Abstract
Long noncoding RNAs (lncRNAs) regulate gene expression by acting with microRNAs (miRNAs). However, the roles of cancer specific lncRNA and its related competitive endogenous RNAs (ceRNA) network in hepatocellular cell carcinoma (HCC) are not fully understood. The lncRNA profiles in 372 HCC patients, including 372 tumor and 48 adjacent non-tumor liver tissues, from The Cancer Genome Atlas (TCGA) and NCBI GEO omnibus (GSE65485) were analyzed. Cancer specific lncRNAs (or HCC related lncRNAs) were identified and correlated with clinical features. Based on bioinformatics generated from miRcode, starBase, and miRTarBase, we constructed an lncRNA-miRNA-mRNA network (ceRNA network) in HCC. We found 177 cancer specific lncRNAs in HCC (fold change ≥ 1.5, P < 0.01), 41 of them were also discriminatively expressed with gender, race, tumor grade, AJCC tumor stage, and AJCC TNM staging system. Six lncRNAs (CECR7, LINC00346, MAPKAPK5-AS1, LOC338651, FLJ90757, and LOC283663) were found to be significantly associated with overall survival (OS, log-rank P < 0.05). Collectively, our results showed the lncRNA expression patterns and a complex ceRNA network in HCC, and identified a complex cancer specific ceRNA network, which includes 14 lncRNAs and 17 miRNAs in HCC.
Introduction
Noncoding RNAs are RNA molecules that are not coding for proteins. They can be divided into several subtypes including long non-coding RNA (lncRNA), microRNA (miRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNA), and transfer RNA according to HUGO Gene Nomenclature Committee (HGNC) (http://www.genenames.org).
After the identification of lncRNA in malignancy diseases, an increasing number of studies on the biological roles of lncRNAs have been conducted in various cancers, including HCC [1], esophageal squamous cell carcinoma [2], colorectal cancer [3], renal cell carcinoma [4] and prostate cancer [5]. The abnormal expression of lncRNAs through interactions with miRNAs or mRNAs is involved in the regulation of tumor progression and tumor biological behaviors in HCC [6–8]. The cancer specific lncRNAs may also impact the invasion and metastasis of HCC [9].
In 2011, Salmena et al. presented a competing endogenous RNA (ceRNA) hypothesis, which unified the transcriptome and formed a regulatory RNA network [10]. The main idea is that all types of RNA transcripts communicate with each other by competing for binding to shared miRNA-binding sites (“miRNA response elements” or “MREs”). This kind of RNA competition crosstalk exists between protein-coding messenger RNAs and non-coding RNAs such as lncRNA, pseudogenes and circular RNAs [11]. Furthermore, artificial miRNA sponges can also participate in this network to regulate gene expression [12].
Zhu et al. reported that the lncRNA expression profile of HCC by microarray analysis from three HCC patients [13]. However, there is lack of studies with large scale sample size and high through detection methods on the expression patterns of cancer specific lncRNA in HCC, and it is unknown whether lncRNAs are correlated with overall survival, gender, or other clinical features or whether the aberrant expression of lncRNAs in HCC has any ceRNA potential. Recently, RNA sequencing data from The Cancer Genome Atlas (TCGA) project or GEO provide the public with lncRNA, miRNA, and mRNA data for HCC. To address the above mentioned questions, we explored lncRNAs in HCC using data sets from TCGA and GEO. These two data sets included RNA sequence results for a total of 372 HCC tumor tissues and 48 adjacent non-tumor liver tissue samples. To the best of our knowledge, this study is the first to make use of large scale sequencing database to investigate the cancer specific lncRNA expression patterns and ceRNA network in HCC. This new approach of predicting cancer specific lncRNA and ceRNA network can help us to understand the function of lncRNAs in HCC.
Methods
Patients and samples
A total of 360 patients with HCC were retrieved from the TCGA data portal. The exclusion criteria were set as follows: 1) histologic diagnosis is not HCC; 2) samples without completed data for analysis; and 3) Overall survival more than 2000 days. Overall, a total of 322 HCC patients were included in our study. Among these 322 HCC patients, the adjacent non-tumor liver tissues were retrieved from 43 subjects. This study meets the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). Another GEO data set (GSE65485) was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/) which included 50 HCC tissues and 5 adjacent non-tumor liver tissues. As the data were obtained from TCGA and GEO, further approval by an ethics committee was not required.
