-test and post hoc class comparison test, we found 345 probesets with significantly changed expression (p < 0.001). The observed differences were also strongly significant in the global test (p < 0.007) (Additional file 2: Table S2).
The evaluation of biological processes represented within the selected genes was done on the basis of the gene ontology over-representation analysis. The most significantly represented ontology classes were connected with neuronal cells building proteins, adhesion molecules, cell junctions, and hormone and neuropeptides activity (Table 2). Within genes with significantly changed expression, there were some that connected with transcriptional processes and acting during embryogenesis and central nervous system differentiation.
Table 2.
Gene ontology (GO) analysis of genes selected from transcripts differentiating the clinical subgroups of pilocytic astrocytomas
| GO ID | GO Term | Observed in the selected subset | Expected in the selected subset | Observed/Expected |
|---|---|---|---|---|
| Cellular components | ||||
| GO:0031225 | Anchored to membrane | 6 | 1.66 | 3.62 |
| GO:0005925 | Focal adhesion | 6 | 1.72 | 3.48 |
| GO:0009897 | External side of plasma membrane | 6 | 1.77 | 3.4 |
| GO:0005924 | Cell-substrate adherens junction | 6 | 1.77 | 3.4 |
| GO:0031253 | Cell projection membrane | 5 | 1.52 | 3.28 |
| GO:0005912 | Adherens junction | 9 | 2.83 | 3.19 |
| GO:0030055 | Cell-substrate junction | 6 | 1.92 | 3.12 |
| GO:0043235 | Receptor complex | 5 | 1.74 | 2.87 |
| GO:0044297 | Cell body | 6 | 2.27 | 2.64 |
| GO:0043025 | Neuronal cell body | 6 | 2.27 | 2.64 |
| Molecular functions | ||||
| GO:0019842 | Vitamin binding | 6 | 1.84 | 3.26 |
| GO:0019900 | Kinase binding | 9 | 2.9 | 3.1 |
| GO:0019901 | Protein kinase binding | 7 | 2.3 | 3.05 |
| GO:0003714 | Transcription corepressor activity | 6 | 2.25 | 2.66 |
| GO:0008234 | Cysteine-type peptidase activity | 6 | 2.34 | 2.56 |
| GO:0042277 | Peptide binding | 5 | 2.3 | 2.18 |
| Biological processes | ||||
| GO:0032677 | Regulation of interleukin-8 production | 6 | 0.37 | 16.38 |
| GO:0032637 | Interleukin-8 production | 6 | 0.4 | 15.12 |
| GO:0003081 | Regulation of systemic arterial blood pressure by renin-angiotensin | 5 | 0.34 | 14.89 |
| GO:0001990 | Regulation of systemic arterial blood pressure by hormone | 5 | 0.43 | 11.7 |
| GO:0002221 | Pattern recognition receptor signaling pathway | 7 | 0.82 | 8.49 |
| GO:0003044 | Regulation of systemic arterial blood pressure mediated by a chemical signal | 5 | 0.61 | 8.19 |
| GO:0002758 | Innate immune response-activating signal transduction | 7 | 0.89 | 7.91 |
| GO:0002218 | Activation of innate immune response | 7 | 0.89 | 7.91 |
| GO:0003073 | Regulation of systemic arterial blood pressure | 5 | 0.64 | 7.8 |
| GO:0050886 | Endocrine process | 5 | 0.73 | 6.83 |
| GO:0021953 | Central nervous system neuron differentiation | 5 | 0.76 | 6.55 |
| GO:0050731 | Positive regulation of peptidyl-tyrosine phosphorylation | 6 | 0.95 | 6.34 |
| GO:0050729 | Positive regulation of inflammatory response | 5 | 0.82 | 6.07 |
| GO:0002768 | Immune response-regulating cell surface receptor signaling pathway | 7 | 1.19 | 5.88 |
| GO:0050864 | Regulation of B cell activation | 6 | 1.07 | 5.62 |
| GO:0050671 | Positive regulation of lymphocyte proliferation | 6 | 1.07 | 5.62 |
| GO:0070665 | Positive regulation of leukocyte proliferation | 6 | 1.1 | 5.46 |
| GO:0048839 | Inner ear development | 8 | 1.47 | 5.46 |
| GO:0032946 | Positive regulation of mononuclear cell proliferation | 6 | 1.1 | 5.46 |
| GO:0002429 | Immune response-activating cell surface receptor signaling pathway | 6 | 1.13 | 5.31 |
| GO:0050730 | Regulation of peptidyl-tyrosine phosphorylation | 7 | 1.47 | 4.78 |
| GO:0050670 | Regulation of lymphocyte proliferation | 8 | 1.71 | 4.68 |
| GO:0070663 | Regulation of leukocyte proliferation | 8 | 1.74 | 4.6 |
| GO:0032944 | Regulation of mononuclear cell proliferation | 8 | 1.74 | 4.6 |
| GO:0042129 | Regulation of T cell proliferation | 5 | 1.13 | 4.43 |
| GO:0032103 | Positive regulation of response to external stimulus | 7 | 1.62 | 4.33 |
| GO:0002706 | Regulation of lymphocyte mediated immunity | 5 | 1.19 | 4.2 |
| GO:0050727 | Regulation of inflammatory response | 6 | 1.47 | 4.1 |
| GO:0043583 | Ear development | 8 | 1.98 | 4.03 |
| GO:0009310 | Amine catabolic process | 8 | 1.98 | 4.03 |
| GO:0090047 | Positive regulation of transcription regulator activity | 5 | 1.31 | 3.81 |
| GO:0051091 | Positive regulation of transcription factor activity | 5 | 1.31 | 3.81 |
| GO:0008217 | Pegulation of blood pressure | 7 | 1.86 | 3.76 |
| GO:0051606 | Detection of stimulus | 6 | 1.65 | 3.64 |
| GO:0021537 | Telencephalon development | 8 | 2.2 | 3.64 |
| GO:0009064 | Glutamine family amino acid metabolic process | 5 | 1.37 | 3.64 |
| GO:0002822 | Regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 5 | 1.37 | 3.64 |
