Abstract
Propionibacterium freudenreichii subsp. freudenreichii DSM 20271T is the type strain of species Propionibacterium freudenreichii that has a long history of safe use in the production dairy products and B12 vitamin. P. freudenreichii is the type species of the genus Propionibacterium which contains Gram-positive, non-motile and non-sporeforming bacteria with a high G + C content. We describe the genome of P. freudenreichii subsp. freudenreichii DSM 20271T consisting of a 2,649,166 bp chromosome containing 2320 protein-coding genes and 50 RNA-only encoding genes.
Keywords: +Propionibacterium, Type strain, Dairy starter, B12 vitamin
Introduction
Strain DSM 20271T (= van Niel 1928T = ATCC 6207) is the type strain of species Propionibacterium freudenreichii, which is the type species of its genus Propionibacterium [1]. There are traditionally two groups described in Propionibacterium genus; the “classical” or “dairy” and the “cutaneous” propionibacteria. P. freudenreichii belongs to the dairy group and is divided into two subspecies on the basis of lactose fermentation and nitrate reductase activity. The DSM 20271T strain represents the P. freudenreichii subsp. freudenreichii distinguished from subsp. shermanii by nitrate reduction and by a lack of lactose fermentation. [1]. Dairy propionibacteria do not belong to human microbiota but can be isolated from various habitats including raw milk, dairy products, soil and fermenting food and plant materials such as silage and fermenting olives [1]. Strains of P. freudenreichii have a long history of safe use in human diet and for instance in the production of Swiss-type cheeses, in which they play central role as ripening starters [1, 2]. Industrial applications of P. freudenreichii include production of vitamin B12 (cobalamin), as well as several other biomolecules like propionic acid, trehalose and conjugated linoleic acid [3]. Recently, there has been growing interest to study P. freudenreichii for its probiotic properties. Complete genome sequence of the type strain P. freudenreichii subsp. shermanii CIRM-BIA1 has been reported [4], but lack of other complete genome sequences has prevented the genomic level comparisons between the two subspecies. Thus, the genomic analysis of DSM 20271T strain should help us in P. freudenrichii subspecies definition that has been under debate [4, 5].
Here we present a summary classification and a set of features for P. freudenreichii DSM 20271T, together with the description of the complete genomic sequence and annotation.
Organism information
Classification and features
P. freudenreichii subsp. freudenreichii DSM 20271 T is a Gram-positive, non-motile, non-sporulating, anaerobic to aerotolerant, mesophilic Actinobacteria belonging to the order Propionibacterium. The strain was originally isolated as one of the three propionic acid-producing strains from Emmental cheese by von Freudenreich and Orla-Jensen as Bacterium acidi propionici a [6] during their work in Bern, Switzerland [7]. The strain was further studied by van Niel and renamed to Propionibacterium freudenreichii [6]. Figure 1 shows the phylogenetic neighborhood of DSM 20271T in a 16S rRNA sequence based tree. Cells of DSM 20271T are short rods with length of approximately 1,5 μm (Fig. 2). According to API 50 CH (Biomerieux, France) carbohydrate fermentation test the growth of DSM 20271T is supported by carbon sources including glucose, fructose, mannose, glycerol, adonitol, inositol, erythritol and galactose (Table 1).
Fig. 1.

Phylogenetic tree showing the relationship of Propionibacterium freudenreichii ssp. freudenreichii DSM 20271T (shown in bold print) to Mycobacterium tuberculosis, Corynebacterium glutamicum, Bifidobacterium longum, Propionibacterium acnes, Propionibacterium acidipropionici and Lactococcus lactis (outgroup). Tree is based on MAFFT [21] aligned 16 s rRNA gene. The tree was built using the Maximum-Likelihood method [22] with GAMMA model. Bootstrap analysis with 500 replicates was performed to assess the support of the tree topology. Tree was visualized with iTOL [23]
Fig. 2.
Optical microscope image. The cells of strain 20271 grown for 72 h, Gram stained. Image from light microspope, magnification 100x
Table 1.