RNA sequence data procession and computational analysis
The RNA expression data (level 3) of the corresponding patients (tumor and/or adjacent non-tumor tissues) were downloaded from TCGA data portal (up to Feb 24, 2015). The lncRNA and mRNA expression profiles were generated from RNA sequencing raw reads by RNASeqV2 post-processing pipelines and demonstrated as RSEM (RNA-Seq by Expectation-Maximization) normalized count data. The miRNA expression profile was performed using the Illumina HiSeq 2000 miRNA sequencing platforms (Illumina Inc, USA) and demonstrated as reads per million miRNA (RPM) mapped data. Because the mRNA, lncRNA, and miRNA expression profile data were already normalized by TCGA, no further normalizations were applied to these data. The GEO data set was also generated from the Illumina HiSeq 2000 platform and normalized as FPKM (fragments per kilo bases of exons for per million mapped reads) data. The lncRNA analyses were performed using BRB-ArrayTools (version 4.4) developed by Dr. Richard Simon and the BRB-ArrayTools Development Team [14].
Construction of the ceRNA network and KEEG Pathway Analysis
The construction of ceRNA network included three steps: (i) cancer specific lncRNA filtration: cancer specific lncRNAs with absolute fold change ≥ 3.0 (either up-regulation or down-regulation) and P < 0.05 were retained. To improve the data reliability, cancer specific lncRNAs have not been annotated by GENCODE (http://www.gencodegenes.org/) were discarded; (ii) lncRNA-miRNA interactions were predicted by miRcode (http://www.mircode.org/) and starBase v2.0 (http://starbase.sysu.edu.cn/); (iii) mRNAs targeted by miRNAs with experimental support were retrieved from miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/). To further enhance the robust of this ceRNA network, the maximal information coefficient (MIC) algorithm and maximal information-based nonparametric exploration (MINE) statistics were used in TCGA data set to filter the pair-wised relationships [15]. A network graph was constructed and visualized using Cytoscape v3.0 [16]. The coding genes involved in ceRNA network were input into the Database for Annotation, Visualization and Integrated Discovery (DAVID) [17] for KEEG pathway enrichment analysis.
Statistical analysis
Data were presented as mean ± SD. Differences among groups were evaluated by two-sample t test and the significance level was set as 0.001 as default to control the false discovery rate (FDR). The univariate Cox proportional hazards regression was conducted to find out the lncRNAs correlated with overall survival [18]. P value less than 0.05 was considered as statistical unless specifically indicated. The statistical analyses were performed by BRB-ArrayTools or R language [19].
Results
Cancer specific lncRNAs in HCC
We identified 604 lncRNAs from TCGA data set and 357 lncRNAs from GSE65485. For any single lncRNA, it appeared in TCGA data set or both TCGA and GEO data sets. We found that 177 lncRNAs and 37 lncRNAs were differentially expressed between HCC tissues and adjacent non-tumor tissues in TCGA data set and GEO data set (absolute fold change ≥ 1.5, P < 0.01, S1 Table), respectively. To further enhance the data reliability, we selected 28 lncRNAs included in GENCODE and these lncRNAs had an absolute fold change ≥ 3.0 from either TCGA or GEO data set to build ceRNA network [20]. Finally, 28 lncRNAs (18 up-regulated; 10 down-regulated) were selected for ceRNA network (Table 1).
Table 1. Twenty eight cancer specific lncRNAs in ceRNA network construction.
This table showed 28 cancer specific lncRNAs for ceRNA network construction with absolute fold change ≥ 3.0, P < 0.01 and included in GENCODE.