| GO:0002703 | Regulation of leukocyte mediated immunity | 5 | 1.37 | 3.64 |
| GO:0050870 | Positive regulation of T cell activation | 7 | 1.95 | 3.58 |
| GO:0002819 | Regulation of adaptive immune response | 5 | 1.4 | 3.56 |
| GO:0043388 | Positive regulation of DNA binding | 5 | 1.43 | 3.49 |
| GO:0051099 | Positive regulation of binding | 6 | 1.74 | 3.45 |
| GO:0009063 | Cellular amino acid catabolic process | 6 | 1.74 | 3.45 |
| GO:0043410 | Positive regulation of MAPKKK cascade | 5 | 1.5 | 3.34 |
| GO:0090046 | Regulation of transcription regulator activity | 8 | 2.47 | 3.24 |
| GO:0051090 | Regulation of transcription factor activity | 8 | 2.47 | 3.24 |
| GO:0048562 | Embryonic organ morphogenesis | 8 | 2.53 | 3.16 |
| GO:0046651 | Lymphocyte proliferation | 8 | 2.53 | 3.16 |
| GO:0042113 | B cell activation | 8 | 2.53 | 3.16 |
| GO:0018108 | Peptidyl-tyrosine phosphorylation | 8 | 2.53 | 3.16 |
| GO:0045927 | Positive regulation of growth | 5 | 1.62 | 3.09 |
| GO:0018212 | Peptidyl-tyrosine modification | 8 | 2.59 | 3.08 |
| GO:0070661 | Leukocyte proliferation | 8 | 2.62 | 3.05 |
| GO:0032943 | Mononuclear cell proliferation | 8 | 2.62 | 3.05 |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | 5 | 1.65 | 3.03 |
| GO:0002237 | Response to molecule of bacterial origin | 6 | 1.98 | 3.02 |
| GO:0046395 | Carboxylic acid catabolic process | 8 | 2.66 | 3.01 |
| GO:0016054 | Organic acid catabolic process | 8 | 2.66 | 3.01 |
| GO:0042098 | T cell proliferation | 5 | 1.68 | 2.98 |
| GO:0042445 | Hormone metabolic process | 7 | 2.38 | 2.94 |
| GO:0019935 | Cyclic-nucleotide-mediated signaling | 6 | 2.04 | 2.93 |
| GO:0009952 | Anterior/posterior pattern formation | 6 | 2.08 | 2.89 |
| GO:0010001 | Glial cell differentiation | 5 | 1.77 | 2.82 |
| GO:0009266 | Response to temperature stimulus | 5 | 1.77 | 2.82 |
| GO:0071375 | Cellular response to peptide hormone stimulus | 6 | 2.14 | 2.81 |
| GO:0030217 | T cell differentiation | 9 | 3.27 | 2.76 |
| GO:0045664 | Regulation of neuron differentiation | 9 | 3.33 | 2.71 |
| GO:0050863 | Regulation of T cell activation | 8 | 2.99 | 2.67 |
| GO:0001934 | Positive regulation of protein amino acid phosphorylation | 6 | 2.29 | 2.62 |
| GO:0051222 | Positive regulation of protein transport | 5 | 1.95 | 2.56 |
| GO:0002460 | Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 7 | 2.87 | 2.44 |
| GO:0043408 | Regulation of MAPKKK cascade | 7 | 2.9 | 2.41 |
| GO:0042327 | Positive regulation of phosphorylation | 6 | 2.5 | 2.4 |
| GO:0034097 | Response to cytokine stimulus | 6 | 2.5 | 2.4 |
| GO:0002250 | Adaptive immune response | 7 | 2.93 | 2.39 |
| GO:0002449 | Lymphocyte mediated immunity | 6 | 2.53 | 2.37 |
| GO:0050953 | Sensory perception of light stimulus | 7 | 3.02 | 2.32 |
| GO:0007601 | Visual perception | 7 | 3.02 | 2.32 |
| GO:0045937 | Positive regulation of phosphate metabolic process | 6 | 2.59 | 2.31 |
| GO:0042063 | Gliogenesis | 5 | 2.17 | 2.31 |
| GO:0010562 | Positive regulation of phosphorus metabolic process | 6 | 2.59 | 2.31 |
| GO:0007411 | Axon guidance | 5 | 2.17 | 2.31 |
| GO:0043010 | Camera-type eye development | 5 | 2.2 | 2.28 |
| GO:0009617 | Response to bacterium | 8 | 3.51 | 2.28 |
| GO:0002697 | Regulation of immune effector process | 5 | 2.2 | 2.28 |
| GO:0032870 | Cellular response to hormone stimulus | 8 | 3.54 | 2.26 |
| GO:0071495 | Cellular response to endogenous stimulus | 8 | 3.57 | 2.24 |
| GO:0030855 | Epithelial cell differentiation | 5 | 2.23 | 2.24 |
| GO:0030258 | Lipid modification | 5 | 2.23 | 2.24 |
| GO:0043122 | Regulation of I-kappaB kinase/NF-kappaB cascade | 6 | 2.78 | 2.16 |
| GO:0008643 | Carbohydrate transport | 5 | 2.32 | 2.16 |
| GO:0048568 | Embryonic organ development | 8 | 3.72 | 2.15 |
| GO:0051239 | Regulation of multicellular organismal process | 72 | 33.82 | 2.13 |
| GO:0008624 | Induction of apoptosis by extracellular signals | 5 | 2.38 | 2.1 |
| GO:0007399 | Nervous system development | 84 | 41.36 | 2.03 |
| GO:0002376 | Immune system process | 80 | 39.37 | 2.03 |
| GO:0003002 | Regionalization | 8 | 3.97 | 2.02 |
The number of genes changed in each category was compared with the number of expected occurrences. Only GO classes and parent classes with at least five observations in the selected subset and with an ’observed vs. expected’ ratio of at least two were shown
The analysis of selected genes’ contribution in the signaling pathways revealed changed regulation of 77 within 2131 curated gene sets, and 14 within 179 motif gene sets. The highest statistical significance was obtained for genes functionally connected with immune response pathways, pathways engaged in silencing suppressors during histone methylation and activation of the NFkB pathway. Interesting group consisted of targets for miR324-5p, miR432, miR299-3P, miR486 and miR499 and genes located near promoter regions of NR6A1, POU3F2, CUTL1, PAX8 and AHR transcription factors (Table 3).
Table 3.