Classification and general features of Propionibacterium freudenreichii subspecies freudenreichii DSM20271 T according to the MIGS recommendations [24]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [25] | |
| Phylum Actinobacteria | TAS [26, 27] | ||
| Class Actinobacteria | TAS [26, 27] | ||
| Order Propionbacteriales | TAS [28] | ||
| Family Propionibacteriaceae | TAS [1] | ||
| Genus Propionibacterium | TAS [1, 29] | ||
| Species Propionibacterium freudenreichii subspecies freudenreichii | TAS [1, 29, 30] | ||
| (Type) strain: van Niel 1928 T, (DSM 20271 T = ATCC 6207) | |||
| Gram stain | Positive | TAS [1] | |
| Cell shape | Rod | TAS [1] | |
| Motility | Non-motile | TAS [1] | |
| Sporulation | Not reported | NAS | |
| Temperature range | Mesophile | TAS [1] | |
| Optimum temperature | 30 °C | TAS [1] | |
| pH range; Optimum | ~4.5–8; ~7 | NAS | |
| Carbon source | Glucose, fructose, mannose, glycerol, adonitol, inositol, erythritol, galactose | IDA | |
| MIGS-6 | Habitat | Swiss cheese | TAS [] |
| MIGS-6.3 | Salinity | Unknown | TAS [] |
| MIGS-22 | Oxygen requirement | Anaerobic | TAS [1] |
| MIGS-15 | Biotic relationship | Free-living | NAS |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Unknown | NAS |
| MIGS-5 | Sample collection | Unknown | NAS |
| MIGS-4.1 | Latitude | Unknown | NAS |
| MIGS-4.2 | Longitude | Unknown | NAS |
| MIGS-4.4 | Altitude | Unknown | NAS |
aEvidence codes - IDA inferred from direct assay, TAS traceable author statement (i.e., a direct report exists in the literature), NAS non-traceable author statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [31]
Genome sequencing information
Genome project
This organism was selected for sequencing on the basis of its importance in food fermentations and in metabolite production.
Growth conditions and genomic DNA preparation
The strain was grown to early stationary growth phase in propionic medium (PPA), composed of 5.0 g. tryptone (Sigma-Aldrich), 10.0 g. yeast extract (Becton, Dickinson), 14.0 ml 60 % w/w DL-sodium lactate (Sigma-Aldrich) per liter and pH adjusted to 6.7. The cells were harvested by centrifugation for 5 min at 21,000 g at 4 °C and washed once with 0.1 M Tris–HCl pH 8.0. The DNA extraction was performed with ILLUSTRA™ bacteria genomicPrep Mini Spin Kit (GE Healthcare) according to the manufacturer’s instruction for Gram-positive bacteria using 100 mg/ml of lysozyme (Sigma-Aldrich) and 30 min incubation time in the lysis step.
Genome sequencing and assembly
The complete finished genome sequence of P. freudenreichii strain DSM 20271T was generated at the Institute of Biotechnology, University of Helsinki, using Pacific Biosciences RS II sequencing platform [8](Table 2) . One standard PacBio 10 kb library was constructed and sequenced using two SMRTCells with 180 min runtime on the RS II instrument, which generated 145,463 reads totaling up to 608.98 Mbp. For the assembly, the data was filtered using default HGAP parameters. The resulting 130,046 reads totaling up to 557.87 Mbp were used to generate the initial genome sequence. 498.74 Mbp of the filtered data mapped to the assembled genome afterwards. The assembled genome sequence was generated using SMRTAnalysis (2.1.0) HGAP2 pipeline [9] with default parameters, excluding the expected genome size and seed cutoff which were set to 2,700,000 and 7000 respectively. The assembly contains one contig which represents the whole genome. The resulting assembly was further improved by two consecutive rounds of mapping the full data on the reference and obtaining a new improved consensus sequence on each run. This was done using the standard SMRTAnalysis resequencing protocol with Quiver algorithm [9]. The circular nature of the final consensus sequence was then confirmed and the start of the sequence manually set to dnaA using Gap4 tool from Staden package [10].
Table 2.