| LncRNA | Gene ID | Chromosome | Expression change (T vs. N) | Data set |
|---|---|---|---|---|
| ASMTL-AS1 | ENSG00000236017 | chrX | Up-regulation | TCGA |
| CDKN2B-AS1 | ENSG00000240498 | chr9 | Up-regulation | TCGA + GEO |
| CECR7 | ENSG00000237438 | chr22 | Up-regulation | TCGA |
| FLVCR1-AS1 | ENSG00000198468 | chr1 | Up-regulation | TCGA |
| FOXD2-AS1 | ENSG00000237424 | chr1 | Up-regulation | TCGA |
| GAS5 | ENSG00000234741 | chr1 | Up-regulation | TCGA |
| IGF2BP2-AS1 | ENSG00000163915 | chr3 | Up-regulation | TCGA |
| LINC00152 | ENSG00000222041 | chr2 | Up-regulation | TCGA |
| LINC00176 | ENSG00000196421 | chr20 | Up-regulation | TCGA |
| LINC00488 | ENSG00000214381 | chr3 | Up-regulation | TCGA |
| LINC00685 | ENSG00000226179 | chrX | Up-regulation | TCGA |
| PVT1 | ENSG00000249859 | chr8 | Up-regulation | TCGA + GEO |
| RUSC1-AS1 | ENSG00000225855 | chr1 | Up-regulation | TCGA |
| SNHG1 | ENSG00000255717 | chr11 | Up-regulation | TCGA |
| SNHG3 | ENSG00000242125 | chr1 | Up-regulation | TCGA + GEO |
| SNHG4 | ENSG00000281398 | chr5 | Up-regulation | TCGA + GEO |
| TSPEAR-AS2 | ENSG00000182912 | chr21 | Up-regulation | TCGA |
| ZNF252P-AS1 | ENSG00000255559 | chr8 | Up-regulation | TCGA |
| DIO3OS | ENSG00000258498 | chr14 | Down-regulation | GEO |
| FAM99A | ENSG00000205866 | chr11 | Down-regulation | TCGA + GEO |
| FAM99B | ENSG00000205865 | chr11 | Down-regulation | TCGA |
| H19 | ENSG00000130600 | chr11 | Down-regulation | TCGA |
| HAND2-AS1 | ENSG00000237125 | chr4 | Down-regulation | TCGA |
| HAR1A | ENSG00000225978 | chr20 | Down-regulation | TCGA |
| LINC00238 | ENSG00000196553 | chr14 | Down-regulation | TCGA |
| LINC01554 | ENSG00000236882 | chr5 | Down-regulation | TCGA |
| PWRN1 | ENSG00000259905 | chr15 | Down-regulation | GEO |
| UCA1 | ENSG00000214049 | chr19 | Down-regulation | TCGA |
The correlations between cancer specific lncRNAs and clinical features
The 177 lncRNAs from the above section were further analyzed according to clinical features including gender, race, tumor grade, AJCC TNM staging system, AJCC pathological stage, vascular invasion, new tumor event, tumor status, and age at diagnosis in TCGA and/or GEO data sets. There were total 41 cancer specific lncRNAs, the levels of which were also significantly different in clinical feature comparisons (P < 0.001, Table 2). Five lncRNAs (LINC01554, LOC255167, A1BG-AS1, LINC00526, and MIR22HG) were differentially expressed in three or four clinical feature comparisons.
Table 2. The correlations between cancer specific lncRNAs and clinical features.
This table showed 41 cancer specific lncRNA which were also differentially expressed in clinical feature comparisons.
| Comparisons | Down-regulated | Up-regulated |
|---|---|---|
| Gender (Female vs. Male) | LOC643837, LINC00526, FAM223B, MIR99AHG, LINC01554, TTTY15 | H19, LINC00092, MIR600HG, LOC645676, COLCA1, LOC723809 |
| Race (White vs. Asian) | GAS5, SNHG7, LOC100128191, SNHG1, HCG18 | HCG11, LINC00242, GLIDR, LINC00261, LINC00638, LINC00574 |
| Tumor grade (G3 + G4 vs. G1 + G2) | LOC255167, FAM99A, LINC00574, LOC399959, A1BG-AS1, LINC00526, LINC00261, MIR22HG, LOC643837 | MALAT1, SNHG12, HCG18, SNHG20, ZFAS1, LOC440944, SNHG7, LOC100128191, MCM3AP-AS1, SNHG1, KCNQ1OT1, DSCR9, GAS5, LOC92659 |
| AJCC TNM staging system (T3 + T4 vs. T1 + T2) | LOC255167, LINC01554, LINC01558, A1BG-AS1 | MCM3AP-AS1 |
| AJCC pathological stage (III + IV vs. I + II) | LOC255167, LINC01554, LINC01558, A1BG-AS1 | UCA1 |
| Vascular invasion (No vs. Yes) | MAPKAPK5-AS1, HNF1A-AS1, DSCR9 | FAM99A |