Selected gene sets differentiated between pilocytic astrocytomas of variable clinical features
| Broad GeneSets | Number of genes | LS | KS | GSA |
|---|---|---|---|---|
| p-value | p-value | p-value | ||
| KONDO_PROSTATE_CANCER_HCP_WITH_H3K27ME3 | 100 | 0.00001 | 0.00001 | <0.005 |
| LI_CISPLATIN_RESISTANCE_UP | 66 | 0.00001 | 0.00019 | <0.005 |
| REACTOME_ACTIVATED_TLR4_SIGNALLING | 25 | 0.00001 | 0.00001 | <0.005 |
| REACTOME_TOLL_LIKE_RECEPTOR_4_CASCADE | 28 | 0.00001 | 0.00001 | <0.005 |
| SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_DN | 7 | 0.00001 | 0.00001 | <0.005 |
| SHARMA_PILOCYTIC_ASTROCYTOMA_LOCATION_UP | 31 | 0.00001 | 0.00001 | <0.005 |
| WATANABE_COLON_CANCER_MSI_VS_MSS_DN | 96 | 0.00001 | 0.00041 | <0.005 |
| ZHAN_MULTIPLE_MYELOMA_PR_DN | 69 | 0.00001 | 0.00001 | <0.005 |
| BIOCARTA_DC_PATHWAY | 13 | 0.00023 | 0.00079 | <0.005 |
| CGGTGTG, MIR-220 | 8 | 0.00001 | 0.09215 | <0.005 |
| V$GNCF_01 | 93 | 0.00002 | 0.00239 | 0.04 |
| GGGATGC, MIR-324-5P | 72 | 0.00003 | 0.01085 | <0.005 |
| CCATCCA, MIR-432 | 64 | 0.00006 | 0.00044 | 0.015 |
| V$POU3F2_01 | 94 | 0.00006 | 0.02333 | 0.025 |
| CCANNAGRKGGC_UNKNOWN | 89 | 0.00017 | 0.00001 | 0.035 |
| V$CDPCR3_01 | 39 | 0.00028 | 0.02874 | 0.065 |
| CCCACAT,MIR-299-3P | 72 | 0.00034 | 0.08243 | 0.07 |
| V$MYOGNF1_01 | 57 | 0.00045 | 0.01553 | 0.045 |
| CTGYNNCTYTAA_UNKNOWN | 73 | 0.00055 | 0.00747 | 0.09 |
| GTACAGG, MIR-486 | 72 | 0.00056 | 0.00176 | 0.01 |
| V$PAX8_01 | 49 | 0.00058 | 0.0018 | <0.005 |
| V$AHR_01 | 86 | 0.00068 | 0.05371 | 0.16 |
| CCAATNNSNNNGCG_UNKNOWN | 65 | 0.00407 | 0.22226 | <0.005 |
| AGTCTTA, MIR-499 | 99 | 0.02398 | 0.00033 | 0.22 |
| V$MEF2_04 | 24 | 0.03997 | 0.01638 | <0.005 |
Three independent tests: LS, KS permutation test, and Efron-Tibshirani’s GSA maxmean test were applied to select significantly affected gene classes
In the next step we analyzed the expression values of genes differentiating clinical subgroups of PA. Genes with highest amplitude were chosen for hierarchical clustering of samples (Fig. 3a). On the basis of such analysis we obtained three distinct clusters showing almost perfect classification of samples, which revealed that the main source of variability is related to the location of the tumors. The cerebellar tumors consist of a homogenic cluster, while the supratentorial samples showed single outlier specimens (Fig. 3b). Within tumors of optic tract and hypothalamus there was also a sample of brain stem PA, which presented a low correlation of gene expression with the supratentorial cases (r = 0,4). This sample was excluded from statistical analyses because of its low RNA quality, and as a consequence both cases of PA located within the brain stem were used only during data visualization.
Fig. 3.
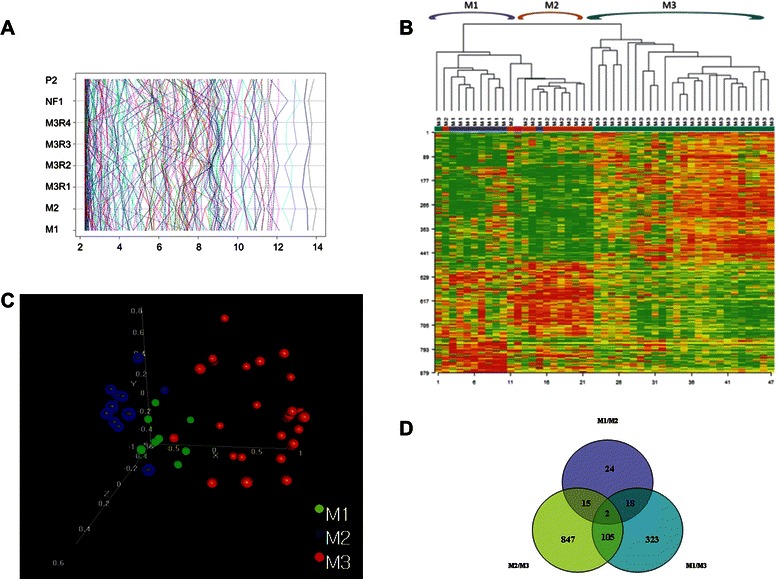
Results of analysis of correlation between gene expression profile and location of childhood pilocytic astrocytomas. a results of probesets expression according to clinical subgroup. b unsupervised hierarchical clustering demonstrated that PA from the three main locations have a unique transcriptome profile. c anatomical location-related subgroups are clearly distinct in fully unsupervised multidimensional scaling analysis. d Venn diagram showing numbers of location-related probesets. M1, cerebral hemispheric tumor; M2, optic tract and hypothalamic tumor; M3, cerebellar tumor; M3R1, cystic cerebellar tumor with an non-enhanced cyst; M3R2, cystic cerebellar tumor with enhanced cyst; M3R3, solid cerebellar tumor with central necrosis; M3R4, solid or mainly solid cerebellar tumor; NF1, tumor linked to the neurofibromatosis type 1; P2, progressive tumor
Bioinformatic analysis of our dataset revealed that 32 probesets showed different expression pattern according to radiological subclasses (p < 0.005). Unfortunately these genes demonstrated weak transcriptome differences (Fig. 4a), with borderline significance in the global test of association (p = 0,88). Hierarchical clustering and PCA analyses taking into account the radiological features of tumors did not show a specific gene expression signature correlated with the radiological features of analyzed PA (Fig. 4b, c).
Fig. 4.
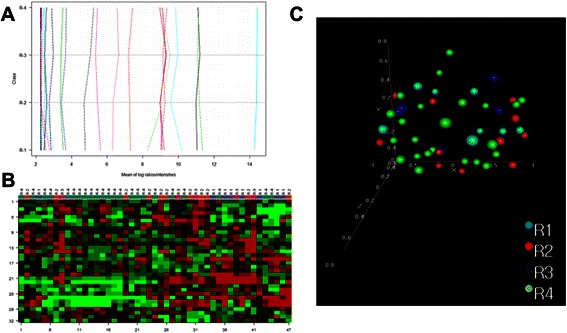
Results of association of radiological features with global gene expression profile. a probesets expression showing a relatively small amplitude of differences radiological subgroups. b unsupervised hierarchical clustering did not show specific gene expression signature correlated with radiological features (32 probesets, FDR adjusted p < 0.005). c radiological-related subgroups are not distinct in fully unsupervised multidimensional scaling analysis. R1, cystic tumor without an enhanced cyst wall; R2, cystic tumor with enhanced cyst; R3, solid tumor with central necrosis; R4, solid or mainly solid tumor
Here we verified the hypothesis that the location of PAs is the major cause of their genomic differences. Analyses of three main anatomical subclasses (M1, M2, M3) using the parametric Welch t-test were very prominent and revealed statistically significant differences for 862 probesets based on the false discovery rate (FDR adjusted p-value of 0.001).