Genome sequencing project information for Propionibacterium freudenreichii DSM 20271T
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | One PacBio 10 kb standard library |
| MIGS 29 | Sequencing platforms | PacBio RS II |
| MIGS 31.2 | Fold coverage | 198x |
| MIGS 30 | Assemblers | SMRTAnalysis (2.1.0), HGAP2 |
| MIGS 32 | Gene calling method | Prodigal v2.50 |
| Locus Tag | RM25 | |
| Genbank ID | CP010341 | |
| GenBank Date of Release | February 1st 2015 | |
| GOLD ID | Gs0113908 | |
| BIOPROJECT | PRJNA269789 | |
| MIGS 13 | Source Material Identifier | DSM 20271T |
| Project relevance | Type strain, dairy starter, B12 vitamin |
Genome annotation
Genes were identified using Prodigal v2.50 tool [11] with manual curation in ARGO Genome Browser [12]. The predicted genes were translated and functionally annotated with description lines, Gene Ontology (GO) classes and Enzyme Commission (EC) numbers with PANNZER program [13] using UniProtKB, Enzyme and GOA databases. PfamA domains were identified using InterProScan 48.0 [14], transmembrane helices and signal peptides were found with TMHMM [15] and SignalP [16], respectively. Clusters of Orthologous Groups (COG) assignments were done by using CD-Search [17]. The tRNAscanSE tool [18] was used to identify tRNA genes and ribosomal RNA were predicted with RNAmmer v1.2 [19].
Genome properties
The circular genome of Propionibacterium freudenreichii subsp. freudenreichii DSM 20271T is 2,649,166 nucleotides with 67.34 % GC content (Table 3) and contains one finalized chromosome with no plasmids. From total number of 2370 genes 2320 (97.9 %) are protein coding and 50 (2.1 %) are RNA genes (Fig. 3). 91.80 % of all proteins were functionally annotated whilst the remaining genes were annotated as “functionally unknown putative proteins”. The distribution of genes into COGs functional categories is presented in Table 4. Three sequence motifs containing methylated bases were also detected in the genome by PacBio sequencing and SMRTAnalysis Modification and Motif detection protocol. Two of these motifs, 5′-GGANNNNNNNCTT-3′ and 5′AAGNNNNNNNTCC-3′, are partner motifs and correspond to same modifications on different strands with m6A as a modified base on 3rd and 2nd position respectively. The modified base is shown in bold. Each motif is found 664 times in the genome and the marked bases are methylated in all of the 1328 motifs. The structure of the motifs and similarity to existing methyltransferases in REBASE [20] suggests that this is a Type I restriction-modification (RM) system. The third modified motif detected in the genome is 5′-TCGWCGA-3′ which partners with itself and is found 4258 times in the genome. In 3676 of these motifs (86.3 %) the 5th nucleotide (C) is found to be modified. The type of this modification could not be reliably identified. However, 319 of the detected modifications are identified as m4C although with low confidence. This finding is supported by the similarity of the recognition site to existing methyltransferases found in REBASE [20] and suggests that there is a Type II RM system acting on this motif in the genome. Therefore also the unidentified modifications are probably m4C bases. This data together suggest that there are two active RM systems present in P. freudenreichii. Comprehensive analysis of these RM systems and corresponding methylations requires further study.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 2,649,166 | 100.00 |
| DNA coding (bp) | 2,321,778 | 87.64 |
| DNA G + C (bp) | 1,783,838 | 67.34 |
| DNA scaffolds | 1 | 100.00 |
| Total genes | 2353 | 100.00 |
| Protein coding genes | 2320 | 97.90 |
| RNA genes | 50 | 2.10 |
| Pseudo genes | NA | NA |
| Genes in internal clusters | NA | NA |
| Genes with function prediction | 2160 | 91.80 |
| Genes assigned to COGs | 1751 | 74.42 |
| Genes with Pfam domains | 1958 | 83.21 |
| Genes with signal peptides | 113 | 4.80 |
| Genes with transmembrane helices | 577 | 24.52 |
| CRISPR repeats | 0 | 0 |
Fig. 3.