| New tumor event (Yes vs. No) | MIR22HG | |
| Tumor status (With tumor vs. Tumor free) | MIR22HG | |
| Age at diagnosis (≥ 61 vs. < 61) | LINC01554, LINC00526, LOC255167 |
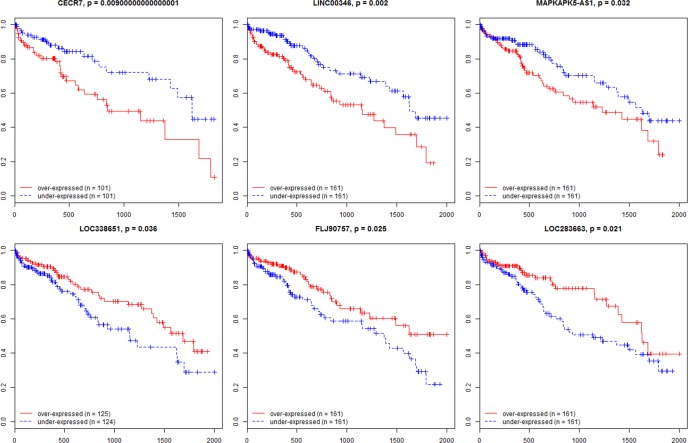
Subsequently, to identify the potential lncRNAs with prognostic characteristics, the levels of 177 lncRNAs in the TCGA data set were profiled using the univariate Cox proportional hazards regression model and six lncRNAs were found to be significantly associated with overall survival (log-rank P < 0.05). Among the six significant lncRNAs, three lncRNA (CECR7, LINC00346, and MAPKAPK5-AS1) were negatively associated with OS, while the remaining three (LOC338651, FLJ90757, and LOC283663) were positively correlated with OS (Fig 1).
Fig 1. Kaplan-Meier survival curves for six lncRNAs associated with overall survival.
(Horizontal axis: overall survival time: days, Vertical axis: survival function).
miRNA targeting lncRNAs predicted by miRcode and starBase
Our previous study has found 207 HCC-associated miRNAs which were differentially expressed between HCC tissues and adjacent non-tumor tissues [21]. We selected 33 miRNAs from 207 HCC-associated miRNAs in current TCGA data set (absolute fold change ≥ 3.0, P < 0.001, S2 Table). Here, we focused on whether these miRNAs would target above 28 cancer specific lncRNAs. In the ceRNA network, miRNAs interact with lncRNAs through MREs, we thus searched for the potential MREs in lncRNAs using miRcode [22] and starBase v2.0 [23]. The results showed that 17 of 33 cancer specific miRNAs might interact with 15 of 28 cancer specific lncRNAs (Table 3).
Table 3. Putative miRNAs that may target cancer specific lncRNAs by MREs.
miR-199A-1 and miR-199A-2 were regards as a single miRNA in our study
| lncRNA | miRNAs |
|---|---|
| LINC00176 | miR-424, miR-184, miR-214, miR-93 |
| PVT1 | miR-195, miR-424, miR-183, miR-199A-1/2, miR-214, miR-383, miR-93 |
| LINC00488 | miR-139 |
| RUSC1-AS1 | miR-10B, miR-135A-1, miR-139, miR-182, miR-199A-1/2, miR-214, miR-96 |
| FLVCR1-AS1 | miR-375 |
| GAS5 | miR-10B, miR-139, miR-182, miR-490, miR-93, miR-96 |
| UCA1 | miR-214, miR-383 |
| SNHG1 | miR-182, miR-195, miR-383, miR-424 |
| SNHG3 | miR-135A-1, miR-139, miR-182 |
| CDKN2B-AS1 | miR-10B |
| LINC00152 | miR-376C |
| CECR7 | miR-199A-1/2, miR-214 |
| DIO3OS | miR-10B, miR-139, miR-199A-1/2, miR-214, miR-34C |
| H19 | miR-93 |
| LINC00238 | miR-33B, miR-375 |
miRNA targets predicted by miRTarBase
To establish lncRNA-miRNA-mRNA network (ceRNA network), the next step was to search for mRNA targeted by miRNAs. Based on the miRNAs described in Table 3, we searched miRNA-targeted mRNA with experimental evidence using miRTarBase [24]. The results identified 17 miRNAs including miR-10B, miR-135A-1, miR-139, miR-182, miR-183, miR-184, miR-195, miR-199A-1/2, miR-214, miR-33B, miR-34C, miR-375, miR-376c, miR-383, miR-424, miR-93, and miR-96 (Table 4). Each miRNA-mRNA pair was experimentally validated by at least two of the following methods including reporter assay, western blot, qPCR, microarray, pSILAC (pulsed stable isotope labelling with amino acids in cell culture) or NGS (CLIP-seq or Degradome-seq). According to the allOnco database (http://www.bushmanlab.org/links/genelists), most of their targets are cancer-associated genes such as SIRT1, VEGFA, RASA1, RAF1, PTEN, MAPK9, MAPK8, MAPK1, MYC, MYB, KRAS, JAK2, IGF1R, IDH2, FOXO3, FOXO1, E2F3, E2F1, MAPK14, CDKN2A, CDKN1A, CDK6, CD44, CCNF, CCNE1, CCND3, CCND2, RUNX2, BCL2, CCND1, APC, AKT2, AKT1, ABCA1, etc.