In the global test the differences were also strongly significant (p < 0.001). The comparisons of pairs (M1vsM3; M2vsM3; M1vsM2) using the post hoc test (BRB ArrayTools) revealed that the majority of genes showed different expression for the M2vsM3 and M1vsM3 (847 and 323 genes respectively), while 105 genes showed differences both for M1vsM3 and M2vsM3 tumors. The most similarities (24 strongly differentiating genes) were noted for two analyzed supratentorial subgroups (Table 4, Fig. 3d, Additional file 3: Table S3 and Additional file 4: Table S4). These comparisons were also repeated with more restricted statistical criteria using the Benjamini-Hochberg multiple comparisons correction, with the criterion of FDR < 1 %. After comparison of the M3 and combined M1 and M2 subgroups, a list of 348 probesets was obtained. The probability of proper classification of tumors on the basis of gene expression profile reached a range of 80 % accuracy.
Table 4.
List of genes differentiated between pilocytic astrocytomas of three different locations
| Probe set | Gene symbol | Description | Parametric p-value | FDR | Geom mean of intensities in class M1 | Geom mean of intensities in class M2 | Geom mean of intensities in class M3 | Pairwise significant |
|---|---|---|---|---|---|---|---|---|
| 228462_at | IRX2 | Iroquois homeobox 2 | <1e-07 | <1e-07 | 7.81 | 36.27 | 470.74 | (1, 2), (1, 3), (2, 3) |
| 231666_at | PAX3 | Paired box 3 | <1e-07 | <1e-07 | 4.97 | 7.59 | 279.28 | (1, 3), (2, 3) |
| 207250_at | SIX6 | SIX homeobox 6 | <1e-07 | <1e-07 | 7.32 | 246.4 | 4.75 | (1, 2), (3, 2) |
| 206140_at | LHX2 | LIM homeobox 2 | <1e-07 | <1e-07 | 1680.77 | 2642.73 | 11.02 | (3, 1), (3, 2) |
| 223582_at | GPR98 | G protein-coupled receptor 98 | <1e-07 | <1e-07 | 50.94 | 146.32 | 15.08 | (1, 2), (3, 1), (3, 2) |
| 230720_at | RNF182 | Ring finger protein 182 | <1e-07 | <1e-07 | 171.48 | 1797.68 | 2061.29 | (1, 2), (1, 3) |
| 210239_at | IRX5 | Iroquois homeobox 5 | <1e-07 | <1e-07 | 7.84 | 25.89 | 100.36 | (1, 2), (1, 3), (2, 3) |
| 213285_at | TMEM30B | Transmembrane protein 30B | <1e-07 | <1e-07 | 7.23 | 26.44 | 5.08 | (1, 2), (3, 2) |
| 230472_at | IRX1 | Iroquois homeobox 1 | <1e-07 | <1e-07 | 10.8 | 66.79 | 205.19 | (1, 2), (1, 3), (2, 3) |
| 227202_at | CNTN1 | Contactin 1 | <1e-07 | <1e-07 | 262.72 | 51.46 | 1717.72 | (2, 1), (1, 3), (2, 3) |
| 214954_at | SUSD5 | Sushi domain containing 5 | <1e-07 | <1e-07 | 125.14 | 1108.04 | 1603.52 | (1, 2), (1, 3) |
| 206018_at | FOXG1 | Forkhead box G1 | <1e-07 | <1e-07 | 1197.42 | 11.25 | 4.93 | (2, 1), (3, 1) |
| 208221_s_at | SLIT1 | Slit homolog 1 (Drosophila) | <1e-07 | <1e-07 | 8.42 | 17.8 | 6 | (1, 2), (3, 2) |
| 238021_s_at | CRNDE | Colorectal neoplasia differentially expressed (non-protein coding) | <1e-07 | <1e-07 | 51.09 | 382.35 | 1216.52 | (1, 2), (1, 3), (2, 3) |
| 226448_at | FAM89A | Family with sequence similarity 89, member A | <1e-07 | <1e-07 | 104 | 625.78 | 808.9 | (1, 2), (1, 3) |
| 229831_at | CNTN3 | Contactin 3 (plasmacytoma associated) | <1e-07 | <1e-07 | 21.57 | 9.97 | 176.8 | (1, 3), (2, 3) |
| 228347_at | SIX1 | SIX homeobox 1 | <1e-07 | <1e-07 | 13.8 | 421.71 | 9.17 | (1, 2), (3, 2) |
| 206634_at | SIX3 | SIX homeobox 3 | <1e-07 | <1e-07 | 4.86 | 49.47 | 4.75 | (1, 2), (3, 2) |
| 238022_at | CRNDE | Colorectal neoplasia differentially expressed (non-protein coding) | <1e-07 | <1e-07 | 14 | 51.83 | 134.25 | (1, 2), (1, 3), (2, 3) |
| 238878_at | ARX | Aristaless related homeobox | <1e-07 | <1e-07 | 102.99 | 6.88 | 4.85 | (2, 1), (3, 1) |
| 227614_at | HKDC1 | Hexokinase domain containing 1 | <1e-07 | <1e-07 | 30.54 | 9.56 | 8.08 | (2, 1), (3, 1) |
| 205858_at | NGFR | Nerve growth factor receptor (TNFR superfamily, member 16) | <1e-07 | <1e-07 | 135.44 | 53.07 | 14.67 | (3, 1), (3, 2) |
M1-cerebral hemispheric tumor; M2-optic tract and hypothalamic tumor; M3-cerebellar tumor
In order to exclude the potential influence of other clinical variables on the obtained results, additional analysis was also performed for infratentorial cases, which included all four radiological types of PA. This approach confirmed our observations.
Our analyses of the transcriptome profile of five cases with progressive disease did not show any correlation with a worse outcome. Only five genes (SIX3, RGS8, FAM82, KIF9, WDR63) reached statistical significance (p = 0.001) when the univariate model was used, but the global test revealed that this association did not meet the criteria of statistical significance (p = 0.83) (Fig. 5a). Cases with neurofibromatosis type 1 had no connection with expression profile.
Fig. 5.
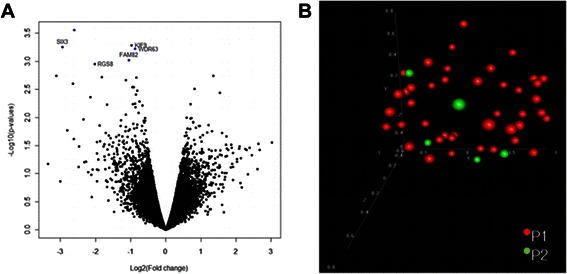
Progression of the disease was not associated with global gene expression profile. a only five genes were down regulated in group of tumors without progression (p < 0.005). b clinical course of the disease was not distinct in fully unsupervised multidimensional scaling analysis. P1, favorable outcome; P2, progressive disease
In the final stage of analysis we applied an unsupervised method (Principal Component Analysis, PCA) to determine the sources of variability in our group of samples according to the clinical data, and that analysis also indicated that the gene expression profile of pilocyticastrocytomas highly depends on the tumor’s location (p = 0.001), but not on other clinical features (Figs. 3c, 4c and 5b).