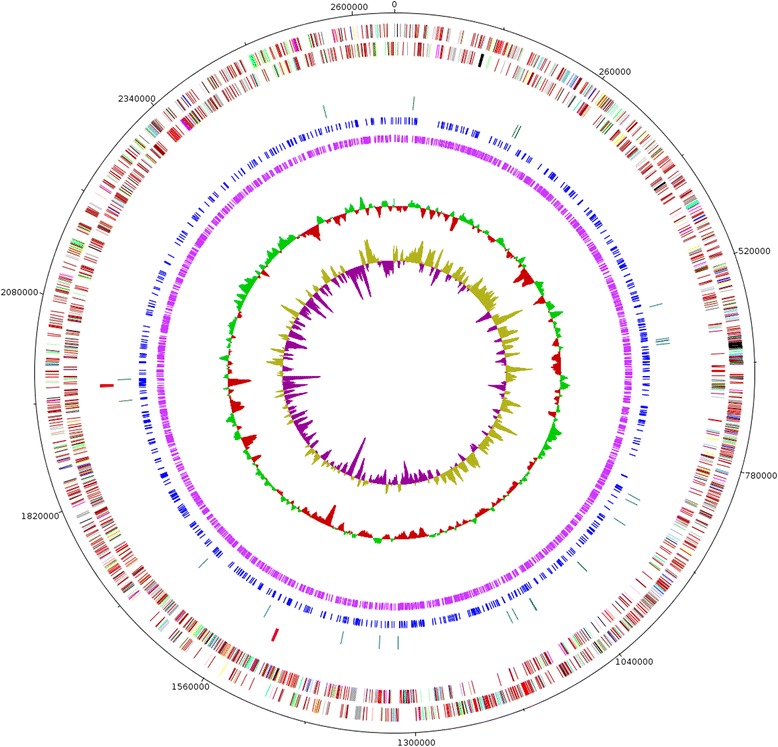
Genome properties of Propionibacterium freudenreichii ssp. freudenreichii DSM 20271T . The circles are from inner to outer order: GC skew, GC%, putative m4C, m6A, tRNA, rRNA, CDS reverse, CDS forward. CDS are colored according to the COG functional categories
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 138 | 5.86 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 152 | 6.46 | Transcription |
| L | 211 | 8.97 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 18 | 0.76 | Cell cycle control, Cell division, chromosome partitioning |
| V | 32 | 1.36 | Defense mechanisms |
| T | 82 | 3.48 | Signal transduction mechanisms |
| M | 77 | 3.27 | Cell wall/membrane biogenesis |
| N | 1 | 0.04 | Cell motility |
| U | 24 | 1.02 | Intracellular trafficking and secretion |
| O | 73 | 3.10 | Posttranslational modification, protein turnover, chaperones |
| C | 138 | 5.86 | Energy production and conversion |
| G | 157 | 6.67 | Carbohydrate transport and metabolism |
| E | 205 | 8.71 | Amino acid transport and metabolism |
| F | 57 | 2.42 | Nucleotide transport and metabolism |
| H | 106 | 4.50 | Coenzyme transport and metabolism |
| I | 58 | 2.46 | Lipid transport and metabolism |
| P | 123 | 5.23 | Inorganic ion transport and metabolism |
| Q | 31 | 1.32 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 219 | 9.31 | General function prediction only |
| S | 80 | 3.40 | Function unknown |
| - | 1017 | 43.22 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Conclusions
Prior to this report only a single genome sequence was available for Propionibacterium freudenreichii, from the type strain of P. freudenreichii subsp. shermanii CIRM-BIA1 [4]. In the present study the first genome sequence of a P. freudenreichii subsp. freudenreichii strain was described. P. freudenreichii is an industrially important species and a rare producer of biologically active form of vitamin B12. Probably the characteristics of P. freudenreichii DNA such as high G + C content and regions of repeated sequences have hampered the unraveling the complete genomes of this species. The results presented here indicate that PacBio RS II sequencing platform is well-suited to overcome these potential obstacles. In this study three DNA sequence motifs containing methylated bases were detected. Our future investigations include using this platform for sequencing of several additional strains for establishing core and pan-genomes as well as methylomes to gain understanding of genome structure and evolution of P. freudenreichii.
Acknowledgements
This study was supported by the Academy of Finland (grant no 257333).