Table 4. Validation of miRNA targets.
| miRNA | mRNAs targeted by miRNAs |
|---|---|
| miR-10B | PPARA, NF1, CDKN2A, HOXD10, KLF4, NCOR2, CDKN1A, TFAP2C, BCL2L11 |
| miR-135A-1 | JAK2, NR3C2, APC, MYC |
| miR-139 | FOS |
| miR-182 | CDKN1A, FOXO1, MITF, FOXO3, RARG, ADCY6, CLOCK, TSC22D3, CYLD, BCL2, CCND2, PDCD4, RECK, EP300, FGF9, NTM |
| miR-183 | SRSF2, PDCD4, AKAP12, FOXO1, ITGB1, KIF2A, BTRC, EZR, IDH2 |
| miR-184 | AKT2, INPPL1, NFATC2 |
| miR-195 | WEE1, E2F3, CDK6, BCL2, CCND1, RUNX2, RAF1, CCL4, BCL2L11, MECP2, CCND3, TBCCD1, VEGFA, SKI, KRT7, SLC2A3, CAB39, BCL2L2 |
| miR-199A-1/2 | CD44, IKBKB, MET, HIF1A, SMARCA2, SMARCA2, MAPK1, DDR1, MAP3K11, FUT4, CAV2, EZH2, CCNL1, MAPK9, AKT1, MAPK8, MAPK14, LIF, JUNB, MED6, MECP2, ETS2, EDN1, TMEM54, SIRT1 |
| miR-214 | PTEN, MAP2K3, MAPK8, PLXNB1, EZH2, POU4F2, GALNT7, XBP1, DAPK1, SRGAP1, TWIST1, BCL2L2, ING4, FLOT1, MAP2K5, HSPD1, AHSA1, CPNE7, RASA1, YWHAQ, ARL2, AP3B1 |
| miR-33B | BCL2, ABCA1 |
| miR-34C | MET, CDK4, NOTCH1, BCL2, E2F3, MYB, CAV1, CCNE2, MYCN, NOTCH4, ULBP2, MYC, SRSF2, SOX2, NANOG |
| miR-375 | ELAVL4, YWHAZ, TIMM8A, PDK1, YAP1, MTDH, RASD1, YY1AP1, RHOA, KCNQ2, MTPN, USP1, JAK2, C1QBP, PLAG1, BCL2L11, RAB10, PARP1, CAB39, DLG4, ITGB1, SETD8, CASP3, CDC42, EIF2AK2, BCL2, NCAM1, LEPROTL1 |
| miR-376C | ACVR1C, IGF1R |
| miR-383 | DIO1, IRF1, VEGFA |
| miR-424 | HIF1A, CUL2, SPI1, PLAG1, CCNE1, CCND3, CDK6, CCND1, MAP2K1, WEE1, NFIA, LGALS3, MYB, SIAH1, CCNF, CDC14A, CDC25A, CHEK1, KIF23, ATF6, ANLN, FGFR1, PIAS1, ITPR1 |
| miR-93 | CDKN1A, E2F1, ITGB8, TUSC2, TP53INP1, KAT2B, PTEN, LATS2, MAPK9, VEGFA |
| miR-96 | CDKN1A, KRAS, FOXO1, FOXO3, ADCY6, REV1, RAD51, MITF, HTR1B, PRMT5 |
ceRNA network construction and KEEG pathway analysis
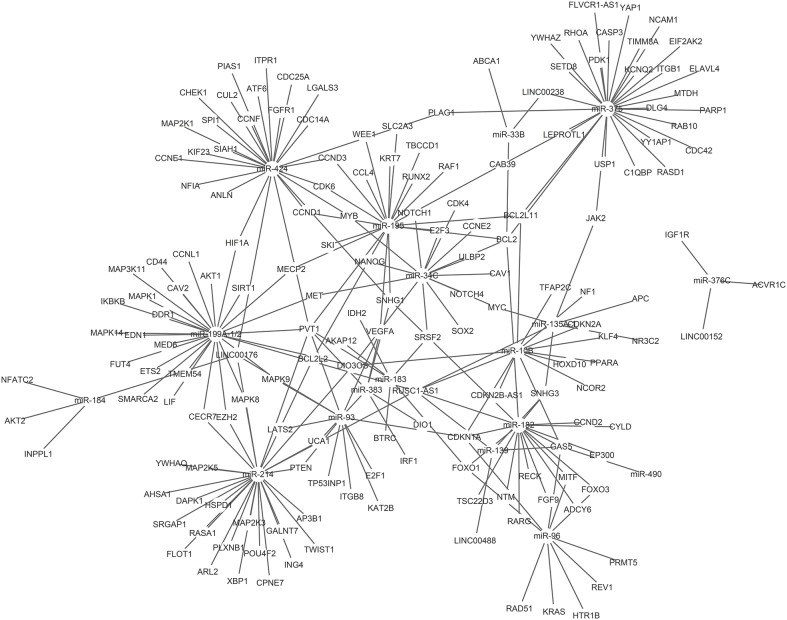
Based on the above data (Tables 3 and 4), we constructed a lncRNA-miRNA-mRNA ceRNA network. To get more robust results, we used the maximal information coefficient (MIC) algorithm to screen the pair-wised relationships based on the expression levels of the lncRNA, miRNA, and mRNA in TCGA data set (MIC > 0.15 and MIC-p2 > 0.15, please refer to Methods). 14 lncRNAs and 17 miRNAs were involved in the proposed ceRNA network (Fig 2).
Fig 2. ceRNA network in HCC.