As the analysis of the most important determinants of gene expression (including location and radiological appearance) include the potential for multivariate associations, we carried out a two-way analysis of variance. In this multivariate approach two variables were taken into account (location: supratentorial vs infratentorial, i.e. M1 + M2 vs M3 + M4, and radiology: cystic vs solid, i.e. R1 + R2 vs R3 + R4). In the multivariate analysis only location seemed to be associated with gene expression (702 probesets with non-corrected p-value < 0.001, 1007 probesets with FDR < 10 %), while radiological appearance was almost without influence on gene expression (5 probesets with non-corrected p < 0.001, no probesets with FDR below 10 %).
During our analysis the most prominent differences connected with the location of the tumor were noted for the IRX2, PAX3, CXCL14, LHX2, SIX6, CNTN1 and SIX1 genes. For all these genes we performed validation results by independent RT-qPCR on 39 cases of PA, which confirmed the data obtained during microarray analysis with similar expression differences. For the PAX3, LHX2, CNCT1 and CXCL14 genes we obtained results which converged with the microarray results and these discriminants were the best to differentiate M1 and M2 from M3 tumors. For PAX3, LHX2, IRX2 and CNTN1 genes similar as in microarray analysis, the major expression differences for M1/M2 and M3 tumors was showen. The gene with a statistically significant level of expression for all three subgroups of tumors was SIX1 (Fig. 6).
Fig. 6.

Results of microarray data validation performed by independent RT-qPCR on 39 samples, equally diversified according to the three main locations. a-g values of PAX3, LHX2, CNCT1, CXCL14, IRX2, SIX6, SIX1 genes expression in pilocytic astrocytoma of different locations, presented as medians with respective inter-quartile ranges. h p-values obtained for site-specific subgroups compared using the Kruskal-Wallis test with post hoc pairwise comparisons using Dwass-Steel-Critchlow-Fligner test. Statistical significance was assumed as p ≤ 0.05. M1, cerebral hemispheric tumor (13 cases); M2, optic tract and hypothalamic tumor (13 cases); M3, cerebellar tumor (13 cases)
The data discussed in this publication have been deposited in the NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE73066 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73066) [20].
Discussion
In the current study we performed expression analysis of PAs diversified according to the main clinical discriminants, from which the tumor location was the most important. We also tried to verify if and how the transcriptional profile is connected with the radiological appearance of the tumor. Moreover we attempted to identify the expression profiles related to the clinical course of the disease.
Unsupervised clustering analysis revealed differences between PAs located in three anatomical regions, which was also well confirmed by decomposition into principal components (Fig. 3b, c). The specific transcriptional profile for hemispheric tumors was mainly characterized by upregulation of FOXG1, NEDD4L, L1CAM and CXCL14 genes. The FOXG1 and NEDD4L have a functional connection with the TGF-beta signaling pathway which is involved in cells’ proliferation and differentiation irregularities noted in other brain tumors, including gliomas, medulloblastomas and supratentorial ependymomas [21–23]. For the following L1CAM, acting during brain development and having an unknowable role in the adult nervous system, the correlation with tumor grade in several other solid human cancers was noted [22, 24]. In turn, expression of CXCL14 chemokine with enigmatic physiological function is connected with the tendency of tumor infiltration described on the basis of in vitro studies [25, 26]. Taken together, these genes are closely related not only to brain development but also to brain tumor growth and expansion. In this subgroup we also found the moderate overexpression of the LHX2 gene. Alterations of LHX2 expression were previously found in all supratentorial or hypothalamo-chiasmatic region tumors [8, 9]. Here we confirmed significantly higher levels of gene expression for PAs located within the hypothalamo-chiasmatic region. The validation of array data reached statistical significance and allowed us to distinguish these two subgroups of supratentorial tumors on the basis of LHX2 activity. The LHX2 gene has a known function in brain development during embryogenesis among different organisms. Previous functional studies on model organisms showed the relationship between lhx2 and two six3 genes in forebrain development. Recent analyses of human embryonic stem cells demonstrated that it also has a crucial role in human brain morphogenesis [27, 28]. There is evidence that LHX2 is expressed in the early neural lineage, which affects signaling pathways fundamental for early lineage differentiation in the human brain via regulation of the neural transcription factors from the SIX gene family [28, 29]. Our results constitute confirmation of this observation. The unique transcriptional profile of our hypothalamo-chiasmatic region tumors was characterized by the overrepresentation of several SIX homeobox transcription factors, SIX1, SIX3 and SIX6.
The upregulation of other homeobox transcription factors was also observed by us in cases of infratentorial tumor, which was shown to be connected with Iroquois homeobox transcription factors (IRX1, IRX2, IRX3, IRX5) and the PAX3 gene. A similar trend was noted by Sharma et al. [9]. These genes play a crucial role in early brain regionalization, including patterning of the anterior-posterior and dorso-ventral axis, as well as the differentiation of its specific regions. On model organisms its homologs overexpression induces the occurrence of ectopic neural tissue and causes inhibition of neuronal differentiation [30, 31].
Contactin family genes (CNTN1, CNTN3), which function as cell adhesion molecules with an essential role in later stages of cerebellar morphogenesis and differentiation, were also significantly overrepresentated in our cohort of cerebellar tumors [32, 33]. Moreover, recent experimental analysis of developing mice cerebellum suggests the cooperation of CNTN proteins in modulating SHH-induced neuronal precursor proliferation and its connection with foliation of the cerebellum [32]. The deregulation of the SHH signaling pathway in cerebellar granule neuron precursors is one of the crucial causes of the SHH subgroup of medulloblastoma, but recent observations also associate this signaling pathway with other central nervous system tumors, including gliomas [34]. This is the first report where CNTN1 and CNTN3 gene expression were shown to be connected with cerebellar pilocytic astrocytomas biology. Also our following observation concerning misregulation of miR-324-5p, engaged in the tumorigenesis of various cancers including glial tumors, suggests SHH contribution in PA biology as probably the consequence of changed progenitor differentiation [35–37].