Abbreviations
- PPA
Propionic medium
- HGAP
Hierarchical genome-assembly process
- RM
Restriction-modification
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PV and VP supplied the strain and background information for this project. PA, PV, LP, KS and PD conceived and designed the experiments. PD performed microbiological experiments and DNA isolation. PK, OPS, FT, JK, LP and PA performed the sequencing, assembly experiments and annotation. PK, OPS, PD, KS and PV wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Patrick S, McDowell A: Family I. Propionibacteriaceae. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s Manual of Systematic Bacteriology Volume Five: The Actinobacteria, Part B. 2. New York: Springer; 2012. pp. 1138–86. [Google Scholar]
- 2.Thierry A, Deutsch S, Falentin H, Dalmasso M, Cousin F, Jan G. New insights into physiology and metabolism of Propionibacterium freudenreichii. Int J Food Microbiol. 2011;149:19–27. doi: 10.1016/j.ijfoodmicro.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 3.Poonam, Pophaly SD, Tomar SK, De S, Singh R: Multifaceted attributes of dairy propionibacteria: a review. World J Microbiol Biotechnol. 2012, 28:3081–95. [DOI] [PubMed]
- 4.Falentin H, Deutsch S, Jan G, Loux V, Thierry A, Parayre S, Maillard M, Dherbécourt J, Cousin F, Jardin J, Siguier P, Couloux A, Barbe V, Vacherie B, Wincker P, Gibrat J, Gaillardin C, Lortal S. The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PloS one. 2010;5:11748. doi: 10.1371/journal.pone.0011748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalmasso M, Nicolas P, Falentin H, Valence F, Tanskanen J, Jatila H, Salusjärvi T, Thierry A. Multilocus sequence typing of Propionibacterium freudenreichii. Int J Food Microbiol. 2011;145:113–20. doi: 10.1016/j.ijfoodmicro.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 6.van Niel CB. The propionic acid bacteria. Haarlem: Uitgeverszaak and Boissevain and Co.; 1928. [Google Scholar]
- 7.Orla-Jensen S. Obituary notice. J Gen Microbiol. 1950;4:107–9. doi: 10.1099/00221287-4-2-107. [DOI] [PubMed] [Google Scholar]
- 8.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, de Winter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 9.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 10.Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucl Acids Res. 1995;23:4992–9. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Argo genome browser [http://www.broadinstitute.org/annotation/argo/].Accessed on 20 Sept 2014.
- 13.Koskinen P, Törönen P, Nokso-Koivisto J, Holm L. PANNZER - high-throughput functional annotation of uncharacterized proteins in an error-prone environment. Bioinformatics. 2015;31:1544–52. doi: 10.1093/bioinformatics/btu851. [DOI] [PubMed] [Google Scholar]
- 14.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, et al. Interpro in 2011: new developments in the family and domain prediction database. Nucl Acids Res. 2011;40(Database issue):D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 16.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 17.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a conserved domain database for the functional annotation of proteins. Nucl Acids Res. 2011;39(suppl 1):225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer rna genes in genomic sequence. Nucl Acids Res. 1997;25:0955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucl Acids Res. 2009;38(Database issue):D234–6. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucl Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 23.Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 24.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnology. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stackebrandt E, Rainey F, Ward-Rainey N. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–91. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 27.Goodfellow M. Phylum XXVI. Actinobacteria phyl. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s Manual of Systematic Bacteriology Volume Five: The Actinobacteria, Part A. 2. New York: Springer; 2012. pp. 33–4. [Google Scholar]
- 28.Patrick S, McDowell A. Order XII. Propionibacteriales ord. nov. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB, editors. Bergey’s Manual of Systematic Bacteriology Volume Five: The Actinobacteria, Part B. 2. New York: Springer; 2012. p. 1137. [Google Scholar]
- 29.Charfreitag O, Stackebrandt E. Inter- and intrageneric relationships of the genus Propionibacterium as determined by 16S rRNA sequences. J Gen Microbiol. 1989;135:2065–70. doi: 10.1099/00221287-135-7-2065. [DOI] [PubMed] [Google Scholar]
- 30.Judicial Commission of the International Committee on Systematics of Prokaryotes Necessary corrections to the Approved Lists of Bacterial Names according to Rule 40d (formerly Rule 46). Opinion 86. Int J Syst Evol Microbiol. 2008;58(Pt 8):1975. doi: 10.1099/ijs.0.2008/006015-0. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