In order to understand the signal pathways involved in ceRNA network, the mRNAs were analyzed by DAVID database. According to number of genes involved, we listed the top 15 KEGG pathways in our study (Table 5). Ten cancer-related pathways including pathways in cancer, pancreatic cancer, melanoma, prostate cancer, chronic myeloid leukemia, colorectal cancer, glioma, small cell lung cancer, bladder cancer, and non-small cell lung cancer, were enriched with the mRNAs, another 5 non-cancer related pathways such as MAPK signaling pathway, focal adhesion, cell cycle, neurotrophin signaling pathway, T cell receptor signaling pathway were also enriched.
Table 5. Top 15 KEEG pathways enriched by the coding genes involved in ceRNA network (P < 0.0001).
The P value was corrected for multiple hypothesis testing using the Benjamini-Hochberg method (Please also refer to S3 Table)
| KEEG pathways | No. of Genes | |
|---|---|---|
| Cancer related | ||
| Pathways in cancer | 41 | |
| Pancreatic cancer | 19 | |
| Melanoma | 19 | |
| Prostate cancer | 19 | |
| Chronic myeloid leukemia | 16 | |
| Colorectal cancer | 16 | |
| Glioma | 15 | |
| Small cell lung cancer | 15 | |
| Bladder cancer | 13 | |
| Non-small cell lung cancer | 13 | |
| Non-cancer related | ||
| MAPK signaling pathway | 22 | |
| Focal adhesion | 21 | |
| Cell cycle | 19 | |
| Neurotrophin signaling pathway | 18 | |
| T cell receptor signaling pathway | 14 |
Discussion
Recently, lncRNAs have been emerged as abundant regulators of cell physiology in HCC and their functions may vary [25, 26]. Only a few studies have tried to reveal the lncRNA expression profiles in HCC by microarray with dozens of or even smaller sample size [13]. LncRNA and mRNA coexpression network was constructed by abnormally expressed lncRNA and mRNA [13]. A few studies described interactions between miRNA and lncRNAs [27, 28] or mRNA and lncRNA [29] in HCC, the results of which indicated that lncRNAs can function as a part of ceRNA network, but such ceRNA network is still poorly explored. In the present study, we identified tumor-specific lncRNAs in HCC and investigated their distributions in different clinical features and their associations with overall survival on the basis of genome-wide RNA profiles of 372 HCC tissues and 48 adjacent non-tumor liver tissues. Furthermore, we constructed a ceRNA network with cancer specific lncRNAs and miRNAs which provides a system biological views of lncRNA-miRNA-mRNA interactions.