Another interesting finding concerns the RUNX1T1 transcript upregulation observed in silico study performed for pilocytic astrocytomas by Deshmukh et al. On the basis of the transcriptional regulatory network, they indicated that RUNX1T1, together with four other transcription factors, is within alteration common for pilocytic astrocytomas and glioblastomas [38]. This is not the only relationship between those histologically different glial tumors. Our group of cerebellar PAs was also characterized by elevated levels of PROM1 expression, which in glioblastoma is considered as a molecular factor of poor prognosis, and in proneural glioblastoma seems to be the potential target of anti-angiogenic therapy [5, 39]. One could consider such observations as the molecular explanation of poor outcomes observed in some cases of PAs, but in light of findings showing gene expression also in normal adult brain astrocytes, such connection should be rather be regarded as additional indication of a gene with a nearly unknown function. Aggressive clinical behavior is more frequently observed in pilocytic astrocytoma located within the hypothalamo-chiasmatic region, and up to now is regarded as the result of limited surgical excision. However, the relation of an unfavorable disease outcome and overexpression of genes contributes to cell growth, proliferation, and differentiation, and hence it could not be excluded that it also promotes the cancerogenesis and tumor metastasis observed in PAs [40–43]. On the other hand, the frequently observed (in our study) alteration of genes involved in brain structures’ development like homeobox transcription factors, contactins and miRNAs inducing neurogenesis, together with the generally benign behavior of PAs, lead to the statement that these tumors could be rather recognized by some scientists as a neurodevelopmental disorder [7, 27, 28, 32, 44]. Another argument for this is the coexistence of neurodevelopmental disorders and pilocytic astrocytomas [45–47]. Such observations, which suggest the plausible association of autism and cerebellar PAs, also raise the question of their similar genetic background.
Until the problem of the biological characteristics of pilocytic astrocytoma will be solved, and the degree of surgical resection remains the only clinically confirmed factor affecting recurrence or dissemination, there is still a need to identify the molecular markers influencing the clinical behavior of this tumor. Taking this into account, we tried to search how gene expression profiling is correlated with the clinical outcomes of children with PA. An additional incentive for us to perform such analyses was the limited studies on this subject [48–50]. Unfortunately in our study we didn’t observe a correlation between a tumor’s global gene expression profile and the clinical course of the disease. In the analysis of the univariate test of progressive and low-risk disease, only five genes differentiated between the subgroups with progressive disease and the others, but in the global test there were no real differences between the analyzed classes.
Hence our unsupervised analysis didn’t disclose any differences between PA of different outcomes. The problem with revealing genes connected with the progressive disease during expression analyses was also noted in other studies, where such correlation were excluded or postulated only for an extremely limited number of genes. Sharma et al. and Rodriguez et al. assumed that the MATN2 and ALDH1L1 genes are connected with the progression of PA, but it seems to be more an accidental occurrence than crucial molecular changes. This observation was confirmed in the following studies [9, 10]. In our opinion this could be an effect of the low biological aggressiveness of Pas, and finding molecular factors usable in clinical practice seems very unlikely. Moreover, malignant transformation of PAs is really a very rare phenomenon and possible histological misdiagnosis cannot be excluded.
We also tried to find any correlations between molecular profile and radiological features, a subject which was completely unknown for PAs. Such plausible connections were noted in other glial brain tumors such as glioblastoma and oligodendroglioma, but this was the first attempt to find them for PAs [8, 9, 51, 52]. Major attention was directed on tumor borders and enhancement patterns after contrast administration. Here we adapted the division of predominant imaging patterns of pilocytic astrocytoma first proposed by Pencalet et al. and modified by others [15, 16]. In our study radiological features presented by pilocytic astrocytoma were not associated with global gene expression profile. In the univariate test only four genes were differentiated between the subgroups, with no differences observed in the global test. Moreover, the genes which best discriminated between different radiological features of tumors had a relatively small amplitude of differences and were not good discriminators between the groups in clustering (Fig. 4). Previous studies indicated that radiological appearance could be a derivative of tumor location; solid tumors were frequently observed infratentorially, and optic tract tumors rarely consist of cystic elements [14–16, 53]. Therefore we performed additional analyses for a cohesive subgroup of infratentorial lesions, which also didn’t reveal any correlations. Similarly, analysis of two subgroups of tumors with mainly solid and mostly cystic features showed no associations with transcriptional pattern. Our results show that different radiological patterns of PA are not determined by trancriptional changes.
Conclusions
In our comprehensive study we showed the differences between pilocytic astrocytoma of different locations and pointed out the heterogeneity of a tumor coming from the supratentorial region (Table 5). We may state that gene expression profile in pilocytic astrocytomas is connected only with tumor location, which suggests a different origin of PA arising within various anatomical brain structures. This observation is complementary to current knowledge of pilocytic astrocytoma’s biology and indicates that for targeted therapy three molecular subgroups of the tumor must be considered, particularly in the case of optimization of the treatment of optic nerve PAs. Our observation, combined with the similar results of a recent methylation study performed by Lambert et al., suggests that the molecular variation of PAs is probably the consequence of region-specific cells of origin’s presence, as is suggested in other brain tumors [54]. We pointed out the difficulties with finding the molecular alterations connected with worse clinical course of the disease, and found no connection between radiological features and global gene expression.
Table 5.
Summary of expression analysis of pilocytic astrocytomas
| Tumor Location | Hemisphere | Optic tracts | Cerebellum |
|---|---|---|---|
| Gene Family | FOXO | LHX SIX | IRX POU |
| Signaling Pathway | TLR | PTEN | |
| PI3/AKT MAPK TGFβ | |||
| miRNA | miR-220 miR-299-3p | miR-486 miR-324-5p | |
Acknowledgements
This work was supported by Ministry of Science and Higher Education Grant No. N401 196 32/4137.
Abbreviations
- FDR
False discovery rate
- GCMRA
GC-robust multi-array
- NF1
Neurofibromatosis type 1
- NUSE
Normalized unsealed standard error
- PA
Pilocytic astrocytoma
- PCA
Principal component analysis
- RT-qPCR
Quantitative real-time polymerase chain reaction
- RIN
RNA integrity number
- RLE
Relative log expression
Additional files
Characteristics of probes used for validation of microarray data (DOCX 15 kb)
List of genes differentiating between pilocytic astrocytomas by different clinical features (DOCX 117 kb)
List of genes differentiating between pilocytic astrocytomas in the three most frequently observed locations. Comparison of three subgroups. FDR, false discovery rate; M1, cerebral hemispheric tumor; M2, optic tract and hypothalamic tumor; M3, cerebellar tumor (DOCX 169 kb)
List of genes differentiating between pilocytic astrocytomas in the three most frequently observed locations. Comparison of all supratentorial and infratentorial tumors. M1/ M2, supratentorial tumors; M3, infratentorial tumors. The microarray data discussed in this publication have been deposited in the NCBI’s Gene Expression Omnibus. The accession number is GSE73066 and the link to freely access is is http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73066 (DOCX 72 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KZ and MZ designed and coordinated the study. KZ provided samples for experiments, obtained clinical data, and drafted and wrote the manuscript. MZ carried out the RNA isolation, RT-qPCR experiments, analyzed data, and interpreted the study. MJ and AP performed the biostatistical analyses and interpreted the obtained data. MO-W carried out the microarray experiments. PPL performed histopathology examinations. BJ participated in the design of the study. All authors participated in analysis and interpretation of the obtained data, and all have read and approved the final manuscript.