Based on the next generation RNA sequence data from TCGA and GEO, we found that 177 cancer specific lncRNAs were differentially expressed in HCC tumor tissues and adjacent non-tumor liver tissues. Then, we revealed that 41 cancer specific lncRNAs were also abnormally expressed in different groups of clinical pathological features such as gender, race, tumor grade, AJCC TNM staging system, AJCC pathological stage, vascular invasion, new tumor event, tumor status, and age at diagnosis. Among the lncRNAs that differentially expressed in three or four groups, MIR22HG was reported to be an indicator of chemical stress responses in human-induced pluripotent stem cells [30]. We concluded that the expression of some lncRNAs is not equally distributed in certain situations. Future studies in this field should be properly designed to cope with this fact. Previous studies reported sexual disparity of HCC incidence [31], the differentially expressed lncRNA between female and male found in this study may contribute to this phenomenon. However, these unevenly distributed lncRNAs may not be significantly associated with overall survival.
With respect to the associations between cancer specific lncRNAs and patients’ survival, we found that six lncRNAs were related to HCC overall survival. Among the three risky lncRNAs, CECR7 is a candidate lncRNA for Cat Eye Syndrome [32]. The functions of the other two risky and three protective novel lncRNAs are still unclear. It is also noted that five of these six lncRNAs (CECR7, LINC00346, LOC338651, FLJ90757, and LOC283663) were not differentially expressed in any clinical feature comparison. Therefore, lncRNAs that do not differentially express in clinical feature comparisons can be correlated with overall survival, whereas lncRNAs that differentially express in clinical feature comparisons may not be necessary to be associated with overall survival.
We believe that there may be some cross-talks between lncRNA, miRNA and mRNA in the progress of HCC. We applied several steps to increase the accuracy of ceRNA network prediction. First, we only included the cancer specific lncRNAs and miRNAs that had an absolute fold change ≥ 3.0 and was annotated by GENCODE. Second, the relationships between lncRNA and miRNA, and miRNA and mRNA were predicted by experiment- supported algorithms or databases such as miRcode, starBase and miRTarBase. These two measurements ensured that the relationships identified would occur not only in silico situations but also by experimental-supported evidences.
To further improve the performance of our prediction, the maximal information coefficient (MIC) algorithm and maximal information-based nonparametric exploration (MINE) statistics were used to filter the pair-wised relationships based on lncRNA-miRNA-mRNA expression correlations. In general gene co-expression network analysis, Pearson’s correlation is a measure for linear regression, but it is very sensitive to outliers. MIC and MINE are able to examine and characterize all potentially interesting relationships in a complex data set [33].
The ceRNA network we built brings to light an unknown ceRNA regulatory network in HCC. In this newly-identified ceRNA network, many oncogenes and tumor suppressors participate in HCC development and treatments. A recent study also identified that lncRNA-miRNA-mRNA interactions were active and might act as potential prognostic biomarkers in cancers [34].
In conclusion, our study has found the cancer specific lncRNAs in HCC using hundreds of candidate lncRNAs and large scale samples, and disclosed abnormal expression pattern of cancer specific lncRNAs under different clinical features. Importantly, we have constructed a ceRNA network to propose a new approach to lncRNA research in HCC. Our findings suggest that cancer specific lncRNAs in HCC may participate in a complex ceRNA network.