Contributor Information
Krzysztof Zakrzewski, Email: krzysztof.zakrzewski_xl@wp.pl.
Michał Jarząb, Email: mjarzab@io.gliwice.pl.
Aleksandra Pfeifer, Email: apfeifer@io.gliwice.pl.
Małgorzata Oczko-Wojciechowska, Email: gosiaoczko@io.gliwice.pl.
Barbara Jarząb, Email: bjarzab@io.gliwice.pl.
Paweł P. Liberski, Email: pawel.liberski@umed.lodz.pl
Magdalena Zakrzewska, Email: magdalena.zakrzewska@umed.lodz.pl.
References
- 1.Scheithauer BW, Hawkins C, Tihan T, VandenBerg SR, Burger PC. Pilocytic astrocytoma. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classification of tumors of the central nervous system. 4. Lyon: IARC; 2007. pp. 14–21. [Google Scholar]
- 2.Schuettpelz LG, McDonald S, Whitesell K, Desruisseau DM, Grange DK, Gurnett CA, et al. Pilocytic astrocytoma in a child with Noonan syndrome. Pediatr Blood Cancer. 2009;53:1147–1149. doi: 10.1002/pbc.22193. [DOI] [PubMed] [Google Scholar]
- 3.Subbiah V, Huff V, Wolff JE, Ketonen L, Lang FF, Jr, Stewart J, et al. Bilateral gonadoblastoma with dysgerminoma and pilocytic astrocytoma with WT1GT-IVS9 mutation: a 46 XY phenotypic female with Frasier syndrome. Pediatr Blood Cancer. 2009;53:1349–1351. doi: 10.1002/pbc.22152. [DOI] [PubMed] [Google Scholar]
- 4.Collins VP, Jones DT, Giannini C. Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:775–788. doi: 10.1007/s00401-015-1410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol. 2010;119:641–649. doi: 10.1007/s00401-009-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong KK, Chang YM, Tsang YT, Perlaky L, Su J, Adesina A, et al. Expression analysis of juvenile pilocytic astrocytomas by oligonucleotide microarray reveals two potential subgroups. Cancer Res. 2005;65:76–84. [PubMed] [Google Scholar]
- 7.Sharma MK, Mansur DB, Reifenberger G, Perry A, Leonard JR, Aldape KD, et al. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 2007;67:890–900. doi: 10.1158/0008-5472.CAN-06-0973. [DOI] [PubMed] [Google Scholar]
- 8.Tchoghandjian A, Fernandez C, Colin C, El Ayachi I, Voutsinos-Porche B, Fina F, et al. Pilocytic astrocytoma of the optic pathway: a tumour deriving from radial glia cells with a specific gene signature. Brain. 2009;132:1523–1535. doi: 10.1093/brain/awp048. [DOI] [PubMed] [Google Scholar]
- 9.Sharma MK, Watson MA, Lyman M, Perry A, Aldape KD, Deák F, et al. Matrilin-2 expression distinguishes clinically relevant subsets of pilocytic astrocytoma. Neurology. 2006;66:127–130. doi: 10.1212/01.wnl.0000188667.66646.1c. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez FJ, Giannini C, Asmann YW, Sharma MK, Perry A, Tibbetts KM, et al. Gene expression profiling of NF-1-associated and sporadic pilocytic astrocytoma identifies aldehyde dehydrogenase 1 family member L1 (ALDH1L1) as an underexpressed candidate biomarker in aggressive subtypes. J Neuropathol Exp Neurol. 2008;67:1194–1204. doi: 10.1097/NEN.0b013e31818fbe1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otero-Rodríguez A, Sarabia-Herrero R, García-Tejeiro M, Zamora-Martínez T. Spontaneous malignant transformation of a supratentorial pilocytic astrocytoma. Neurochirugia (Austr) 2010;21:245–252. doi: 10.1016/s1130-1473(10)70084-1. [DOI] [PubMed] [Google Scholar]
- 12.Parsa CF, Givrad S. Pilocytic astrocytomas as hamartomas: implications for treatment. Br J Ophthalmol. 2008;92:3–6. doi: 10.1136/bjo.2007.124941. [DOI] [PubMed] [Google Scholar]
- 13.Rozen WM, Joseph S, Lo PA. Spontaneous regression of low-grade gliomas in pediatric patients without neurofibromatosis. Pediatr Neurosurg. 2008;44:324–328. doi: 10.1159/000134925. [DOI] [PubMed] [Google Scholar]
- 14.Villarejo F, De Diego JM, de la Riva AG. Prognosis of cerebellar astrocytomas in children. Childs Nerv Syst. 2008;24:203–210. doi: 10.1007/s00381-007-0449-8. [DOI] [PubMed] [Google Scholar]
- 15.Pencalet P, Maixner W, Sainte-Rose C, Lellouch-Tubiana A, Cinalli G, Zerah M, et al. Benign cerebellar astrocytomas in children. J Neurosurg. 1999;90:265–273. doi: 10.3171/jns.1999.90.2.0265. [DOI] [PubMed] [Google Scholar]
- 16.Koeller KK, Rushing EJ. From the archives of the AFIP: pilocytic astrocytoma: radiologic-pathologic correlation. Radiographics. 2004;24:1693–1708. doi: 10.1148/rg.246045146. [DOI] [PubMed] [Google Scholar]
- 17.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adesina AM, Nguyen Y, Mehta V, Takei H, Stangeby P, Crabtree S, et al. FOXG1 dysregulation is a frequent event in medulloblastoma. J Neurooncol. 2007;85:111–122. doi: 10.1007/s11060-007-9394-3. [DOI] [PubMed] [Google Scholar]
- 22.Zeimet AG, Reimer D, Huszar M, Winterhoff B, Puistola U, Azim SA, et al. L1CAM in early-stage type I endometrial cancer: results of a large multicenter evaluation. J Natl Cancer Inst. 2013;105:1142–1150. doi: 10.1093/jnci/djt144. [DOI] [PubMed] [Google Scholar]
- 23.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J Oncol. 2012;2012:376894. doi: 10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Augsten M, Hägglöf C, Olsson E, Stolz C, Tsagozis P, Levchenko T, et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc Natl Acad Sci U S A. 2009;106:3414–3419. doi: 10.1073/pnas.0813144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabrusiewicz K, Ellert-Miklaszewska A, Lipko M, Sielska M, Frankowska M, Kaminska B. Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS One. 2011;6:e23902. doi: 10.1371/journal.pone.0023902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando H, Kobayashi M, Tsubokawa T, Uyemura K, Furuta T, Okamoto H. Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev Biol. 2005;287:456–468. doi: 10.1016/j.ydbio.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Hou PS, Chuang CY, Kao CF, Chou SJ, Stone L, Ho HN, et al. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 2013;41:7753–7770. doi: 10.1093/nar/gkt567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez C, Dastot-Le Moal F, Collot N, Legendre M, Abadie I, Bertrand AM, et al. Screening of LHX2 in patients presenting growth retardation with posterior pituitary and ocular abnormalities. Eur J Endocrinol. 2012;167:85–91. doi: 10.1530/EJE-12-0026. [DOI] [PubMed] [Google Scholar]