Supporting Information
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank Mr. Rocky Ho for their technical assistance. The results published or shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This study was supported by SRFDP and RGC ERG Joint Research (No: M-CUHK406/13) and the National Natural Science Foundation of China (No.81472339).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Specialized Research Fund for the Doctoral Program of Higher Education and Research Grants Council Earmarked Research Grants Joint Research Scheme (No. M-CUHK406/13) and the National Natural Science Foundation of China (No. 81472339). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60(4):1278–1290. Epub 2014/07/22. 10.1002/hep.27239 . [DOI] [PubMed] [Google Scholar]
- 2. Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. Epub 2014/02/14. 10.1136/gutjnl-2013-305806 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, et al. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5(8):2230–2242. Epub 2014/05/09. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fachel AA, Tahira AC, Vilella-Arias SA, Maracaja-Coutinho V, Gimba ER, Vignal GM, et al. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: emerging functional associations. Molecular cancer. 2013;12(1):140 Epub 2013/11/19. 10.1186/1476-4598-12-140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. Epub 2014/02/13. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer research. 2015;75(5):846–857. Epub 2015/01/17. 10.1158/0008-5472.CAN-14-1192 . [DOI] [PubMed] [Google Scholar]
- 7. Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. Epub 2013/10/12. 10.1002/hep.26740 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia (New York, NY). 2015;17(1):79–88. Epub 2015/01/28. 10.1016/j.neo.2014.11.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gao Y, Chen G, Zeng Y, Zeng J, Lin M, Liu X, et al. Invasion and metastasis-related long noncoding RNA expression profiles in hepatocellular carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015. Epub 2015/04/23. 10.1007/s13277-015-3408-0 . [DOI] [PubMed] [Google Scholar]
- 10. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. Epub 2011/08/02. 10.1016/j.cell.2011.07.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. Epub 2014/01/17. 10.1038/nature12986 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5(3):317–333. Epub 2014/01/01. 10.1002/wrna.1213 . [DOI] [PubMed] [Google Scholar]
- 13. Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, et al. The long noncoding RNA expression profile of hepatocellular carcinoma identified by microarray analysis. PloS one. 2014;9(7):e101707 Epub 2014/07/16. 10.1371/journal.pone.0101707 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer informatics. 2008;6:9–15. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, et al. Detecting novel associations in large data sets. Science (New York, NY). 2011;334(6062):1518–1524. Epub 2011/12/17. 10.1126/science.1205438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–2504. Epub 2003/11/05. 10.1101/gr.1239303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. Epub 2009/01/10. 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 18. Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS biology. 2004;2(4):E108 10.1371/journal.pbio.0020108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2014.
- 20. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22(9):1760–1774. Epub 2012/09/08. 10.1101/gr.135350.111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J, Chong CC, Chen GG, Lai PB. A Seven-microRNA Expression Signature Predicts Survival in Hepatocellular Carcinoma. PloS one. 2015;10(6):e0128628 10.1371/journal.pone.0128628 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics (Oxford, England). 2012;28(15):2062–2063. 10.1093/bioinformatics/bts344 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids research. 2014;42(Database issue):D92–97. Epub 2013/12/04. 10.1093/nar/gkt1248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic acids research. 2014;42(Database issue):D78–85. 10.1093/nar/gkt1266 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54(5):1679–1689. Epub 2011/07/20. 10.1002/hep.24563 . [DOI] [PubMed] [Google Scholar]
- 26. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. The Journal of biological chemistry. 2012;287(31):26302–26311. Epub 2012/06/12. 10.1074/jbc.M112.342113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver international: official journal of the International Association for the Study of the Liver. 2015;35(5):1597–1606. Epub 2014/11/27. 10.1111/liv.12746 . [DOI] [PubMed] [Google Scholar]
- 28. Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899–7917. Epub 2015/03/12. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25(5):666–681. Epub 2014/04/29. 10.1016/j.ccr.2014.03.010 . [DOI] [PubMed] [Google Scholar]
- 30. Tani H, Onuma Y, Ito Y, Torimura M. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PloS one. 2014;9(8):e106282 Epub 2014/08/30. 10.1371/journal.pone.0106282 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148(1–2):72–83. Epub 2012/01/24. 10.1016/j.cell.2011.11.026 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bridgland L, Footz TK, Kardel MD, Riazi MA, McDermid HE. Three duplicons form a novel chimeric transcription unit in the pericentromeric region of chromosome 22q11. Hum Genet. 2003;112(1):57–61. Epub 2002/12/17. 10.1007/s00439-002-0827-y . [DOI] [PubMed] [Google Scholar]
- 33. Weiler S, Ademokun JA, Norton JD. ID helix-loop-helix proteins as determinants of cell survival in B-cell chronic lymphocytic leukemia cells in vitro. Molecular cancer. 2015;14(1):30 10.1186/s12943-014-0286-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z, et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic acids research. 2015;43(7):3478–3489. 10.1093/nar/gkv233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.