- 30.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrios N, González-Pérez E, Hernández R, Campuzano S. The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields. PLoS Genet. 2015;11(8):e1005463. doi: 10.1371/journal.pgen.1005463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xenaki D, Martin IB, Yoshida L, Ohyama K, Gennarini G, Grumet M, et al. F3/contactin and TAG1 play antagonistic roles in the regulation of sonic hedgehog-induced cerebellar granule neuron progenitor proliferation. Development. 2011;138:519–529. doi: 10.1242/dev.051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bizzoca A, Virgintino D, Lorusso L, Buttiglione M, Yoshida L, Polizzi A, et al. Transgenic mice expressing F3/contactin from the TAG-1 promoter exhibit developmentally regulated changes in the differentiation of cerebellar neurons. Development. 2003;130:29–43. doi: 10.1242/dev.00183. [DOI] [PubMed] [Google Scholar]
- 34.Ju B, Chen W, Spitsbergen JM, Lu J, Vogel P, Peters JL, et al. Activation of Sonic hedgehog signaling in neural progenitor cells promotes glioma development in the zebrafish optic pathway. Oncogenesis. 2014;3:e96. doi: 10.1038/oncsis.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, et al. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PLoS One. 2013;8:e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu HS, Zong HL, Shang M, Ming X, Zhao JP, Ma C, et al. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur Rev Med Pharmacol Sci. 2014;18:828–832. [PubMed] [Google Scholar]
- 37.Rush SZ, Abel TW, Valadez JG, Pearson M, Cooper MK. Activation of the Hedgehog pathway in pilocytic astrocytomas. Neuro Oncol. 2010;12:790–798. doi: 10.1093/neuonc/noq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deshmukh H, Yu J, Shaik J, MacDonald TJ, Perry A, Payton JE, et al. Identification of transcriptional regulatory networks specific to pilocytic astrocytoma. BMC Med Genomics. 2011;4:57. doi: 10.1186/1755-8794-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding BS, James D, Iyer R, Falciatori I, Hambardzumyan D, Wang S, et al. Prominin 1/CD133 endothelium sustains growth of proneural glioma. PLoS One. 2013;8:e62150. doi: 10.1371/journal.pone.0062150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Barón AE, Harrell JC, et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rorive S, Maris C, Debeir O, Sandras F, Vidaud M, Bièche I, et al. Exploring the distinctive biological characteristics of pilocytic and low-grade diffuse astrocytomas using microarray gene expression profiles. J Neuropathol Exp Neurol. 2006;65:794–807. doi: 10.1097/01.jnen.0000228203.12292.a1. [DOI] [PubMed] [Google Scholar]
- 43.Anagnostopoulos AK, Dimas KS, Papathanassiou C, Braoudaki M, Anastasiadou E, Vougas K, et al. Proteomics studies of childhood pilocytic astrocytoma. J Proteome Res. 2011;10:2555–2565. doi: 10.1021/pr200024m. [DOI] [PubMed] [Google Scholar]
- 44.Das E, Bhattacharyya NP. MicroRNA-432 contributes to dopamine cocktail and retinoic acid induced differentiation of human neuroblastoma cells by targeting NESTIN and RCOR1 genes. FEBS Lett. 2014;588:1706–1714. doi: 10.1016/j.febslet.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Minhas HM, Pescosolido MF, Schwede M, Piasecka J, Gaitanis J, Tantravahi U, et al. An unbalanced translocation involving loss of 10q26.2 and gain of 11q25 in a pedigree with autism spectrum disorder and cerebellar juvenile pilocytic astrocytoma. Am J Med Genet A. 2013;161A:787–791. doi: 10.1002/ajmg.a.35841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adachi K, Murai Y, Teramoto A. Infantile cerebellar pilocytic astrocytoma with autism spectrum disorder. J Nippon Med Sch. 2012;79:228–231. doi: 10.1272/jnms.79.228. [DOI] [PubMed] [Google Scholar]
- 47.Chen YH, Gutmann DH. The molecular and cell biology of pediatric low-grade gliomas. Oncogene. 2014;33:2019–2026. doi: 10.1038/onc.2013.148. [DOI] [PubMed] [Google Scholar]
- 48.Horbinski C, Hamilton RL, Lovell C, Burnham J, Pollack IF. Impact of morphology, MIB-1, p53 and MGMT on outcome in pilocytic astrocytomas. Brain Pathol. 2010;20:581–588. doi: 10.1111/j.1750-3639.2009.00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tibbetts KM, Emnett RJ, Gao F, Perry A, Gutmann DH, Leonard JR. Histopathologic predictors of pilocytic astrocytoma event-free survival. Acta Neuropathol. 2009;117:657–665. doi: 10.1007/s00401-009-0506-3. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez C, Figarella-Branger D, Girard N, Bouvier-Labit C, Gouvernet J, Paz Paredes A, et al. Pilocytic astrocytomas in children: prognostic factors-a retrospective study of 80 cases. Neurosurgery. 2003;53:544–553. doi: 10.1227/01.NEU.0000079330.01541.6E. [DOI] [PubMed] [Google Scholar]
- 51.Strong JA, Hatten HP, Jr, Brown MT, Debatin JF, Friedman HS, Oakes WJ, et al. Pilocytic astrocytoma: correlation between the initial imaging features and clinical aggressiveness. AJR Am J Roentgenol. 1993;161:369–372. doi: 10.2214/ajr.161.2.8333380. [DOI] [PubMed] [Google Scholar]
- 52.Beni-Adani L, Gomori M, Spektor S, Constantini S. Cyst wall enhancement in pilocytic astrocytoma: neoplastic or reactive phenomena. Pediatr Neurosurg. 2000;32:234–239. doi: 10.1159/000028944. [DOI] [PubMed] [Google Scholar]
- 53.Hirst DG, Robson T. Molecular biology: the key to personalised treatment in radiation oncology? Br J Radiol. 2010;83:723–7228. doi: 10.1259/bjr/91488645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambert SR, Witt H, Hovestadt V, Zucknick M, Kool M, Pearson DM, et al. Differential expression and methylation of brain developmental genes define location-specific subsets of pilocytic astrocytoma. Acta Neuropathol. 2013;126:291–301. doi: 10.1007/s00401-013-1124-7. [DOI] [PubMed] [Google Scholar]


